Abstract
The genomic DNA from four species of ixodid ticks in western Canada was tested for the presence of Rickettsiella by PCR analyses targeting the 16S rRNA gene. Eighty-eight percent of the Ixodes angustus (n = 270), 43% of the I. sculptus (n = 61), and 4% of the I. kingi (n = 93) individuals examined were PCR positive for Rickettsiella, whereas there was no evidence for the presence of Rickettsiella in Dermacentor andersoni (n = 45). Three different single-strand conformation polymorphism profiles of the 16S rRNA gene were detected among amplicons derived from Rickettsiella-positive ticks, each corresponding to a different sequence type. Furthermore, each sequence type was associated with a different tick species. Phylogenetic analyses of sequence data of the 16S rRNA gene and three other genes (rpsA, gidA, and sucB) revealed that all three sequence types were placed in a clade that contained species and pathotypes of the genus Rickettsiella. The bacterium in I. kingi represented the sister taxon to the Rickettsiella in I. sculptus, and both formed a clade with Rickettsiella grylli from crickets (Gryllus bimaculatus) and “R. ixodidis” from I. woodi. In contrast, the Rickettsiella in I. angustus was not a member of this clade but was placed external to the clade comprising the pathotypes of R. popilliae. The results indicate the existence of at least two new species of Rickettsiella: one in I. angustus and another in I. kingi and I. sculptus. However, the Rickettsiella strains in I. kingi and I. sculptus may also represent different species because each had unique sequences for all four genes.
INTRODUCTION
Terrestrial invertebrates harbor a diverse range of endosymbiotic bacteria (1–4), some of which are mutualists, assisting their invertebrate partners in metabolic processes or increasing their ability to resist infection by pathogens (5, 6). Other endosymbionts may reduce the fitness of their invertebrate hosts (7, 8) and/or have pathogenic effects on vertebrate hosts used by hematophagous arthropods (e.g., mosquitoes and ticks) (9, 10). For example, there are some species of Rickettsiella that are intracellular pathogens of arthropods (11–16), whereas other species may provide some benefit to their arthropod hosts (17).
Bacteria within the genus Rickettsiella were first described in 1952 as “small Rickettsia” (18) and assigned to the order Rickettsiales (Alphaproteobacteria) (19). However, phylogenetic analyses of sequence data of the 16S rRNA gene subsequently revealed that Rickettsiella grylli represented a sister taxon to two genera of Gammaproteobacteria: Coxiella and Legionella (12). As a consequence, the genus Rickettsiella was transferred from the Rickettsiales to the family Coxiellaceae within the order Legionellales (20), a placement that has been supported by other molecular studies (11, 21–29). Rickettsiella have been reported in a diverse range of arthropod hosts, including insects (e.g., beetles, flies, crickets, locusts, cockroaches, wasps, midges, moths, and aphids), collembolans, crustaceans (e.g., isopods and crabs), and arachnids (e.g., spiders, scorpions, ticks, and mites) (20, 30). Currently, only four species of Rickettsiella are recognized: R. popilliae, R. grylli, R. chironomi, and R. stethorae (27). However, for some species, several pathotypes (e.g., “R. tipulae,” “ R. costelytrae,” and “R. melolonthae”) have been described based on the host species they infect, their pathogenic effects on their hosts, and genetic comparisons to other Rickettsiella spp. (28, 30). Some pathotypes have been shown, using genetic comparisons, to have identical sequences of the 16S rRNA gene (e.g., “R. costelytrae” and “R. pyronotae” [28]) and are considered synonyms of one of the four recognized species (22, 27–29). Several other pathotypes are considered unassigned species (30).
Ticks have been shown to be suitable hosts for Rickettsiella. For example, a Rickettsiella genetically similar to R. grylli has been isolated in the ovarian tissues and malpighian tubules of unfed female Ixodes woodi (31), while female Dermacentor reticulatus have been experimentally infected with “R. phytoseiuli” isolated from the mite Phytoseiulus persimilis (32). Rickettsiella DNA has also been detected by PCR in I. tasmani and I. ricinus (33–35), and in a few individuals of I. sculptus during a molecular study of the bacterial diversity in this tick species (36). Therefore, the aim of the present study was to develop a PCR-based assay to screen for the presence of Rickettsiella in four species of tick from western Canada and to compare their DNA sequences to those of different species and pathotypes of Rickettsiella.
MATERIALS AND METHODS
DNA extraction and PCR.
A total of 469 ticks (Table 1) representing four species—Ixodes angustus, I. kingi, I. sculptus, and Dermacentor andersoni—were collected from small mammals at three localities in western Canada (37–39). Total genomic DNA (gDNA) was extracted and purified from each tick using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany), but with the modifications described previously (40, 41). PCR analyses were conducted to test for the presence of Rickettsiella DNA in the total gDNA of each tick. Initially, PCRs were conducted using the primers (RCL16S-211F and RCL16S-470R) and conditions described by Tsuchida et al. (17); however, no amplicons were produced for any sample. PCRs were then conducted using RCL16S-211F and a universal primer for the bacterial 16S rRNA gene (i.e., primer 802r [5′-ACTACCAGGGTATCTAATCCTG-3′]) (42), and the conditions of Tsuchida et al. (17), but with modifications to the number of cycles (n = 30) and annealing temperature (58°C). This PCR assay produced amplicons from the ticks tested; however, subsequent sequencing of representative PCR products revealed the presence of multiple bacterial species within each amplicon. The same problem was encountered when PCRs were conducted using the universal eubacterial primers (i.e., fD1 and rP2) (43) that have been used to amplify the 16S rRNA gene of Rickettsiella in other studies (see, for example, references 22, 31, and 33). As a consequence, two new primers, Rickella-F (5′-GTAGGAATCTGTCCTGGAG-3′) and Rickella-R2 (5′-TGCTTATTCTGTGGGTACCG-3′), were designed based on a comparison of all available nucleotide sequences available on GenBank, to specifically amplify ∼380 bp of the 16S rRNA gene of Rickettsiella. PCRs were performed in 25-μl volumes containing 2.5 μl of 10× iTaq PCR buffer (Bio-Rad), 3 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 25 pmol (1 μM) of each primer, 0.5 U of iTaq DNA polymerase (Bio-Rad)/μl, and 1.5 μl of gDNA template. A negative control (i.e., without gDNA) sample was included in each set of PCR assays. PCRs were performed in a thermocycler (iCycler; Bio-Rad, Hercules, CA) under the following conditions: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 5 min. Amplicons were subjected to electrophoresis on SYBR Safe-stained 1.5% agarose–TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.3]) gels, and their banding patterns were visualized by UV transillumination.
