Abstract
Escherichia coli ATCC 12806 was exposed to increasing subinhibitory concentrations of three biocides widely used in food industry facilities: trisodium phosphate (TSP), sodium nitrite (SNI), and sodium hypochlorite (SHY). The cultures exhibited an acquired tolerance to biocides (especially to SNI and SHY) after exposure to such compounds. E. coli produced biofilms (as observed by confocal laser scanning microscopy) on polystyrene microtiter plates. Previous adaptation to SNI or SHY enhanced the formation of biofilms (with an increase in biovolume and surface coverage) both in the absence and in the presence (MIC/2) of such compounds. TSP reduced the ability of E. coli to produce biofilms. The concentration of suspended cells in the culture broth in contact with the polystyrene surfaces did not influence the biofilm structure. The increase in cell surface hydrophobicity (assessed by a test of microbial adhesion to solvents) after contact with SNI or SHY appeared to be associated with a strong capacity to form biofilms. Cultures exposed to biocides displayed a stable reduced susceptibility to a range of antibiotics (mainly aminoglycosides, cephalosporins, and quinolones) compared with cultures that were not exposed. SNI caused the greatest increase in resistances (14 antibiotics [48.3% of the total tested]) compared with TSP (1 antibiotic [3.4%]) and SHY (3 antibiotics [10.3%]). Adaptation to SHY involved changes in cell morphology (as observed by scanning electron microscopy) and ultrastructure (as observed by transmission electron microscopy) which allowed this bacterium to persist in the presence of severe SHY challenges. The findings of the present study suggest that the use of biocides at subinhibitory concentrations could represent a public health risk.
INTRODUCTION
Escherichia coli strains are common contaminants in the food industry, especially in foods of animal origin (1). This bacterium is part of the common microbiota of the gastrointestinal tracts of animals and human beings, and although most isolates are nonpathogenic and are considered merely indicators of poor hygiene and sanitation conditions (fecal contamination), some 10% to 15% of E. coli strains are opportunistic and pathogenic serotypes capable of causing food-borne disease (2, 3). This bacterium is also of concern in the health care system, where it is a frequent nosocomial pathogenic microorganism (4). Escherichia coli and Enterococcus spp. are considered indicators of antibiotic resistance (5).
Biofilm formation is a major cause for concern in the food system, resulting in serious operational and maintenance costs (decreasing heat transfer and operational efficacy in heat exchange equipment, causing filter plugging and blockages of tubes in water distribution systems, increasing energy consumption, and accelerating damage to surfaces) (6). Moreover, biofilms provide a reservoir of microorganisms, increasing the risk of contamination in food processing plants and leading to critical problems in terms of public health and financial losses (7). Food-borne outbreaks of pathogens associated with biofilms have been related to the presence of Salmonella, Listeria monocytogenes, Yersinia enterocolitica, Campylobacter jejuni, Staphylococcus aureus, and E. coli (8). Moreover, biofilm formation is considered an important virulence factor in human infections, and it has been reported that approximately 80% of all bacterial infections are associated with biofilms (9, 10).
It is a well-established fact that the metabolic changes and protective effects of biofilm matrices create a conducive environment for biofilm inhabitants to adapt to higher concentrations of antimicrobial compounds (11). However, there are hardly any reports discussing the reverse phenomenon, i.e., the effect of antimicrobial adaptation on the potential of bacteria to form biofilms. Such a study would provide useful information from a food safety as well as clinical point of view if such adaptations confer a survival advantage upon microorganisms.
Antibiotic resistance is a global public health threat that involves all major microbial pathogens and antimicrobial drugs (12). Recent scientific evidence suggests that the selective pressure exerted by the use of biocides at sublethal concentrations could contribute to the expression and dissemination of antibiotic resistance mechanisms (5). In view of the large and increasing use of biocides, the risk of inappropriate use of such compounds, leading to the selection and spread of antibiotic-resistant bacteria, is of increasing concern, and the need for new research on this topic has been highlighted (5, 13).
An increase in biocide tolerance is also a major public health issue, as it could be expected to contribute to the increased persistence of pathogenic and spoilage bacteria in the food chain. It has been suggested that exposure to sublethal concentrations of biocides could potentially have an impact on the responses of bacteria to commonly used food processes, enabling microorganisms to survive challenges such as the concentrations of biocides currently permitted for use in food environments (14). However, at present, there is a limited understanding of the mechanisms which contribute to biocide tolerance.
This study aimed to examine whether exposure of E. coli ATCC 12806 to subinhibitory concentrations of three food-grade biocides (trisodium phosphate [TSP], sodium nitrite [SNI], and sodium hypochlorite [SHY]), and consequent adaptation, could influence its ability to form biofilms, resistance to antibiotics, and capacity to survive subsequent severe challenge by such compounds. In addition, the roles of associated changes in the growth kinetics, surface hydrophobicity, morphology, and ultrastructure of E. coli cells were examined.
MATERIALS AND METHODS
E. coli strain and culture conditions.
Escherichia coli ATCC 12806 was maintained in tryptone soy broth (TSB; Oxoid Ltd., Hampshire, United Kingdom) supplemented with 20% (vol/vol) glycerol at −30°C. Prior to experiments, frozen cells were subcultured twice in TSB at 37°C. Working cultures were kept at 4°C ± 1°C on plates of tryptone soy agar (TSA; Oxoid) and were subcultured monthly.
Biocides.
Three compounds were tested: TSP (Merck, Darmstadt, Germany), SNI (Sigma-Aldrich, Steinheim, Germany), and SHY (Sigma-Aldrich). Sterile solutions were prepared in distilled water immediately before each experiment.
Determination of MICs.
The MIC values were established using a microdilution broth method in accordance with CLSI guidelines (15). Five colonies of E. coli were taken from TSA plates, inoculated into 10 ml of Mueller-Hinton (MH) broth (Oxoid), and incubated at 37°C. Previous experiments showed that after 24 h of incubation, these bacterial cultures contained approximately 108 CFU/ml. For the experiment, 100-well polystyrene microwell plates (Oy Growth Curves Ab Ltd., Helsinki, Finland) were used. Wells were filled with 20 μl of chemical solution (a range of concentrations was used for each biocide) and 180 μl of an appropriate dilution (in MH broth) of inoculum in order to give a final concentration in the well of 5 × 105 CFU/ml. The inoculum concentration was confirmed by plating. The microwell plates were incubated at 37°C in a Bioscreen C MBR incubator (Oy Growth Curves Ab). Both positive (200 μl of inoculum at 5 × 105 CFU/ml) and negative (180 μl of MH broth plus 20 μl of chemical solution) controls were included in each experiment. The experiments were replicated five times on separate days.
The MIC was established as the lowest biocide concentration necessary to prevent growth after 48 h of incubation. A calibration equation was performed in order to establish a cutoff for bacterial growth. The growth of the strain in MH broth was determined in the following two ways for this purpose: by measuring the optical density at 420 nm to 580 nm (OD420–580) in a Bioscreen C MBR instrument and by enumerating viable cells on plate count agar (PCA; Oxoid) in duplicate, using 0.1% (wt/vol) peptone water (PW; Oxoid) for decimal dilutions, and counting visible colonies after 48 h of incubation at 37°C. Blank sample wells with uninoculated broth were included as a control for contamination. Five replications were carried out on different days. The degree of correlation between optical densities and microbial counts was investigated by linear regression of continuous data.
Adaptation to increasing concentrations of biocides.
The test was performed in the same manner as that described for determining the MIC. The starting concentration of biocide was MIC/2. When growth was observed, 20 μl of the suspension were aseptically transferred to the next well, which contained 160 μl of MH broth and 20 μl of the chemical solution. After the transfer, each well contained a concentration of biocide 1.5 times stronger than that in the previous well. This procedure was continued until no growth was observed after 72 h of incubation at 37°C. The suspension in the last well with recorded growth was streaked on TSA plates with biocides (one-half the maximum concentration of biocide that supported microbial growth was added to TSA). Nonexposed cells were grown in TSB and subsequently streaked on TSA plates without biocides. After incubation at 37°C for 48 h, agar plates were kept at 4°C ± 1°C for no longer than 1 week. For the experiments described below, TSB with biocides (a concentration equivalent to MIC/2 was added) was used for growth of previously adapted cells and TSB without biocides was used in the case of nonadapted cells. Culture broths were inoculated from the cultures kept on TSA (nonadapted cells) or TSA with biocides (adapted cells). All groups of cells (nonadapted and adapted to TSP, SNI, or SHY) were tested simultaneously on the same day of storage on agar plates.
Biofilm determination.
The study of the formation and structure of biofilms was performed using a previously described method (16), with minimal modifications. Cultures were grown at 37°C for 18 h, and appropriate dilutions in TSB were prepared to obtain a concentration of approximately 106 CFU/ml. TSB with biocides (MIC/2) was used for growth and dilution of previously adapted cells, and TSB without biocides was used in the case of nonadapted cells. A volume of 250 μl was added to the wells of sterile Matrix 96-well polystyrene flat-bottom microplates (Thermo Scientific, NH) having high optical quality, a low fluorescence background, and overall flatness, which allowed for high-resolution imaging. After 1 h of adhesion at 37°C, the wells were rinsed with 150 mM NaCl in order to eliminate any nonadherent bacteria before being refilled with 250 μl of culture broth. TSB with biocides (MIC/2) and without biocides was added to all groups of wells (containing cells that were nonadapted, adapted to TSP, adapted to SNI, or adapted to SHY). Thus, a total of 16 different conditions were tested. The plate was then incubated for 24 h at 37°C. After the development of biofilms, the cells were rinsed with 150 mM NaCl and refilled with TSB containing 5 μM Syto9 (1:1,000 dilution from a Syto9 stock solution at 5 mM in dimethyl sulfoxide [DMSO]; Invitrogen, Barcelona, Spain), a cell-permeating green fluorescent marker of the nucleic acids of cells. The plate was then incubated in the dark at 30°C for 20 min to enable fluorescence labeling of the bacteria.
