Abstract
Here, we describe data obtained from transcriptome profiling of human cell lines and intestinal cells of a murine model upon exposure and colonization, respectively, with Bifidobacterium bifidum PRL2010. Significant changes were detected in the transcription of genes that are known to be involved in innate immunity. Furthermore, results from enzyme-linked immunosorbent assays (ELISAs) showed that exposure to B. bifidum PRL2010 causes enhanced production of interleukin 6 (IL-6) and IL-8 cytokines, presumably through NF-κB activation. The obtained global transcription profiles strongly suggest that Bifidobacterium bifidum PRL2010 modulates the innate immune response of the host.
INTRODUCTION
During the last decade, bifidobacteria have attracted a lot of scientific attention due to their perceived role as health-promoting or probiotic bacteria. Although there is suggestive proof to corroborate some of these functional claims, the molecular mechanisms behind such probiotic activities remain largely unknown. The decoding and functional analysis of genome sequences of probiotic bacteria, i.e., probiogenomics, offer the possibility of accelerating research into the mechanisms of probiotic action (1, 2). Probiogenomic investigations have highlighted a plethora of genetic features that may explain how bifidobacteria have so well adapted to the human gut. Prominent examples are represented by the genes and their products that allow bifidobacteria to interact with the human host, either through different morphological structures, such as pili (3, 4) or exopolysaccharides (5), or through the utilization of host-derived glycans, such as mucin or human milk oligosaccharides (6, 7). Nevertheless, so far, relatively little is known about the changes that occur in the host's transcriptome in response to bifidobacterial colonization/exposure (8, 9). Among the various human bifidobacteria, a special mention should be reserved for Bifidobacterium bifidum PRL2010, whose genome has been subjected to sequencing and functional analysis, which has revealed a number of key traits that render this bacterium a model organism for the investigation of human-microbe coevolution (6, 10, 11). These analyses of B. bifidum PRL2010 have provided insights that explain not only the genomic strategy adopted by this strain to metabolize host-derived glycans (6, 10) but also the process of host colonization through sortase-dependent pili (12).
B. bifidum strains are claimed to exert a key role in the evolution and maturation of the immune system of the host, which is still rather undeveloped following birth (9). The interaction of B. bifidum with the host immune system has recently been explored by analyzing the impact of B. bifidum Z9 in combination with a second human gut commensal, Lactobacillus acidophilus, on the transcriptome of dendritic cells (DCs) (8). This study revealed that B. bifidum Z9 inhibits expression of genes related to the adaptive immune system in murine dendritic cells. These findings are in line with those of other publications that employed in vitro assays involving various bifidobacterial strains belonging to different species, showing a clear and distinct cytokine profile induced by bifidobacteria (9, 13).
The purpose of the current study was to investigate the particular host gene expression profile following exposure to and/or colonization with B. bifidum PRL2010 by employing microarray-based transcriptome analyses coupled with in vitro (colonic cell line) and in vivo (murine-colonization) approaches.
MATERIALS AND METHODS
Growth conditions.
B. bifidum PRL2010 was cultivated in an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2) in a chamber (Concept 400; Ruskin) at 37°C for 32 h in de Man-Rogosa-Sharpe (MRS; Scharlau Chemie, Barcelona, Spain) medium, supplemented with 0.05% (wt/vol) l-cysteine hydrochloride.
Stimulation of Caco-2 monolayers and ELISA measurement of cytokine production.
A predetermined number of Caco-2 cells was seeded into 24-well plates and grown as described previously (14). B. bifidum PRL2010 cells (final concentration of 108 bacterial cells ml−1) were added to monolayers of Caco-2 cells at a multiplicity of infection (MOI) of 100 in 0.5 ml of fresh antibiotic-free Dulbecco's modified Eagle's medium (DMEM) and incubated for 18 h at 37°C in 5% CO2. This timing was optimized as described previously (14). The cytokine interleukin 1β (IL-1β) (1 ng/ml), casein (10 μg/ml), and bovine serum albumin (BSA) (10 μg/ml) were used as controls. Microtiter plates were kept at −70°C for 4 h to completely disrupt the Caco-2 cells. The supernatant and disrupted Caco-2 cells were then collected, and phenylmethylsulfonyl fluoride (20 mM final concentration) was added. After centrifugation at a relative centrifugal force of 16,000 × g for 1 min, levels of IL-2, IL-4, IL-6, IL-8, IL-10, gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) in the supernatants were determined by enzyme-linked immunosorbent assay (ELISA) using Bio-Plex human cytokine multiplex panel (Bio-Rad Laboratories, Segrate, Italy) as instructed by the manufacturer. Each sample was processed in duplicate in two independent experiments.
Evaluation of NF-κB activation.
A stable recombinant Caco-2 cell line was obtained by transfecting cells with the plasmid pNiFty2-Luc (InvivoGen, Labogen, Rho, Italy) as described previously (15). This plasmid combines five nuclear factor κB (NF-κB)-dependent transcriptional activation sites upstream of the insect luciferase reporter gene luc. The presence of active NF-κB molecules in the cell activates the promoter, resulting in expression of the luciferase gene. Following growth in the presence of 50 μg/ml zeocin, cell monolayers (approximately 3 × 105 cells/well) were carefully washed with 0.1 M Tris-HCl buffer (pH 8.0). Subsequently, 50 μl of a bacterial suspension containing 2.5 × 108 cells was added to 0.45 ml of fresh DMEM containing 100 mM HEPES (pH 7.4). The resulting 500 μl was finally pipetted into the microtiter plate well containing the Caco-2 cell layer, resulting in an MOI of 100. Stimulation was conducted both in the presence and the absence of 2 ng/ml of IL-1β. After incubation at 37°C for 4 h, the samples were treated and the bioluminescence was measured as described previously (16). All conditions were analyzed in triplicate in at least two independent experiments.
Tissue culture experiments.
About 2 × 105 HT29 cells in 1.5 ml of DMEM (high glucose, HEPES) medium supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies, Italy), penicillin (100 U/ml), streptomycin (0.1 mg/ml), amphotericin B (0.25 μg/ml), and 4 mM l-glutamine were seeded into the upper compartments of a six-well transwell plate. The lower compartments contained 3.0 ml of the same medium. HT29 cells were incubated at 37°C in a 5% CO2 atmosphere until they reached 3 days postconfluence. Cells were then washed with Hank's solution and stepped down in DMEM supplemented with l-glutamine (4 mM), sodium selenite (0.2 μg/ml), and transferrin (5 μg/ml) for 24 h. These transwell inserts were transferred to an anaerobic culture box within an MACS-MG-1000 anaerobic workstation at 37°C, and each insert was filled with anaerobic DMEM. A culture of B. bifidum PRL2010 grown to exponential phase was harvested by centrifugation at 3,500 × g for 5 min and washed with 10 ml of anaerobic DMEM. The obtained pellet was resuspended in 0.8 ml of the same medium. One hundred microliters of bacterial suspension (108 CFU/ml) was added to wells, with control wells receiving the same amount of medium without bacterial cells. An additional control included bacterial cells incubated without HT29 cells.