TABLE 1.
Numbers of larvae, nymphs, and adults of four tick species collected at different localities in western Canada that were PCR positive for Rickettsiella
| Locality and tick speciesa | Life cycle stage | No. tested | No. (%) PCR positive |
|---|---|---|---|
| Kootenay N.P., BC | |||
| Ixodes angustus | Larvae | 178 | 163 (92) |
| Nymphs | 68 | 55 (81) | |
| Adults | 24 | 20 (83) | |
| Dermacentor andersoni | Adults | 2 | 0 (0) |
| Beechy, SK | |||
| I. sculptus | Larvae | 33 | 16 (49) |
| Nymphs | 21 | 8 (38) | |
| Adults | 3 | 0 (0) | |
| I. kingi | Larvae | 1 | 0 (0) |
| Nymphs | 4 | 1 (25) | |
| Adults | 1 | 1 (100) | |
| D. andersoni | Nymphs | 20 | 0 (0) |
| Adults | 20 | 0 (0) | |
| Clavet, SK | |||
| I. sculptus | Nymphs | 4 | 2 (50) |
| I. kingi | Larvae | 82 | 0 (0) |
| Nymphs | 2 | 0 (0) | |
| Adults | 3 | 2 (66) | |
| D. andersoni | Larvae | 3 | 0 (0) |
BC, British Columbia, Canada; SK, Saskatchewan, Canada; N.P., National Park.
SSCP analyses.
All PCR-positive samples were prescreened for genetic variation using single-strand conformation polymorphism (SSCP) analyses (44) according to the protocol described previously (40). This mutation scanning technique can be used to distinguish among DNA sequences of 150 to 450 bp that differ by one or more nucleotides (44). Amplicons from eight PCR-positive ticks (i.e., two I. angustus, four I. kingi, and two I. sculptus), representing the three different SSCP banding patterns (i.e., profiles), were purified (40) and subjected to automated DNA sequencing using the primers Rickella-F and Rickella-R2 in separate reactions. This was performed to confirm that amplicons with the same SSCP profile have identical DNA sequences and that amplicons with different SSCP profiles differ in sequence by one or more nucleotides.
Characterization and phylogenetic analyses of the 16S rRNA gene.
The Rickettsiella spp. in the gDNA of two I. angustus, two I. kingi, and two I. sculptus were further characterized by amplifying a larger (∼1,270 bp) fragment of the 16S rRNA gene using the primers Rickella-F and 1387R-mod (5′-GGGCGGTGTGTACAAGGC-3′) (45). The same temperature conditions were used for the PCR as described above except that the duration of each phase was increased to 60 s. In addition, the MgCl2 concentration was reduced to 2.5 mM and the volume of gDNA template was increased to 2 μl. All amplicons were purified prior to DNA sequencing with the primers Rickella-F and 1387R-mod in separate reactions. BLAST searches (GenBank) were performed on the DNA sequence data. The DNA sequences of the Rickettsiella in each species of tick were aligned manually with the sequences of Rickettsiella available on GenBank (for accession numbers see Table S1 in the supplemental material). Phylogenetic analyses were performed by using the neighbor-joining (NJ) and maximum-parsimony (MP) methods in PAUP (46). For the MP analyses, characters were treated as unordered and were equally weighted, whereas alignment gaps were treated as “missing” characters. The sequence of the 16S rRNA gene of Coxiella burnetii was used as the outgroup for the MP analyses. Heuristic searches with TBR branch swapping were used to infer the shortest trees. The length, the consistency index (CI) excluding uninformative characters, and the retention index (RI) of each most parsimonious tree were recorded. Bootstrap analyses (1,000 replicates for the NJ analyses and 100 replicates for MP analyses) were conducted to determine the relative support for clades in the consensus trees.
Characterization and phylogenetic analyses of three protein-encoding genes.
The sequences of three additional genes, rpsA (30S ribosomal protein A), gidA (glucose inhibited cell division protein A), and sucB (dihydrolipoamide succinyl-transferase component E2), were also determined for the Rickettsiella in the gDNA of two I. angustus, two I. kingi, and two I. sculptus. PCRs were performed using the conditions and oligonucleotide primers described by Leclerque et al. (26). For each gene, the nucleotide and amino acid sequences were aligned manually with those of the other taxa within the genus Rickettsiella (for the GenBank accession numbers, see Tables S2 to S7 in the supplemental material), C. burnetii and Legionella pneumophila (accession numbers NC_002971 and NC_002942, respectively). Phylogenetic analyses (NJ and MP) were performed on both the nucleotide and amino acid sequence data for each gene and the concatenated data sets, using the sequences of Escherichia coli as the outgroup for the MP analyses.
Ethics statement.
This study was approved by the University of Saskatchewan's Animal Research Ethics Board and adhered to the Canadian Council on Animal Care guidelines for humane animal use.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene, rpsA, gidA, and sucB of the Rickettsiella spp. in I. angustus, I. kingi, and I. sculptus have been deposited in GenBank under accession numbers HF912419 to HF912421 and HG792868 to HG792876.
RESULTS
No amplicons were obtained for any of the 45 D. andersoni or the negative-control samples, whereas 268 (63%) of the 424 Ixodes individuals tested were PCR positive (Table 1). On TBE-agarose gels, all PCR-positive samples had a single band of the expected size (∼380 bp) for the partial fragment of the 16S rRNA gene amplified using primers Rickella-F and Rickella-R2. There was a significant difference (χ22 = 224.9, P < 0.001) in the proportions of I. angustus, I. sculptus, and I. kingi (i.e., 88%, 43%, and 4%, respectively) that were PCR positive for Rickettsiella. For I. angustus, there was no significant difference (χ22 = 5.94, P > 0.05) among life cycle stages in the proportions of individuals that were PCR positive for Rickettsiella (Table 1). Although none of the three I. sculptus adults were infected with Rickettsiella, there was no significant difference (χ22 = 0.37, P > 0.05) in the proportions of I. sculptus larvae and nymphs that were PCR positive for Rickettsiella (Table 1). Of the 95 I. kingi individuals screened for Rickettsiella DNA, some of the adult and nymphal ticks were PCR positive, whereas none of the 81 larvae was PCR positive. Furthermore, individuals of I. kingi and I. sculptus collected from both Clavet and Beechy (Saskatchewan [SK], Canada) were PCR positive for Rickettsiella.