Confocal laser scanning microscopy (CLSM) image acquisition was performed using a Nikon Eclipse TE 2000-U confocal laser scanning microscope with EZ-C13.60 software (Nikon Instruments Inc., NY). All biofilms were scanned at 400 Hz, using a 40× objective lens with a 488-nm argon laser set at 90% intensity. Emitted fluorescence was recorded within the range of 500 nm to 600 nm in order to visualize Syto9 fluorescence. Three stacks of horizontal plane images (512 by 512 pixels, corresponding to 119 by 119 μm) with a z step of 1 μm were acquired for each biofilm, using different areas in the well. Three independent experiments were performed for each condition, on different days.
For image analysis, three-dimensional projections of the structures of the biofilms were reconstructed using the Easy3D function of IMARIS 7.5 software (Bitplane, Zurich, Switzerland). The quantitative structural parameters of the biofilms, such as their biovolume, substratum coverage, and roughness, were calculated using the computer program COMSTAT (17, 18). The biovolume represented the overall volume of cells (μm3) in the observation field (14.161 μm2) and provided an estimate of the biomass in the biofilm. Substratum coverage (%) reflected the efficiency of substratum colonization by the population of bacteria. Roughness provided a measure of how much the thickness of the biofilm varied and is an indicator of biofilm heterogeneity. The maximum thickness (μm) of biofilms was determined directly from the confocal stack images.
Growth curves.
E. coli cells were grown in TSB (nonadapted cells) or TSB with biocides (MIC/2; adapted cells) for 18 h at 37°C. Volumes of 30 ml were centrifuged (4,000 × g, 10 min, 4°C), washed with phosphate-buffered saline (PBS; Oxoid), and resuspended in TSB. The wells of microwell plates (Oy Growth Curves Ab) were filled with 200 μl of inoculum (final concentration, 105 CFU/ml) or with 180 μl of inoculum and 20 μl of chemical solution at an appropriate concentration in order to give a final concentration of biocide in the well of MIC/2. Bacterial growth was monitored before incubation (at hour 0) and every hour until 120 h had elapsed. Growth was determined by measuring the OD420–580 in a Bioscreen C MBR instrument. The microwell plates were shaken for 1 min prior to the measurement of turbidity. The experiment was replicated three times, on separate days.
The model used to fit growth curves to the data obtained was the modified Gompertz equation (19): ODt = A + B × exp{−exp[2.71828183 × μ × (L − t)/B + 1]}, where t is the time, in hours, that has elapsed since inoculation, ODt is the OD420–580 at time t, L is the lag time (hours), μ is the maximum growth rate achieved (ΔOD420–580/h), B is the increase in OD420–580 between the inoculation and the stationary phase (E), and A is the upper asymptote curve (OD420–580 in the stationary phase [E]) − B. The OD420–580 value at 24 h (OD24h) was obtained for each strain and replication by fitting a sigmoidal curve to the data set, using a Marquardt algorithm that calculated the parameter values which gave the minimum residual sum of squares. The goodness of fit was evaluated using the coefficient of determination (R2). The calibration equation performed was used to transform the OD420–580 at 24 h into plate counts (log10 CFU/ml).
Determination of CSH.
Microbial cell surface hydrophobicity (CSH) was determined by the microbial adhesion to solvents (MATS) test based on affinity for nonpolar solvents (20). Xylene was used as the hydrocarbon phase. E. coli cells were grown in TSB (nonadapted cells) or TSB with biocides (MIC/2; adapted cells) for 18 h at 37°C. Cells were harvested by centrifugation (4,000 × g, 10 min, 4°C) and washed with sterile PBS. All groups of cells were resuspended in TSB and in TSB with biocides (MIC/2) at an initial concentration of 105 CFU/ml. Thus, a total of 16 conditions were tested. After 18 h at 37°C, cells were centrifuged, washed twice with PBS, and resuspended in 150 mM NaCl at a concentration of approximately 108 CFU/ml. The cell suspension (2.4 ml) was vortexed with 0.4 ml of the solvent for 60 s and then allowed to stand for 15 min at room temperature, which resulted in the complete separation of the two phases. An aqueous-phase sample (1 ml) was obtained, and the absorbance at 400 nm was determined (Bioscreen C MBR). The percentage of cells present in the solvent was calculated with the following equation: % affinity = 100 × [1 − (A/A0)], where A0 is the absorbance of the original suspension at 400 nm prior to mixing and A is the absorbance of the aqueous phase. Each experiment was performed in triplicate with three independently prepared cultures on separate days. Cell surface hydrophobicity was grouped into three categories: weak (<21%), moderate (21% to 50%), and strong (>50%) affinities for xylene (11).
Determination of antibiotic susceptibility.
E. coli cells were screened for susceptibility, before and after adaptation to the chemicals, to a panel of 29 antibiotics on Mueller-Hinton agar (Oxoid) by a disc diffusion method described by the CLSI (15). Adapted cells were grown in TSB with biocides (MIC/2), and nonadapted cells were grown in TSB without biocides. The following antibiotic discs (Oxoid) were used: spectinomycin (SH; 100 μg), amikacin (AK; 30 μg), gentamicin (CN; 10 μg), kanamycin (K; 30 μg), streptomycin (S; 10 μg), tobramycin (TOB; 10 μg), rifampin (RD; 5 μg), ampicillin (AMP; 10 μg), ticarcillin (TIC; 75 μg), amoxicillin-clavulanic acid (AMC; 30 μg), ampicillin-sulbactam (SAM; 20 μg), piperacillin-tazobactam (TZP; 110 μg), cephalothin (KF; 30 μg), cefazolin (KZ; 30 μg), cefoxitin (FOX; 30 μg), cefotaxime (CTX; 30 μg), ceftazidime (CAZ; 30 μg), cefepime (FEP; 30 μg), imipenem (IPM; 10 μg), aztreonam (ATM; 30 μg), sulfonamides (S3; 300 μg), trimethoprim-sulfamethoxazole (SXT; 25 μg), chloramphenicol (C; 30 μg), nalidixic acid (NA; 30 μg), ciprofloxacin (CIP; 5 μg), enrofloxacin (ENR; 5 μg), tetracycline (TE; 30 μg), phosphomycin (FOS; 50 μg), and nitrofurantoin (F; 300 μg). The inhibition zones were measured and scored as sensitive, intermediate, or resistant according to the CLSI guidelines (15). Cultures of Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 with known antimicrobial resistance patterns were used as reference strains for antibiotic disc control. The stability of resistance to therapeutic antimicrobials was determined by measuring susceptibility to antibiotics for both nonadapted and adapted strains after storage (4°C ± 1°C) for 3 months on TSA plates without biocides, with monthly transfers.
Determination of resistance to high concentrations of biocides.
E. coli cells were grown in TSB (nonadapted cells) or TSB with biocides (MIC/2; adapted cells) for 18 h at 37°C. Volumes of 30 ml were centrifuged (4,000 × g, 10 min, 4°C), washed with PBS, and resuspended in TSB or TSB with biocides (at 1.5 times the maximum concentration of biocide that allowed bacterial growth after adaptation) at an initial concentration of 107 CFU/ml. Initial bacterial concentrations were confirmed by plating. Experimental samples were incubated at 37°C for 60 min. After incubation, 0.75 ml of 100 mM sodium thiosulfate (Na2SO3; Sigma-Aldrich Química, Madrid, Spain) was added to 30 ml of bacterial suspension to arrest the activity of the biocides prior to sampling.
To determine cell survival, each sample was serially diluted with sterile 0.1% (wt/vol) PW, using 10-fold serial dilutions. After dilution, 0.1-ml samples were taken and spread plated on PCA. Two plates per dilution were prepared, inverted, and incubated at 37°C for 24 h. Each experiment was repeated three times, on separate days. The average number of colonies from the duplicate plates was recorded for each sample. Results are expressed as percentages of CFU after exposure to biocides relative to the initial (preexposure) CFU (percentage of survivors).
Determination of bacterial morphology and ultrastructure.
E. coli cells were tested before adaptation to SHY (nonadapted cells were inoculated in TSB and incubated at 37°C for 18 h) and after adaptation to SHY (cells adapted to SHY were incubated for 18 h at 37°C in TSB with SHY at MIC/2). Both groups of cells were examined before and after exposure to SHY (cells were incubated at 37°C for 60 min in TSB with 1.5 times the maximum concentration of SHY that allowed bacterial growth after adaptation). Thus, a total of four conditions were tested. The cells were harvested by centrifugation (4,000 × g, 10 min, 4°C) and were washed twice by suspension and centrifugation in PBS. E. coli cells were fixed in PBS with 2.5% glutaraldehyde (TAAB Laboratories Equipment Ltd., Berkshire, United Kingdom) for 1 h at 4°C. The bacteria were then centrifuged (4,000 × g, 10 min, 4°C) and washed by suspension and centrifugation in PBS. This washing step was repeated twice. For transmission electron microscopy (TEM), cells were treated with 1% osmium tetroxide (TAAB Laboratories Equipment) in PBS for 45 min at room temperature in darkness, followed by three new washing steps. Cells were pelleted in bacteriological agar (Oxoid), and the pellets were thereafter dehydrated in ethanol solutions of increasing concentrations and embedded in an epoxy resin (Epon 812; Tousimis Research Corp., Rockville, MD), which was polymerized by incubation for 48 h at 60°C. Ultrathin sections were collected on copper grids and stained with uranyl acetate and lead citrate. Observations were made using a JEM 1010 (JEOL Ltd., Tokyo, Japan) microscope at 80 kV. Samples for scanning electron microscopy (SEM) were dried under vacuum, mounted on aluminum stubs, and sputter coated with gold. Samples were viewed with a JSM 6480LV (JEOL) scanning electron microscope operating at 20 kV. Electron microscope observations were documented through the acquisition of representative microphotographs.