Tissue culture cells were harvested for analyses after 1 h, 2 h, and 4 h of incubation (with bacterial cultures) and 1 h (unexposed cell control placed in contact with phosphate-buffered saline [PBS] for 1 h) for time course-dependent gene expression studies. This study design is similar to those employed in two previous studies, one involving B. bifidum PRL2010 and a Caco-2 human cell monolayer (6) and the second involving human gut commensal bacteria and human epithelial cells (17). Nonadherent bacteria were carefully aspirated from the wells and pooled, while the adherent fraction was collected after washing of the inserts with anaerobic media and also pooled. Each fraction was collected into 1.5-ml tubes and centrifuged at 3,500 × g for 5 min, and the resulting pellet resuspended in 400 μl of RNAlater and subjected to RNA extraction according to the protocol described below. Caco-2 cells or HT29 cells were harvested from the wells, pooled, and stored in RNAlater at 4°C.
Mouse colonization.
All procedures were approved by the University of Parma, as represented by the Institutional Animal Care and Use Committee (Dipartimento per la Sanità Pubblica Veterinaria, la Nutrizione e la Sicurezza degli Alimenti Direzione Generale della Sanità Animale e del Farmaco Veterinario). Two groups of mice were orally inoculated with either 109 CFU of PRL2010 cells (test strain) or water (control), according to previously described protocols (12, 18). Each group contained five animals of 3-month-old female BALB/c mice. Bacterial colonization was established by five consecutive daily administrations, during which each animal received 20 μl of 109 cells using a micropipette tip placed immediately behind the incisors (19). Bifidobacterial inocula were prepared as described previously (18).
In order to estimate the number of B. bifidum PRL2010 cells per gram of feces, individual fecal samples were serially diluted and cultured on selective agar (MRS agar) using mupirocin as described previously (20). Following enumeration of B. bifidum PRL2010 cells in fecal samples, 100 random colonies were further tested to verify their identity using PCR primers targeting the pil-2 and pil-3 loci (3).
Animals were sacrificed by cervical dislocation, and individual gastrointestinal tracts were removed and used for RNA extraction.
Eukaryotic RNA isolation.
Human cell lines as well as cells from murine gastrointestinal tracts in RNAlater were diluted 1:1 in an equal volume of sterile PBS, followed by centrifugation at 5,000 × g for 10 min at 4°C. Total RNA from the pellet was isolated using the RNeasy minikit (Qiagen, Italy) according to the manufacturer's instructions, including an RNase-free DNase I (Qiagen, Italy) digestion step. Eukaryotic RNA integrity was determined using the Experion automated electrophoresis system (Bio-Rad, Italy).
Acceptable RNA purity values (optical density at 260 nm [OD260]/OD280) had to be ≥1.8, with corresponding RNA integrity numbers (RIN) ranging from 7 to 9.
Microarray, description, labeling, and hybridizations.
Microarray analysis was performed with an oligonucleotide array based on the human genome as well as the murine genome from CombiMatrix. Oligonucleotides were present in triplicate on a 90k CombiMatrix array (CombiMatrix, Mukilteo, WA, USA). Replicates were distributed on the chip at random, nonadjacent positions. A set of 74 negative-control probes designed on the basis of phage and plant sequences were also included on the chip.
Reverse transcription (RT) and amplification of 500 ng of total RNA was performed with ImProm-II reverse transcriptase (Promega, Madison, USA) according to the manufacturer's instructions. Five micrograms of cDNA was then labeled with a ULS labeling kit for CombiMatrix arrays with Cy5 (Kreatech, The Netherlands). Hybridizations were performed according to a previously described protocol (21).
Microarray data acquisition and treatment.
Fluorescence scanning was performed on an InnoScan 710 microarray scanner (Innopsys, France). Signal intensities for each spot were determined using GenePix Pro 7 software (Molecular Devices, USA). Signal background was calculated as the mean of results for negative controls plus 2 times the standard deviation (22). A global quantile normalization analysis was performed (23), and log2 ratios between the reference sample and the test sample results were calculated. The distribution of the log2-transformed ratios was separately calculated for each hybridization reaction. The fold change cutoff value for transcriptomic data analyses was ≥2 for upregulated genes and ≤0.5 for downregulated genes. Only those genes whose transcription levels were shown to be significantly different (24) were further taken into consideration in our analyses.
Statistical analysis.
Statistical significance between means was analyzed using the unpaired Student t test, with a threshold P value of <0.0005. Values are expressed as the means ± the standard errors of the means of results from three experiments. Statistical calculations were performed using the software program GraphPad (La Jolla, CA, USA) Prism 5.
Microarray accession number.
The transcriptional array data have been deposited in the GEO database under accession number GSE48533.
RESULTS AND DISCUSSION
Modulation of cytokine gene expression by PRL2010 in Caco-2 cells is mediated by NF-κB activation.
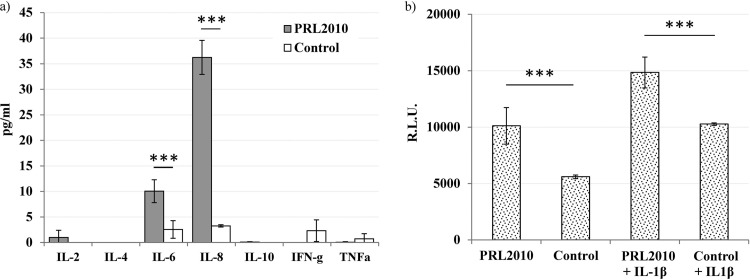
Caco-2 cells are human colonic adenocarcinoma cells that are able to express differentiation features typical of mature intestinal cells and that, consequently, are one of the most commonly employed cell lines for in vitro studies related to intestinal cell function and differentiation (25). In a previous study, we demonstrated that, using this Caco-2 in vitro model, B. bifidum PRL2010 can modulate the expression of genes encoding certain interleukins and cytokine transcriptional regulators (6). Specifically, 4-h incubation of PRL2010 with Caco-2 cells was shown to cause a significant rise in the transcription (4.19-fold; P < 0.0005) of the genes for the nuclear factor κB (NF-κB) p105 subunit (NFKB1) and interleukin 8 (IL-8) (6). Particularly, the increased transcription of NFKB1 is expected to cause enhanced activation of the transcriptional regulator NF-κB, which is a well-known transcriptional inducer of IL-8, thus explaining its observed enhanced transcription, as well as of other cytokines. To experimentally confirm this notion, we quantified several cytokines in the broth culture of Caco-2 cells by ELISA analysis. Differentiated Caco-2 layers were prepared as previously described (14) and stimulated with B. bifidum PRL2010 cells for 18 h at a bifidobacterial-cell/Caco-2-cell ratio of 100. The results clearly show that strain PRL2010 induced an 11-fold (P < 0.001) and a 4-fold (P < 0.001) increase in secreted IL-6 and IL-8, respectively (Fig. 1a), while broth concentrations of IL-2, IL-4, IL-10, IFN-γ, and TNF-α were not significantly above the detection limit (Fig. 1a). The ability to directly stimulate secretion of similar cytokines by Caco-2 cells has not received much attention. For instance, B. bifidum MIMBb75 has been reported to trigger IL-8 production, whereas IL-6 was shown to be unaffected (14). In contrast, the enhanced simultaneous stimulation of IL-6 and IL-8 was reported for other bifidobacterial species, such as B. breve (26).
FIG 1.