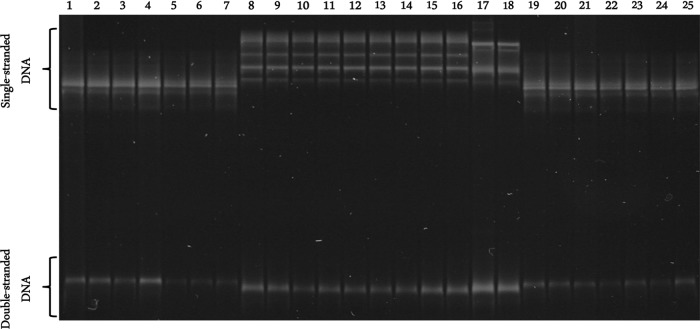
There were three different SSCP profiles among the 268 amplicons of the bacterial 16S rRNA gene (Fig. 1). The SSCP profiles of the amplicons from four I. kingi samples (two from Beechy and two from Clavet) were identical to one another but differed from those of all 26 amplicons derived from I. sculptus (24 from Beechy and two from Clavet). Similarly, there was no variation in SSCP profiles among the 238 amplicons from I. angustus. However, the profiles of each sample differed from those derived from I. sculptus and I. kingi. A comparison of the DNA sequences (340 bp) of representative samples of each SSCP profile type revealed that samples with identical banding patterns had identical sequences of the 16S rRNA gene, whereas those that differed in banding pattern differed by 3 to 22 bp in sequence. BLAST searches of the three sequence types revealed that they were most similar to the 16S rRNA gene sequences of species within the genus Rickettsiella, but each was unique compared to these sequences.
FIG 1.
SSCP profiles of representative amplicons of the 16S rRNA gene of Rickettsiella from the total gDNA of Ixodes angustus (lanes 1 to 7 and lanes 19 to 25), I. kingi (lanes 17 and 18), and I. sculptus (lanes 8 to 16).
Given the novel sequences of the Rickettsiella from I. angustus, I. sculptus, and I. kingi, comparisons were made for the sequence of a larger fragment (1,272 bp) of the 16S rRNA gene for six Rickettsiella-infected ticks (i.e., two I. angustus, two I. kingi, and two I. sculptus). There were 55 variable positions in the sequence alignment of the three taxa, representing 36 transitional (23 purine and 13 pyrimidine) changes, 15 transversional changes, two multiple mutational changes, and two indels (Table 2). The DNA sequences of the Rickettsiella in the two I. angustus were identical to one another but differed by 3.8% (i.e., 49 bp) from the Rickettsiella in the two I. kingi and from the Rickettsiella in the two I. sculptus. The DNA sequences of the Rickettsiella in I. kingi differed by 1.1% (i.e., 14 bp) from the Rickettsiella in the I. sculptus (Table 2). The DNA sequences of the Rickettsiella in I. angustus, I. kingi, and I. sculptus differed by 2.0 to 6.6% (i.e., 25 to 82 bp), 2.6 to 6.2% (i.e., 33 to 78 bp), and 2.5 to 6.0% (i.e., 32 to 75 bp), respectively, compared to the sequences of taxa within the genus Rickettsiella (see Table S1 in the supplemental material).
TABLE 2.
Variable nucleotide positions in the aligned 16S rRNA gene sequences of “Rickettsiella kingi,” “R. sculptus,” and “R. angustus” detected within three species of Ixodes in western Canada
| Species | Nucleotide positiona |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 35 | 52 | 64 | 68 | 82 | 84 | 87 | 116 | 124 | 165 | 223 | 240 | 281 | 320 | 322 | 328 | 330 | 331 | 332 | 335 | 337 | 350 | 405 | 452 | 509 | 512 | 696 | 706 | 715 | 719 | 725 | 726 | 871 | 875 | 876 | 877 | 878 | 888 | 889 | 894 | 908 | 909 | 985 | 997 | 1007 | 1009 | 1051 | 1056 | 1060 | 1115 | 1177 | 1135 | 1156 | |
| “R. kingi” | A | C | G | T | A | C | C | A | T | G | G | G | G | A | C | A | T | A | T | G | A | A | T | A | G | T | A | A | A | A | T | G | - | C | G | G | A | A | A | C | T | G | T | G | T | G | A | A | A | G | C | A | G | T | A |
| “R. sculptus” | . | T | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | G | . | . | A | C | G | . | . | C | . | A | - | . | A | . | . | G | . | . | . | A | . | . | . | A | . | . | . | A | . | G | . | . | . |
| “R. angustus” | C | T | A | C | - | A | A | G | G | T | A | A | A | C | A | G | C | T | C | T | G | G | C | G | A | C | . | C | G | T | C | . | A | A | . | T | G | . | G | T | C | T | C | A | C | . | T | T | G | . | T | G | A | C | T |
A dot indicates the same nucleotide as in the sequence of the “R. kingi,” and a hyphen indicates a nucleotide deletion.
The NJ analysis of the sequence data for the 16S rRNA gene revealed that the Rickettsiella in I. kingi represented the sister taxon to the Rickettsiella in I. sculptus with 100% statistical support (Fig. 2). There was also some statistical support (i.e., a bootstrap value of 77%) for both taxa belonging to a clade that contained “R. ixodidis” and R. grylli (ex Gryllus bimaculatus). In contrast, the Rickettsiella in I. angustus was placed external, with strong statistical support (80% bootstrap value), to a group comprising two clades; the first containing seven pathotypes of R. popilliae, and the second containing Rickettsiella ex Eisenia fetida and Rickettsiella ex Folsomia candida (Fig. 2). The single most-parsimonious tree (not shown) produced by the MP analysis of the sequence data (i.e., 158 cladistically informative characters) had a length of 607, a CI of 0.58 and a RI of 0.60. As with the NJ analysis, there was very strong statistical support (i.e., bootstrap value of 99%) for a sister taxa relationship between the Rickettsiella in I. kingi and I. sculptus in the MP tree, but no support for these taxa forming a clade with R. grylli (ex G. bimaculatus) and “R. ixodidis.” There was also no statistical support in the MP tree for the inclusion of the Rickettsiella in I. angustus in a clade with other taxa in the genus.