Statistical analysis.
The quantitative structural parameters of biofilms, percentages of affinity for xylene, and bacterial concentrations (OD420–580 or log10 CFU/ml) in culture broth were compared for statistical significance by using analysis of variance techniques. Mean separations were obtained using Duncan's multiple-range test. The percentages of cultures with increased resistance to antibiotics after exposure to biocides were compared by means of Fisher's exact test. All data processing in this study was carried out using the Statistica 6.0 software package (Statsoft Ltd., Chicago, IL). Significance was determined at the 5% (P < 0.05) level.
RESULTS
Adaptation of E. coli to biocides.
Optical densities (OD420–580) gave a linear relationship to microbial counts in the range of 0.200 to 0.700 (7.4 to 8.3 log10 CFU/ml). An R2 value of 0.842 was obtained over this range. Thus, an OD420–580 of 0.200 was considered the cutoff for bacterial growth in order to calculate the MICs. The regression equation for the relationship between microbial counts (log10 CFU/ml) and OD420–580 values was as follows: log10 CFU/ml = 7.061 + (1.809 × OD420–580).
The MICs of TSP, SNI, and SHY for Escherichia coli prior to its exposure to subinhibitory concentrations of biocides were 10.428, 14.876, and 0.239 mg/ml, respectively. After several passages through gradually higher concentrations of the compounds, the maximum concentrations of biocides that allowed bacterial growth were 11.731 mg/ml (TSP), 37.655 mg/ml (SNI), and 0.403 mg/ml (SHY). Adaptive tolerance to SNI and SHY was stable after repeated subculture in nonselective broth without biocides (the strain was passed through biocide-free TSB every 24 h for 7 days). On the other hand, in the absence of selective pressure, cells that had adapted to TSP returned to their preadaptation sensitivity.
Biofilm formation.
E. coli cells formed biofilms on polystyrene surfaces under the experimental conditions tested. To determine whether previous exposure to subinhibitory concentrations of biocide (TSP, SNI, or SHY), and consequent adaptation, would influence biofilm production, the architecture of biofilms produced by adapted and nonadapted E. coli cells in the absence or presence of sublethal concentrations (MIC/2) of such compounds was studied.
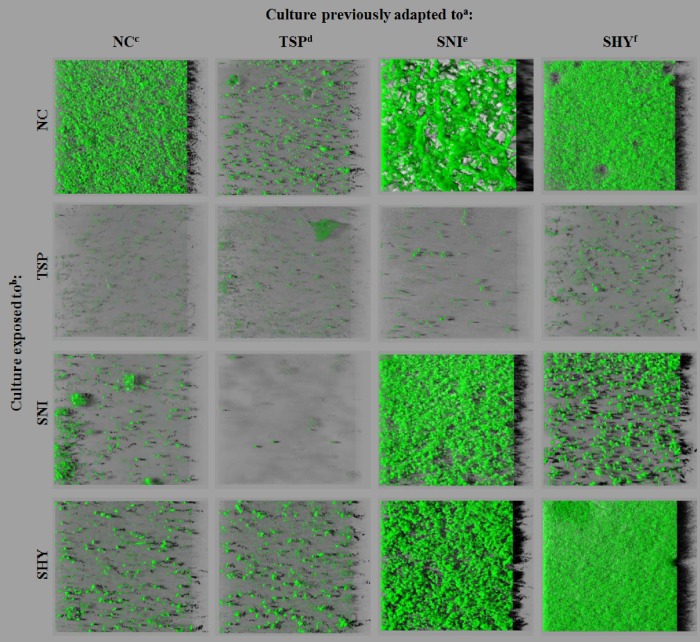
Representative 24-h biofilm structures observed using CLSM for the cells under study are presented in Fig. 1. The images correspond to three-dimensional reconstructions obtained from confocal stack images by use of IMARIS 7.5. software, including virtual shadow projections on the right. A marked variability in the three-dimensional biofilm architecture was noted between conditions. When cells were grown in the absence of biocides, cells adapted to TSP formed biofilms containing several small aggregates. Nonadapted cells and those adapted to SHY produced rough biofilms with a patchy coverage and confluent growth areas where the bacteria formed clumps. Cells adapted to SNI produced a specific spatial arrangement, forming snake-shaped structures of variable thickness. In the presence of TSP, E. coli cells formed only a few, small scattered cell clusters. In the presence of SNI or SHY, small scattered cell clusters were also observed for nonadapted cultures and for cells adapted to TSP, while cells adapted to SNI or SHY displayed a marked ability to form biofilms under these experimental conditions. It should be noted that cells adapted to SHY produced compact structures that covered most of the surface available in the presence of this compound.
FIG 1.
Three-dimensional projections of biofilm structures of E. coli ATCC 12806 under 16 different conditions, with shadow projections on the right, obtained from confocal z stacks by use of IMARIS software. a, for adaptation, cultures were previously exposed to increasing subinhibitory concentrations of biocides; b, exposed cultures were grown in the presence of biocides (MIC/2); c, no compound; d, trisodium phosphate; e, sodium nitrite; f, sodium hypochlorite.
Biovolume, substratum coverage, maximum thickness, and roughness parameters were extracted from confocal stack images in order to quantify biofilm structures with numerical data that would allow statistical analysis (Tables 1 to 4). Numerical data confirmed the visual observations. In TSB without biocides, cells adapted to TSP produced the smallest biofilms, with low biovolume (biomass) and surface coverage figures. Cultures adapted to SNI or SHY formed stronger biofilms (with a higher percentage of covered surface and larger biovolume) than did nonadapted strains. In the presence of TSP, E. coli cells formed weak biofilms, irrespective of adaptation. In the presence of SNI and SHY, strains adapted to these compounds produced biofilms with a larger biovolume and more surface coverage than those of nonadapted strains or those adapted to TSP.
TABLE 1.
Biovolume values observed for E. coli ATCC 12806 adapted and/or exposed to subinhibitory concentrations of food-grade biocides
| Culture exposed tob: | Biovolume (μm3) of culture previously adapted toa: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 23,664.6 ± 2,703.9Aa | 3,174.1 ± 613.4Ba | 80,467.2 ± 4,593.5Ca | 39,434.1 ± 1,393.6Da |
| TSP | 509.9 ± 275.4Ab | 857.9 ± 148.5ABb | 435.3 ± 50.5Ab | 1,485.6 ± 822.3Bb |
| SNI | 2,363.3 ± 445.6Ab | 227.4 ± 328.7Ab | 42,183.0 ± 3,980.1Bc | 6,114.7 ± 5,480.7Ab |
| SHY | 2,164.0 ± 296.7Ab | 2,798.7 ± 755.4Aa | 45,419.8 ± 7,564.5Bc | 74,474.1 ± 7,307.5Cc |
For adaptation, cultures were previously exposed to increasing subinhibitory concentrations of biocides. Data are means ± SD for nine determinations. Means in the same row with no capital letters in common are significantly different (P < 0.05). Means in the same column with no lowercase letters in common are significantly different (P < 0.05). NC, no compound; TSP, trisodium phosphate; SNI, sodium nitrite; SHY, sodium hypochlorite.
Cultures were grown in the presence of biocides (at MIC/2).
TABLE 4.
Roughness values observed for E. coli ATCC 12806 adapted and/or exposed to subinhibitory concentrations of food-grade biocidesa
| Culture exposed to: | Roughness value of culture previously adapted to: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 0.47 ± 0.01Aa | 0.47 ± 0.02Aab | 0.53 ± 0.14Aa | 0.46 ± 0.01Aab |
| TSP | 0.35 ± 0.07Ab | 0.39 ± 0.06Aa | 0.49 ± 0.07Aa | 0.39 ± 0.09Aa |
| SNI | 0.48 ± 0.02Aa | 0.61 ± 0.14Ab | 0.47 ± 0.01Aa | 0.52 ± 0.02Ab |
| SHY | 0.51 ± 0.03Aa | 0.55 ± 0.09Aab | 0.49 ± 0.02Aa | 0.40 ± 0.04Ba |
For information on interpretation, see the footnotes to Table 1.
TABLE 3.
Maximum thicknesses observed for E. coli ATCC 12806 adapted and/or exposed to subinhibitory concentrations of food-grade biocidesa
| Culture exposed to: | Maximum thickness (μm) of biofilms of culture previously adapted to: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 19.7 ± 0.6ABa | 17.3 ± 1.5Aa | 36.0 ± 2.0Ca | 20.3 ± 0.6Bab |
| TSP | 21.0 ± 2.7Aa | 21.3 ± 1.5Aa | 14.3 ± 3.2Bb | 21.0 ± 1.0Aab |
| SNI | 20.7 ± 4.0Aa | 8.0 ± 1.0Bb | 20.0 ± 1.0Ac | 18.3 ± 2.3Aa |
| SHY | 23.0 ± 1.0Aa | 19.0 ± 1.0Ba | 23.0 ± 2.0Ac | 23.3 ± 2.1Ab |
For information on interpretation, see the footnotes to Table 1.
Strong correlations (P < 0.001) were observed between biovolume and substratum coverage (r = 0.906), between biovolume and maximum thickness (r = 0.634), and between substratum coverage and maximum thickness (r = 0.471). No significant correlations (P > 0.05) were observed between roughness and the remaining structural parameters of biofilms (biovolume, surface coverage, and maximum thickness), thus suggesting similar levels of heterogeneity of biofilm thickness under the different conditions tested.