Effect of B. bifidum PRL2010 on cytokine production by Caco-2 cells. (a) ELISA quantification of secreted IL-6 and IL-8 following an 18-h stimulation at a bifidobacterial-cell/Caco-2-cell ratio of 100. (b) Effect of PRL2010 on Caco-2 cells stably transfected with an NF-κB/luciferase reporter vector, with or without stimulation with IL-1β (2 ng/ml). R.L.U., relative luminescence units. Numerical results from two experiments conducted in duplicate are given as arithmetic means ± standard deviations. The vertical lines indicate standard deviations. ***, statistically significant difference from control value (untreated Caco-2 cells) according to an unpaired Student t test (P < 0.001).
To further explore the immune-modulatory properties of PRL2010, we tested the effect of this bifidobacterial strain on NF-κB activation using a recombinant cell line, obtained by transfecting Caco-2 cells with a vector containing an NF-κB-dependent luciferase reporter (15). In this model, 4-h incubation with strain PRL2010 showed a stimulatory effect on NF-κB-dependent production of bioluminescence (+81%) (Fig. 1b). The increase of NF-κB activation was also evident for strain PRL2010 when Caco-2 cells were incubated with bacteria and IL-1β, which has been used as a proinflammatory stimulus. These results demonstrate that B. bifidum PRL2010 can actively modulate epithelial cell responses at a transcriptional level. A similar scenario has previously been noticed for a number of other bacteria, which were shown to either increase (in the case of Streptococcus salivarius ST3 [27]) or decrease (in the case of Lactobacillus helveticus MIMLh5 [15]) NF-κB activation in epithelial cells. Further examples concerning bifidobacteria are available in the literature; secreted factors from Bifidobacterium animalis subsp. lactis were shown to inhibit NF-κB-mediated IL-8 gene expression in Caco-2 cells (28), while increased NF-κB expression in Caco-2 cells was observed upon stimulation with B. breve DSMZ 20091-conditioned media (29). Therefore, modulation of the NF-κB signal transduction pathway can be considered a general mechanism of cross talk between host epithelial cells and various (commensal and/or probiotic) microorganisms. In addition, the observed ability of B. bifidum PRL2010 to induce chemokine production of IL-8 by intestinal epithelial cells may contribute to the enhancement of the innate immune response.
Modulation of HT29 gene expression by PRL2010.
The human HT29 cell line was originally isolated from a human colonic carcinoma, and it is commonly considered a valid human colonic mucosa model for study of human-microbe interactions (30–32). We therefore decided to use an HT29 monolayer as an in vitro model to investigate the transcriptomic impact of B. bifidum PRL2010 on these human intestinal cells. HT29 monolayers cultivated at 15 days postconfluence were placed in contact with PRL2010 cells for various lengths of time, namely, 0 h, 1 h (T1), and 2 h (T2) to 4 h (T4), as well as with PBS for 1 h (representing an unexposed cell monolayer control).
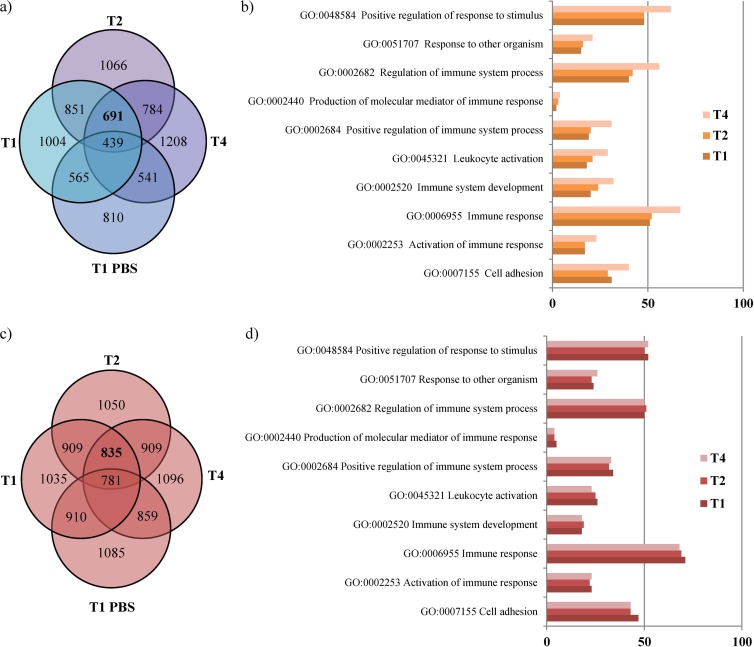
Microarray analysis was performed on samples of tissue for each time point with the use of a CombiMatrix human array, which contains 62,078 oligonucleotides representing >24,311 cDNAs. Interestingly, a change in the transcription profile occurred when different times of exposure to PRL2010 cells were used; 1,526 genes exhibited a significant change in their transcription for all time points (691 genes were upregulated and 835 genes were downregulated) (Fig. 2a and c). The number of genes whose transcription exhibited significant changes when exposed to PRL2010 cells for different lengths of time are also shown in Fig. 2a and c. Functional classification of the up- and downregulated PRL2010-affected human genes according to the Gene Ontology (GO) database using GO terms (AmiGo) is shown in Fig. 2b and d. Notably, the majority of the host genes whose transcription was influenced by the presence of PRL2010 cells were shown to be involved in immune response and positive regulation of response to stimulus, which confirmed previously published data (6). Interestingly, we noticed that after 4 h of treatment of HT29 with PRL2010 (time point T4), the number of genes for each GO assignment was consistently higher than those of the other time points assayed, thus suggesting that PRL2010 cells are most effective in modulating the transcriptome of HT29 human cell monolayers upon 4 h of contact. These results also reinforce what was previously noticed for similar experiments performed on a different type of human cell monolayer (6). In general, analysis of the HT29 monolayer transcriptome data suggests that PRL2010 elicits a host innate immune response. This idea is supported by the significant downregulation of several genes coding for cytokines and cytokine receptors. Among these, we observed a decrease in the transcription of genes specifying cytokines and a related receptor belonging to the IL-10 family (33), namely, IL-20, IL-24, and the receptor for IL-22 (Fig. 3b). For instance, IL-20 has been shown to induce expression of proinflammatory cytokines, such as TNF-α in HaCat keratinocytes (34), and to act as a proinflammatory mediator in rheumatoid and experimental arthritis (35). Furthermore, the IL-22 receptor, which is found on cells of nonhematopoietic origin in the skin, kidney, liver, lung, and gut, allows regulation of local epithelial cell responses after infection or exposure to inflammatory stimuli mediated by IL-22 (36). IL-22 is a pleiotropic cytokine that has been shown to either enhance maintenance of the epithelial barrier (37) or exert proinflammatory/pathological properties (38) following infection. In addition, PRL2010 was shown to downregulate transcription of the gene specifying IL-25, a member of the IL-17 family known to be expressed in epithelial cells and involved in triggering Th2-type cytokines; it is thus implied to play a role in allergy (39).
FIG 2.
Venn diagram illustrating the numbers of shared and unique human genes upregulated (a) and downregulated (c) between each time point of sampling and the corresponding GO functional categories of the upregulated human genes (b) and downregulated human genes (d). The PRL2010 cells were placed in contact with an HT29 monolayer for different lengths of time, i.e., 1 h (T1) or 2 h (T2) to 4 h (T4); as a control, HT29 monolayer cells were incubated with PBS for 1 h (T1 PBS). Each circle in the Venn diagram represents the number of genes upregulated (a) or downregulated (b) at different time points. The number of genes within each GO functional category observed to be upregulated (b) or downregulated (d) for various time points is indicated on the x axis. The code for each GO category described is provided.