FIG 2.
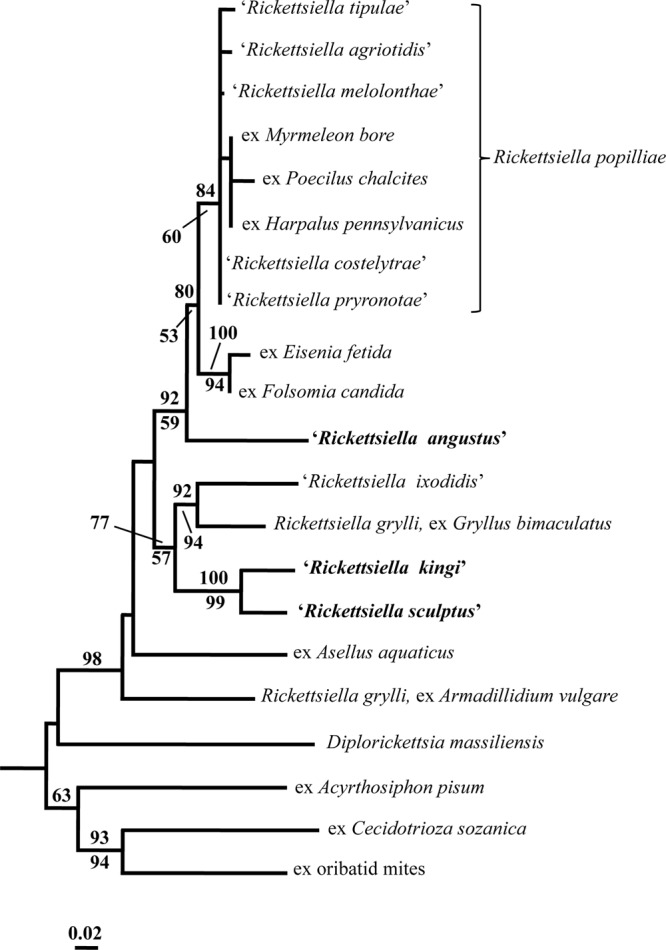
Neighbor-joining tree depicting the relationships of the 16S rRNA gene sequences of “Rickettsiella angustus,” “R. kingi,” “R. sculptus,” and other species and pathotypes of the genus Rickettsiella. The scale bar represents the inferred substitutions per nucleotide site. The relative support for clades in the tree produced from the NJ and MP analyses are indicated above and below branches, respectively.
The Rickettsiella in I. angustus, I. sculptus, and I. kingi each had novel nucleotide and amino acid sequences for three additional genes; rpsA, gidA, and sucB (see Tables S2 to S7 in the supplemental material). The nucleotide sequences of all three genes for the Rickettsiella in I. kingi and I. sculptus were more similar to one another (96.0 to 98.1%) than they were to the nucleotide sequences of the Rickettsiella in I. angustus (82.6 to 90.2%) (Table 3). The extent of the nucleotide differences in DNA sequence of the three genes between the Rickettsiella in I. angustus and the Rickettsiella in I. kingi and I. sculptus were greater than those between different pathotypes of R. popilliae and were of a similar magnitude between different species of Rickettsiella (i.e., between R. popilliae and the Rickettsiella in I. woodi) (see Tables S2, S3, and S5 in the supplemental material). The magnitude of the nucleotide differences in DNA sequence of all three genes between the Rickettsiella in I. kingi and I. sculptus were similar to that among some pathotypes of R. popilliae (see Tables S2, S3, and S5 in the supplemental material). Similarly, the differences in amino acid sequence for all three genes between the Rickettsiella in I. angustus and the Rickettsiella in I. kingi and I. sculptus (Table 4) were of a similar magnitude between all pathotypes of R. popilliae and the Rickettsiella in I. woodi (see Tables S3, S5, and S7 in the supplemental material). The amino acid differences in sequence of gidA and sucB between the Rickettsiella in I. kingi and I. sculptus were of a similar magnitude to the differences among some pathotypes of R. popilliae, whereas the number of amino acid differences in the sequences of rpsA for the Rickettsiella in I. kingi and I. sculptus (n = 6) was greater than that among pathotypes of R. popilliae (n = 0 to 2) (see Tables S3, S5, and S6 in supplemental material).
TABLE 3.
Pairwise comparisons of the numbers of nucleotide differences and percent similarities in DNA sequences of three genes (rpsA, gidA, and sucB) between Rickettsiella spp. in three species of ixodid tick: Ixodes angustus, I. kingi, and I. sculptus
| Tick host | Pairwise comparisonsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
rpsA |
gidA |
sucB |
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| 1. I. angustus | 85.6 | 90.2 | 86.7 | 86.3 | 82.6 | 83.0 | |||
| 2. I. kingi | 93 | 97.5 | 105 | 98.1 | 162 | 96.0 | |||
| 3. I. sculptus | 88 | 22 | 108 | 15 | 158 | 37 | |||
Pairwise comparisons show the numbers of amino acid differences (lower diagonals for each gene) and the percent similarities (upper diagonals for each gene) in the DNA sequences of three genes (rpsA [894 bp], gidA [787 bp], and sucB [928 to 931 bp]). The numbers in column 1 correspond to the comparison number subheadings.
TABLE 4.
