Abstract
The fermentation of lignocellulose-derived sugars, particularly xylose, into ethanol by the yeast Saccharomyces cerevisiae is known to be inhibited by compounds produced during feedstock pretreatment. We devised a strategy that combined chemical profiling of pretreated feedstocks, high-throughput phenotyping of genetically diverse S. cerevisiae strains isolated from a range of ecological niches, and directed engineering and evolution against identified inhibitors to produce strains with improved fermentation properties. We identified and quantified for the first time the major inhibitory compounds in alkaline hydrogen peroxide (AHP)-pretreated lignocellulosic hydrolysates, including Na+, acetate, and p-coumaric (pCA) and ferulic (FA) acids. By phenotyping these yeast strains for their abilities to grow in the presence of these AHP inhibitors, one heterozygous diploid strain tolerant to all four inhibitors was selected, engineered for xylose metabolism, and then allowed to evolve on xylose with increasing amounts of pCA and FA. After only 149 generations, one evolved isolate, GLBRCY87, exhibited faster xylose uptake rates in both laboratory media and AHP switchgrass hydrolysate than its ancestral GLBRCY73 strain and completely converted 115 g/liter of total sugars in undetoxified AHP hydrolysate into more than 40 g/liter ethanol. Strikingly, genome sequencing revealed that during the evolution from GLBRCY73, the GLBRCY87 strain acquired the conversion of heterozygous to homozygous alleles in chromosome VII and amplification of chromosome XIV. Our approach highlights that simultaneous selection on xylose and pCA or FA with a wild S. cerevisiae strain containing inherent tolerance to AHP pretreatment inhibitors has potential for rapid evolution of robust properties in lignocellulosic biofuel production.
INTRODUCTION
Although petroleum displacement by lignocellulose-derived biofuels is a priority around much of the world, technological and economic barriers still remain that prevent a wide-scale deployment of these technologies. For technologies based on the biological conversion of sugars derived from plant cell wall polysaccharides into biofuels such as ethanol, one challenge, among many others, is that a sizable fraction of the sugars in plant cell wall-derived hydrolysates consists of pentoses, including xylose. Xylose is the second most abundant sugar in the cell walls of angiosperms, which include most of the promising bioenergy plants such as high-productivity, short-rotation woody dicots (e.g., hybrid poplar), agricultural residues (e.g., corn stover), and dedicated energy grasses (e.g., switchgrass). Relative to hexoses such as glucose, the conversion of xylose to ethanol by wild-type bacteria and yeasts at high product yields, titers, and productivities has remained a refractory problem (1).
As a consequence of this first problem of limited substrate range, significant research has been invested into metabolic engineering for the cofermentation of glucose and xylose to ethanol in bacteria, including Escherichia coli (2) and Zymomonas mobilis (3, 4), and yeasts such as Saccharomyces cerevisiae (5, 6). Xylose fermentation by native S. cerevisiae is restricted by insufficient biochemical activities to convert xylose into xylulose-5-phosphate, which can be fermented into ethanol via the nonoxidative pentose phosphate pathway (PPP) and glycolysis (7–9). To overcome this metabolic barrier, others have engineered S. cerevisiae to express NADPH-coupled xylose reductases (XR) together with NAD+-coupled xylitol dehydrogenases (XDH), or xylose isomerases for the conversion of xylose into xylulose (7, 9–14). Subsequent phosphorylation of xylulose into xylulose-5-phosphate by engineered xylulokinases (XK) then allowed for complete xylose catabolism. Even with the minimal enzymes needed for conversion, xylose fermentation often remains inefficient. In particular, xylose metabolism can be impaired by cofactor imbalance generated with the use of NADPH and NAD+-coupled enzymes (15, 16). This imbalance is further exacerbated during anaerobic fermentation during which oxygen is unavailable for respiratory generation the NAD+ cofactor for xylitol dehydrogenase activity (17). Furthermore, native S. cerevisiae lack high-affinity xylose transporters and instead rely on low affinity transport through glucose transporters, resulting in slower diauxic sugar consumption (15, 16). To overcome these barriers, adaptation and directed evolution are commonly employed to significantly improve the rate of xylose fermentation from laboratory media (13, 17–19). While some of these engineered and evolved strains are efficient at fermenting xylose in innocuous laboratory media, most strains are significantly inhibited by compounds present in pretreated lignocellulosic hydrolysates that drain cellular ATP levels and alter cofactor balance.
Pretreatment is an essential step for processes that utilize enzymatic hydrolysis of the plant cell wall polysaccharides for biological conversion into a biofuel (20). The wide range of pretreatment chemistries differ in their impact on the plant cell wall polymers, the higher-order cell wall structure, and the generation of cell wall degradation products that may inhibit both enzymatic hydrolysis and microbial conversion of monomer sugars into biofuels (21, 22). This introduces a second challenge to the fermentation of lignocellulosic hydrolysates. Unlike the sugar streams used in starch- or sucrose-based fermentations, lignocellulosic hydrolysates contain a complex assortment of inhibitory compounds derived from biomass pretreatment (23–25), as well as chemicals used for pretreatment and/or pH adjustment, including inorganics such as sulfate or sodium (26). The identification of compounds derived from a number of pretreatment methods and biomass sources, their inhibitory effects on cell growth and fermentation, and approaches to detoxify lignocellulose-derived hydrolysates have been well covered in the literature (23, 26–34).
Alkaline hydrogen peroxide (AHP) pretreatment has several potential advantages compared to other pretreatment processes, including minimal loss of polysaccharides, operation at low temperature and pressure, and potentially high enzymatic digestibilities in grasses (2, 35, 36). Previous studies performed AHP pretreatment and hydrolysis at relatively low solid concentrations, and therefore, the yeast strains were not challenged to produce high ethanol titers in industrially relevant sugar and inhibitor concentrations (35, 36). Other work found that removal of Na+, which is added during pH adjustment by NaOH, from AHP hydrolysates via cost- and energy-prohibitive electrodialysis was required for Clostridium beijerinckii fermentations (25, 37),. To date, the spectrum of inhibitors in AHP hydrolysates produced from high-solid pretreatment and hydrolysis have not been published; thus, their specific impacts on xylose fermentation have not been systematically assessed. The soluble fermentation inhibitors in AHP hydrolysates may be comparable to those generated from alkaline wet oxidation (23), which is a pretreatment performed at substantially higher temperatures. Specifically, we propose that the key classes of inhibitors may include aliphatic oxidation products of cell wall polymers (e.g., one- to six-carbon aliphatic alcohols, aldehydes, and carboxylic acids), acetate produced by saponification of xylan, phenolic compounds (acids, aldehydes, and alcohols) derived primarily from acylated p-hydroxycinnamates in grasses (38), and inorganics such as Na+ and SO42− introduced via NaOH and H2SO4 during pH adjustment. Furans, which are significant inhibitors in acid hydrolysates, are notably absent in the hydrolysates of alkaline pretreatments (23, 27), as these are produced by the acid-catalyzed dehydration of sugar monomers.