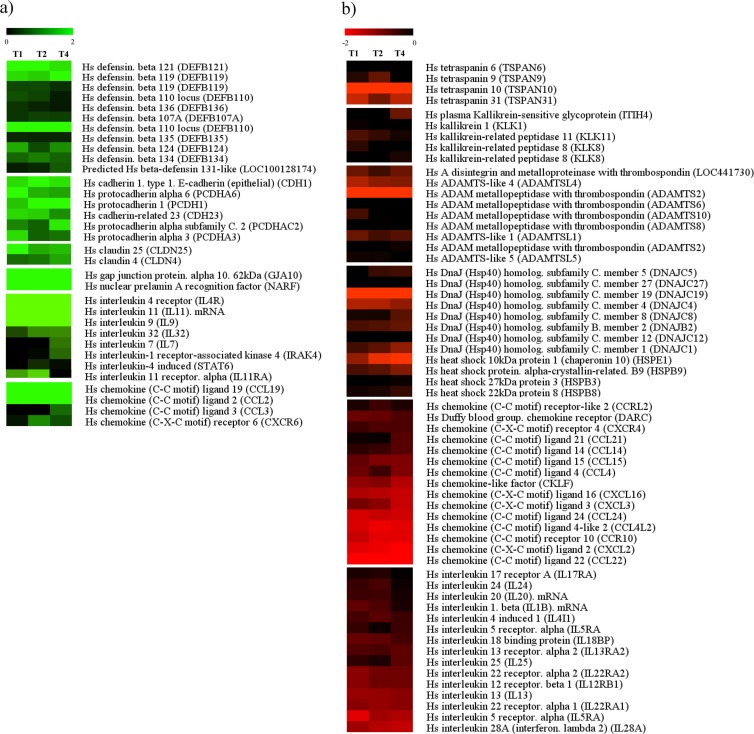
FIG 3.
Identification of the most statistically significantly differentially expressed Homo sapiens (Hs) genes upon treatment with PRL2010 by microarray analyses. (a) Upregulated human genes; (b) downregulated human genes. Each row represents a separate transcript, and each column represents a separate sample (T1, T2, and T4). The color legend is at the top of the microarray plot; green indicates increased transcription levels compared to those of the reference sample.
The effect of PRL2010 cells on the HT29 transcriptome also positively affected the transcription of certain interleukin-encoding genes. In particular, PRL2010 exposure induced a marked increase (4-fold) in the transcription of the IL-11-encoding gene and a modest increase (2.1-fold) in the transcription of the gene specifying its corresponding receptor, IL11RA (Fig. 3a). Interestingly, preclinical in vivo and in vitro studies have demonstrated that IL-11 inhibits the secretion of a number of proinflammatory cytokines, such as TNF, IL-12, IL-1β, and nitric oxide, from activated macrophages as well as gamma interferon (IFN-γ) and IL-12 from activated T cells (40–42).
Intestinal epithelial cells are important players in gut homeostasis due to their role in regulating inflammation also through the expression of chemokines and related receptors and the consequential interaction with circulating leukocytes (43). Chemokines are a superfamily of chemotactic cytokines which are involved in attraction and recruitment of leukocytes to the site of inflammation, while they also play a role in angiogenesis and carcinogenesis (44). Our data show that the stimulation of HT29 cells with B. bifidum PRL2010 induced significant transcriptional downregulation of genes that encode several chemokines and chemokine receptors (Fig. 3b). For instance, PRL2010 was shown to downregulate the genes coding for chemokines of the CXC family, such as CXCL2 and CXCL3. These two chemokines (also known as growth-related oncogenes GRO2 and GRO3) have been found to be overexpressed in colorectal carcinoma compared to healthy tissue (45). CXCL2 and CXCL3 are in fact considered biomarkers for the angiogenesis of tumors, and their expression in HT29 was demonstrated to be triggered by proinflammatory stimulation (44). Nevertheless, in vivo studies of humans involving PRL2010 should be performed in order to further investigate the relevance and importance of these findings. Furthermore, PRL2010 was shown to markedly downregulate the transcription of the CXCL16-encoding gene. Interestingly, CXCL16 is considered a marker for inflammation due to its perceived role in the development of inflammatory bowel disease (46). In addition, expression of CXCL16 has been found to be upregulated in several types of primary and metastatic cancer tissue and cancer cell lines and has been proposed as a prognostic marker for colorectal cancer (CRC) (47).
The chemokine whose corresponding gene was most strongly repressed by PRL2010 exposure was CCL22 (Fig. 3b). Also known as macrophage-derived chemokine (MDC), CCL22 has been found to be expressed in the small intestine (48) and was demonstrated to be a potent chemoattractant of immune cells, in particular, immature dendritic cells (DCs). High levels of CCL22 have been found following stimulation with proinflammatory stimuli like TNF-α, IL-1β, IFN-γ, and lipopolysaccharide (LPS) (49, 50) and in specimens of inflamed mucosae (51).
Finally, PRL2010 exposure induced a strong increase (5-fold) in the transcription of the gene encoding CCL19, which is a chemokine that attracts both DCs and T lymphocytes. CCL19 expression has been found to be enhanced during inflammation by cells properly associated with the immune system (e.g., DCs) (52). Nonetheless, it is noteworthy that CCL19 has been shown to serve as a potential immune stimulator that can reduce tumor burden in a model of advanced lung cancer (53). Taken together, these results indicate an opposing effect of PRL2010 exposure on the production of certain chemokines, such as CCL22 and CCL19, with apparently similar functions, which supports the idea that PRL2010 has a balanced effect on chemokine-mediated immune functions.
It has been well established that the intestinal mucosa plays a pivotal role in rejecting pathogens, dampening inflammatory responses, and discriminating between “friends and foes,” thus orchestrating tolerance to or mounting immune responses against commensals or pathogens, respectively. Consequently, the overall ability of B. bifidum PRL2010 to modulate cytokine/chemokine gene expression in HT29 cells suggests that this bacterium can actively exert tolerogenic and potentially immunomodulatory effects.
The ability of PRL2010 to modulate inflammatory responses is corroborated by the observed transcriptional downregulation of the genes encoding various heat shock proteins (HSPs) of the host (Fig. 3b). Although best described as protein chaperones during cellular stress (54), recent evidence suggests that HSPs are also important for activation of the innate immune response (55). Release of inducible HSPs from necrotic cells is interpreted as a danger signal by antigen-presenting cells and as a result causes cellular activation and cytokine production (56, 57). Furthermore, peptides binding to HSPs act as sources of antigen and affect the maturation of dendritic cells (56, 58). Upregulation of HSP expression is the general response to all types of general stresses, including infection (59). Consequently, downregulation of HSPs by B. bifidum PRL2010 can be interpreted as a means to affect the host inflammatory response to pathogens. This scenario is completely different than that caused by many enteropathogens, which have been shown to increase the levels of expression of HSPs (60). Furthermore, according to a recent model regarding tumor development, HSPs have been implicated in tumorigenesis resulting from both infection and chronic inflammation (60).