Pairwise comparisons of the numbers of amino acid differences and percent similarities in DNA sequences of three genes (rpsA, gidA, and sucB) between Rickettsiella in three species of ixodid tick: Ixodes angustus, I. kingi, and I. sculptus
| Tick host | Pairwise comparisonsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
rpsA |
gidA |
sucB |
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| 1. I. angustus | 95.3 | 96.6 | 91.6 | 92.0 | 84.8 | 81.2 | |||
| 2. I. kingi | 14 | 98.0 | 22 | 98.1 | 47 | 93.9 | |||
| 3. I. sculptus | 10 | 6 | 20 | 5 | 46 | 19 | |||
Pairwise comparisons show the numbers of amino acid differences (lower diagonals for each gene) and the percent similarity (upper diagonals for each gene) in the DNA sequences of three genes (rpsA [encoding 298 amino acids], gidA [encoding 262 amino acids], and sucB [encoding 309 to 310 amino acids]). The numbers in column 1 correspond to the comparison number subheadings.
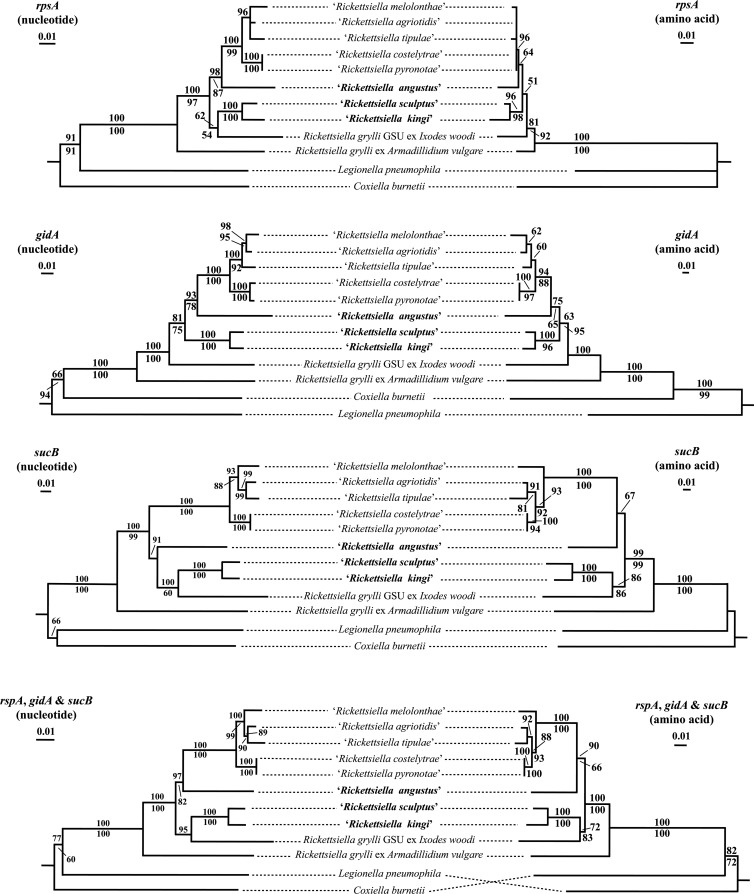
The results of the phylogenetic analyses conducted on the nucleotide and amino acid sequence data of three genes (rpsA, gidA and sucB) and on the concatenated sequence data of these genes are shown in Fig. 3. In all trees, except for the MP tree of the rpsA amino acid sequence data, there was strong to total statistical support (i.e., bootstrap values of 88 to 100%) for the five pathotypes of R. popilliae (i.e., “R. melolonthae,” “R. agriotidis,” “R. tipulae,” “R. costelytrae,” and “R. pyronotae”) forming a monophyletic clade (Fig. 3). In addition, NJ and MP analyses of the nucleotide and amino acid sequence data for each gene and for the concatenated data revealed a sister taxa relationship between the Rickettsiella in I. kingi and I. sculptus with 96 to 100% statistical support. In some phylogenetic analyses, there was some statistical support (i.e., bootstrap values of 72 to 100%) for the Rickettsiella in I. kingi and I. sculptus forming a clade with R. grylli GSU ex I. woodi, whereas in other analyses they were placed on a branch external to the different pathotypes of R. popilliae and the Rickettsiella in I. angustus. In most NJ and MP analyses, there was strong statistical support (i.e., bootstrap values 87 to 98%) for a sister taxa relationship between the Rickettsiella in I. angustus and the different pathotypes of R. popilliae. The exception to this was in the NJ tree produced from the nucleotide sequence data of sucB, where there was strong support (i.e., bootstrap value of 91%) for the Rickettsiella in I. angustus representing the sister taxon to the Rickettsiella in I. kingi and I. sculptus (Fig. 3).
FIG 3.
Phylogenetic trees depicting the relationships of “‘Rickettsiella angustus,” “R. kingi,” “R. sculptus,” and other species and pathotypes of the genus Rickettsiella based on NJ analyses of the nucleotide and amino acid sequences of the rpsA, gidA, and sucB genes, and the concatenation of the sequence data for these three genes. The scale bar represents the inferred substitutions per nucleotide site. The relative support for clades in the tree produced from the NJ and MP analyses are indicated above and below branches, respectively.
DISCUSSION
A PCR assay, utilizing genus-specific primers, was used to test for the presence of the Rickettsiella 16S rRNA gene in the total gDNA of 469 individual ticks representing four species collected from three different localities in western Canada: Kootenay National Park (British Columbia), Beechy (SK), and Clavet (SK). A total of 268 ticks (i.e., 23 adults, 66 nymphs, and 179 larvae), all Ixodes sp., were PCR positive for Rickettsiella using our PCR assay. None of the 45 D. andersoni tested were infected with Rickettsiella, even though many of these ticks were found parasitizing the same host individuals as I. sculptus (at Beechy) or I. angustus (at Kootenay National Park) that were found to contain Rickettsiella. This suggests that there was no transfer of Rickettsiella to D. andersoni individuals feeding on the same small mammal host as ticks infected with Rickettsiella. Interestingly, those Rickettsiella detected in ticks thus far have only been found in members of the genus Ixodes: I. ricinus in the Palearctic (34, 35), I. woodi in the Nearctic (31) and I. tasmani in Australia (33). The results of the present study also detected Rickettsiella DNA in three species of Ixodes: I. angustus, I. sculptus, and I. kingi. All feeding life cycle stages (i.e., larvae, nymphs, and adults) of I. angustus were infected with Rickettsiella, whereas for I. kingi, all but the larval stage contained the bacterium, and for I. sculptus, only the larvae and nymphs contained Rickettsiella. However, this finding may be a consequence of the small sample sizes examined for some of the life cycle stages and/or a low prevalence of Rickettsiella in the ticks (e.g., in I. kingi). There were significant differences among the three species of Ixodes in the proportions of individuals infected with Rickettsiella. A majority (88%) of the 270 I. angustus individuals were infected with Rickettsiella, compared to 43% of the 61 I. sculptus individuals and four (4%) of the 95 I. kingi individuals tested. The PCR-positive ticks included I. sculptus and I. kingi individuals collected from both Beechy and Clavet.