In addition to hydrolysate detoxification, numerous studies have shown that genetic approaches for S. cerevisiae strain development can permit sufficient growth and fermentation in the presence of pretreatment inhibitors. For example, the overexpression or deletion of genes that enhance cellular detoxification of furfural (39, 40), acetic (41, 42), p-coumaric (43) and ferulic (44) acids in laboratory strains of S. cerevisiae have enhanced growth on glucose. In addition, experimental evolution has allowed for the selection of tolerant strains by adaptation on glucose in the presence of sodium (45) and acetic acid (46, 47). In contrast, fewer studies have investigated genetic approaches that improve xylose conversion in combination with inhibitor tolerance. Examples include targeted deletion of the PHO13 gene, which appeared to improve xylose fermentation in the presence of acetic acid and furfural (48) and evolutionary selection of engineered S. cerevisiae in the presence of acetic acid (49, 50).
Another approach to cope with inhibitory stress is to utilize native strain backgrounds with tolerant properties. Yeast strains employed in industrial applications are subject to continuous selection, often resulting in the development of robust stress-tolerant properties (1). Some of these industrial strains that have been metabolically engineered for xylose utilization outperform laboratory strains in the fermentation of pretreated lignocellulosic hydrolysates (19, 51, 52). In two specific cases, faster xylose fermentation has been independently obtained by engineered industrial strains evolved on defined media containing a cocktail of inhibitors found in dilute acid-pretreated spruce hydrolysate (53) or selected on SO2-impregnated steam explosion-pretreated spruce hydrolysate (54). While industrial yeast strains display a wide range of robust tolerance properties, wild S. cerevisiae strains isolated from a range of ecological niches also exhibit a range of phenotypic properties, including tolerance to various stressors (55–57). Moreover, this range in tolerance phenotypes can be attributed to the significant degree of natural genetic variation between strains (57, 58). Despite their observed abilities to tolerate environmental stress, wild S. cerevisiae strains have not been systematically explored for lignocellulosic biofuel production and may offer alternative mechanisms of stress tolerance that are not present in industrial strains.
Given that xylose fermentation by engineered S. cerevisiae is highly sensitive to perturbation, particularly by compounds generated from lignocellulose pretreatment, we designed and tested an experimental approach to develop strains with improved xylose fermentation of inhibitor-laden AHP-pretreated lignocellulose. First, we detected and quantified the inhibitors in three AHP-pretreated lignocellulosic hydrolysates and then selected an S. cerevisiae strain among more than 100 genetically diverse strains with innate tolerance to the most abundant inhibitors, specifically acetate, sodium, and phenolic acids. We then engineered and experimentally evolved that strain for improved xylose fermentation in the presence of p-coumaric (pCA) and ferulic (FA) acids, the phenolic acids at the highest concentrations found in AHP hydrolysates. The resulting strain, named GLBRCY87, displayed higher xylose uptake rates in laboratory media containing FA, as well as during anaerobic fermentation of undetoxified AHP-pretreated switchgrass. Finally, genome sequencing comparisons between the evolved GLBRCY87 and parental GLBRCY73 strains strikingly revealed that two major genetic events, aneuploidy and loss of heterozygosity (LOH), occurred during the phenotypic evolution. Together, our data show that simultaneous selection of engineered S. cerevisiae strains on xylose and high pCA or FA concentrations, in a strain background selected initially for high tolerance, can allow for accelerated fermentation of xylose in AHP hydrolysate to ethanol.
MATERIALS AND METHODS
Hydrolysate preparation.
Corn stover (Zea mays L. Pioneer hybrid 36H56) and switchgrass (Panicum virgatum cv. Cave-In-Rock) were the same material as used in our previous work (59). AHP pretreatment was performed as described previously (35), with the difference that the biomass insoluble-solid concentrations (200, 250, or 350 g initial insoluble solids/kg of total slurry) were utilized as listed in Table 1 and pretreatment was performed at a scale of 70-g biomass (dry basis) in sealable 500-ml polyethylene boxes. No portion of the pretreatment liquor (prehydrolysate) was discarded, and hydrolysates represent all the compounds solubilized during both pretreatment and enzymatic hydrolysis. Without removing material from the containers, the pH of hydrolysates was adjusted to 4.8 for the subsequent enzymatic hydrolysis using 18.36 M H2SO4. Enzyme cocktails included either Accelerase 1500 and Multifect xylanase (Genencor, Palo Alto, CA) or Cellic C-Tec 2 and H-Tec 2 (Novozymes, Davis, CA) at an enzyme ratio of 7:3 cellulase-xylanase on a protein basis (Bradford assay; Sigma-Aldrich) and a total enzyme loading of 30 mg protein/g original cellulose with conditions for individual hydrolysates indicated in Table 1. During hydrolysis, the containers holding the biomass were incubated with shaking at 180 rpm at 50°C for 72 h, with additional manual stirring every 2 h for the first 8 h. The hydrolysate was centrifuged at 17,700 × g for 15 min and the liquid phase was decanted. Urea (Sigma-Aldrich) and yeast nitrogen base (YNB) without amino acids (Sigma-Aldrich) were added into the raw hydrolysate to final concentrations of 2.27 g/liter and 1.67 g/liter, respectively. The pH was adjusted to 5.5 using NaOH pellets (Sigma-Aldrich). Prior to fermentation, the hydrolysates were sterilized by filtration through a 0.22-μm filter (Millipore Stericup; Fisher Scientific).
TABLE 1.
Compositions, yields, and conditions used to generate hydrolysates
| Parameter | Value or ingredients for AHP hydrolysate |
||
|---|---|---|---|
| SG1 | CS1 | CS2 | |
| Solids to pretreatment (g/kg slurry) | 200 | 250 | 350 |
| Equivalent initial solids to hydrolysis (g/kg slurry) | 193 | 239 | 329 |
| Enzyme cocktail | Cellic H-Tec + C-Tec | Accelerase 1500 + Multifect Xylanase | Cellic H-Tec + C-Tec |
| Enzyme loading (mg/g original glucan) | 30 | 30 | 30 |
| Sugars | |||
| Glc (g/liter) | 63.6 | 77.6 | 89.7 |
| Xyl (g/liter) | 39.1 | 35.7 | 44.4 |
| Ara (g/liter) | 4.4 | 2.5 | 3.9 |
| Glc yield (%) | 59 | 75 | 64 |
| Xyl yield (%) | 36 | 71 | 61 |
| Na+ (mM) | 580 | 718 | 988 |
| Formate (g/liter; mM) | 1.2; 26 | 0.8; 17 | 0.8; 17 |
| Acetate (g/liter; mM) | 5.8; 97 | 6.9; 115 | 8.9; 148 |
| Ferulic acid (g/liter; mM) | 0.25; 1.3 | 0.25; 1.3 | 0.44; 2.3 |
| p-Coumaric acid (g/liter; mM) | 0.59; 3.6 | 0.57; 3.5 | 1.01; 6.2 |
Growth phenotyping of yeast strains in 96-well plates.
