Abstract
Single-cell oil (SCO) represents a sustainable alternative for the oil industry. Accordingly, the identification of microorganisms with either higher lipidogenic ability or novel capacities for the transformation of raw materials constitutes a major challenge for the field of oil biotechnology. With this in mind, here, we were prompted to address the lipidogenic profile of the filamentous hemiascomycete Ashbya gossypii, which is currently used for the microbial production of vitamins. We found that A. gossypii mostly accumulates unsaturated fatty acids (FAs), with more than 50% of the total FA content corresponding to oleic acid. In addition, we engineered A. gossypii strains both lacking the beta-oxidation pathway and also providing ATP-citrate lyase (ACL) activity to block the degradation of FA and to increase the cytosolic acetyl-coenzyme A (CoA) content, respectively. The lipidogenic profile of the newly developed strains demonstrates that the mere elimination of the beta-oxidation pathway in A. gossypii triggers a significant increase in lipid accumulation that can reach 70% of cell dry weight. The use of A. gossypii as a novel and robust tool for the production of added-value oils is further discussed.
INTRODUCTION
The oil industry sustains the increasing demand for most carbon-based compounds, such as fuels, lubricants, functional polymers, and other high-value fine chemicals. In addition, some feed supplements, colorants, and nutraceuticals, such as carotenoids and polyunsaturated fatty acids (PUFAs), are also derived from lipids (1). Crude oil, as well as plant oil and animal fat, is the main source of oil for industry. However, fossil oil supplies are limited, and their use involves negative environmental consequences. Moreover, the use of oilseeds and animal fat for nonfood applications results in competition with food, higher prices, and other economic and environmental concerns (2–4).
In this context, microbial oil—also referred to as single-cell oil (SCO)—represents a sustainable alternative feedstock for the oleochemical industry. SCO accumulated by oleaginous microorganisms may have several advantages over other oil resources: the fermentative processes are independent of climate and, more importantly, the use of waste products as substrates avoids competition with edible resources and makes the process environmentally friendly (4, 5). In addition, the application of “white biotechnology” (i.e., industrial biotechnology) to fermentation processes allows strain design for the production of many different high-value oleochemicals (2, 4, 6).
An oleaginous microorganism is defined by its ability to accumulate more than 20 to 25% of the cell dry weight as lipid content. Additionally, it has been reported than some oleaginous yeasts are able to reach a level of lipid accumulation of up to 70 to 80% of their cell dry weight in oil-containing media (4, 7, 8).
The concept of lipid accumulation denotes a metabolic imbalance between lipid biosynthesis and degradation. Fatty-acid (FA) biosynthesis starts with the carboxylation of acetyl-coenzyme A (CoA) by acetyl-CoA carboxylase (ACC) to form malonyl-CoA. Next, the FA synthase (FAS) multienzymatic complex catalyzes the elongation of the acyl-CoA chain by condensing malonyl-CoA molecules and acetyl-CoA (8). Therefore, acetyl-CoA is the essential donor molecule for FA biosynthesis, and it has been reported that a continuous supply of cytosolic acetyl-CoA is crucial for lipid accumulation. Indeed, the presence of the enzyme ATP-citrate lyase (ACL), which catalyzes the formation of acetyl-CoA from citrate in the cytosol, is considered a common hallmark of all oleaginous microorganisms (8, 9). Unlike FA biosynthesis, beta-oxidation is the principal catabolic pathway for the degradation of FA and hence represents a competing pathway for lipid accumulation (10, 11). Beta-oxidation comprises a four-step oxidative cycle that removes two carbons (an acetyl-CoA molecule) from the acyl-CoA chain in each cycle. The pathway is present in all eukaryotes, although there is significant diversity in terms of substrate specificity, enzyme architecture, and subcellular localization among the different organisms (11).
Several strategies have been developed to enhance the accumulation of lipids in oleaginous microorganisms (10, 12, 13). Metabolic flux channeling toward lipid accumulation has mainly been succeeded by two different strategies: (i) increasing the cytosolic pool of acetyl-CoA by the overexpression of genes encoding FA synthesis-promoting activities, such as the ACL genes, and (ii) blocking the principal FA-degrading activities by deleting the genes controlling FA beta-oxidation (10, 12–15).
Ashbya gossypii is a filamentous hemiascomycete that belongs to the family Saccharomycetaceae and that naturally overproduces riboflavin (vitamin B2). A. gossypii has been considered a paradigm of sustainable white biotechnology through its use in the industrial overproduction of riboflavin and other vitamins (16). Indeed, we have previously described metabolically engineered strains of A. gossypii that are able to produce 10-fold more riboflavin than a wild-type (WT) strain (17–19), and similar results have been achieved for the production of folic acid (J. L. Revuelta, unpublished results). The fermentation processes using A. gossypii have several advantages, such as the mycelial autolysis that occurs in late stationary phase, which avoids costly recovery steps, and its ability to grow in different oil sources and oil-containing wastes (16, 20).
According to general criteria, A. gossypii is considered to be a nonoleaginous microorganism (9). However, it has been reported that A. gossypii is able to accumulate between 10 and 20% of the cell dry weight as lipid bodies and, moreover, 54% of the total FA composition corresponded to the essential unsaturated omega-6 linoleic acid when soybean oil was used as the sole carbon source (21). In contrast to most bona fide oleaginous microorganisms, A. gossypii lacks any ACL-coding gene, and hence, cytosolic acetyl-CoA is provided solely by acetyl-CoA synthase (ACS) activity. Furthermore, A. gossypii has only one FA beta-oxidation pathway with a unique acyl-CoA oxidase gene (AER358C-POX1) that controls the first beta-oxidation reaction. Conversely, most oleaginous microorganisms, such as Yarrowia lipolytica or Mucor circinelloides, show two or three beta-oxidation pathways in several different cellular compartments and up to six different acyl-CoA oxidase-encoding genes (9, 15).