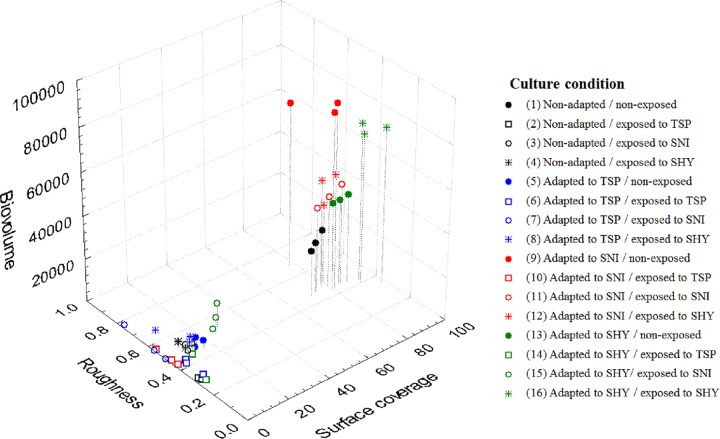
A three-dimensional scatterplot was drawn up to show the relationship between biovolume, substratum coverage, and roughness (Fig. 2). As can be seen, cells may be classified into two main groups (conditions 1, 9, 11 to 13, and 16 versus conditions 2 to 8, 10, 14, and 15).
FIG 2.
Relationships between biovolume (μm3), surface coverage (%), and roughness of biofilms produced by E. coli ATCC 12806 adapted and/or exposed to sublethal concentrations of food-grade biocides. Data are the averages for three determinations. For additional interpretation, see the legend to Fig. 1.
The possible influences of growth kinetics (concentration of bacteria in the culture broth in contact with the polystyrene surface at 24 h of incubation, the time at which biofilm formation was determined) and of CSH on biofilm formation were tested. Including the replicates, a total of 48 OD420–580 curves were generated for the E. coli cells tested and fitted to the modified Gompertz equation. The R2 values for the Gompertz model fit were high (>0.90). The OD420–580 values at 24 h (OD24h) were extracted from these curves. The regression equation obtained between microbial counts (log10 CFU/ml) and OD420–580 values (log10 CFU/ml = 7.061 + 1.809 × OD420–580) was used to estimate microbial counts after 24 h of incubation. Thus, OD24h values obtained throughout the experiment were transformed into microbial counts in accordance with this equation. The estimated OD24h values and microbial counts (log10 CFU/ml) recorded after 24 h are shown in Table 5. In the absence of biocides, similar cell densities were found in adapted and parent (not previously adapted) cultures. In TSB with TSP, SNI, or SHY, the highest cell density under each condition was found for cells adapted to TSP, SNI, or SHY, respectively.
TABLE 5.
Optical densities and bacterial concentrations observed for E. coli ATCC 12806 in tryptone soy broth after 24 h of incubation at 37°Ca
| Culture exposed to: | OD24h (log10 CFU/ml) of culture previously adapted to: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 0.68 ± 0.07Aa (8.30 ± 0.13Aa) | 0.66 ± 0.04Aa (8.26 ± 0.08Aa) | 0.59 ± 0.02Aa (8.14 ± 0.03Aa) | 0.61 ± 0.07Aa (8.17 ± 0.12Aa) |
| TSP | 0.44 ± 0.02ABb (7.85 ± 0.03ABb) | 0.47 ± 0.05Ab (7.92 ± 0.09Ab) | 0.46 ± 0.01ABb (7.89 ± 0.01ABb) | 0.40 ± 0.04Bb (7.78 ± 0.06Bb) |
| SNI | 0.20 ± 0.01Ac (7.42 ± 0.01Ac) | 0.24 ± 0.04ABc (7.49 ± 0.08ABc) | 0.28 ± 0.05Bc (7.56 ± 0.09Bc) | 0.20 ± 0.01Ac (7.41 ± 0.02Ac) |
| SHY | 0.30 ± 0.01Ad (7.60 ± 0.02Ad) | 0.31 ± 0.02Ac (7.62 ± 0.04Ac) | 0.26 ± 0.00Bc (7.53 ± 0.01Bc) | 0.43 ± 0.04Cb (7.84 ± 0.07Cb) |
OD24h, OD420–580 after 24 h. OD420–580 values were transformed to log10 CFU/ml according to the regression equation obtained [log10 CFU/ml = 7.061 + (1.809 × OD420–580)]. Data are means ± SD for three determinations. For additional information on interpretation, see the footnotes to Table 1.
Important differences in hydrophobicity values (ranging from 4.5 ± 0.3 to 66.2 ± 2.4) were found between cells, depending on the conditions tested (Fig. 3). Weak (<20% affinity for xylene), moderate (21% to 50% affinity), and strong (>50% affinity) reactions were observed for 6 (37.5%), 4 (25.0%), and 6 (37.5%) conditions, respectively. The highest values for cell surface hydrophobicity were shown by cells adapted and/or exposed to SNI, followed by those adapted and/or exposed to SHY. The lowest figures were observed for cultures adapted and/or exposed to TSP.
FIG 3.
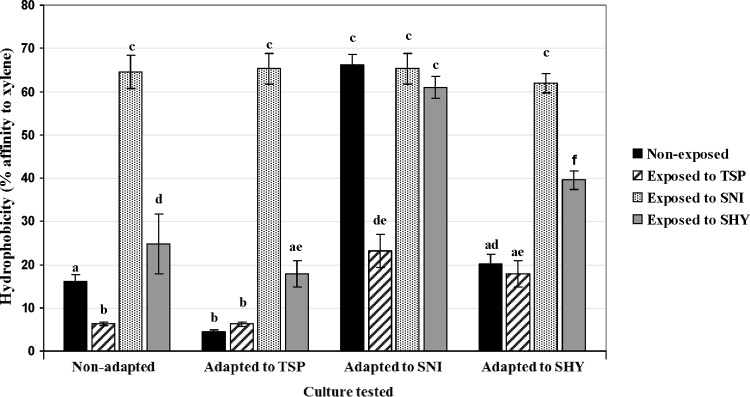
Hydrophobicity values observed for cultures of E. coli ATCC 12806 tested under 16 different conditions. Data are means ± standard deviations (SD) for three determinations. Mean values with no letters in common are significantly different (P < 0.05). For additional interpretation, see the legend to Fig. 1.
The relationship between surface coverage of the biofilm (%), estimated concentration of bacteria in TSB at 24 h (log10 CFU/ml), and hydrophobicity (percentage of adhesion to xylene) was examined. In the case of cells growing in the absence of biocides, similar concentrations of bacteria in TSB were observed for the four conditions tested (adapted and nonadapted cells) after 24 h of incubation at 37°C (Table 5). No substantial differences were found between the hydrophobicity values of parent cells and cells previously adapted to either TSP or SHY (Fig. 3). However, cultures adapted to TSP showed the least ability to form biofilms (low surface coverage) (Table 2). Cells adapted to SNI showed both the highest hydrophobicity values and the greatest potential for forming biofilms.
TABLE 2.
Surface coverage values observed for E. coli ATCC 12806 adapted and/or exposed to subinhibitory concentrations of food-grade biocidesa
| Culture exposed to: | Surface coverage (%) of culture previously adapted to: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 74.0 ± 3.7Aa | 14.0 ± 2.2Ba | 80.2 ± 1.88Ca | 85.1 ± 3.1Ca |
| TSP | 3.1 ± 1.5Ab | 4.5 ± 0.5ABb | 1.1 ± 0.2Ab | 7.3 ± 3.5Bb |
| SNI | 9.9 ± 1.7Ac | 0.4 ± 0.5Bc | 80.4 ± 5.3Ca | 26.8 ± 1.8Dc |
| SHY | 8.7 ± 0.1Ac | 10.6 ± 3.3Aa | 80.9 ± 2.1Ba | 91.2 ± 8.0Ca |
For information on interpretation, see the footnotes to Table 1.
In the presence of TSP, minimal figures were observed for both hydrophobicity and surface coverage, even though moderate concentrations of bacteria in the culture broth were found. When cells were grown in the presence of SNI, a high hydrophobicity and a low bacterial concentration were observed for both nonadapted and adapted cells. The highest surface coverage of the biofilm was found for cells adapted to SNI, followed by cells adapted to SHY. Nonadapted cells and those adapted to TSP showed a low surface coverage. In the presence of SHY, no substantial differences were found between conditions with regard to the bacterial concentration at 24 h. However, cells adapted to SNI and SHY showed the highest values for surface coverage and hydrophobicity.
Antibiotic susceptibility.
E. coli cultures were screened for susceptibility to 29 antibiotics before and after exposure to sublethal concentrations of biocides. Before adaptation, E. coli was susceptible to all antibiotics tested by the disc diffusion method. The exposure of E. coli to increasing subinhibitory concentrations of biocides was associated with reductions in the susceptibility to several antibiotics (Table 6).
TABLE 6.
Antibiotic resistance patterns of E. coli cultures tested

SH, spectinomycin (100 μg); AK, amikacin (30 μg); CN, gentamicin (10 μg); K, kanamycin (30 μg); S, streptomycin (10 μg); TOB, tobramycin (10 μg); RD, rifampin (5 μg); AMP, ampicillin (10 μg); TIC, ticarcillin (75 μg); AMC, amoxicillin-clavulanic acid (30 μg); SAM, ampicillin-sulbactam (20 μg); TZP, piperacillin-tazobactam (110 μg); KF, cephalothin (30 μg); KZ, cefazolin (30 μg); FOX, cefoxitin (30 μg); CTX, cefotaxime (30 μg); CAZ, ceftazidime (30 μg); FEP, cefepime (30 μg); IPM, imipenem (10 μg); ATM, aztreonam (30 μg); S3, sulfonamides (300 μg); SXT, trimethoprim-sulfamethoxazole (25 μg); C, chloramphenicol (30 μg); NA, nalidixic acid (30 μg); CIP, ciprofloxacin (5 μg); ENR, enrofloxacin (5 μg); TE, tetracycline (30 μg); FOS, phosphomycin (50 μg); F, nitrofurantoin (300 μg). Resistances are indicated as follows: R, resistant strain; I, strain of intermediate susceptibility; and S, susceptible strain. Data for exposed strains with increased resistance to antibiotics (relative to unexposed strains) are shaded; values for exposed strains not exhibiting increased susceptibility to antibiotics are not shaded. An increase in resistance was defined as a change from S (before exposure) to R (after exposure).