Native S. cerevisiae strains used in this study were obtained from Cletus Kurtzmann (USDA ARS, Peoria, IL), National Collection of Yeast Cultures (Norwich, United Kingdom), and Justin Fay (Washington University, St. Louis, MO) and are described in Table S1 in the supplemental material. Strains were prepared for inoculation as previously described (60). Strains arrayed in 96-well plates were inoculated from 10-μl volumes of saturated cultures into the interior wells of a standard 96-well plate (Nunc) containing 190 μl of a single type of yeast extract-peptone-dextrose (YPD) containing inhibitor media, while outer wells contained 200 μl sterile water. To study the inhibitory effects of Na+ and acetate, which requires a counterion, either in an acetate salt or from base adjustment (e.g., NaOH) of glacial acetic acid, sodium acetate (NaAc) was selected since both ionic compounds were present in all AHP hydrolysates and also prevented the introduction of a new potential inhibitor (e.g., elevated concentration of K+ from potassium acetate). YPD containing 20 g/liter NaAc was made by dissolving solid sodium acetate (Sigma-Aldrich) in YPD with stirring, followed by adjustment to pH 5.5. YPD medium containing 0.242 M Na2SO4 media was made by dissolving solid Na2SO4 decahydrate (Sigma-Aldrich) in YPD with stirring, followed by adjustment to pH 5.5. Stock solutions of p-coumaric and ferulic acids (Sigma-Aldrich) dissolved in ethanol were prepared just prior to use and added to YPD for a final concentration of 2.4 g/liter pCA and 1.2 g/liter FA. For the control medium, an equivalent volume of 95% ethanol was added to YPD.
To monitor growth of native yeast strains in YPD with inhibitors, inoculated 96-well plates were placed in Tecan F500 or M1000 multimode plate readers maintaining an interior temperature of 30°C. Plates were shaken for 10 s, and absorbance at 595 nm was measured from each well every 10 min for approximately 24 h, with no shaking in between absorbance readings. Background subtracted cell density readings for each strain were analyzed using an automated program called GCAT (J. Shao, T. K. Sato, and Y. Bukhman, submitted for publication; GCAT is available for download at http://www.glbrc.org/gcat-vm/), which uses nonlinear regression (61) or a logistic function (62), to report individual specific growth rates. Averaged specific growth rates for each strain from three independent biological replicates in YPD at pH 5.5 alone or with 20 g/liter NaAc, or YPD with 2.4 g/liter pCA and 1.2 g/liter FA, were ranked ordered from 1 to 112 (108 unique strains with 3 sets of control strains—BY4741, CEN.PK113-5D, and CEN.PK2-1D—on two 96-well plates) for highest average specific growth to lowest, respectively. For all strains with no detectable specific growth rates, strains were assigned a rank of 112. Strain ranks in each media condition were hierarchically clustered and displayed with Spotfire (TIBCO).
Directed evolution of yeast strains.
Engineering of the GLBRCY2A strain integrated with a single copy of a cassette for expressing xylose reductase (XYL1), xylitol dehydrogenase (XYL2), and xylulokinase (XYL3) from Scheffersomyces (Pichia) stipitis has been described previously (14). Strain GLBRCY73 was selected from serial subcultures of GLBRCY2A adapted aerobically in shake flasks or culture tubes with YP media containing 20 g/liter xylose and 1 g/liter glucose for 7.8 generations, 20 g/liter xylose for 8.8 generations, 30 g/liter xylose for 8 generations, and 40 g/liter xylose for 22.5 generations. At the end of adaptation, the culture was diluted and spread on agar plates with YP plus 20 g/liter xylose (YPX). Single colonies were selected and phenotyped for growth on YPX in 96-well plate assays described previously (14), with the GLBRCY73 isolate being one with significantly faster growth on YPX relative to the GLBRCY2A parent and other adapted isolates. Strain GLBRCY87 was isolated from a culture of GLBRCY73 adapted aerobically in shake flasks or culture tubes with YP and 10 g/liter xylose containing various concentrations of pCA and FA in a 2:1 ratio. Quantities of 1.6 g/liter pCA and 0.8 g/liter FA (Sigma-Aldrich) were dissolved in ethanol for 28.6 generations, 2.0 g/liter pCA and 1.0 g/liter FA for 10.4 generations, 2.2 g/liter pCA and 1.1 g/liter FA for 33.3 generations, and 2.28 g/liter pCA and 1.14 g/liter FA for 29.8 generations (see Fig. S1 in the supplemental material). Single isolates were screened as described for GLBRCY73 but with YPX with 2.4 g/liter pCA and 1.2 g/liter FA, and one isolate, designated GLBRCY87, with significantly faster growth was selected for further characterization.
Growth and cell viability assays.
For determining cell growth, viability, and sugar consumption, a single colony of an engineered yeast strain was inoculated in 5 ml YPD with 200 μg/ml Geneticin (Life Technologies) in a culture tube and incubated with shaking at 30°C overnight. Saturated cultures were then diluted to an optical density at 600 nm (OD600) of 0.3 in fresh YPD with Geneticin and incubated with shaking at 30°C. When the culture reached mid-log-phase growth (OD600 of 1 to 2), cells equivalent to an OD600 of 8 were centrifuged at 3,000 × g for 5 min and the supernatant was aspirated. The harvested cell pellets were washed gently with 5 ml YP medium and harvested again by centrifugation and aspiration. Cells were then resuspended in 8 ml YPD or YPX medium containing pCA and FA made as described above, transferred into sterile culture tubes, and incubated with shaking at 30°C. At 0, 20, 25, 44, and 49 h after resuspension of cells in the new YPD medium with inhibitors, 25 μl cell culture diluted to OD600 of 0.3 in YP media was incubated in 475 μl viability staining buffer (10 mM HEPES, 20 g/liter glucose [pH 7.2]) containing 1.1 μl/ml LIVE/DEAD FungaLight viability dyes for flow cytometry (Invitrogen) at room temperature for 20 to 30 min. Stained cells were then transferred to vials and loaded into a Guava EasyCyte Plus flow cytometer. With Guava ExpressPro software, the numbers of live and dead cells were determined based on red and green fluorescence levels per cell relative to red and green fluorescence levels found in isopropanol-killed cells.
Hydrolysate and defined medium fermentations.
Hydrolysate and anaerobic minimal medium studies were performed in sterile 250-ml shake flasks (100-ml working volume) fitted with fermentation locks (Bacchus & Barleycorn Ltd., Shawnee, KS). Seed cultures of parental control GLBRCY2A, xylose-adapted GLBRCY73 (evolved from GLBRCY2A), and xylose-pCA-FA-adapted GLBRCY87 (evolved from GLBRCY73) strains were used for inoculation to an initial OD600 of 1.0 in minimal media (urea, 2.27 g/liter; YNB without amino acids, 1.67 g/liter; glucose, 50 g/liter; and xylose, 50 g/liter). To determine the effects of pCA and FA adaptation on the fermentation of xylose in AHP hydrolysates, seed cultures of parental control GLBRCY73 and pCA- and FA-adapted GLBRCY87 strains were inoculated to an initial OD600 of 1.0 into filter-sterilized hydrolysate. Flasks were purged with N2 initially and after sampling to maintain anaerobic conditions and were incubated at 30°C and with orbital shaking at 150 rpm. Fermentations were performed in duplicate flasks, and approximately 300-μl samples were taken periodically to measure cell density (OD600) and glucose, xylose, ethanol, glycerol, acetate, formate, and xylitol.
Analysis of hydrolysates and fermentations.