Other genes involved in bacterial-host interaction are genes encoding integral transmembrane proteins identified in tight junctions (TJ) called cadherin and claudin (61). Cadherins and claudins play an important role in the maintenance of cell-cell adhesion and recognition, and we found an upregulation especially for cadherin type 1 (3.5-fold; P < 0.0005), type 23 (3-fold; P < 0.0005), protocadherin 6 (2.8-fold; P < 0.0005), claudin 25 (2.5-fold; P < 0.0005), and claudin 4 (2.3-fold; P < 0.0005) (Fig. 3a). It has previously been shown that an opposite expression effect is observed for pathogens, as they are known to cause a downregulation of cadherin, thereby linking it to early gastric carcinogenesis events (62).
Remarkably, a series of β-defensin genes were also upregulated by PRL2010. The β-defensin family is one of the major antimicrobial protein families that can be expressed in the large intestine, acting against Gram-positive and Gram-negative bacteria (63), being efficacious against Enterobacteriaceae (e.g., Escherichia coli, Salmonella enterica serovar Typhimurium, Listeria monocytogenes), Streptococcus pyogenes, Staphylococcus aureus, Pseudomonas aeruginosa, and Bacteroides vulgatus (64). This finding, together with the observed overexpression of genes coding for TJ proteins, supports the notion of B. bifidum PRL2010 being an active contributor to the strengthening of the intestinal epithelium barrier as part of the innate immune response. Interestingly, transcription of the genes encoding tetraspanin membrane proteins, which are used by pathogens to cause infection (65, 66), is downregulated following contact with PRL2010, especially for cadherin types 6, 9, 10, and 31 (≤0.5-fold; P < 0.0005).
We also found transcriptional downregulation of kallikrein genes (≤0.5-fold; P < 0.0005). Many bacterial pathogens interfere with the contact system (kallikrein-kinin system) in human plasma, especially with the release of proinflammatory kinin (67). The downregulation of these genes may reflect an anti-inflammatory activity exerted by beneficial microorganisms (Fig. 3b).
The ADAM (A disintegrin and metalloproteinase) family proteins represent membrane-anchored cell surface glycoproteins that may have important roles as modulators of inflammation (68, 69). Pathogens cause upregulation of expression of ADAM membrane glycoproteins in epithelial cells at mucosal sites, and this upregulation may be instrumental in regulating local mucosal immune responses, growth factor-mediated epithelial proliferation, and tissue remodeling associated with chronic mucosal infections (70). We found transcriptional downregulation of a small number of ADAM genes, especially ADAMTS2 and ADAMTS8 (≤0.5-fold; P < 0.0005) (Fig. 3b).
The microarray results based on HT29 cells were verified by RT-PCR for the most significant HT29 up- or downregulated genes. Such analyses corroborated upregulation of IL-32 and IL-9 and downregulation of IL-13, CXCL3, CXCL2, and CXCL16 in PRL2010-treated HT29 cells (see Fig. S1a in the supplemental material).
Transcriptomic profile of PRL2010 colonization in the murine model.
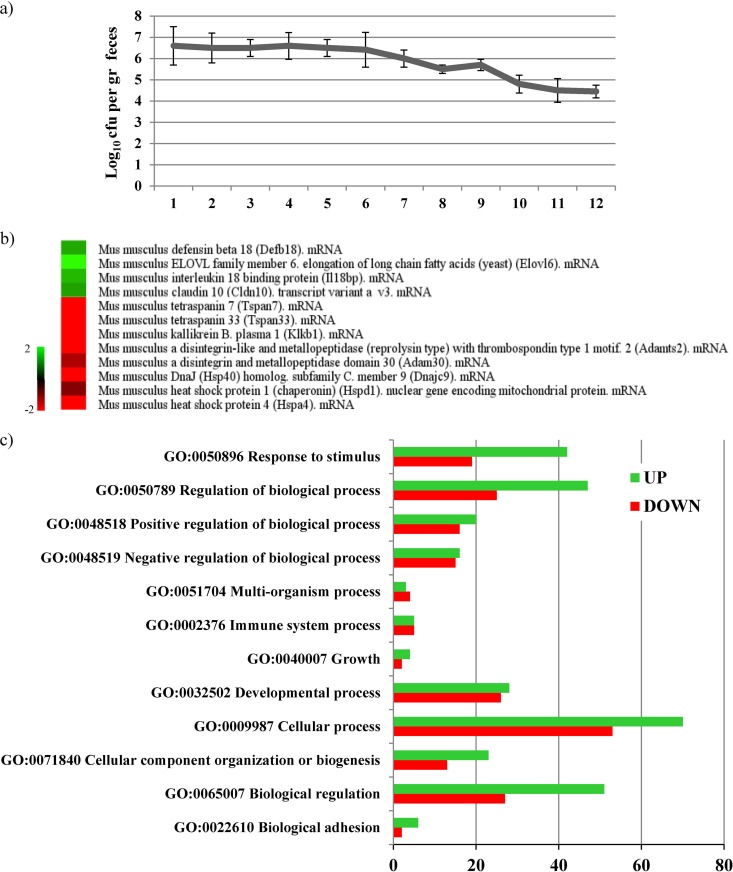
In order to substantiate the global transcription data achieved when exposing PRL2010 to human cell lines, we extended our investigations using BALB/c mice as an in vivo host-microbe model. Colonization and clearance of PRL2010 was monitored during the period of bacterial administration as well as for 1 week of posttreatment by determining viable counts of PRL2010 cells recovered from fecal samples. Levels of PRL2010 colonization remained stable, at approximately 106 CFU/g of fecal material during the course of the intervention period and the subsequent 2 days and at around 105 CFU/g during the washout period (Fig. 4a). After the mice were sacrificed by cervical dislocation, their individual gastrointestinal tracts were removed and used for RNA extraction. Total RNA was extracted from cecal tissue samples and hybridized on a CombiMatrix mouse array representing 24,667 murine cDNAs (NCBI source).
FIG 4.
Host response upon in vivo mice colonization of B. bifidum PRL2010. (s) Population numbers of B. bifidum PRL2010 organisms colonizing the intestines of BALB/c mice upon five consecutive daily administrations of 109 CFU of PRL2010 followed by a washout period of 1 week. Each point represents the average of the log-transformed population sizes ± standard deviation for five mice. The days of treatment and washout are indicated on the x axis. (b) Selection of differentially expressed murine genes upon colonization with PRL2010 as determined by transcriptome analyses. Green indicates upregulated genes and red downregulated genes upon treatment with PRL2010 cells. (c) GO functional categories of the differentially expressed mouse genes upon PRL2010 colonization. Numbers of genes within each GO functional category resulting in upregulation (represented by green bars) or downregulation (indicated by red bars) according to the different time points are indicated on the x axis. The code for each GO category described is provided.
Transcriptome analysis showed that upon colonization by PRL2010, a total of 288 murine genes were transcribed differentially from those of the murine control group (Fig. 4b), which is numerically less than those identified in the transcriptome of the human cell monolayer. This could be explained by the fact that, compared to experiments in human cell monolayers, in vivo trials in a murine model are based on a more complex biological system (e.g., a complete eukaryotic organism with a complex gut microbiota) and could ultimately be more buffered in terms of gene expression. One hundred forty-three genes were significantly upregulated by more than 2-fold, while 91 genes were significantly downregulated upon PRL2010 colonization. Functional classification of the murine genes that were upregulated by PRL2010 mediation indicated that the majority encode proteins involved in cellular processes and regulation, as well as in responses to stimuli (Fig. 4b).