The amplicons of the 16S rRNA gene (∼380 bp) for the 268 PCR-positive samples were compared using SSCP. This mutation scanning technique has been used effectively to differentially display genetic variation between DNA sequences that are 150 to 450 bp in size and that differ by one or more nucleotides (see, for example, references 40, 41, 47, and 48). Three different SSCP profiles were detected among the 268 Rickettsiella amplicons, each profile type associated with a different tick species. DNA sequencing of representative samples confirmed that those with the same SSCP profile had identical sequences of the 16S rRNA gene, whereas those with different SSCP profiles differed in sequence. The sequences of the partial (340 bp) 16S rRNA gene for the Rickettsiella in I. kingi differed from the Rickettsiella in the I. sculptus by 3 bp, while both taxa differed from the Rickettsiella in I. angustus by 22 bp. A larger number of nucleotide differences (i.e., 14 to 49 bp; 1.1 to 3.8%) were detected among the Rickettsiella from the three species of Ixodes when a much larger fragment (1,272 bp) of the 16S rRNA gene was analyzed. BLAST searches of the sequence data showed that each taxon was closest in sequence to a member of the genus Rickettsiella; however, the Rickettsiella in I. angustus, I. sculptus, and I. kingi each had novel sequences of the 16S rRNA gene compared to those of all recognized species and pathotypes of Rickettsiella. Similarly, the Rickettsiella in I. angustus, I. sculptus, and I. kingi each had novel nucleotide and amino acid sequences for three additional genes: rpsA, gidA, and sucB. Given this, we propose to provisionally name the Rickettsiella spp. in I. angustus, I. sculptus, and I. kingi as “Rickettsiella angustus,” “Rickettsiella sculptus,” and “Rickettsiella kingi,” respectively, in accordance with the nomenclature used in other studies (see, for example, references 26 and 27).
Phylogenetic analyses of the sequence data for the 16S rRNA gene revealed that “R. angustus” was placed external to a clade, comprising two groups; the first of which contained seven pathotypes of R. popilliae (i.e., “R. tipulae,” “R. agriotidis,” “R. melolonthae,” “R. costelytrae,” Rickettsiella in Myrmeleon bore, Rickettsiella in Poecilus chalcites, and Rickettsiella in Harpalus pennsylvanicus), and the second of which contained the Rickettsiella spp. in the earthworm, Eisenia fetida, and in the springtail, Folsomia candida. The magnitude of sequence differences between “R. angustus” and other members of the genus, including “R. sculptus” and “R. kingi,” ranged from 25 to 82 bp (2.0 to 6.6%), which is greater than the differences (i.e., 1 to 7 bp; 0.1 to 0.6%) among seven pathotypes of R. popilliae. In addition, phylogenetic analyses of the nucleotide and amino acid sequence data for the rpsA, gidA, and sucB genes revealed a similar topology, with high bootstrap support for both the NJ and MP analyses. For each of these genes, the magnitude of the sequence differences between “R. angustus” and “R. sculptus”/“R. kingi” were greater than those among isolates from recognized species of Rickettsiella. This suggests that “R. angustus” represents a new species of Rickettsiella based on the results of the phylogenetic analyses and the magnitude of differences in the sequences of the four genes compared to other members of the genus.
The results of the phylogenetic analyses for all four genes (i.e., 16S rRNA, rpsA, gidA, and sucB) revealed that “R. kingi” and “R. sculptus” were sister taxa. In some analyses these two taxa formed a clade with R. grylli (a pathogen of the cricket, Gryllus bimaculatus) and/or “R. ixodidis” in the tick I. woodi, whereas in others they were positioned on a branch external to the different pathotypes of R. popilliae and “R. angustus.” These results suggest that “R. sculptus” and “R. kingi” are not pathotypes of any recognized species of Rickettsiella. However, given that species delineation within the genus Rickettsiella is controversial (30), either “R. sculptus” and “R. kingi” may represent different pathotypes of a single new species or each may represent a distinct Rickettsiella species. There was a significant difference in the proportions of I. sculptus and I. kingi individuals infected with Rickettsiella. Furthermore, “R. sculptus” and “R. kingi” were only detected in individuals of I. sculptus and I. kingi (respectively), and there was no evidence of cross-transmission of the Rickettsiella in I. sculptus to I. kingi, or vice versa, even though there were instances of infected ticks of both species feeding on the same small mammal hosts at two localities (∼200 km apart) in Saskatchewan. The sequences of the 16S rRNA gene for “R. sculptus” and “R. kingi” differed from those of other Rickettsiella by 32 to 78 bp (2.5 to 6.2%), which is similar to or exceeds the sequence differences (i.e., ∼3%) among closely related species of bacteria (49). Moreover, the number of differences in the sequences of the 16S rRNA gene of “R. sculptus” and “R. kingi” (i.e., 14 bp over an alignment length of 1,255 bp) was greater than that among different pathotypes of R. popilliae (i.e., 1 to 7 bp). Although “R. sculptus” and “R. kingi” had novel nucleotide and amino acid sequences for three other genes (rpsA, gidA, and sucB), the magnitude of the sequence differences between these two taxa for two of these genes (gidA and sucB) were of a similar magnitude to that among pathotypes of R. popilliae. Nonetheless, the number of differences in amino acid sequence of rpsA between “R. sculptus” and “R. kingi” (n = 6) was greater than that among the different pathotypes of R. popilliae (n = 0 to 2). These combined results suggest that “R. sculptus” and “R. kingi” may each represent a distinct species within the genus Rickettsiella; however, this requires further investigation.