For adaptation, cultures were previously exposed to increasing subinhibitory concentrations of biocides. For additional information on interpretation, see the footnotes to Table 1.
The number of increased resistances caused by SNI (14 [48.28% of the antibiotics tested]) was larger (P < 0.05) than those for TSP (1 [3.44%]) and SHY (3 [10.34%]). The impact of biocide exposure on reduced susceptibility to antibiotics depended on the antibiotic family tested, with aminoglycosides (especially spectinomycin), cephalosporins, and quinolones (especially nalidixic acid) showing the greatest increases in resistance. The reduced susceptibility to antibiotics as a result of adaptation to biocides remained stable after 3 months in the absence of biocides (E. coli cells were maintained on TSA plates and subcultured monthly).
Resistance to strong concentrations of biocides.
E. coli cultures (initial concentration of 107 CFU/ml) were incubated at 37°C for 1 h in biocide-free TSB or in TSB with biocides (in this case, 1.5 times the maximum doses that supported microbial growth after adaptation) prior to plating to determine viability.
No differences between groups of cells were observed in terms of the morphology of the colonies on TSA. In the absence of biocides or in the presence of SNI, the population density after incubation was essentially stable. E. coli cells were completely killed by the presence of TSP. Lastly, when E. coli cells were exposed to SHY, the bacterial population decreased substantially during the 1-h test period (Table 7).
TABLE 7.
Survival of E. coli ATCC 12806 after incubation for 1 h at 37°C in TSB or TSB with biocides
| Culture incubated in the presence ofb: | % survival of culture previously adapted toa: |
|||
|---|---|---|---|---|
| NC | TSP | SNI | SHY | |
| NC | 133.87 ± 35.07Aa | 103.89 ± 57.19Aa | 99.94 ± 40.40Aa | 108.44 ± 26.84Aa |
| TSP | 0.00 ± 0.00Ab | 0.00 ± 0.00Ab | 0.00 ± 0.00Bb | 0.00 ± 0.00Bb |
| SNI | 118.35 ± 30.95ABa | 111.47 ± 48.72ABa | 101.73 ± 34.15Aa | 145.95 ± 27.67Bc |
| SHY | 0.08 ± 0.08Ab | 0.22 ± 0.15Ab | 0.00 ± 0.00Ab | 56.20 ± 25.56Bd |
Values represent the concentrations of bacteria after incubation with regard to initial concentrations (%). For adaptation, cultures were previously exposed to increasing subinhibitory concentrations of biocides. Data are means ± SD for three determinations. For additional information on interpretation, see the footnotes to Table 1.
Cultures were incubated at 37°C for 60 min in TSB with 1.5 times the maximum concentration of biocide that allowed bacterial growth after adaptation.
In the absence of biocides, the bacterial concentrations during the study period were similar for adapted and nonadapted cells. The influence of previous adaptation on bacterial resistance to TSP or SNI was slight. On the other hand, extreme differences in susceptibility of bacteria to inhibition by SHY were found with respect to their previous adaptation. Thus, after exposure to high concentrations of SHY for 1 h, cells adapted to SHY showed a >250-fold higher percentage of survivors than nonadapted cells or those adapted to TSP or SNI. Thus, the morphological and ultrastructural changes in E. coli cells associated with adaptation and/or exposure to SHY were investigated in order to determine the possible mechanisms of acquired resistance to this compound.
Determination of bacterial morphology and ultrastructure in the presence of SHY.
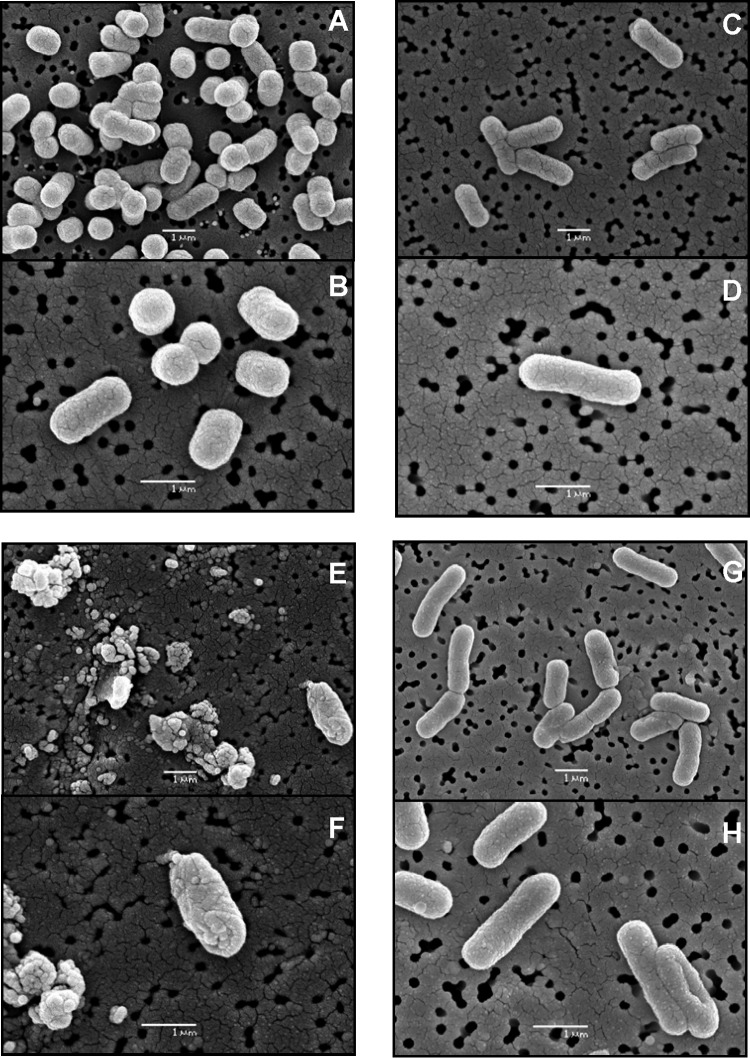
To elucidate why bacterial adaptation to SHY decreases the deleterious effect of high concentrations of this compound, morphological and ultrastructural changes in E. coli occurring after adaptation and/or exposure to SHY were examined by SEM and TEM.
The SEM analysis (Fig. 4) revealed that after adaptation to SHY, ridges appeared on the outer surfaces of the cells. Moreover, the cells appeared thinner and longer than control (nonadapted) cells. After exposure to high concentrations of SHY, the adapted cultures displayed few or no morphological changes relative to nonexposed cultures. However, after challenge by SHY, nonadapted cultures exhibited considerable morphological modifications in comparison with control (nonexposed) cultures. Modifications ranged from deformed cells, with individual bumps, buds, grooves, and cavities, to overall cell surface wrinkling. Cell wall disruption was observed in the form of cracks, holes, or even cell lysis. In some places, clusters of numerous lysed and fused cells were seen.
FIG 4.
Scanning electron micrograph sections of E. coli ATCC 12806 cells. (A and B) Nonadapted untreated cells; (C and D) adapted untreated cells; (E and F) nonadapted treated cells; (G and H) adapted treated cells. For adaptation, cultures were previously exposed to increasing subinhibitory concentrations of sodium hypochlorite. Treated cells were exposed to sodium hypochlorite at 1.5 times the maximum concentration of biocide that allowed bacterial growth after adaptation.
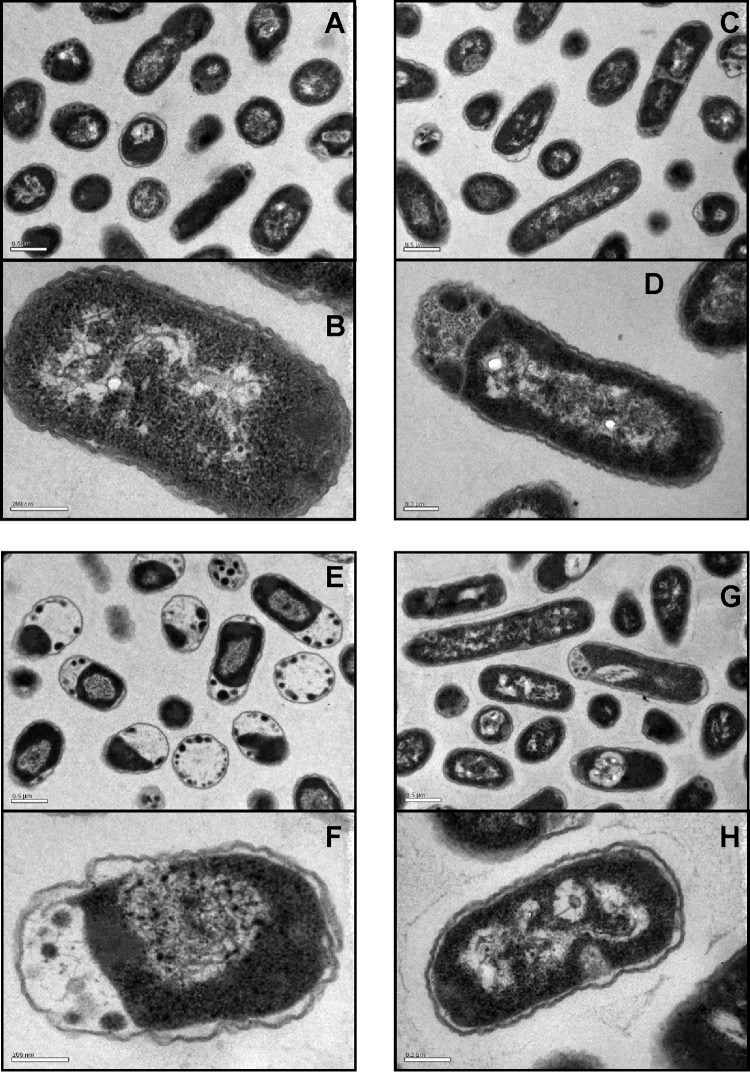
Transmission electron microscopy examination of cells adapted to SHY confirmed the changes (elongation) in the nature of the cellular suspension compared with nonadapted cultures (Fig. 5). In addition, E. coli adapted to SHY showed electron-dense regions and evidence of cytoplasmic condensation. In E. coli cells exposed to high concentrations of SHY (both adapted and nonadapted cells), the cytoplasm was disorganized and the inner and outer membranes were separated. No uniform spacing between the E. coli cytoplasmic contents and the cell envelope was observed. Exposure to SHY caused major structural changes in nonadapted cells. Thus, in contrast to adapted bacteria after exposure to SHY, nonadapted E. coli cells showed coagulated material, especially in the periphery of the cells, and a very wide space between the cytoplasm and the cell membranes. Cultures exhibited wavy cell envelope structures, with the presence of blebs in the outer membrane, and the integrity of the outer membrane was not maintained in some cases, demonstrating that the E. coli cells were lysed after exposure to SHY.