In the present study, we wished to investigate the potential ability of A. gossypii to accumulate significant amounts of SCOs in a biotechnologically feasible process. We show that the metabolic engineering of A. gossypii allows the isolation of oleaginous strains that are able to accumulate up to 70% of their biomass as lipid content. Additionally, the role of ACL genes as an essential feature of oleaginous microorganisms is examined. In sum, we report a novel microbial tool for the production of higher-added-value lipids by the rational design of metabolic engineering manipulations.
MATERIALS AND METHODS
A. gossypii strains, media, and growth conditions.
The A. gossypii ATCC 10895 strain was used and was considered a wild-type strain. The other A. gossypii strains used in the study are listed in Table S1 in the supplemental material. A. gossypii was cultured at 28°C using MA2 rich medium, synthetic complete (SC) medium, or synthetic minimal medium lacking leucine (SC−leu) (17). For lipid accumulation analyses, the carbon source of the MA2 medium was either 8% glucose or 1% glucose plus 2% oleic acid (Sigma), previously emulsified by sonication in the presence of 0.02% Tween 40 (22). Cultures with oil were centrifuged at 10,000 × g for 10 min, and the resulting cell pellet was washed three times with equal volumes of SB solution (9 g/liter NaCl in 0.5% bovine serum albumin [BSA]) (23). A. gossypii transformation, sporulation conditions, and spore isolation have been described elsewhere (24). Briefly, DNA was introduced into A. gossypii germlings by electroporation, and primary heterokaryon transformants were isolated in selective medium. Homokaryon transformant clones were obtained by sporulation of the primary transformants and isolation on antibiotic-containing plates. Genomic DNA and RNA isolation was carried out as previously described (17). Concentrations of 250 mg/liter for Geneticin (G418) (Gibco-BRL) and 100 mg/liter for nourseothricin (cloNAT; Werner Bioagents) were used where indicated. Liquid cultures were initiated either from mycelial overnight preinocula or with 1 × 106 spores per liter of medium and were performed on a rotary shaker at 200 rpm.
Lipid extraction and quantification by GC-MS.
Fatty acid methyl esters (FAMEs) were extracted and transmethylated from freeze-dried A. gossypii biomass using a modification of the method described by Bligh and Dyer (25). One hundred micrograms of lyophilized mycelia was mixed with 1 ml of hexane (Sigma) and 750 μl of 2% sulfuric acid in methanol under a nitrogen atmosphere. The samples were incubated at 100°C for 90 min. Then, the samples were cooled on ice, and the transmethylation reaction was stopped by the addition of 0.5 ml of distilled water. The upper phase was recovered by centrifugation, and the extraction step was repeated by adding 0.5 ml of hexane to the lower phase. The two hexane-soluble phases were mixed and evaporated with nitrogen. The FAMEs were resuspended in 100 μl of hexane and used for gas chromatography-mass spectrometry (GC-MS) analysis.
GC-MS was carried out using an Agilent 7890A gas chromatograph with an Agilent MS200 mass spectrometer. A VF50 column (30 m long, 0.25-mm internal diameter, and 25-μm film) was used. The conditions for analysis were as follows: helium was used as the carrier at 1 ml/min, with a split ratio of 1:20. The oven program was as follows: an initial temperature of 90°C for 5 min, a ramp of 12°C/min up to 190°C, and a ramp of 4°C/min up to 290°C. Mass spectrometric detection was from 50 to 400 Da. FAs were identified by comparison with commercial fatty acid methyl ester standards (FAME32; Supelco) and quantified by an internal-standard method using 50 μg of C17:0 (Sigma).
Gene deletion and gene overexpression.
For gene deletions, a loxP-KanMX-loxP or a loxP-NatMX-loxP replacement cassette flanked by recombinogenic sequences for the corresponding target gene was used (data not shown). The loxP repeated inverted sequences enabled the selection marker to be eliminated and subsequently reused by expressing a Cre recombinase, as described elsewhere (26, 27). The selection markers with recombinogenic flanks for the deletion of POX1 (loxP-KanMX-loxP module for G418 resistance) and FOX2 (loxP-NatMX-loxP module for nourseothricin resistance) were obtained using the primers listed in Table S2 in the supplemental material. Genomic integration of the deletion modules was confirmed by analytical PCR and DNA sequencing (data not shown). For the heterologous overexpression of the ACL genes from Y. lipolytica (http://genolevures.org/yali.html#), each open reading frame (ORF) (YALI0E34793g and YALI0D24431g, here referred to as ACL1 and ACL2, respectively) was PCR amplified from Y. lipolytica genomic DNA (the primer sequences are listed in Table S2 in the supplemental material) and verified by DNA sequencing. Each ORF was fused to the promoter and terminator sequences of the A. gossypii GPD (AgGPD) gene (for ACL2, the terminator of the AgPGK1 gene was used) following a previously described cloning strategy (18, 28). The ACL2 overexpression module comprising the recombinogenic flanks for the AgSTE12 locus, the selection marker loxP-KanMX-loxP, the ACL2 ORF, the AgGPD promoter, and the AgPGK1 terminator sequences was assembled following a one-pot DNA-shuffling method using the sequence of the BsaI restriction enzyme in the acceptor vector, as previously described (28). For the ACL1 overexpression module, the ACL1 ORF was PCR amplified as an NdeI-BamHI fragment using the corresponding primers (see Table S2 in the supplemental material) and inserted between the AgGPD promoter and terminator sequences in an overexpression cassette containing the integration and selection modules, as previously described (18). The overexpression modules were isolated and purified by enzymatic restriction (SapI and PmeI for the ACL2 and ACL1 modules, respectively) and were used to transform spores of the corresponding A. gossypii strain. The ACL2 cassette was integrated into the STE12 locus, and the ACL1 cassette was inserted into the LEU2 locus of the genome (data not shown). For the construction of double mutants, the selection marker was eliminated in the single mutants by expressing a Cre recombinase, and the double mutant was obtained using the same selection marker (data not shown). The genomic integration of the overexpression modules was confirmed by analytical PCR and DNA sequencing (data not shown).