In conclusion, three novel Rickettsiella were detected in the total gDNA of three species of Ixodes in North America that use small mammals as hosts. More work is needed to determine whether these putative new species of Rickettsiella have pathogenic or beneficial effects on their tick hosts, as has been shown for other members of the genus (see, for example, references 13 and 17) and whether other species of Ixodes in North America are hosts for Rickettsiella.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this study was provided (to N.B.C.) by the Natural Sciences and Engineering Research Council of Canada and the Canadian Foundation for Innovation. A University of Saskatchewan Graduate Scholarship provided financial support to C.A.A.
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03564-13.
REFERENCES
- 1.Jeyaprakash A, Hoy MA, Allsopp MH. 2003. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J. Invertebr. Pathol. 84:96–103. 10.1016/j.jip.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 2.Campbell CL, Mummey DL, Schmidtmann ET, Wilson WC. 2004. Culture-independent analysis of midgut microbiota in the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae). J. Med. Entomol. 41:340–348. 10.1603/0022-2585-41.3.340 [DOI] [PubMed] [Google Scholar]
- 3.Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudo T. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590–6599. 10.1128/AEM.71.11.6590-6599.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinert LA, Tinsley MC, Temperley M, Jiggins FM. 2007. Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biol. Lett. 3:678–681. 10.1098/rsbl.2007.0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgdorfer W, Hayes SF, Mavros AJ. 1981. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii, p 585–594 In Burgdorfer W, Anacker RL. (ed), Rickettsiae and rickettsial diseases. Academic Press, Inc, New York, NY [Google Scholar]
- 6.Graf J, Kikuchi Y, Rio RV. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365–371. 10.1016/j.tim.2006.06.009 [DOI] [PubMed] [Google Scholar]
- 7.McGraw EA, Merritt DJ, Droller JN, O'Neill SL. 2002. Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. 99:2918–2923. 10.1073/pnas.052466499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turley AP, Moreira LA, O'Neill SL, McGraw EA. 2009. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 3:e516. 10.1371/journal.pntd.0000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilly K, Rosa PA, Stewart PE. 2008. Biology of infection with Borrelia burgdorferi. Infect. Dis. Clin. N. Am. 22:217–234. 10.1016/j.idc.2007.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, Llinás M. 2009. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5:191–199. 10.1016/j.chom.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordaux R, Paces-Fessy M, Raimond M, Michel-Salzat A, Zimmer M, Bouchon D. 2007. Molecular characterization and evolution of arthropod-pathogenic Rickettsiella bacteria. Appl. Environ. Microbiol. 73:5045–5047. 10.1128/AEM.00378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux V, Bergoin M, Lamaze N, Raoult D. 1997. Reassessment of the taxonomic position of Rickettsiella grylli. Int. J. Syst. Bacteriol. 47:1255–1257. 10.1099/00207713-47-4-1255 [DOI] [PubMed] [Google Scholar]
- 13.Dutky SR, Gooden EL. 1952. Coxiella popilliae, n. sp., a Rickettsia causing blue disease of Japanese beetle larvae. J. Bacteriol. 63:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federici BA. 1980. Reproduction and morphogenesis of Rickettsiella chironomi, an unusual intracellular procaryotic parasite of midge larvae. J. Bacteriol. 143:995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federici BA. 1984. Diseases of terrestrial isopods. Symp. Zool. Soc. Lond. 53:233–245 [Google Scholar]
- 16.Adamo SA. 1998. The specificity of behavioral fever in the cricket Acheta domesticus. J. Parasitol. 84:529–533. 10.2307/3284717 [DOI] [PubMed] [Google Scholar]
- 17.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon J-C, Fukatsu T. 2010. Symbiotic bacterium modifies aphid body color. Science 330:1102–1104. 10.1126/science.1195463 [DOI] [PubMed] [Google Scholar]
- 18.Philip CB. 1956. Comments on the classification of the order Rickettsiales. Can. J. Microbiol. 2:261–270. 10.1139/m56-030 [DOI] [PubMed] [Google Scholar]
- 19.Weiss E, Dasch GA, Chang K-P. 1984. Genus VIII. Rickettsiella Philip 1956, p 713–717 In Krieg NR, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 1 The Williams & Wilkins Co, Baltimore, MD [Google Scholar]
- 20.Fournier P-E, Raoult D. 2005. Genus II. Rickettsiella Philip 1956, 267AL, p 241–247 In Garrity GM, Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2, part B Springer, New York, NY [Google Scholar]
- 21.Leclerque A. 2008. Whole genome-based assessment of the taxonomic position of the arthropod pathogenic bacterium Rickettsiella grylli. FEMS Microbiol. Lett. 283:117–127. 10.1111/j.1574-6968.2008.01158.x [DOI] [PubMed] [Google Scholar]
- 22.Leclerque A, Kleespies RG. 2008. 16S rRNA-, GroEL-, and MucZ-based assessment of the taxonomic position of ‘Rickettsiella melolonthae' and its implications for the organization of the genus Rickettsiella. Int. J. Syst. Evol. Microbiol. 58:749–755. 10.1099/ijs.0.65359-0 [DOI] [PubMed] [Google Scholar]
- 23.Leclerque A, Kleespies RG. 2008. Genetic and electron-microscopic characterization of Rickettsiella tipulae, an intracellular bacterial pathogen of the crane fly, Tipula paludosa. J. Invertebr. Pathol. 98:329–334. 10.1016/j.jip.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 24.Mediannikov O, Sekeyová Z, Birg M-L, Raoult D. 2010. A novel obligate intracellular gamma-proteobacterium associated with ixodid ticks, Diplorickettsia massiliensis, gen. nov., sp. nov. PLoS One 7:e11478. 10.1371/journal.pone.0011478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleespies RG, Marshall SDG, Schuster C, Townsend RJ, Jackson TA, Leclerque A. 2011. Genetic and electron-microscopic characterization of Rickettsiella bacteria from the manuka beetle, Pyronota setosa (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 107:206–211. 10.1016/j.jip.2011.05.017 [DOI] [PubMed] [Google Scholar]