FIG 5.
Transmission electron micrograph sections of E. coli ATCC 12806 cells. (A and B) Nonadapted untreated cells; (C and D) adapted untreated cells; (E and F) nonadapted treated cells; (G and H) adapted treated cells. For adaptation, cultures were previously exposed to increasing subinhibitory concentrations of sodium hypochlorite. Treated cells were exposed to sodium hypochlorite at 1.5 times the maximum concentration of biocide that allowed bacterial growth after adaptation.
DISCUSSION
Adaptation of E. coli to biocides.
E. coli ATCC 12806 exhibited acquired tolerance to biocides (especially SNI and SHY) after exposure to increasing subinhibitory concentrations of such compounds. The increased tolerance to SNI or SHY remained after 7 successive passages in biocide-free TSB, suggesting that in E. coli ATCC 12806, adaptive tolerance to both compounds could be the result of heritable mutations. These findings are in agreement with reports by other authors, where adaptation of E. coli to both chemical and physical sublethal stresses has been demonstrated (21). The increased tolerance observed suggests that the use in food environments of compounds which may provide sublethal exposure when used inappropriately (improper use, inappropriate storage, or the presence of excessive amounts of organic matter, known to inactivate several biocides) represents a real risk for the development of adaptation to biocides. Caution should also be exercised when food preservation treatments are used at low intensities. It is a frequent practice worldwide to use combined preservative factors (hurdles) at sublethal doses because of their synergistic antimicrobial effect. Hurdle technology aims to improve the microbial status and the sensory quality of foods, as well as their nutritional properties (22). The potential link between preservation treatments at low intensities and microbial acquisition of new or modified characteristics, such as more resistance to antimicrobials, has been suggested previously (12, 23).
Biofilm formation.
The ability of E. coli to attach to, colonize, and form biofilms on different surfaces is well documented (24). However, only limited studies have thus far been conducted to determine the influences of adaptation and/or exposure to sublethal doses of biocides on the potential to form biofilms, and it appears that the effect of contact with TSP, SNI, or SHY on the ability of E. coli to form biofilms may have been tested for the first time in this work.
E. coli cells that had neither been adapted nor exposed to biocides produced rough biofilms on polystyrene, with clusters and irregular coverage, as previously described both under static conditions (16) and in continuous-flow systems (25). These findings are a matter for concern, because plastic materials are used extensively in surfaces and equipment throughout the food chain (10). Moreover, several studies have shown a correlation between biofilm production on polystyrene microwells and biofilm formation on the surfaces of different materials in food facilities (26, 27).
The finding of substantially improved biofilm formation ability after adaptation to SNI or SHY reported here, which is in agreement with similar observations in earlier studies performed with different bacterial species and chemical compounds (1, 28–30), suggests that the use of antimicrobial compounds at inappropriate doses in the food industry may increase the ability of bacteria to produce biofilms, thus imposing a food safety threat because the presence of biofilms on food processing surfaces favors the contamination of foodstuffs.
The concentration of bacteria in the culture broth in contact with the polystyrene surface at 24 h of incubation (biofilm formation was evaluated at this time point), as well as the cell surface hydrophobicity of E. coli cultures, was studied in order to improve our understanding of the differences in the ability to form biofilms between cultures subjected to different conditions. The cell densities of the culture broths were found not to have any apparent relationship to their biofilm-producing capability. Thus, for TSB with and without compounds, important differences in biofilm formation ability were observed between groups of cells (nonadapted or adapted to TSP, SNI, or SHY), even though no substantial differences in bacterial concentrations were found. These results agree with findings from other authors, who found that the number of attached cells remained constant over the period of incubation (31, 32). It has been suggested that after a few hours of incubation, the surface reaches a saturation level where larger numbers of planktonic cells do not entail larger numbers of attached cells (7). This is one possible explanation for the absence of any relationship between biofilm formation and the density of suspended cells after 24 h of incubation in the present study. It must be pointed out, however, that these results should be considered with caution, because the different sizes of E. coli cells under different growth conditions (Fig. 4 and 5) could bring into question the relationship between cell density (determined in this study) and microbial counts in some circumstances.
As an adaptive response, bacterial cell surfaces may undergo chemical modifications to repel water-soluble compounds. These modifications may result in a change in cell surface hydrophobicity. The hydrophobicity of the microbial cell surface plays a critical role in the adherence of bacteria to a wide variety of hydrophobic surfaces (e.g., polystyrene) (33, 34). Because adherence of bacteria to the surface is the first step in biofilm formation, it is expected that an increase in cell surface hydrophobicity is accompanied by an increase in bacterial ability to form biofilms on polystyrene surfaces (11, 26, 35). However, controversial results on the effect of cell surface hydrophobicity on biofilm formation on cell contact surfaces have been reported (24).
The hydrophobicity values observed for parent E. coli cells grown in the absence of biocides (16.1 ± 1.6) are in agreement with figures reported by other authors for different E. coli serovars, which showed that E. coli strains exhibit low hydrophobicity (4). However, comparisons between reports should be considered with caution because several factors (e.g., characteristics of the culture medium, growth conditions, variability of the bacterial strains, presence of antimicrobials, or methods used in the study) may modify the hydrophobic surface properties of bacterial cells (26, 36).
Parent (nonadapted) E. coli cells growing in the absence of biocides showed a weak hydrophobicity (16.1 ± 1.6), a high estimated bacterial concentration (8.30 ± 0.13 log10 CFU/ml), and a considerable surface coverage by the biofilm (74.0% ± 3.7%). Adaptation and/or exposure to biocides significantly modified these figures (Tables 2 and 5; Fig. 3).
The increase in cell surface hydrophobicity observed after contact (adaptation and/or exposure) with sublethal concentrations of SHY and, especially, SNI could be related to a reduction in the charged (hydrophilic) binding sites for the biocides (37). Changes in hydrophobicity do not explain the differences observed in the ability to form biofilms in some cases. Thus, similar levels of surface coverage were observed in TSB without biocides for cells adapted to SNI (which showed high values for hydrophobicity) and for those adapted to SHY or nonadapted (with lower hydrophobicity figures), thus suggesting that biofilm formation in E. coli ATCC 12806 is regulated by different factors.
The cell surface hydrophobicity of E. coli cells increased in the presence of SNI, with figures in the same range being obtained for both nonadapted and adapted strains (Fig. 3). Despite these similar values (figures of a like nature were also observed for parent and adapted cultures in the case of cell density), cells adapted to SHY and SNI had a greater ability to produce biofilms than nonadapted cells and those adapted to TSP. Thus, factors other than hydrophobicity and cell density might influence the biofilm-forming potential of E. coli in the presence of SNI. Along these lines, several reports have shown that bacterial adhesion to surfaces is the first step in biofilm formation. Once adhered to a surface, bacteria aggregate to form microcolonies organized in the three-dimensional structure of a biofilm. However, high levels of initial adherence (associated with high cell surface hydrophobicity) do not necessarily lead to the formation of thick biofilms (38). On the other hand, the hydrophobicity values seen had a strong influence on biofilm formation in the presence of SHY, as cells with the highest hydrophobicity values showed the greatest potential to form biofilms (Table 2; Fig. 3).
Lastly, cells adapted and/or exposed to subinhibitory concentrations of TSP showed little potential to form biofilms, together with low values for hydrophobicity, highlighting the positive effects of TSP in the context of food safety. Several authors (39, 40) have suggested that changes in cell surface properties with alkaline and/or other environmentally induced damage can significantly disrupt cell metabolism and structure, preventing effective interactions between bacterial cells and their environment. This fact might explain the limited biofilm formation ability of cells adapted and/or exposed to TSP. In addition to the high pH and ionic strength of the TSP solutions, the ability of TSP to remove fat films and to have a surfactant or detergent effect (41) may be responsible for the limited ability of E. coli to form biofilms after contact with this compound.
Antibiotic susceptibility.
Concerns about a possible linkage between adaptation to biocides and antibiotic resistance have been growing for a number of years and have been raised at both the national and international levels (12, 42). If pathogenic bacteria are resistant to commonly used antibiotics, then this causes an added problem for people acquiring infections. It is noteworthy that the study presented here revealed an increase in resistance to quinolones after adaptation to biocides. Fluoroquinolones are critically important drugs for treating serious E. coli infections in humans, and continued surveillance is required to detect emerging fluoroquinolone-resistant phenotypes (43). The movement of strains from the category of “sensitive” to that of “resistant” observed for aminoglycosides and cephalosporins after exposure to subinhibitory concentrations of biocides (marked for aminoglycosides) is also a matter for concern in view of the fact that these compounds are the front-line antimicrobials for treating serious bacterial infections in humans (44, 45). It should be pointed out, however, that clinical levels of resistance were not reached and that data indicating public health relevance are lacking.