Acetyl-CoA quantification.
A. gossypii mycelium (1 to 2 g) was lyophilized and pulverized mechanically. Then, samples (60 to 100 mg cell dry weight) were deproteinized by the addition of 6 μl of 1 N perchloric acid and sonicated on ice at medium intensity to avoid excessive heating. The homogenized extract was centrifuged at 13,000 × g to remove insoluble material and kept on ice. The supernatant (200 μl) was neutralized (pH 6 to 8) with three aliquots (20 μl each) of 3 M potassium bicarbonate solution, adding the aliquots while vortexing. The samples were cooled on ice and centrifuged to pellet the potassium bicarbonate. Twenty to 50 μl of the samples was used to quantify the acetyl-CoA content using an Acetyl-Coenzyme A Assay Kit (Sigma) following the manufacturer's instructions.
RNA extraction and quantitative real-time PCR.
Previously frozen A. gossypii mycelium (200 to 300 mg) was homogenized mechanically in liquid nitrogen using TRIzol reagent (Invitrogen, Carlsbad, CA), and total RNA was isolated as described by the manufacturer. RNA was incubated with 20 U of RNase-free DNase I (Roche, Basel, Switzerland). The cDNA samples were prepared using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative real-time PCR was performed with a LightCycler 480 real-time PCR instrument (Roche), using SYBR green I master mix (Roche) following the manufacturer's instructions. Primer sequences are listed in Table S2 in the supplemental material. All real-time PCRs were performed in duplicate and in at least two independent experiments. Quantitative analyses were carried out using the LightCycler 480 software.
Fluorescence microscopy.
For the visualization of lipid bodies, A. gossypii mycelium was suspended and washed in SB solution. The Bodipy lipid probe (2.5 mg/ml in ethanol; Invitrogen) was added to the hyphal suspension, and the solution was washed three times with SB solution. The lipid bodies were visualized using an Axio Imager.M2 (Zeiss, Le Pecq, France) fluorescence microscope at 495 nm (green fluorescent protein [GFP] filter) with a 100× oil immersion objective. AxioVision release 4.8 software was used to record the images.
RESULTS
Accumulation of FA in A. gossypii.
A. gossypii is considered to be a model organism for the biotechnological production of vitamins, nucleotides, and proteins (19, 29, 30). Furthermore, it has been reported that A. gossypii is able to accumulate a significant amount of lipid bodies in its multinucleated hyphae (21) (Fig. 1A). However, A. gossypii has typically been regarded as a nonoleaginous microorganism (9).
FIG 1.
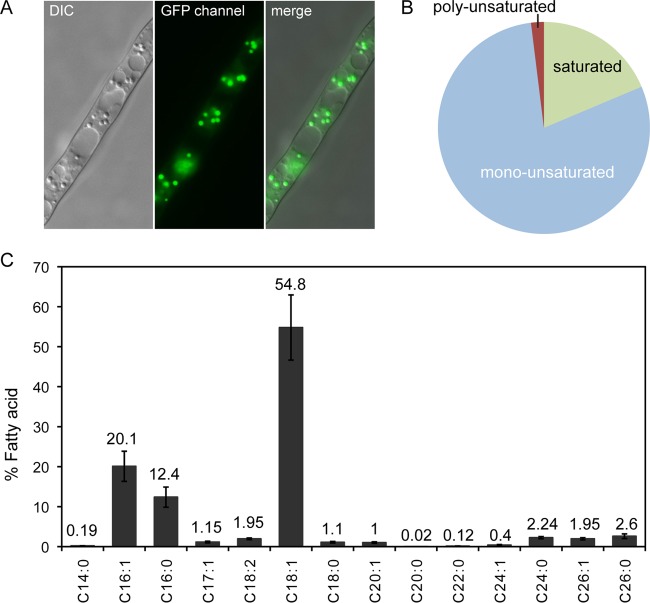
Lipid profile of a wild-type strain of A. gossypii. (A) Lipid bodies inside the A. gossypii mycelia were stained with Bodipy and visualized under fluorescence microscopy. DIC, differential interference contrast. (B) FA content of a WT strain of A. gossypii grown in 8% glucose. (C) Characterization of the FA composition of a WT strain of A. gossypii grown in 8% glucose; the numbers indicate the percentage of each FA. The results are the means of two independent experiments performed in duplicate. The error bars represent the standard deviations.