- 26.Leclerque A, Kleespies RG, Ritter C, Schuster C, Feiertag S. 2011. Genetic and electron-microscopic characterization of ‘Rickettsiella agriotidis', a new Rickettsiella pathotype associated with wireworm, Agriotes sp. (Coleoptera: Elateridae). Curr. Microbiol. 63:158–163. 10.1007/s00284-011-9958-5 [DOI] [PubMed] [Google Scholar]
- 27.Leclerque A, Kleespies RG. 2012. A Rickettsiella bacterium from the hard tick, Ixodes woodi: molecular taxonomy combining multilocus sequence typing (MLST) with significance testing. PLoS One 7:e38062. 10.1371/journal.pone.0038062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leclerque A, Kleespies RG, Schuster C, Richards NK, Marshall SDG, Jackson TA. 2012. Multilocus sequence analysis (MLSA) of ‘Rickettsiella costelytrae' and ‘Rickettsiella pyronotae', intracellular bacterial entomopathogens from New Zealand. J. Appl. Microbiol. 113:1128–1237. 10.1111/j.1365-2672.2012.05419.x [DOI] [PubMed] [Google Scholar]
- 29.Schuster C, Kleespies RG, Ritter C, Feiertag S, Leclerque A. 2012. Multilocus sequence analysis (MLSA) of ‘Rickettsiella agriotidis', an intracellular bacterial pathogen of Agriotes wireworms. Curr. Microbiol. 66:1–9. 10.1007/s00284-012-0219-z [DOI] [PubMed] [Google Scholar]
- 30.Bouchon D, Cordaux R, Grève P. 2012. Chapter 7: Rickettsiella, intracellular pathogens of arthropods, p 127–148 In Zchori-Fein E, Bourtzis K. (ed), Manipulative tenants: bacteria associated with arthropods. CRC Press, Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- 31.Kurtti TJ, Palmer AT, Oliver JH., Jr 2002. Rickettsiella-like bacteria in Ixodes woodi (Acari: Ixodidae). J. Med. Entomol. 39:534–540. 10.1603/0022-2585-39.3.534 [DOI] [PubMed] [Google Scholar]
- 32.Šut'áková G, Řeháček J. 1990. Mixed infection of Rickettsiella phytoseiuli and Coxiella burnetii in Dermacentor reticulatus female ticks: electron microscope study. J. Invertebr. Pathol. 55:407–416. 10.1016/0022-2011(90)90085-K [DOI] [PubMed] [Google Scholar]
- 33.Vilcins I-ME, Old JM, Deane E. 2009. Molecular detection of Rickettsia, Coxiella, and Rickettsiella DNA in three native Australian tick species. Exp. Appl. Acarol. 49:229–242. 10.1007/s10493-009-9260-4 [DOI] [PubMed] [Google Scholar]
- 34.Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, Tomsho LP, Drautz DI, Rizzoli A, Schuster SC. 2011. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6:e25604. 10.1371/journal.pone.0025604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tveten A-K, Sjåstad KK. 2011. Identification of bacteria infecting Ixodes ricinus ticks by 16S rDNA amplification and denaturing gradient gel electrophoresis. Vector Borne Zoonotic Dis. 11:1329–1334. 10.1089/vbz.2011.0657 [DOI] [PubMed] [Google Scholar]
- 36.Anstead CA. 2013. Comparison of the ticks and tick-borne bacteria of small mammals in western Canada. Ph.D. thesis University of Saskatchewan, Saskatoon, Saskatchewan, Canada [Google Scholar]
- 37.Anstead CA, Chilton NB. 2011. Ticks feeding on northern pocket gophers (Thomomys talpoides) in central Saskatchewan and the unexpected detection of Ixodes scapularis larvae. J. Vector Ecol. 36:355–360. 10.1111/j.1948-7134.2011.00176.x [DOI] [PubMed] [Google Scholar]
- 38.Anstead CA, Hwang YT, Chilton NB. 2013. Ticks (Acari: Ixodidae) on small mammals in Kootenay National Park, British Columbia, Canada. J. Med. Entomol. 50:1208–1214. 10.1603/ME13067 [DOI] [PubMed] [Google Scholar]
- 39.Anstead CA, Wallace S, Chilton NB. 2014. Mutation scanning-based identification of larval and nymphal ticks (Acari: Ixodidae) from Richardson's ground squirrels (Spermophilus richardsonii). Mol. Cell. Probes 28:6–9. 10.1016/j.mcp.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 40.Dergousoff SJ, Chilton NB. 2007. Differentiation of three species of ixodid tick, Dermacentor andersoni, D. variabilis, and D. albipictus, by PCR-based approaches using markers in ribosomal DNA. Mol. Cell. Probes 21:343–348. 10.1016/j.mcp.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 41.Anstead CA, Chilton NB. 2013. Detection of a novel Rickettsia (Alphaproteobacteria: Rickettsiales) in rotund ticks (Ixodes kingi) from Saskatchewan, Canada. Ticks Tick-Borne Dis. 4:202–206. 10.1016/j.ttbdis.2012.11.013 [DOI] [PubMed] [Google Scholar]
- 42.Dergousoff SJ. 2011. Comparison of the bacteria within ticks from allopatric and sympatric populations of Dermacentor andersoni and Dermacentor variabilis near their northern distributional limits in Canada. Ph.D. thesis University of Saskatchewan, Saskatoon, Saskatchewan, Canada [Google Scholar]
- 43.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasser RB, Hu M, Chilton NB, Campbell BE, Jex AJ, Otranto D, Cafarchia C, Beveridge I, Zhu X. 2006. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 1:3121–3128 [DOI] [PubMed] [Google Scholar]
- 45.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 47.Dergousoff SJ, Gajadhar AJA, Chilton NB. 2009. Prevalence of Rickettsia species in Canadian populations of Dermacentor andersoni and D. variabilis. Appl. Environ. Microbiol. 75:1786–1789. 10.1128/AEM.02554-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dergousoff SJ, Chilton NB. 2012. Association of different genetic types of Francisella-like organisms with the Rocky Mountain wood tick (Dermacentor andersoni) and the American dog tick (Dermacentor variabilis) in localities near their northern distributional limits. Appl. Environ. Microbiol. 78:965–971. 10.1128/AEM.05762-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition of bacteriology. Int. J. Syst. Bacteriol. 44:846–849. 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.