Glucose, xylose, arabinose, ethanol, formate, acetate, glycerol, furans, and xylitol concentrations in hydrolysates and fermentation samples were determined by high-performance liquid chromatography (HPLC; Agilent 1100 series) using an Aminex HPX-87H column (Bio-Rad) operating at 65°C, a mobile phase of 0.005 M H2SO4, a flow rate of 0.6 ml/min, and detection by refractive index (35). FA and pCA were analyzed by HPLC using a C18 column (Discovery, 5-μm particle size, 5-cm length by 2.1-mm internal diameter; Sigma-Aldrich) operating at 40°C at 0.25 ml/min using a gradient elution with mobile phases consisting of 1% (vol/vol) acetic acid in water and 100% methanol. Beginning with only acetic acid mobile phase, after 2 min the fraction of methanol was increased stepwise by 2.5% methanol/min up to 40% and held for 2 min. The column was then flushed by increasing to 50% methanol for 3 min before being returned to 100% acetic acid. All chromatograms were quantitated using an external set of calibration standards containing each compound. Na+ was estimated based on the mass of NaOH used and known solution volumes used during pretreatment. Samples for cell density were diluted to a range within the sensitivity of the spectrophotometer, and the absorbance at 600 nm was recorded. The absorbances of hydrolysate samples were corrected for experiment variability by subtraction of the absorbance of the supernatant of the sample after centrifugation (3,780 × g). The OD600 was converted to dry cell weight (DCW) through an experimentally determined correlation generated through a series of serial dilutions of a high-cell-density sample. After measurement of the suspended cell OD600, the samples were centrifuged and the absorbance of the supernatant was subtracted to obtain the OD600 due to the cells alone. The cells were then oven dried overnight to determine the experimental DCW.
Determination of yields, rates, and measurement errors.
The yields of metabolites and biomass from either glucose or xylose were determined by regression of the measured consumption of substrate versus either the metabolite or the cell mass generated. Metabolite yields on xylose were calculated using data points after glucose consumption was complete. The data used for the xylose rates were taken from the time period after glucose consumption was complete but before xylose concentrations decreased to less than 10 g/liter in order to prevent interference by the relatively high Ks values for xylose uptake. The xylose rates were calculated using between 4 and 6 data pairs of xylose concentration, cell concentration, and time. Specific xylose uptake rates (qXyl) were determined by linear least-squares (LLS) regression of the xylose concentration (g/liter) divided by the cell concentration (g/liter) against time (h), while the absolute xylose uptake rates (QXyl) were determined by LLS regression of the xylose concentration (g/liter) against time (h). The standard errors for parameters determined by regressions were calculated as the parametric error associated with the LLS fit (63), while the significances of parameter differences were assessed by testing the null hypothesis that the parameters were equal using “pooled” parameter error variance (64). The biomass yields on glucose (YX/Glc) for individual hydrolysates were determined using only the initial time point and after 24 h, when all the glucose was consumed. Errors presented for these parameters were determined by propagation of the standard errors associated with the initial glucose concentration, and the final biomass concentration and statistical differences were assessed using the Student t test (63).
High-throughput genomic sequencing and analysis.
Unamplified libraries were generated from 1.0 μg GLBRCY73 or GLBRCY87 genomic DNA using a modified version of the Illumina standard protocol. The DNA was sheared using a sonicator (Covaris, Inc.) to generate fragments of 100 to 500 bp in length. The fragments were size selected by SPRI to ∼200 to 500 bp. Selected fragments either were end repaired, phosphorylated, and A tailed using the polymerase activity of Klenow fragment and then ligated with Illumina paired-end sequencing adapters (Illumina Inc.) or end repaired, A tailed, and then ligated with Illumina-compatible paired-end sequencing adapters (IDT, Inc.). The prepared sample library was quantified using a KAPA library quantification kit (KAPA Biosystems) and run on a Roche LightCycler 480 real-time PCR instrument. The quantified sample library was then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit and Illumina cBot instrument to generate clustered flow cells for sequencing. Sequencing of the flow cells was performed on the Illumina HiSeq2000 sequencer using a TruSeq SBS sequencing kit for 200 cycles by following a paired-end 100-bp indexed run recipe.
Genome mapping and analysis.
Read quality analysis was performed in Galaxy (65). Short reads were examined for quality and trimmed at the 3′ end when average base quality in a 3-nucleotide (nt) window fell below Q30. Short reads were mapped to the standard S. cerevisiae reference genome, strain S288c (obtained from the NCBI RefSeq repository, February 2011), using Burrows-Wheeler alignment (BWA, version 1.2.2 [66]) with default parameters except the fraction of missing alignments threshold was 0.08 (−n in “bwa aln”). Single nucleotide polymorphism (SNP) and indel detection was performed with the Genome Analysis Toolkit (GATK, version 1.4 [67]) following their “best-practice” variant-calling work flow (www.broadinstitute.org/gatk/). Duplicate reads were marked, followed by base quality recalibration using an SNP database designed for S. cerevisiae. To minimize false-positive variant calls, stringent parameters were used: namely, the minimum base quality required to consider a base for calling was 30, and the minimum phred-scaled confidence threshold for genotype calling was 50 (−mbq and −scc in the UnifiedGenotyper tool). Custom Perl scripts were used to further filter calls on the basis of read depth, mapping quality, and strand bias. Filtered SNP calls were used to analyze heterozygosity with custom Perl scripts. In order to estimate ploidy, output from the GATK DepthOfCoverage tool was analyzed using custom Perl scripts.
RESULTS
Detection and quantification of inhibitors in hydrolysates.
Three AHP hydrolysates (one from switchgrass and two from corn stover) were used to identify potential inhibitors for assessing their impact on yeast fermentation. One AHP-pretreated switchgrass (SG1) and two corn stover (CS1 and CS2) hydrolysates were generated by pretreatment and hydrolysis at different solid concentrations, so they were expected to contain a range of final sugar titers, sugar yields, and inhibitor concentrations. With high-solid AHP pretreatment (200 to 350 g insoluble solids/kg slurry) and hydrolysis (193 to 329 g insoluble solids/kg slurry), all three hydrolysates contained high sugar titers, which could potentially generate more than 40 g/liter ethanol based on a theoretical yield of 0.51 g ethanol/g sugar. Of the expected spectrum of compounds in these hydrolysates, methanol, lactate, succinate, glycolate, oxalate, furfural, and 5-hydroxymethylfurfural (HMF) were not quantified at levels above the detection limits of the refractive index and UV detectors in analysis. Acetate and formate were present at significant concentrations in all hydrolysates (Table 1). Two phenolic acids, p-coumaric (pCA) and ferulic (FA) acids, were identified in initial screening as the most abundant phenolics in AHP hydrolysates (data not shown), and for this set of hydrolysates, these compounds were quantified at concentrations approaching their solubility limit in water (Table 1).
Phenotyping of yeast strains for inhibitor tolerance.