In order to characterize the lipidogenic (FA) profile of A. gossypii, we first wished to determine the total FA content of a wild-type strain of A. gossypii grown in medium containing glucose as the sole carbon source. Our results revealed that A. gossypii mostly accumulated unsaturated fatty acids, which comprised 81% of the total FA content (Fig. 1B). In good agreement with published results (21), we found that oleic acid (C18:1) was the most abundant FA in A. gossypii (54.8%), followed to a lesser extent by palmitoleic acid (C16:1) and palmitic acid (C16:0), which represented 20.1% and 12.4% of total FAs, respectively (Fig. 1C). In addition, we quantified significant levels (representing more than 1% of the total FA content) of other long-chain and very long-chain FAs, including the uncommon FA ginkgolic acid (C17:1), and we also found trace amounts (less than 0.5% of the total FA content) of myristic (C14:0), arachidic (C20:0), behenic (C22:0), and nervonic (C24:1) acids (Fig. 1C).
Lipid accumulation in A. gossypii mutants lacking the beta-oxidation pathway.
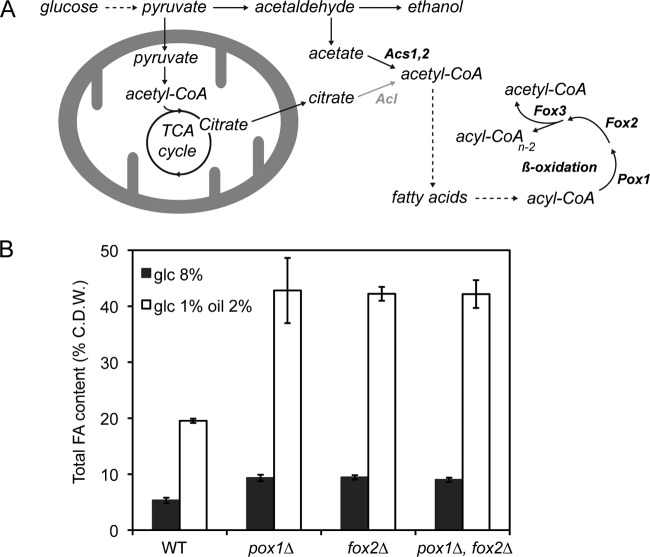
An increase in lipid accumulation has been achieved previously in engineered microorganisms lacking the FA beta-oxidation pathway, which is the main metabolic pathway that competes with lipid accumulation (10, 14, 15, 22). In contrast to other microorganisms, A. gossypii has only one beta-oxidation pathway localized to the peroxisome (9). According to the Ashbya Genome Database (http://agd.vital-it.ch/index.html), the genes AER358C (POX1), AGL060W (FOX2), and AFR302W (FOX3) are syntenic homologs of the S. cerevisiae genes POX1, FOX2, and POT1, respectively, which encode the enzymatic activities of the beta-oxidation pathway (Fig. 2A).
FIG 2.
Abolition of the FA beta-oxidation pathway increases lipid accumulation in A. gossypii. (A) Simplified diagram of FA biosynthesis and degradation in A. gossypii. ACL activity (gray) is not present in A. gossypii. The dashed arrows indicate a multistep pathway. (B) Total FA quantification of A. gossypii mutant strains lacking beta-oxidation genes. Analyses were carried out with cultures grown in 8% glucose (glc) or 1% glucose plus 2% oleic acid for 3 days. The results are the means of two independent experiments performed in duplicate. The error bars represent the standard deviations. C.D.W., cell dry weight.
In order to evaluate the effect of blocking the beta-oxidation process on the accumulation of lipids in A. gossypii, we carried out single and double deletions of both the AER358C (POX1) and AGL060W (FOX2) genes, which code for acyl-CoA oxidase and the multifunctional beta-oxidation enzyme, respectively. The pox1Δ and fox2Δ single mutants and the pox1Δ fox2Δ double mutant did not show any growth defect in glucose-containing medium; however, their growth was impaired when oleic acid was used as the sole carbon source (data not shown). Accordingly, we measured the lipid accumulation of the mutants in glucose-containing medium. As shown in Fig. 2B, abolition of the beta-oxidation pathway triggered a 2-fold increase (up to 9.4% of cell dry weight) in the accumulation of lipids in 8% glucose-containing medium. Furthermore, when oleic acid (2%) was added to the culture medium, the FA content rose to 19.5% of cell dry weight in the WT strain, and after 3 days of culture, the three beta-oxidation mutants displayed a level of lipid accumulation of about 40% of their cell dry weight.
Heterologous expression of Y. lipolytica ACL genes in A. gossypii.
The immediate precursor for the biosynthesis of FA is cytosolic acetyl-CoA, which can be synthesized from both acetate and citrate through two enzymatic pathways (Fig. 2A): the ACS (from acetate) and the ACL (from mitochondrial citrate) pathways (9, 31). In this regard, a correlation between lipid accumulation in oleaginous yeasts and the presence of ACL activity has been reported (32).