As with the in vitro experiments involving HT29, the transcriptomes of colonized mice depicted a clear transcriptional downregulation of murine HSP-encoding genes, such as those for Hspd1, Hspa4, and Hsp40 (Fig. 4b).
Furthermore, the transcriptomes of mice colonized with PRL2010 were shown to exhibit enhanced transcription of the ctnNal1 and cldn10 genes, encoding catenin alpha-like 1 and claudin 10, respectively, which are proteins located at the tight junctions (TJ) of epithelial cells. These results are in accordance with our in vitro observations using the HT29 model with regard to the upregulation of tight junction-associated proteins. Catenins, cadherins, and claudins, in fact, play a key structural role in the maintenance of the intestinal barrier against infection with enteropathogens like Listeria, which are known to infect the host by penetrating the intestinal epithelium through a paracellular route (71).
Transcriptional analyses also provided evidence that PRL2010 increases the transcription of genes encoding antimicrobial proteins such as β-defensin 18 (Fig. 4b). This protein belongs to the defensin family proteins that are expressed in neutrophils and on mucosal surfaces, where they are thought to play key roles in innate host defense (72).
Another interesting indication of the immune-modulatory role exerted by PRL2010 in the gut is represented by the downregulation of the transmembrane protein tetraspanin 33. Tetraspanins are used by intracellular pathogens as a means of entering and replicating within human cells. Although previous investigations have focused mainly on viruses such as hepatitis C and HIV, it has now become evident that other microorganisms may also associate with tetraspanins, using tetraspanin-enriched microdomain as a gateway to infection (73).
The gene encoding kallikrein B, plasma 1 (Klkb1) was shown to undergo transcriptional downregulation upon exposure to PRL2010, which further reinforces the hypothesis that this microorganism might invoke a potential anti-inflammatory activity. Kallikrein cleaves kininogens to release kinins, which in turn act as inflammation mediators. Intestinal kallikrein may degrade growth factors and peptides, whereas kinins are responsible for capillary permeability, pain, synthesis of cytokines, and the adhesion molecule neutrophil cascade. Recent studies have shown that kallikrein is present in intestinal goblet cells and is released into interstitial space during inflammation (74).
Other interesting proteins mediating host inflammation are represented by disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif 2 (ADAMTSL4). Notably, the transcriptomes of mice colonized by PRL2010 show a downregulation of the genes encoding Adamts, which confirmed what was observed with in vitro assays involving HT29 human cell lines (see above and Fig. S2 in the supplemental material).
Finally, microarray analysis revealed that B. bifidum PRL2010 was able to highly induce (5-fold) the expression of host-metabolism-related genes, such as those encoding ELOVL family member 6, elongation of long-chain fatty acids (Elovl6). This enzyme catalyzes the synthesis of saturated and monounsaturated fatty acids, as well as the conversion of palmitate to stearate (75).
Interestingly, mice deficient for Elovl6 have been shown to become obese and develop hepatosteatosis when fed a high-fat diet, identifying Elovl6 as a promising target for the treatment of metabolic disorders (76).
The transcription levels of the most significantly downregulated murine genes as identified by DNA microarray experiments were also assayed by quantitative RT-PCR. These analyses indeed confirmed downregulation of various hsp genes, such as cspd1, dnaj9, hspd1, and hspa4 in the PRL2010-treated animals (Fig. S1b).
Conclusions.
A large body of data pertaining to modulated genes in B. bifidum PRL2010 upon contact with human cell lines (6) or during colonization of animal models (12) was previously obtained. However, very little was known about the transcriptional response of the host as a consequence of the presence of PRL2010 cells. Here, through the use of high-throughput gene expression technology and by employing both an in vitro cell line model and a murine model, we assayed the host gene response triggered by B. bifidum PRL2010 cells. Comparison of transcriptome changes following B. bifidum PRL2010 exposure with the identified murine transcriptome changes upon colonization of mice with PRL2010 cells revealed intriguing similarities with regard to transcription profiles involving HSP-encoding genes and genes involved in tight junctions, as well as Adamts-encoding genes. Such findings involving two different models (e.g., a rather simple HT29 model and a complete eukaryotic organism) clearly support the possibility of a specific effect of B. bifidum PRL2010 on the human immune system. The data presented also confirm and extend previous published results achieved from Caco-2 cell monolayers. Interestingly, the overall host response scenario driven by PRL2010 cells may be summarized as a stimulatory response to prime the immune system, while it, at the same time, attenuates the proinflammatory response by downregulating certain chemokines and HSPs and upregulating defensin and tight junction genes (Fig. S2). Overall, these results suggest that PRL2010 exerts an immunomodulatory activity as well as a reinforcement of the innate defense toward its host during gut colonization. Future comparative studies involving B. bifidum PRL2010 as well as other bifidobacterial strains belonging to different species will be carried out in order to evaluate the existence of specific strain/species immune-modulatory activities.
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. This work was financially supported by Fondazione Cariplo (grant 2010-0678 to S.G. and V.T.) and by a FEMS Jensen Award to F.T. This publication has emanated from research supported in part by a research grant from Science Foundation Ireland (SFI) under grant SFI/12/RC/2273. The grant ILINK2010-0122, funded by CSIC, is also acknowledged.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03313-13.