Since acetate, Na+, ferulate, and p-coumarate have been previously shown to individually impair growth and fermentation by S. cerevisiae, it was expected that these compounds together in the three AHP hydrolysates would have significant impacts on fermentation, particularly for conversion of xylose. We opted to evaluate wild and domesticated yeast strains with a wide range of genetic variation for innate tolerance to these inhibitors, which could then be genetically manipulated for xylose consumption in AHP hydrolysates. A panel of 112 wild, industrial, and laboratory S. cerevisiae strains (108 unique strains with duplicate control strains) were phenotyped for their abilities to grow in YPD media at pH 5.5 alone or containing either 2.4 g/liter pCA and 1.2 g/liter FA, 0.24 M Na2SO4, or 20 g/liter sodium acetate (0.24 M NaAc), which evaluated strain growth during simultaneous inhibition from Na+ and acetate. At pH 5.5, 15.3% of acetate was in the inhibitory, protonated form; acetate has been shown to specifically inhibit yeast growth beyond equal or higher molar concentrations of NaCl (41, 42). The effect of acetate alone could not be determined, due to the requirement for a counterion in any acetate salt or raising the pH of acetic acid with base. Assuming that inhibitor tolerance can be reflected by strain growth rate (i.e., sensitive strains grow more slowly in the presence of an inhibitor than do tolerant strains), we continuously monitored the cell densities of each strain over 24 h and determined their specific growth rates in each of the four medium conditions by curve fitting models. Specific growth rates for each strain cultured in medium containing the single AHP inhibitor were normalized to their growth rate in YPD medium alone and rank ordered fastest to slowest. Interestingly, only 34 of the 112 strains could grow in the presence of 2.4 g/liter pCA and 1.2 g/liter FA, suggesting that this particular condition was the most inhibitory. Additionally, poor correlation in growth rate rank between strains grown in 0.24 M NaAc and 0.24 M Na2SO4 (R2 = 0.26) was observed, with many strains growing relatively well in one medium but not the other. For example, CLIB382, K13, and UC1 grew relatively fast in 0.24 M Na2SO4 but not in 0.24 M NaAc, while YJM427, YPS1009, and YJM326 grew relatively fast in NaAc but not in Na2SO4 (see Table S1 in the supplemental material). Similar to previous observations that acetate at pH 5.5 can impact cell growth (41, 42), the differences in wild yeast strain responses to NaAc and Na2SO4 at pH 5.5 also suggested that the growth phenotypes induced by NaAc were not a general response to osmotic stress.
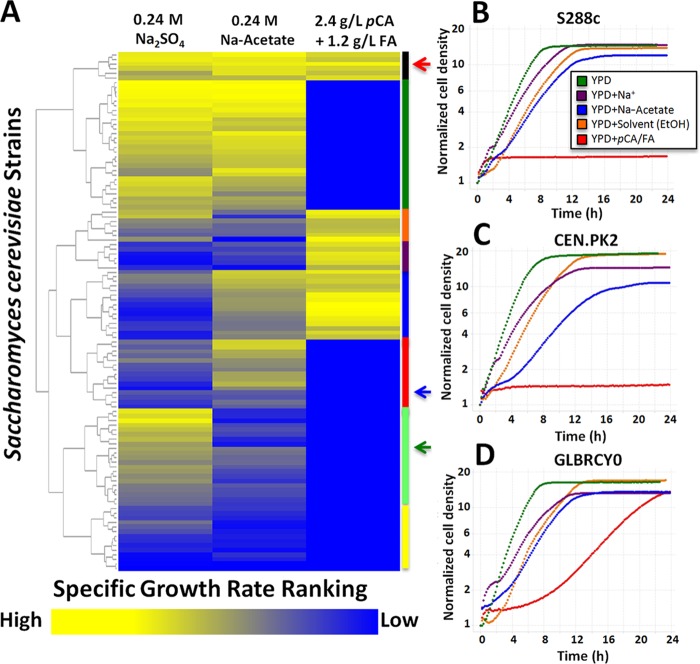
Hierarchical clustering of growth rate rank identified 8 major phenotypic clusters of strains with combinations of tolerance and sensitivity to each inhibitor (Fig. 1A). Commonly used laboratory strains, S288c (Fig. 1A, green arrow) and CEN.PK2 (Fig. 1A, blue arrow), clustered in groups of strains that grew relatively fast in only 0.48 M Na+ and 0.24 M NaAc or neither, respectively. Most of the strains ranked highly for growth rate in the presence of one or two different inhibitors, suggesting that genetic backgrounds with tolerance to multiple inhibitors are a rare occurrence in wild strains. Six wild strains isolated from distinct environmental sources ranked highest for growth rate in all three inhibitor conditions: I14 (vineyard soil), Y389 (mushroom), Y9 (ragi wine), GLBRCY0 (banana), YIIc17_E5 (wine), and YJM454 (clinical). The variety of ecological niches that these strains were isolated from suggests that selection of strains isolated from a single environment (e.g., industrial fermentation tanks) may not necessarily yield the optimal strain for engineering and evolution, but rather, selection from an environmentally diverse collection may yield more ideal strains. Given the interest in selecting a strain with the highest tolerance to all three sets of inhibitors found in AHP hydrolysates, we focused on the cluster of strains that ranked highest in relative growth rate for all three inhibitory conditions, including a wild strain assigned as GLBRCY0 (Fig. 1C, red arrow), for further analysis.
FIG 1.
Phenotyping of S. cerevisiae strains grown in laboratory media containing AHP inhibitors. (A) The 112 wild, domesticated, or industrial S. cerevisiae strains cultured in YPD media containing the indicated AHP inhibitors hierarchically cluster into 8 phenotypically distinct groups. The cell densities of strains grown at 30°C in 96-well plates were monitored for 24 h. From the growth data, average specific growth rates from 3 biological replicates for each strain were calculated, rank ordered from fastest (1, yellow) to slowest (112, blue) in each medium condition and then hierarchically clustered. Colored bars indicate groups of strains with relative tolerance to specific inhibitory conditions (black, relatively tolerant to all three inhibitor conditions; green, Na2SO4 and NaAc; orange, Na2SO4 and pCA plus FA; purple, pCA or FA only; blue, NaAc and pCA or FA; red, NaAc only; light green, Na2SO4 only; yellow, relatively sensitive to all inhibitors). Arrows indicate positions of GLBRCY0 (red), CEN.PK2 (blue), and S288c (green) strains in the heat map. Also shown are representative growth curves of S288c (B), CEN.PK2 (C), and GLBRCY0 (D) cultured in YPD alone (green) or YPD with Na2SO4 (purple), NaAc (blue), solvent-ethanol (orange), or pCA or FA in ethanol (red).
Directed evolution for improved xylose utilization in the presence of p-hydroxycinnamic acids.
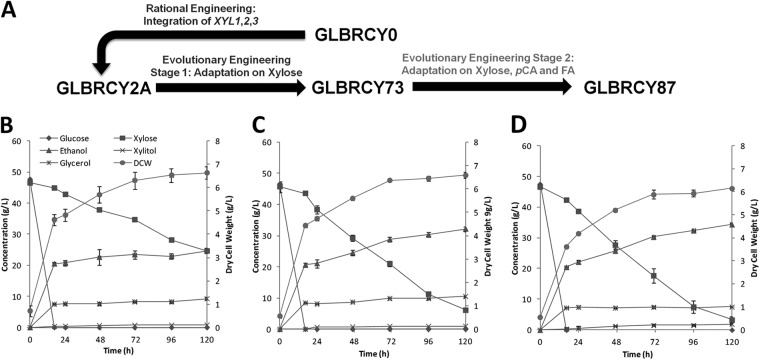
Since the native GLBRCY0 wild strain displayed relatively high tolerance to sodium, acetate, and p-hydroxycinnamic acids, we utilized a GLBRY0 strain previously engineered for xylose utilization by expression of the XR, XDH, and XK enzymes from the xylose-fermenting yeast S. stipitis (14). Unfortunately, the engineered GLBRCY0 strain, renamed GLBRCY2A, displayed very limited growth on xylose from laboratory media, making it a poor starting point for adaptation on xylose in the presence of inhibitors (see Fig. S1A in the supplemental material). Therefore, a two-stage directed evolution approach was employed to improve growth on xylose in the presence of inhibitory concentrations of p-hydroxycinnamic acids (Fig. 2A). First, the GLBRCY2A strain was adapted by aerobic serial subculturing in YP media containing increasing xylose concentrations for a total of 47.1 generations. A single clone from this adaptation was isolated and determined to have faster growth on xylose than the GLBRCY2A parent strain (see Fig. S1A in the supplemental material). The GLBRCY73 strain was then subjected to a second round of coadaptation by aerobic serial selection in YP media containing 20 g/liter xylose (YPX) and increasing concentrations of pCA and FA (see Fig. S1B in the supplemental material). From this adapted culture, a single clone named GLBRCY87 was isolated that displayed faster growth in YPX (see Fig. S1C in the supplemental material) and YPD (see Fig. S1D in the supplemental material) media containing 2.4 g/liter pCA and 1.2 g/liter FA relative to both GLBRCY73 and GLBRCY2A strains.