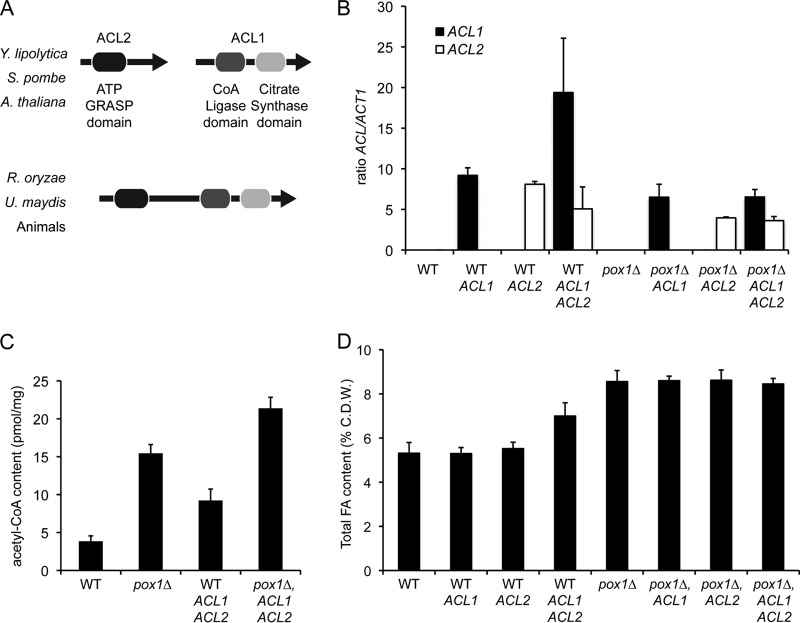
The ACL enzyme of plants and most yeasts and fungi is a heteromeric protein that comprises different subunits encoded by the ACL2 and ACL1 genes. However, animals and other fungi, such as zygomycetes and basidiomycetes, have homomeric ACL enzymes consisting of a large subunit encoded by a single ACL gene, which was originated by gene fusion of ACL2 and ACL1 (Fig. 3A) (31, 33). Analysis of the A. gossypii genome did not retrieve any predicted ACL-coding gene; accordingly, A. gossypii must generate acetyl-CoA for FA biosynthesis through ACS activity (Fig. 2A), as has also been described for Saccharomyces cerevisiae (34). In contrast, Y. lipolytica contains the ACL2 and ACL1 genes encoding the two subunits of the heteromeric ACL enzyme (33). Therefore, we decided to investigate the effect of the heterologous expression of both the ACL2 and ACL1 genes from Y. lipolytica on the accumulation of FA in A. gossypii.
FIG 3.
Heterologous expression of the Y. lipolytica ACL genes in A. gossypii. (A) Domain organizations of ACL-encoding genes in different species. S. pombe, Schizosaccharomyces pombe; A. thaliana, Arabidopsis thaliana; R. oryzae, Rhizopus oryzae; U. maydis, Ustilago maydis. (B) Relative transcription levels of the ACL2 and ACL1 genes in our A. gossypii mutant strains. Transcription levels were normalized using the A. gossypii ACT1 gene as a reference. Relative quantitative analyses were performed using LightCycler 480 software. The results are the means of two independent experiments performed in duplicate and are expressed as a ratio of the cDNA abundances of the target genes with respect to the ACT1 mRNA levels. (C) Acetyl-CoA quantification in A. gossypii strains. The results are the means of two independent experiments performed in duplicate. (D) Total FA quantification in A. gossypii strains. Analyses were carried out with cultures grown in 8% glucose for 3 days. The results are the means of two independent experiments performed in duplicate. The error bars represent the standard deviations.
The ACL2 and ACL1 ORFs were amplified from Y. lipolytica genomic DNA and used to generate two integrative cassettes for their heterologous overexpression in A. gossypii under the control of the strong promoter pAgGPD (see Materials and Methods). We carried out all the different combinations for the expression of the Y. lipolytica ACL2 and ACL1 genes in both the WT and pox1Δ backgrounds of A. gossypii. The expression cassette for ACL2 was targeted to the STE12 locus, while the expression cassette for ACL1 was inserted into the LEU2 locus. Integration of the overexpression cassettes in the target genomic locations was confirmed by analytical PCR, and the mRNA levels of both the ACL2 and ACL1 genes in the engineered strains of A. gossypii were determined by quantitative PCR (Fig. 3B). Next, we measured the acetyl-CoA contents in the mutant strains that overexpressed the two subunits of the ACL enzyme. As shown in Fig. 3C, overexpression of the ACL genes induced a strong increase in the acetyl-CoA pool in both the WT and pox1Δ backgrounds of A. gossypii. Furthermore, we found that the acetyl-CoA content was significantly higher in the pox1Δ background than in the WT background (Fig. 3C).
The ACL-expressing mutants were cultured in 8% glucose-containing medium, and lipid accumulation was determined as described in Materials and Methods. We detected an increase in the lipid content in the strain that overexpressed both the ACL2 and ACL1 genes in the WT background (Fig. 3D). However, the increased levels of acetyl-CoA in the poxΔ mutant that overexpressed the ACL genes did not correlate with any significant change in the accumulation of lipids (Fig. 3D).
Engineered strains of A. gossypii as novel SCOs.
As mentioned above, A. gossypii has been considered a nonoleaginous organism on the basis of two principal features: (i) a lipid accumulation capacity below 20% of the total cell dry weight and (ii) the absence of ACL-coding genes. However, we observed that the lipid content of a pox1Δ mutant of A. gossypii was able to exceed 40% of the mycelial dry weight when oleic acid was present in the culture medium (Fig. 2B).