SNI caused the largest number of increased resistances (14), followed by SHY (3). Adaptation to TSP caused only one increased resistance (to AMP). These results are not surprising because among the compounds tested, SNI showed the largest increases in the maximum concentration of biocides that allowed bacterial growth after adaptation. According to Braoudaki and Hinton (46), a relationship between insensitivity to biocides and resistance to antibiotics is generally seen. The decreases simultaneously observed in susceptibility to SNI and antibiotics, which have different modes of action, suggest a nonspecific resistance mechanism. It has been suggested that the linkage between biocide adaptation and resistance to antibiotics in Gram-negative bacteria might be due to nonspecific efflux pumps. In fact, the classes of antibiotics showing a marked increase in resistance after exposure to biocides (aminoglycosides, cephalosporins, and quinolones) are known efflux pump substrates (47). Changes in the cell surface which induce a reduction in cell permeability do not allow chemically unrelated molecules into the resistant cells and also play a vital role in the establishment of resistance to a variety of antimicrobial agents (48). In the present study, cultures with high cell surface hydrophobicity (adapted to SNI) also showed an increased resistance to antibiotics. Similarly, Braoudaki and Hinton (46) reported that increased cell surface hydrophobicity was associated with antibiotic resistance in Salmonella strains.
Resistance to strong concentrations of biocides.
This work demonstrated that E. coli ATCC 12806 has the capacity to resist inactivation by SHY after exposure to sublethal concentrations of such a compound. This finding is of particular concern because SHY is commonly used in food processing facilities and other environments to reduce or eliminate pathogenic and spoilage microorganisms. Even through SHY is generally used at concentrations above those employed in this research, suboptimal concentrations of biocide could occur as a consequence of improper use (erroneous concentration or inadequate distribution), inappropriate storage of the formulations (resulting in a decrease in the effective biocide concentration), or the presence of excessive amounts of organic matter (known to inactivate several chlorinated compounds) (5).
An increase in E. coli tolerance to biocides after exposure to sublethal concentrations has been demonstrated widely for triclosan, benzalkonium chloride, and chlorhexidine (14, 49). In order to avoid an increase in biocide tolerance, several biocidal agents (with various modes of action on their microbial target organisms) should be combined in the development of commercial biocides (50). Moreover, the consequences of adaptation can be limited by alternating the formulations used over time (14).
Determination of bacterial morphology and ultrastructure in the presence of SHY.
One major way in which microorganisms cope with their environment is to alter their morphology, and various morphological adaptations have been reported and reviewed (51). The elongation observed after adaptation, probably due to disturbances in cell division, is in agreement with previous findings on tolerance to environmental stresses (20, 39, 52). Moreover, the changes in the surfaces of adapted cells, which were more undulating and rougher than those of parent cells, suggest the involvement of modifications of the outer membrane of E. coli in its tolerance to SHY. Outer membrane remodeling has been shown to be partially responsible for the adaptive resistance of Gram-negative bacteria to several antimicrobials (53, 54).
The electron-dense regions observed in TEM examinations of E. coli cells exposed to SHY are likely due to the accumulation of glycogen inclusion bodies (55). Other authors (56, 57) reported similar electron-dense granules in TEM examinations following E. coli exposure to low concentrations of biocides (silver ions and mineral leachates, respectively). Coagulated cytoplasmic material was observed near the cell wall of treated E. coli. This coagulated material is thought to be a precipitate of abnormal proteins or denatured membrane (58, 59). The treated cells also showed numerous breaks in the cell membranes. Similar observations have been reported as evidence of outer membrane damage (60, 61). Notably, these ultrastructural alterations were not evident in the previously adapted cells, thus indicating that adaptation to SHY increases bacterial resistance to high concentrations of this compound.
A characteristic effect of SHY on the ultrastructure of E. coli cells seems to be the formation of blebs on the outer membrane of the cell envelope. These findings are consistent with corresponding studies of Gram-negative bacteria, in which it was also found that bacteria treated with antimicrobials present these structures (62). As in the study presented here, these reports described thin-section projections that appeared to consist of a unit of membrane clearly continuous with the whole outer membrane, without participation of the entire cell envelope. The physical cause for these changes in outer membrane morphology may be due to the loss of certain outer membrane proteins and/or lipopolysaccharides (63).
The findings of the present study have improved our understanding of the mechanism of SHY's antibacterial action. Upon exposure to SHY, membrane integrity is compromised and structural integrity is not retained. We suggest that the morphological and structural changes induced by sublethal doses of SHY are among the possible mechanisms of resistance of E. coli to high concentrations of this compound.
ACKNOWLEDGMENTS
This work was supported by the Junta de Castilla y León (project LE013A10-2) and the Ministerio de Economía y Competitividad (project AGL2011-29645).
Footnotes
Published ahead of print 6 December 2013
REFERENCES
- 1.Pagedar A, Singh J, Batish VK. 2012. Adaptation to benzalkonium chloride and ciprofloxacin affects biofilm formation potential, efflux pump and haemolysin activity of Escherichia coli of dairy origin. J. Dairy Res. 79:383–389. 10.1017/S0022029912000295 [DOI] [PubMed] [Google Scholar]
- 2.Barnes HJ, Gross WB. 1997. Colibacillosis, p 131–139 In Calnek BW, Barnes HJ, Beard CW, Mcdougald LR. (ed), Diseases of poultry, 10th ed. Mosby-Wolfe Medical Publication Ltd, London, United Kingdom [Google Scholar]
- 3.EFSA 2013. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 11:3129–3378. 10.2903/j.efsa.2013.3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamadi F, Latrache H, Zahir H, Elghmari A, Timinouni M, Ellouali M. 2008. The relation between Escherichia coli surface functional groups composition and their physicochemical properties. Braz. J. Microbiol. 39:10–15. 10.1590/S1517-83822008000100003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SCENIHR 19 January 2009. Assessment of the antibiotic resistance effects of biocides. Scientific Committee on Emerging and Newly Identified Health Risks, European Commission, Brussels, Belgium [Google Scholar]
- 6.Myszka K, Czaczyk K. 2011. Bacterial biofilms on food contact surfaces—a review. Pol. J. Food Nutr. Sci. 61:73–180. 10.2478/v10222-011-0018-4 [DOI] [Google Scholar]
- 7.Díez-García M, Capita R, Alonso-Calleja C. 2012. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 31:173–180. 10.1016/j.fm.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 8.Shi X, Zhu X. 2009. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 20:407–413. 10.1016/j.tifs.2009.01.054 [DOI] [Google Scholar]
- 9.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114–122. 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 10.Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ. 2012. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45:502–531. 10.1016/j.foodres.2011.01.038 [DOI] [Google Scholar]
- 11.Norouzi F, Mansouri S, Moradi M, Razavi M. 2010. Comparison of cell surface hydrophobicity and biofilm formation among ESBL- and non-ESBL-producing Pseudomonas aeruginosa clinical isolates. Afr. J. Microbiol. 4:1143–1147 [Google Scholar]
- 12.Capita R, Alonso-Calleja C. 2013. Antibiotic-resistant bacteria: a challenge for the food industry. Crit. Rev. Food Sci. Nutr. 53:11–48. 10.1080/10408398.2010.519837 [DOI] [PubMed] [Google Scholar]
- 13.OJEU 2009. Council Decision 2009/121/EC of 18 December 2008 rejecting the proposal from the Commission for a Council Regulation Implementing Regulation (EC) no. 853/2004 of the European Parliament and of the Council as regards the use of antimicrobial substances to remove surface contamination from poultry carcasses. Off. J. Eur. Union L42:13–15 [Google Scholar]
- 14.Sheridan A, Lenahan M, Duffy G, Fanning S, Burgess C. 2012. The potential for biocide tolerance in Escherichia coli and its impact on the response to food processing stresses. Food Control 26:98–106. 10.1016/j.foodcont.2012.01.018 [DOI] [Google Scholar]
- 15.CLSI 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals. Approved standard M31-A3. CLSI, Wayne, PA [Google Scholar]
- 16.Bridier A, Dubois-Brissonnet F, Boubetra A, Thomas V, Briandet R. 2010. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J. Microbiol. Methods 82:64–70. 10.1016/j.mimet.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 17.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 18.Heydorn A, Ersbøll BK, Hentzer M, Parsek MR, Givskov M, Molin S. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409–2415 [DOI] [PubMed] [Google Scholar]
- 19.Garthright WE. 1991. Refinements in the prediction of microbial growth curves. Food Microbiol. 8:239–248. 10.1016/0740-0020(91)90056-8 [DOI] [Google Scholar]