FIG 2.
The strategy for strain development employed in this work, which includes a combination of rational and evolutionary engineering (A) along with representative kinetic profiles of growth, sugar consumption, and end product formation from fermentations by these strains, including GLBRCY2A (B), GLBRCY73 (C), and GLBRCY87 (D) in minimal yeast media containing glucose and xylose. Error bars represent data ranges in biological duplicates.
Fermentation in minimal media and AHP-pretreated hydrolysates.
To assess whether our selection and evolution approach generally improved xylose fermentation to favor ethanol production, we compared the anaerobic fermentation performances of the GLBRCY2A, -73 and -87 in defined minimal media without inhibitors. All three strains were inoculated into N2-sparged medium containing yeast nitrogen base (YNB), urea as a nitrogen source, and 50 g/liter glucose and xylose at pH 5.5 (Fig. 2B to D). While the parental GLBRCY2A strain could only ferment approximately 50% of the xylose within 120 h, the two strains evolved on xylose, GLBRCY73 and -87, both consumed over 80% of the xylose from the medium. Moreover, both GLBRCY73 and -87 had significantly higher specific (GLBRCY73, 0.102 ± 0.007 g xylose consumed/g [dry cell weight]/h, and GLBRCY87, 0.132 ± 0.012 g xylose consumed/g [dry cell weight]/h [Fig. 3A]) and absolute (GLBRCY73, 0.369 ± 0.021 g xylose consumed/liter/h, and GLBRCY87, 0.446 ± 0.03 g xylose consumed/liter/h [Fig. 3B]) xylose uptake rates in minimal YNB media than the parental GLBRCY2A strain (0.053 ± 0.004 g xylose consumed/g [dry cell weight]/h and 0.191 ± 0.007 g xylose consumed/liter/h, respectively). Interestingly, strain GLBRCY87, which was evolved on xylose, pCA, and FA, showed a higher specific xylose uptake rate than GLBRCY73 in the absence of inhibitors. This may be partly due the significantly lower biomass yield on glucose for GLBRCY87 relative to GLBRCY2A and -73 (Fig. 3C). These increased xylose consumption rates resulted in corresponding increases in ethanol titers after 120 h (GLBRCY2A, 24.6 ± 0.86 g EtOH/liter; GLBRCY73, 32.2 ± 0.70 g EtOH/liter; and GLBRCY87, 34.3 ± 0.50 g EtOH/liter), suggesting that the aerobic directed evolution steps also improved anaerobic xylose conversion into ethanol. Differences in ethanol yield were not likely to contribute to these differences, as ethanol yield was unaffected by evolution or medium (hydrolysate or minimal media), with relatively constant ethanol yields on glucose (0.43 g/g) and xylose (0.30 g/g).
FIG 3.
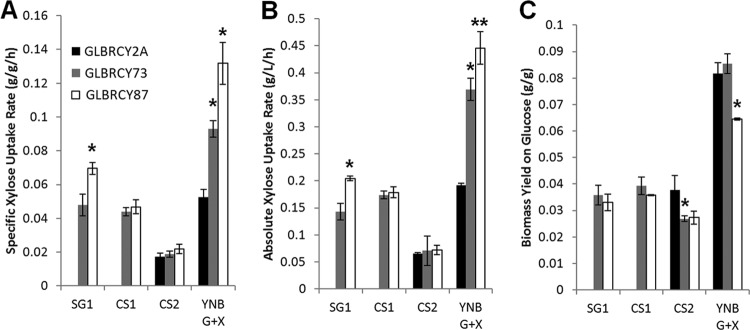
Specific (A) and absolute (B) xylose uptake rates and biomass yields on glucose (C) for the 3 strains in the 3 hydrolysates and YNB minimal media. Fermentation using GLBRCY2A was not performed on hydrolysates SG1 and CS1. Asterisks indicate statistically significant differences between the strain and its parent based on testing the null hypothesis that the parameters are equal (Student t test: *, P < 0.01; **, P < 0.025).
We next examined whether the evolution of GLBRCY87 on xylose and pCA-FA also allowed for improved anaerobic fermentation of xylose from the characterized AHP hydrolysates, which contained high concentrations of inhibitors. From fermentation of AHP pretreated switchgrass hydrolysate (SG1) at pH 5.5, the GLBRCY87 strain evolved on xylose and pCA or FA displayed significantly higher specific (Fig. 3A) and absolute (Fig. 3B) xylose uptakes than the GLBRCY73 ancestor, resulting in greater overall xylose consumed within 144 h (Fig. 4A and B). In the SG1 hydrolysate, the evolved GLBRCY87 strain displayed higher specific (0.07 ± 0.004 g/g/h) and absolute (0.205 ± 0.005 g/liter/h) xylose consumption rates than the GLBRCY73 strain (0.048 ± 0.006 g/g/h and 0.143 ± 0.016 g/liter/h, respectively). In contrast, no significant differences in xylose uptake rates between strains in AHP-pretreated corn stover hydrolysates (CS1 and CS2) were observed (Fig. 3A and B). In all three AHP hydrolysates, the biomass yields from glucose (YX/Glc) were similar between GLBRCY73 and GLBRCY87, while the biomass yield was lower for GLBRCY87 in defined media. Since pH is known to play a significant role in the fermentation of hydrolysates through protonation of weak acids (32), we tested the effect of pH on the fermentation of hydrolysate. Although the GLBRCY87 strain could ferment the xylose in the CS1 AHP-pretreated corn stover hydrolysate to completion at pH 5.5 (Fig. 4C), we observed significant inhibition of cell growth and sugar consumption during anaerobic fermentation of CS1 at pH 5.0 (Fig. 4D). Increasing the pH to 6.0 or 6.5 did not result in any differences compared to results obtained at pH 5.5 (data not shown). Taken together, these results indicate that the directed evolution on xylose and pCA or FA produced the GLBRCY87 strain with faster anaerobic xylose consumption from the AHP switchgrass hydrolysate.
FIG 4.
Representative kinetic profiles of growth, sugar consumption, and end product formation from fermentations of undetoxified switchgrass hydrolysate SG1 by GLBRCY73 (A) or GLBRCY87 (B). Also shown is fermentation by GLBRCY87 of undetoxified corn stover hydrolysate (CS1) at pH 5.5 (C) or pH 5.0 (D). Error bars represent data ranges in biological replicates.