A. gossypii shows two differentiated growth stages that strongly affect the production of metabolites such as riboflavin: a trophic phase, when the growth rate increases exponentially and riboflavin production is minimal, and a productive phase, when the growth rate decreases and riboflavin is overproduced (19). Accordingly, we wished to check whether the accumulation of lipids in our engineered strains changed during both the trophic and the productive phases. We measured the lipid contents at different time points of both the trophic and productive phases during 7 days of culture in oleic acid-containing media (Fig. 4A) and observed that the accumulation of lipids was maximal in the pox1Δ strain at the end of the productive phase, yielding a total FA content of more than 70% of cell dry weight. To our knowledge, this represents one of the highest lipid accumulation rates described so far (4, 7, 8). However, we also found that overexpression of both the ACL2 and ACL1 genes from Y. lipolytica induced an increase in the FA content in a WT strain only during the trophic phase (after 3 days of culture). In addition, during the productive phase (after 7 days of culture), the expression of ACL activity did not change the accumulation of lipids or even induce a decrease in the FA content in the WT and the pox1Δ strains, respectively (Fig. 4A). As shown in Fig. 4B, the accumulation of lipids in the pox1Δ strain could be visualized directly by staining of the intracellular lipid bodies, which were significantly larger in the pox1Δ strain than in the wild type.
FIG 4.
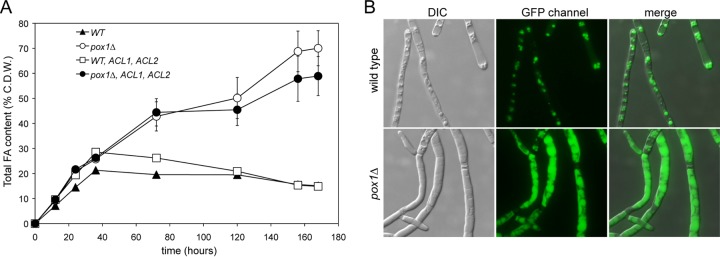
Lipid accumulation in the engineered A. gossypii strains. (A) Lipid quantification in A. gossypii strains at different time points of cultures grown in 1% glucose plus 2% oleic acid at 0 to ∼50 h (trophic phase) and 50 to 168 h (productive phase). The results are the means of two independent experiments performed in duplicate. The error bars represent the standard deviations. (B) Micrographs of the WT and pox1Δ strains of A. gossypii grown in 1% glucose plus 2% oleic acid for 7 days. Lipid bodies were stained with Bodipy and visualized under fluorescence microscopy.
DISCUSSION
The development of novel microbial factories with the ability to transform low-value lipids into different fine chemicals of industrial significance is an important issue for biotechnology. The presence of lipid bodies in A. gossypii hyphae has been reported previously (21); however, the biotechnological potential of A. gossypii for SCO production has remained unexplored. Here, we analyzed the FA content of a wild-type strain of A. gossypii, and we found that unsaturated FAs represented more than 80% of the total FA composition, suggesting an effective system of FA desaturation in A. gossypii. Indeed, a BLAST search for A. gossypii orthologs of the OLE1 gene, which codes for a delta-9 desaturase in S. cerevisiae (35), retrieved two different ORFs (AAL078W and AAR153C) that were predicted to encode desaturases. We also identified significant amounts of C24 and C26 FAs, which indicates the presence of enzymatic activities that catalyze the elongation of the acyl-CoA chain toward the synthesis of very long-chain FAs in A. gossypii.
According to general criteria regarding biomass, an oleaginous microorganism must be able to accumulate a minimum lipid content of 20% (4, 7, 8). Also, it has been proposed that lipid accumulation mostly depends on a continuous supply of both cytosolic acetyl-CoA and NADPH as the reducing power for FA biosynthesis (8), and by chance, two enzymes have been suggested to be essential for these metabolic processes, namely, ATP-citrate lyase and malic enzyme (8, 36). In this regard, it has been reported that the presence of an active ACL enzyme is a general feature of all oleaginous species (32, 36), although we observed that a wild-type strain of A. gossypii that lacks ACL-coding genes was able to accumulate lipids at a high concentration (19.5% of dry biomass), although still below the level of an oleaginous microorganism. However, when the ACL enzyme from Y. lipolytica was overexpressed in A. gossypii, the total FA yield reached 26% of the cell dry weight in the early phases of growth, indicating that ACL genes are likely essential for the lipidogenic ability of oleaginous microorganisms. Nevertheless, the presence of ACL activity did not correlate with such an increase in lipid accumulation in a pox1Δ mutant of A. gossypii, supporting a different hypothesis. First, the pox1Δ mutant must undergo metabolic flux redirection that triggers an increase in the cytosolic acetyl-CoA pool independently of ACL activity. Indeed, alternative pathways derived from carbohydrate and amino acid metabolism have also been described for the synthesis of acetyl-CoA (9). Second, a high concentration of acetyl-CoA in the pox1Δ ACL2 ACL1 strain could induce pleiotropic effects that would mask the effect of the overexpression of the ACL enzyme on lipid accumulation in the pox1Δ background. For example, the pox1Δ ACL2 ACL1 strain might undergo a depletion of the coenzyme A pool induced by the increased ACL activity that finally triggers inhibition of the activation of extracellular FAs. In addition, the nucleocytosolic pool of acetyl-CoA is also used for histone acetylation, which is crucial for the epigenetic mechanisms of transcriptional regulation (37). Therefore, the ACS activity and other metabolic reactions that provide cytosolic acetyl-CoA must be regulated to avoid drawbacks in the global gene expression pattern. In any case, the high level of acetyl-CoA in our engineered strains might serve for future biotechnological approaches involving the manipulation of acetyl-CoA metabolism, such as the production of biobutanol (38).