REFERENCES
- 1.Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91:351–372. 10.1007/s10482-006-9122-6 [DOI] [PubMed] [Google Scholar]
- 2.Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O'Toole PW. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61–71. 10.1038/nrmicro2047 [DOI] [PubMed] [Google Scholar]
- 3.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O'Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10(Suppl 1):S16. 10.1186/1475-2859-10-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222. 10.1073/pnas.1105380108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U. S. A. 109:2108–2113. 10.1073/pnas.1115621109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964–18969. 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss G, Rasmussen S, Nielsen Fink L, Jarmer H, Nohr Nielsen B, Frokiaer H. 2010. Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS One 5:e11065. 10.1371/journal.pone.0011065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez P, Gonzalez-Rodriguez I, Gueimonde M, Margolles A, Suarez A. 2011. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One 6:e24776. 10.1371/journal.pone.0024776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. 10.4161/gmic.2.3.16105 [DOI] [PubMed] [Google Scholar]
- 11.Turroni F, Foroni E, Montanini B, Viappiani A, Strati F, Duranti S, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2011. Global genome transcription profiling of Bifidobacterium bifidum PRL2010 under in vitro conditions and identification of reference genes for quantitative real-time PCR. Appl. Environ. Microbiol. 77:8578–8587. 10.1128/AEM.06352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U. S. A. 110:11151–11156. 10.1073/pnas.1303897110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez P, Gueimonde M, Margolles A, Suarez A. 2010. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 138:157–165. 10.1016/j.ijfoodmicro.2009.12.023 [DOI] [PubMed] [Google Scholar]
- 14.Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74:4695–4702. 10.1128/AEM.00124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hamalainen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. 2013. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl. Environ. Microbiol. 79:1221–1231. 10.1128/AEM.03056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuknyte M, De Noni I, Guglielmetti S, Minuzzo M, Mora D. 2011. Potential immunomodulatory activity of bovine casein hydrolysates produced after digestion with proteinases of lactic acid bacteria. Int. Dairy J. 21:763–769. 10.1016/j.idairyj.2011.04.011 [DOI] [Google Scholar]
- 17.O'Flaherty S, Klaenhammer TR. 2012. Influence of exposure time on gene expression by human intestinal epithelial cells exposed to Lactobacillus acidophilus. Appl. Environ. Microbiol. 78:5028–5032. 10.1128/AEM.00504-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serafini F, Turroni F, Guglielmetti S, Gioiosa L, Foroni E, Sanghez V, Bartolomucci A, Motherway MO, Palanza P, van Sinderen D, Ventura M. 2012. An efficient and reproducible method for transformation of genetically recalcitrant bifidobacteria. FEMS Microbiol. Lett. 333:146–152. 10.1111/j.1574-6968.2012.02605.x [DOI] [PubMed] [Google Scholar]
- 19.Sleator RD, Gahan CG, Hill C. 2001. Mutations in the listerial proB gene leading to proline overproduction: effects on salt tolerance and murine infection. Appl. Environ. Microbiol. 67:4560–4565. 10.1128/AEM.67.10.4560-4565.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serafini F, Bottacini F, Viappiani A, Baruffini E, Turroni F, Foroni E, Lodi T, van Sinderen D, Ventura M. 2011. Insights into physiological and genetic mupirocin susceptibility in bifidobacteria. Appl. Environ. Microbiol. 77:3141–3146. 10.1128/AEM.02540-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, Ventura M. 2010. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 76:3206–3219. 10.1128/AEM.02938-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilban M, Buehler LK, Head S, Desoye G, Quaranta V. 2002. Defining signal thresholds in DNA microarrays: exemplary application for invasive cancer. BMC Genomics 3:19. 10.1186/1471-2164-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- 24.Flores M, Hsiao TH, Chiu YC, Chuang EY, Huang Y, Chen Y. 2013. Gene regulation, modulation, and their applications in gene expression data analysis. Adv. Bioinformatics 2013:360678. 10.1155/2013/360678.23573084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engle MJ, Goetz GS, Alpers DH. 1998. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell. Physiol. 174:362–369. [DOI] [PubMed] [Google Scholar]
- 26.Pozo-Rubio T, Mujico JR, Marcos A, Puertollano E, Nadal I, Sanz Y, Nova E. 2011. Immunostimulatory effect of faecal Bifidobacterium species of breast-fed and formula-fed infants in a peripheral blood mononuclear cell/Caco-2 co-culture system. Br. J. Nutr. 106:1216–1223. 10.1017/S0007114511001656 [DOI] [PubMed] [Google Scholar]
- 27.Guglielmetti S, Taverniti V, Minuzzo M, Arioli S, Stuknyte M, Karp M, Mora D. 2010. Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl. Environ. Microbiol. 76:3948–3958. 10.1128/AEM.00109-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Wang J, Cheng Y, Liu X, Huang Y. 2011. Secreted factors from Bifidobacterium animalis subsp. lactis inhibit NF-kappaB-mediated interleukin-8 gene expression in Caco-2 cells. Appl. Environ. Microbiol. 77:8171–8174. 10.1128/AEM.06145-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakhdari O, Tap J, Beguet-Crespel F, Le Roux K, de Wouters T, Cultrone A, Nepelska M, Lefevre F, Dore J, Blottiere HM. 2011. Identification of NF-kappaB modulation capabilities within human intestinal commensal bacteria. J. Biomed. Biotechnol. 2011:282356. 10.1155/2011/282356.21765633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene JD, Klaenhammer TR. 1994. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl. Environ. Microbiol. 60:4487–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck BL, Altermann E, Svingerud T, Klaenhammer TR. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344–8351. 10.1128/AEM.71.12.8344-8351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D. 2009. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 59:167–172. 10.1007/s00284-009-9415-x [DOI] [PubMed] [Google Scholar]
- 33.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. 2004. Interleukin-10 and related cytokines and receptors. Annu. Rev. Immunol. 22:929–979. 10.1146/annurev.immunol.22.012703.104622 [DOI] [PubMed] [Google Scholar]
- 34.Rich BE. 2003. IL-20: a new target for the treatment of inflammatory skin disease. Expert Opin. Ther. Targets 7:165–174. 10.1517/14728222.7.2.165 [DOI] [PubMed] [Google Scholar]
- 35.Hsu YH, Li HH, Hsieh MY, Liu MF, Huang KY, Chin LS, Chen PC, Cheng HH, Chang MS. 2006. Function of interleukin-20 as a proinflammatory molecule in rheumatoid and experimental arthritis. Arthritis Rheum. 54:2722–2733. 10.1002/art.22039 [DOI] [PubMed] [Google Scholar]
- 36.Sonnenberg GF, Nair MG, Kirn TJ, Zaph C, Fouser LA, Artis D. 2010. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J. Exp. Med. 207:1293–1305. 10.1084/jem.20092054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14:275–281. 10.1038/nm1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz M, Heimesaat MM, Danker K, Struck D, Lohmann U, Plickert R, Bereswill S, Fischer A, Dunay IR, Wolk K, Loddenkemper C, Krell HW, Libert C, Lund LR, Frey O, Holscher C, Iwakura Y, Ghilardi N, Ouyang W, Kamradt T, Sabat R, Liesenfeld O. 2009. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J. Exp. Med. 206:3047–3059. 10.1084/jem.20090900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licona-Limon P, Kim LK, Palm NW, Flavell RA. 2013. TH2, allergy and group 2 innate lymphoid cells. Nat. Immunol. 14:536–542. 10.1038/ni.2617 [DOI] [PubMed] [Google Scholar]