Functional and genetic comparisons of the evolved strains.
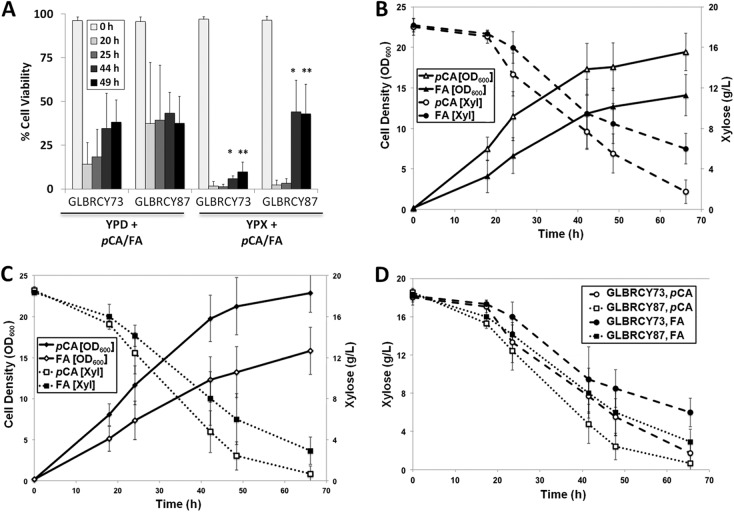
The growth and fermentation studies in both laboratory media and AHP hydrolysates indicate that the evolved GLBRCY87 displays a functionally relevant phenotype. We sought to better understand the mechanism by which the GLBRCY87 strain acquired greater resistance to p-hydroxycinnamic acids relative to GLBRCY73. To accomplish this, the viabilities of the GLBRCY73 and GLBRCY87 strains aerobically cultured in YPD or YPX with p-hydroxycinnamic acids were monitored by flow cytometry. While there were no significant differences in the percentages of viable cells grown in YPD media with pCA and FA, statistically significant differences between GLBRCY73 and GLBRCY87 in YPX with pCA and FA were observed at the later time points (Fig. 5A). In contrast, no differences in the viabilities between the GLBRCY73 and GLBRCY87 strains after 49 h in YPD and YPX media containing 0.48 M Na+ or 20 g/liter acetate as well as CS1 hydrolysates were observed in comparison to media without inhibitors (data not shown).
FIG 5.
GLBRCY87 displays greater tolerance to ferulic acid than GLBRCY73 during xylose consumption. In panel A, the cell viabilities of GLBRCY73 or GLBRCY87 grown in culture tubes at 30°C with the indicated media were measured. At the indicated times, the number of live or dead cells were counted flow cytometry, and the average percent cell viability and standard deviations from 3 independent biological replicates were calculated. Asterisks indicate statistically significant differences (Student t test: *, P < 0.03; **, P < 0.04). In panels B to D, representative growth profiles of xylose uptake by engineered and evolved yeast strains in YPX media containing 2.4 g/liter pCA or FA for strain GLBRCY73 (B) and strain GLBRCY87 (C) and comparison of xylose consumption by the 2 strains (D) are shown.
Although strain GLBRCY87 was evolved from adaptation on xylose in the presence of both p-coumaric and ferulic acids, it is possible that the strain may have evolved tolerance to one of the p-hydroxycinnamic acids, especially if one acid was more inhibitory. We explored this possibility by comparing the individual impacts of equimolar concentrations of p-coumaric or ferulic acid on xylose utilization by GLBRCY73 and GLBRCY87, which showed higher xylose consumption rates than GLBRCY73 in AHP hydrolysates that contained both inhibitors (Fig. 3A and B). GLBRCY73 and -87 were grown aerobically on YPX media at pH 5.5 in the presence of either pCA (1.5 g/liter) or FA (1.75 g/liter) at 9.0 mM, which is the molar equivalent to the total p-hydroxycinnamic acid concentration in the CS2 hydrolysate. Specifically, cell growth and xylose consumption by the GLBRCY73 strain were significantly more inhibited in 9.0 mM FA than pCA at the 68-h time point (Fig. 5B). In contrast, while cell growth by the GLBRCY87 strain was significantly more inhibited, there was no statistical difference in xylose consumption between FA and pCA (Fig. 5C). Moreover, in directly comparing the xylose consumption between strains and inhibitor conditions, we found no difference between strains for xylose consumption in pCA but significantly greater xylose consumption for GLBRCY87 than GLBRCY73 in the presence of FA (Fig. 5D). Together, these results suggest that GLBRCY87 evolved the improved xylose uptake phenotype primarily through tolerance to FA, which also appears to be more inhibitory for the GLBRCY0 strain background than pCA at equimolar concentrations. This higher growth inhibition by FA has also been observed with winemaking yeast strains (68).
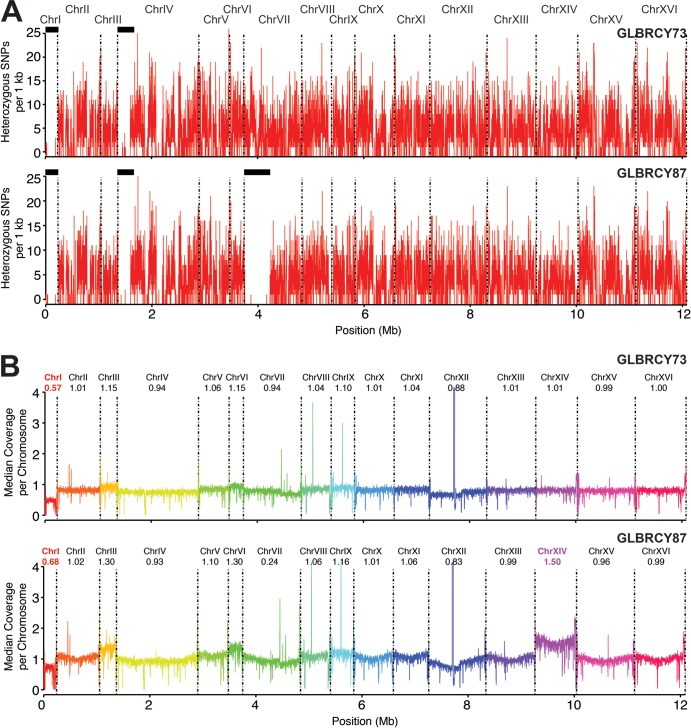
Our physiological data indicate that the evolved GLBRCY87 strain acquired stable genetic changes that confer improved xylose fermentation in the presence of ferulic and p-coumaric acids compared to the GLBRCY73 parent. We first performed standard Sanger sequencing of PAD1 and FDC1, two genes responsible for decarboxylation and detoxification of pCA and FA (44, 69), respectively, but found no coding sequence differences between GLBRCY73 and -87 (data not shown). Thus, in order to obtain a full picture of the strain-specific genetic differences, we performed high-throughput genome sequencing of both strains. Libraries of 200- to 500-bp genomic DNA fragments from the diploid GLBRCY73 and GLBRCY87 strains were prepared and sequenced using Illumina instruments (see Materials and Methods). Given that both GLBRCY73 and -87 are diploid strains, we first examined both strains for the prevalence of heterozygous SNPs (e.g., more than one nucleotide call for a single base sequence). GLBRCY87 had a significant reduction in the number of heterozygous SNP calls within regions of multiple chromosomes, including a large region within the left arm of chromosome VII (ChrVII) spanning open reading frames from YGL008C to YGL259W (Fig. 6A). In contrast, GLBRCY73 and the parental GLBRCY2A (D. J. Wohlbach, T. K. Sato, and A. P. Gasch, unpublished data) strains had numerous heterozygous SNPs within this region, suggesting that the GLBRCY87 strain underwent loss of heterozygosity (LOH) in ChrVII.