We also found a significant difference in the lipid accumulation pattern between the WT and pox1Δ backgrounds during the trophic and productive phases of A. gossypii growth. Indeed, as shown in Fig. 4A, the pox1Δ mutants increased lipid accumulation after 3 days of culture and throughout the productive phase. In contrast, the POX1 strains (WT backgrounds) underwent a decrease in the FA content after the trophic phase. This difference is probably due to activation of the beta-oxidation pathway during the productive phase.
Metabolic engineering for SCO production has been carried out in many microorganisms using different approaches that generally involve either the overexpression of lipid biosynthesis genes or the inactivation of FA-degrading activities by gene deletion (10, 12). With the aim of increasing the lipid accumulation in A. gossypii, we took a multigene approach in A. gossypii by exploiting both the overexpression of the ACL genes from Y. lipolytica and the inactivation of the beta-oxidation pathway by POX1 gene deletion. As mentioned above, the heterologous expression of ACL genes in A. gossypii afforded an FA content of 26% of the cell dry weight, thus transforming A. gossypii into an oleaginous fungus. Nevertheless, we found that blockade of the beta-oxidation pathway in the pox1Δ mutant was the most efficient modification for increasing lipid accumulation in A. gossypii up to 70% of the dry biomass, which makes our pox1Δ strain an attractive tool for oil biotechnology. The lack of FA degradation in the pox1Δ strain in combination with high levels of acetyl-CoA (4-fold more than the wild type) may explain the strong increase in the FA content measured in the strain. Recent studies have described the metabolic engineering of lipid biosynthesis in both oleaginous and nonoleaginous microorganisms, with rates of lipid accumulation not exceeding 60% of cell dry weight (12, 14, 15, 39, 40). Such modifications include increasing intracellular citrate levels, the overexpression of ACL genes, the overexpression of genes to enhance TAG biosynthesis, and the abolition of the beta-oxidation pathway and other competing pathways (12, 14, 15, 22, 23, 40).
A. gossypii is currently used as a microbial factory for several bioprocesses utilizing both low-cost and waste oil to cultivate the fungus (16). Therefore, it is possible to take advantage of the ability of A. gossypii to accumulate lipids for its use in biotransformation processes. Further characterization and modification of the A. gossypii genes that are predicted to encode desaturases and elongases may allow the design, through metabolic engineering, of new strains that accumulate valuable high-cost oils. This approach to the practical economic use of waste oil is emerging as a sustainable opportunity for the oleochemical industry (5, 41, 42). In this context, A. gossypii, which naturally produces mostly unsaturated FAs, may represent a novel tool for the production of high-cost oils, such as PUFAs, considered to be nutraceuticals for food fortification.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant BIO2011-23901 from the Spanish Ministerio de Economía y Competitividad. R.L.-A. was the recipient of an FPU predoctoral fellowship from the Spanish Ministerio de Economía y Competitividad.
We thank M. D. Sánchez and S. Domínguez for excellent technical help and N. Skinner for correcting the manuscript.
Footnotes
Published ahead of print 6 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03560-13.
REFERENCES
- 1.Sabirova JS, Haddouche R, Van Bogaert IN, Mulaa F, Verstraete W, Timmis KN, Schmidt-Dannert C, Nicaud JM, Soetaert W. 2011. The LipoYeasts project: using the oleaginous yeast Yarrowia lipolytica in combination with specific bacterial genes for the bioconversion of lipids, fats and oils into high-value products. Microb. Biotechnol. 4:47–54. 10.1111/j.1751-7915.2010.00187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. 10.1038/nature08721 [DOI] [PubMed] [Google Scholar]
- 3.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. 2006. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. U. S. A. 103:11206–11210. 10.1073/pnas.0604600103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beopoulos A, Nicaud JM, Gaillardin C. 2011. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 90:1193–1206. 10.1007/s00253-011-3212-8 [DOI] [PubMed] [Google Scholar]
- 5.Tuck CO, Pérez E, Horváth IT, Sheldon RA, Poliakoff M. 2012. Valorization of biomass: deriving more value from waste. Science 337:695–699. 10.1126/science.1218930 [DOI] [PubMed] [Google Scholar]
- 6.Lennen RM, Pfleger BF. 2013. Microbial production of fatty acid-derived fuels and chemicals. Curr. Opin. Biotechnol. 24:1044–1053. 10.1016/j.copbio.2013.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. 2011. Oily yeasts as oleaginous cell factories. Appl. Microbiol. Biotechnol. 90:1219–1227. 10.1007/s00253-011-3200-z [DOI] [PubMed] [Google Scholar]
- 8.Ratledge C. 2004. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86:807–815. 10.1016/j.biochi.2004.09.017 [DOI] [PubMed] [Google Scholar]
- 9.Vorapreeda T, Thammarongtham C, Cheevadhanarak S, Laoteng K. 2012. Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: involvement in the lipid production of oleaginous yeast and fungi. Microbiology 158:217–228. 10.1099/mic.0.051946-0 [DOI] [PubMed] [Google Scholar]
- 10.Courchesne NM, Parisien A, Wang B, Lan CQ. 2009. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 141:31–41. 10.1016/j.jbiotec.2009.02.018 [DOI] [PubMed] [Google Scholar]
- 11.Shen YQ, Burger G. 2009. Plasticity of a key metabolic pathway in fungi. Funct. Integr. Genomics 9:145–151. 10.1007/s10142-008-0095-6 [DOI] [PubMed] [Google Scholar]