- 20.To MS, Favrin S, Romanova N, Griffiths MW. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5258–5264. 10.1128/AEM.68.11.5258-5264.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung HJ, Bang W, Drake MA. 2006. Stress response of Escherichia coli. Compr. Rev. Food Sci. Food Saf. 5:52–64. 10.1111/j.1541-4337.2006.00002.x [DOI] [Google Scholar]
- 22.Leistner L. 2000. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 55:181–186. 10.1016/S0168-1605(00)00161-6 [DOI] [PubMed] [Google Scholar]
- 23.Rodrigo D, Sampedro F, Silva A, Palop A. 2010. New food processing technologies as a paradigm of safety and quality. Br. Food J. 112:467–475. 10.1108/00070701011043727 [DOI] [Google Scholar]
- 24.Dourou D, Beauchamp CS, Yoon Y, Geornaras I, Belk KE, Smith GC, Nychas G-JE, Sofos JN. 2011. Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. Int. J. Food Microbiol. 149:262–268. 10.1016/j.ijfoodmicro.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 25.Reisner A, Haagensen JAJ, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933–946. 10.1046/j.1365-2958.2003.03490.x [DOI] [PubMed] [Google Scholar]
- 26.Patel J, Sharma M, Ravishakar S. 2011. Effect of curli expression and hydrophobicity of Escherichia coli O157:H7 on attachment to fresh produce surfaces. J. Appl. Microbiol. 110:737–745. 10.1111/j.1365-2672.2010.04933.x [DOI] [PubMed] [Google Scholar]
- 27.Vestby LK, Møretrø T, Langsrud S, Heir E, Nesse L. 2009. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal and feed factories. BMC Vet. Res. 5:20–25. 10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. 10.1038/416740a [DOI] [PubMed] [Google Scholar]
- 29.Machado I, Lopes SP, Sousa AM, Pereira MO. 2012. Adaptive response of single and binary Pseudomonas aeruginosa and Escherichia coli biofilms to benzalkonium chloride. J. Basic Microbiol. 52:43–52. 10.1002/jobm.201100137 [DOI] [PubMed] [Google Scholar]
- 30.Milisavljevic V, Tran LP, Batmalle C, Bootsma H. 2008. Benzyl alcohol and ethanol can enhance the pathogenic potential of clinical Staphylococcus epidermidis strains. Am. J. Infect. Control 36:552–558. 10.1016/j.ajic.2007.10.025 [DOI] [PubMed] [Google Scholar]
- 31.Stepanović S, Ćirković I, Mijać V, Švabić-Vlahović M. 2003. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 20:339–343. 10.1016/S0740-0020(02)00123-5 [DOI] [Google Scholar]
- 32.Tondo EC, Machado TRM, Malheiros PDS, Padrão DK, de Carvalho AL, Brandelli A. 2010. Adhesion and biocides inactivation of Salmonella on stainless steel and polyethylene. Braz. J. Microbiol. 41:1027–1037. 10.1590/S1517-83822010000400022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallardo-Moreno AM, González-Martín ML, Pérez-Giraldo C, Garduno E, Bruque JM, Gómez-García AC. 2002. Thermodynamic analysis of growth temperature dependence in the adhesion of Candida parapsilosis to polystyrene. Appl. Environ. Microbiol. 68:2610–2613. 10.1128/AEM.68.5.2610-2613.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallardo-Moreno AM, González-Martín ML, Pérez-Giraldo C, Bruque JM, Gómez-García AC. 2002. Serum as a factor influencing adhesion of Enterococcus faecalis to glass and silicone. Appl. Environ. Microbiol. 68:5784–5787. 10.1128/AEM.68.11.5784-5787.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi H, Suda T, Tanaka Y, Kimura B. 2010. Cellular hydrophobicity of Listeria monocytogenes involves initial attachment and biofilm formation on the surface of polyvinyl chloride. Lett. Appl. Microbiol. 50:618–625. 10.1111/j.1472-765X.2010.02842.x [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Baño J, Pascual A. 2008. Clinical significance of extended-spectrum beta-lactamases. Expert Rev. Anti Infect. Ther. 6:671–683. 10.1586/14787210.6.5.671 [DOI] [PubMed] [Google Scholar]
- 37.Loughlin MF, Jones MV, Lambert PA. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631–639. 10.1093/jac/49.4.631 [DOI] [PubMed] [Google Scholar]
- 38.Cerca N, Pier GB, Vilanova M, Olivera R, Azeredo J. 2005. Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res. Microbiol. 22:996–1006. 10.1016/j.resmic.2005.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giotis ES, Blair IS, McDowell DA. 2009. Effects of short-term alkaline adaptation on surface properties of Listeria monocytogenes 10403S. Open Food Sci. J. 3:62–65. 10.2174/1874256400903010062 [DOI] [Google Scholar]
- 40.Somers EB, Schoeni JL, Wong ACL. 1994. Effect of trisodium phosphate on biofilm and planktonic cells of Campylobacter jejuni, Escherichia coli O157:H7, Listeria monocytogenes and Salmonella Typhimurium. Int. J. Food Microbiol. 22:269–276. 10.1016/0168-1605(94)90178-3 [DOI] [PubMed] [Google Scholar]
- 41.Capita R, Alonso-Calleja C, García-Fernández MC, Moreno B. 2002. Review: trisodium phosphate treatment for decontamination of poultry. Food Sci. Technol. Int. 8:11–24. 10.1106/108201302023118 [DOI] [Google Scholar]
- 42.Alonso-Hernando A, Capita R, Prieto M, Alonso-Calleja C. 2009. Comparison of antibiotic resistance patterns in Listeria monocytogenes and Salmonella enterica strains pre-exposed and exposed to poultry decontaminants. Food Control 20:1108–1111. 10.1016/j.foodcont.2009.02.011 [DOI] [Google Scholar]
- 43.Van TTH, Chin J, Chapman T, Tran LT, Coloe PJ. 2008. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 124:217–223. 10.1016/j.ijfoodmicro.2008.03.029 [DOI] [PubMed] [Google Scholar]
- 44.Lei T, Tian W, He L, Huang X-H, Sun Y-X, Deng Y-T, Sun Y, Lv D-H, Wu C-M, Huang L-Z, Shen J-Z, Liu J-H. 2010. Antimicrobial resistance in Escherichia coli isolates from food animals, animal food products and companion animals in China. Vet. Microbiol. 146:85–89. 10.1016/j.vetmic.2010.04.025 [DOI] [PubMed] [Google Scholar]
- 45.Pallett A, Hand K. 2010. Complicated urinary tract infections: practical solutions for the treatment of multiresistant Gram-negative bacteria. J. Antimicrob. Chemother. 65(Suppl 3):iii25–iii33. 10.1093/jac/dkq298 [DOI] [PubMed] [Google Scholar]
- 46.Braoudaki M, Hilton AC. 2005. Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan. Int. J. Antimicrob. Agents 25:31–37. 10.1016/j.ijantimicag.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 47.Condell O, Iversen C, Cooney S, Power KA, Walsh C, Burgess C, Fanning S. 2012. Efficacy of biocides used in the modern food industry to control Salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl. Environ. Microbiol. 78:3087–3097. 10.1128/AEM.07534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikaido H. 1996. Multidrug efflux pumps of Gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braoudaki M, Hilton AC. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42:73–78. 10.1128/JCM.42.1.73-78.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell AD. 2001. Mechanisms of bacterial insusceptibility to biocides. Am. J. Infect. Control 29:259–261. 10.1067/mic.2001.115671 [DOI] [PubMed] [Google Scholar]
- 51.Young KD. 2007. Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 10:596–600. 10.1016/j.mib.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shalamanov DS. 2005. Chlorhexidine gluconate-induced morphological changes in Gram negative microorganisms. Biotechnol. Biotechnol. Equip. 19:121–124 [Google Scholar]
- 53.Ernst RK, Guina T, Miller SI. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327–1334. 10.1016/S1286-4579(01)01494-0 [DOI] [PubMed] [Google Scholar]
- 54.Sallum UW, Chen TT. 2008. Inducible resistance of fish bacterial pathogens to the antimicrobial peptide cecropin B. Antimicrob. Agents Chemother. 52:3006–3012. 10.1128/AAC.00023-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makinoshima H, Aizawa S, Hayashi H, Miki T, Nishimura A, Ishihama A. 2003. Growth phase-coupled alterations in cell structure and function of Escherichia coli. J. Bacteriol. 185:1338–1345. 10.1128/JB.185.4.1338-1345.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. 2000. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 52:662–668. [DOI] [PubMed] [Google Scholar]
- 57.Otto CC, Cunningham TM, Hansen MR, Haydel SE. 2010. Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin-resistant Staphylococcus aureus. Ann. Clin. Microb. Antimicrob. 9:26–38. 10.1186/1476-0711-9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becerril R, Gómez-Lus R, Goñi P, López P, Nerón C. 2007. Combination of analytical and microbiological techniques to study the antimicrobial activity of a new active food packaging containing cinnamon or oregano against E. coli and S. aureus. Anal. Bioanal. Chem. 388:1003–1011. 10.1007/s00216-007-1332-x [DOI] [PubMed] [Google Scholar]
- 59.Gustafson JE, Liew YC, Markham J, Bell HC, Wyllie SG, Warmington JR. 1998. Effects of tea tree oil on Escherichia coli. Lett. Appl. Microbiol. 26:194–198. 10.1046/j.1472-765X.1998.00317.x [DOI] [PubMed] [Google Scholar]
- 60.Bouhdid S, Abrini J, Amensour M, Zhiri A, Espuny MJ, Manresa A. 2010. Functional and ultrastructural changes in Pseudomonas aeruginosa and Staphylococcus aureus cells induced by Cinnamomum verum essential oil. J. Appl. Microbiol. 109:1139–1149. 10.1111/j.1365-2672.2010.04740.x [DOI] [PubMed] [Google Scholar]
- 61.Kadurugamuwa JL, Beveridge TJ. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615–621. 10.1093/jac/40.5.615 [DOI] [PubMed] [Google Scholar]
- 62.Belguith H, Kthiri F, Ben Ammar A, Jaafoura H, Ben Hamida J, Landoulsi A. 2009. Morphological and biochemical changes of Salmonella Hadar exposed to aqueous garlic extract. Int. J. Morphol. 27:705–713. 10.4067/S0717-95022009000300013 [DOI] [Google Scholar]
- 63.Irvin RT, Chatterjee AK, Sanderson KE, Costerton JW. 1975. Comparison of the cell envelope structure of a lipopolysaccharide-defective (heptose-deficient) strain and a smooth strain of Salmonella typhimurium. J. Bacteriol. 124:930–941 [DOI] [PMC free article] [PubMed] [Google Scholar]