- 40.Trepicchio WL, Ozawa M, Walters IB, Kikuchi T, Gilleaudeau P, Bliss JL, Schwertschlag U, Dorner AJ, Krueger JG. 1999. Interleukin-11 therapy selectively downregulates type I cytokine proinflammatory pathways in psoriasis lesions. J. Clin. Invest. 104:1527–1537. 10.1172/JCI6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leng SX, Elias JA. 1997. Interleukin-11 inhibits macrophage interleukin-12 production. J. Immunol. 159:2161–2168 [PubMed] [Google Scholar]
- 42.Opal SM, Keith JC, Palardy JE, Parejo N. 2000. Recombinant human interleukin-11 has anti-inflammatory actions yet does not exacerbate systemic Listeria infection. J. Infect. Dis. 181:754–756. 10.1086/315247 [DOI] [PubMed] [Google Scholar]
- 43.Manousou P, Kolios G, Valatas V, Drygiannakis I, Bourikas L, Pyrovolaki K, Koutroubakis I, Papadaki HA, Kouroumalis E. 2010. Increased expression of chemokine receptor CCR3 and its ligands in ulcerative colitis: the role of colonic epithelial cells in in vitro studies. Clin. Exp. Immunol. 162:337–347. 10.1111/j.1365-2249.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doll D, Keller L, Maak M, Boulesteix AL, Siewert JR, Holzmann B, Janssen KP. 2010. Differential expression of the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and their impact on metastatic disease and survival. Int. J. Colorectal Dis. 25:573–581. 10.1007/s00384-010-0901-1 [DOI] [PubMed] [Google Scholar]
- 45.Friederichs J, Rosenberg R, Mages J, Janssen KP, Maeckl C, Nekarda H, Holzmann B, Siewert JR. 2005. Gene expression profiles of different clinical stages of colorectal carcinoma: toward a molecular genetic understanding of tumor progression. Int. J. Colorectal Dis. 20:391–402. 10.1007/s00384-004-0722-1 [DOI] [PubMed] [Google Scholar]
- 46.Lehrke M, Konrad A, Schachinger V, Tillack C, Seibold F, Stark R, Parhofer KG, Broedl UC. 2008. CXCL16 is a surrogate marker of inflammatory bowel disease. Scand. J. Gastroenterol. 43:283–288. 10.1080/00365520701679249 [DOI] [PubMed] [Google Scholar]
- 47.Messerini L, Cianchi F, Cortesini C, Comin CE. 2006. Incidence and prognostic significance of occult tumor cells in lymph nodes from patients with stage IIA colorectal carcinoma. Hum. Pathol. 37:1259–1267. 10.1016/j.humpath.2006.04.023 [DOI] [PubMed] [Google Scholar]
- 48.Mantovani A, Gray PA, Van Damme J, Sozzani S. 2000. Macrophage-derived chemokine (MDC). J. Leukoc. Biol. 68:400–404 [PubMed] [Google Scholar]
- 49.Berin MC, Dwinell MB, Eckmann L, Kagnoff MF. 2001. Production of MDC/CCL22 by human intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G1217–G1226 [DOI] [PubMed] [Google Scholar]
- 50.Sonnier DI, Bailey SR, Schuster RM, Gangidine MM, Lentsch AB, Pritts TA. 2012. Proinflammatory chemokines in the intestinal lumen contribute to intestinal dysfunction during endotoxemia. Shock 37:63–69. 10.1097/SHK.0b013e31823cbff1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jugde F, Alizadeh M, Boissier C, Chantry D, Siproudhis L, Corbinais S, Quelvennec E, Dyard F, Campion JP, Gosselin M, Bretagne JF, Semana G, Heresbach D. 2001. Quantitation of chemokines (MDC, TARC) expression in mucosa from Crohn's disease and ulcerative colitis. Eur. Cytokine Netw. 12:468–477 [PubMed] [Google Scholar]
- 52.Middel P, Raddatz D, Gunawan B, Haller F, Radzun HJ. 2006. Increased number of mature dendritic cells in Crohn's disease: evidence for a chemokine mediated retention mechanism. Gut 55:220–227. 10.1136/gut.2004.063008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hillinger S, Yang SC, Batra RK, Strieter RM, Weder W, Dubinett SM, Sharma S. 2006. CCL19 reduces tumour burden in a model of advanced lung cancer. Br. J. Cancer 94:1029–1034. 10.1038/sj.bjc.6603061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 55.Basu S, Srivastava PK. 2000. Heat shock proteins: the fountainhead of innate and adaptive immune responses. Cell Stress Chaperones 5:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava P. 2002. Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2:185–194. 10.1038/nri749 [DOI] [PubMed] [Google Scholar]
- 57.Todryk SM, Gough MJ, Pockley AG. 2003. Facets of heat shock protein 70 show immunotherapeutic potential. Immunology 110:1–9. 10.1046/j.1365-2567.2003.01725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milani V, Noessner E, Ghose S, Kuppner M, Ahrens B, Scharner A, Gastpar R, Issels RD. 2002. Heat shock protein 70: role in antigen presentation and immune stimulation. Int. J. Hyperthermia 18:563–575. 10.1080/02656730210166140 [DOI] [PubMed] [Google Scholar]
- 59.Kregel KC. 2002. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J. Appl. Physiol. 92:2177–2186. 10.1152/japplphysiol.01267.2001.11960972 [DOI] [PubMed] [Google Scholar]
- 60.Lecuit M, Sonnenburg JL, Cossart P, Gordon JI. 2007. Functional genomic studies of the intestinal response to a foodborne enteropathogen in a humanized gnotobiotic mouse model. J. Biol. Chem. 282:15065–15072. 10.1074/jbc.M610926200 [DOI] [PubMed] [Google Scholar]
- 61.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361–367. 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- 62.Yu J, Ebert MP, Miehlke S, Rost H, Lendeckel U, Leodolter A, Stolte M, Bayerdorffer E, Malfertheiner P. 2000. alpha-catenin expression is decreased in human gastric cancers and in the gastric mucosa of first degree relatives. Gut 46:639–644. 10.1136/gut.46.5.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallo RL, Hooper LV. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12:503–516. 10.1038/nri3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai Y, Gallo RL. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30:131–141. 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monk PN, Partridge LJ. 2012. Tetraspanins: gateways for infection. Infect. Disord. Drug Targets 12:4–17. 10.2174/187152612798994957 [DOI] [PubMed] [Google Scholar]
- 66.van Spriel AB, Figdor CG. 2010. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 12:106–112. 10.1016/j.micinf.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 67.Murphy EC, Morgelin M, Cooney JC, Frick IM. 2011. Interaction of Bacteroides fragilis and Bacteroides thetaiotaomicron with the kallikrein-kinin system. Microbiology 157:2094–2105. 10.1099/mic.0.046862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuno K, Kanada N, Nakashima E, Fujiki F, Ichimura F, Matsushima K. 1997. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 272:556–562. 10.1074/jbc.272.1.556 [DOI] [PubMed] [Google Scholar]
- 69.Apte SS. 2004. A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motifs: the ADAMTS family. Int. J. Biochem. Cell Biol. 36:981–985. 10.1016/j.biocel.2004.01.014 [DOI] [PubMed] [Google Scholar]
- 70.Yoshimura T, Tomita T, Dixon MF, Axon AT, Robinson PA, Crabtree JE. 2002. ADAMs (A disintegrin and metalloproteinase) messenger RNA expression in Helicobacter pylori-infected, normal, and neoplastic gastric mucosa. J. Infect. Dis. 185:332–340. 10.1086/338191 [DOI] [PubMed] [Google Scholar]
- 71.Burkholder KM, Bhunia AK. 2010. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect. Immun. 78:5062–5073. 10.1128/IAI.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bevins CL, Martin-Porter E, Ganz T. 1999. Defensins and innate host defence of the gastrointestinal tract. Gut 45:911–915. 10.1136/gut.45.6.911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hassuna N, Monk PN, Moseley GW, Partridge LJ. 2009. Strategies for targeting tetraspanin proteins: potential therapeutic applications in microbial infections. BioDrugs 23:341–359. 10.2165/11315650-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stadnicki A. 2011. Intestinal tissue kallikrein-kinin system in inflammatory bowel disease. Inflamm. Bowel Dis. 17:645–654. 10.1002/ibd.21337 [DOI] [PubMed] [Google Scholar]
- 75.Green CD, Olson LK. 2011. Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic beta-cells by stearoyl-CoA desaturase and Elovl6. Am. J. Physiol. Endocrinol. Metab. 300:E640–E649. 10.1152/ajpendo.00544.2010 [DOI] [PubMed] [Google Scholar]
- 76.Shimamura K, Takahashi H, Kitazawa H, Miyamoto Y, Nagumo A, Tang C, Dean D, Nagase T, Sato N, Tokita S. 2009. Identification and characterization of a selective radioligand for ELOVL6. J. Biochem. 146:429–437. 10.1093/jb/mvp088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.