- 12.Liang MH, Jiang JG. 2013. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 52:395–408. 10.1016/j.plipres.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 13.Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C, Nicaud JM. 2009. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48:375–387. 10.1016/j.plipres.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 14.Tang X, Feng H, Chen WN. 2013. Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab. Eng. 16C:95–102. 10.1016/j.ymben.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 15.Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, Chardot T, Nicaud JM. 2008. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74:7779–7789. 10.1128/AEM.01412-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahmann KP, Revuelta JL, Seulberger H. 2000. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53:509–516. 10.1007/s002530051649 [DOI] [PubMed] [Google Scholar]
- 17.Jiménez A, Santos MA, Pompejus M, Revuelta JL. 2005. Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 71:5743–5751. 10.1128/AEM.71.10.5743-5751.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiménez A, Santos MA, Revuelta JL. 2008. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol. 8:67. 10.1186/1472-6750-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateos L, Jiménez A, Revuelta JL, Santos MA. 2006. Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii. Appl. Environ. Microbiol. 72:5052–5060. 10.1128/AEM.00424-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park E, Kato A, Ming H. 2004. Utilization of waste activated bleaching earth containing palm oil in riboflavin production by Ashbya gossypii. J. Am. Oil Chem. Soc. 81:57–62. 10.1007/s11746-004-0857-z [DOI] [Google Scholar]
- 21.Stahmann KP, Kupp C, Feldmann SD, Sahm H. 1994. Formation and degradation of lipid bodies found in the riboflavin-producing fungus Ashbya gossypii. Appl. Microbiol. Biotechnol. 42:121–127. 10.1007/BF00170234 [DOI] [Google Scholar]
- 22.Dulermo T, Nicaud JM. 2011. Involvement of the G3P shuttle and beta-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 13:482–491. 10.1016/j.ymben.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 23.Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, Nicaud JM. 2012. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 93:1523–1537. 10.1007/s00253-011-3506-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos MA, Mateos L, Stahmann KP, Revuelta JL. 2004. Insertional mutagenesis in the vitamin B2 producer fungus Ashbya gossypii. Methods Biotechnol. 18:283–300. 10.1385/1-59259-847-1:283 [DOI] [Google Scholar]
- 25.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 26.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524. 10.1093/nar/24.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez A, Lisa-Santamaria P, Garcia-Marino M, Escribano-Bailon MT, Rivas-Gonzalo JC, Revuelta JL. 2010. The biological activity of the wine anthocyanins delphinidin and petunidin is mediated through Msn2 and Msn4 in Saccharomyces cerevisiae. FEMS Yeast Res. 10:858–869. 10.1111/j.1567-1364.2010.00679.x [DOI] [PubMed] [Google Scholar]
- 28.Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553. 10.1371/journal.pone.0005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledesma-Amaro R, Jiménez A, Santos MA, Revuelta JL. 2013. Biotechnological production of feed nucleotides by microbial strain improvement. Proc. Biochem. 48:1263–1270. 10.1016/j.procbio.2013.06.025 [DOI] [Google Scholar]
- 30.Ribeiro O, Magalhaes F, Aguiar TQ, Wiebe MG, Penttila M, Domingues L. 2013. Random and direct mutagenesis to enhance protein secretion in Ashbya gossypii. Bioengineered 4:322–331. 10.4161/bioe.24653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hynes MJ, Murray SL. 2010. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot. Cell 9:1039–1048. 10.1128/EC.00080-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boulton CA, Ratledge C. 1981. Correlation of lipid accumulation in yeasts with possession of ATP: citrate lyase. J. Gen. Microbiol. 127:169–176 [Google Scholar]
- 33.Fatland BL, Ke J, Anderson MD, Mentzen WI, Cui LW, Allred CC, Johnston JL, Nikolau BJ, Wurtele ES. 2002. Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol. 130:740–756. 10.1104/pp.008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Siewers V, Nielsen J. 2012. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS One 7:e42475. 10.1371/journal.pone.0042475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stukey JE, McDonough VM, Martin CE. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537–16544 [PubMed] [Google Scholar]
- 36.Ratledge C. 2002. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. Trans. 30:1047–1050. 10.1042/bst0301047 [DOI] [PubMed] [Google Scholar]
- 37.Galdieri L, Vancura A. 2012. Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 287:23865–23876. 10.1074/jbc.M112.380519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krivoruchko A, Serrano-Amatriain C, Chen Y, Siewers V, Nielsen J. 2013. Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J. Ind. Microbiol. Biotechnol. 40:1051–1056. 10.1007/s10295-013-1296-0 [DOI] [PubMed] [Google Scholar]
- 39.Wang ZP, Xu HM, Wang GY, Chi Z, Chi ZM. 2013. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim. Biophys. Acta 1831:675–682. 10.1016/j.bbalip.2012.12.010 [DOI] [PubMed] [Google Scholar]
- 40.Kamisaka Y, Kimura K, Uemura H, Yamaoka M. 2013. Overexpression of the active diacylglycerol acyltransferase variant transforms Saccharomyces cerevisiae into an oleaginous yeast. Appl. Microbiol. Biotechnol. 97:7345–7355. 10.1007/s00253-013-4915-9 [DOI] [PubMed] [Google Scholar]
- 41.Huang C, Chen XF, Xiong L, Chen XD, Ma LL, Chen Y. 2013. Single cell oil production from low-cost substrates: the possibility and potential of its industrialization. Biotechnol. Adv. 31:129–139. 10.1016/j.biotechadv.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 42.Nicol RW, Marchand K, Lubitz WD. 2012. Bioconversion of crude glycerol by fungi. Appl. Microbiol. Biotechnol. 93:1865–1875. 10.1007/s00253-012-3921-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.