FIG 6.
pCA- and FA-tolerant GLBRCY87 displays large chromosomal differences, including chromosome duplication and LOH, compared to GLBRCY73. Both GLBRCY73 and -87 genomic DNAs were subjected to Illumina sequencing and individual reads mapped (see Materials and Methods). (A) Total number of heterozygous SNPs in 1-kb windows with a 100-bp step size for the indicated strains. Regions of LOH are indicated by the black bars at the top of the plot. LOH in ChrI is due to the single copy (hemizygosity) of this chromosome in both strains. (B) The median coverage of all reads in 1 kb windows with a 100 bp step size relative to the total genomic coverage for each strain indicated. Individual chromosomes are demarked by color and dashed vertical lines. ChrI (red) and ChrXIV (purple) are highlighted to denote aneuploidies.
In addition to determining changes in heterozygosity, we examined alterations in chromosome copy number, which can also result in phenotypic changes. To do this, we determined the relative sequence read coverage across all chromosomes and discovered that ChrI read coverage in both GLBRCY73 and -87 was approximately half of the median read coverage for all chromosomes (Fig. 6B), suggesting that ChrI is hemizygous in GLBRCY73 and -87. We further found that the read coverage for ChrXIV in strain GLBRCY87 was 1.5-fold higher than that of the diploid GLBRCY73 parent, suggesting that the evolved GLBRCY87 has three copies of ChrXIV, while GLBRCY73 has two copies. These results suggest that amplification of ChrXIV occurred during adaptation of GLBRCY87 and may contribute toward the pCA- and FA-tolerant phenotype.
DISCUSSION
Identification of AHP hydrolysate inhibitors for evolution of a tolerant, xylose-fermenting yeast strain.
Microbial fermentation of mixed-sugar lignocellulose-derived hydrolysates to industrially relevant ethanol titers (i.e., >50 g/liter) is challenging due to a number of inhibitory compounds derived from plant cell wall polymers, extractives, and pretreatment chemicals. Compared to defined media or detoxified hydrolysates, most ethanologens ferment undetoxified lignocellulosic hydrolysates with slower growth, sugar utilization, and product formation rates (7, 70, 71), ultimately prolonging the bioreactor residence time for fermentation and translating into higher capital costs for process equipment. In particular, xylose consumption rates by XR-XDH-expressing strains are much slower than glucose consumption rates due to an imbalance in regenerating NADH and NADPH cofactors (6). In our AHP-pretreated hydrolysates, this impact on xylose consumption rate was presumably due to the combinatorial effects of inhibitors on metabolism.
Since we did not detect significant levels of furfural and HMF, two inhibitors that are known to alter cofactor balance through their detoxification by NADH and NADPH-coupled reductases (72–75), it appears less likely that redox imbalance is the primary source of poor xylose fermentation by GLBRCY73 and -87 in the three AHP hydrolysates. Consistent with this, neither of these strains excreted significant amounts of xylitol into the media (Fig. 4), which can be indicative of suboptimal NAD+ levels and cofactor imbalance. Instead, high concentrations of Na+, in the range of 0.5 to 1.0 M (Table 1), introduced during high-solid AHP pretreatment and hydrolysis may have impaired xylose fermentation by inducing osmotic stress and impacting cell membrane potential, thereby limiting ATP production (76). Acetate is another well-known fermentation inhibitor that is commonly released from hemicellulose xylan under alkaline conditions (38). From the three AHP hydrolysates examined (Table 1), acetate was detected at concentrations (5.8 to 8.9 g/liter) that could dramatically impact fermentation. Acetate and other weak acids are known to inhibit yeast biomass yield from sugars (32) as well as glucose and xylose uptake rates (71, 77–80) by diffusing into and acidifying the cytosol (81). To counteract this, yeast responds by increasing the expression of ATPase-driven proton symport proteins (42), which results in reduced ATP availability. In addition to acetate and Na+, we detected high concentrations of two phenolic compounds, pCA and FA, that are known to inhibit yeast fermentation and growth (22, 43, 68, 82) in the AHP-pretreated corn stover and switchgrass hydrolysates (Table 1). In grasses, AHP pretreatment is known to release pCA, which is acylated to the side chain region of lignin, and FA, which cross-links arabinoxylans and lignins (38, 83, 84). Both pCA and FA appear to enter into the yeast cell, where they can be decarboxylated into 2-vinylphenol and 4-vinylguaiacol, respectively, by the Fdc1p and Pad1p enzymes (43, 44). To date, the mechanisms by which these phenolic acids and their vinyl derivatives impact yeast physiology and fermentation have not been precisely determined, although they have been proposed to act through partitioning and disrupting the integrity of biological membranes (33, 85).
While previous work focused on engineering and evolution of a selected number industrial strains, we comprehensively searched for a strain with robust innate tolerance to individual AHP inhibitors among a panel of wild S. cerevisiae strains isolated from a variety of environmental niches. Surprisingly, only six wild strains were found to rank highest in relative growth rates across laboratory media containing acetate, sodium, or pCA and FA (Fig. 1A and B). In contrast, commonly used laboratory strains S288c and CEN.PK2 exhibited less robust growth in the presence of inhibitors, particularly pCA and FA (Fig. 1C and D). Importantly, all 108 of these strains can be publically acquired, so the phenotypic data can be used for the selection of S. cerevisiae strains for metabolic engineering for the fermentation of other hydrolysates containing the tested inhibitors.
Through a series of genetic engineering and directed evolution, we obtained two strains, GLBRCY73 and GLBRCY87, with higher growth rates relative to their ancestor in aerobic xylose only and xylose containing p-hydroxycinnamic acids, respectively (see Fig. S1A, C, and D in the supplemental material), as well as faster anaerobic xylose consumption rates in minimal laboratory media and AHP switchgrass hydrolysate (SG1), respectively (Fig. 2, 3A and B, and 4A and B). Importantly, the higher rate of specific xylose uptake by GLBRCY87 in AHP-pretreated SG1 hydrolysate was not due to lower biomass yields, and the specific xylose uptake rates (qXyl = 0.05 to 0.14 g/g/h) for these strains and medium conditions are comparable to others reported for XR-XDH-XK-engineered S. cerevisiae strains (1, 17). Although there were significant differences in SG1, it was unclear why GLBRCY87 did not perform better than GLBRCY73 in both AHP corn stover hydrolysates, but this could be due to higher concentrations of acetate and Na+, which together synergistically suppressed the evolved tolerance to p-hydroxycinnamic acids. Evidence for the higher acetate concentrations impacting xylose fermentation is suggested by the strong inhibition of growth and fermentation of xylose in AHP CS1 at pH 5.0 (Fig. 4D), which is closer to the dissociation constant of acetic acid (pKa = 4.75) than pH 5.5. More notable is our achievement of complete fermentation of highly concentrated sugars from undetoxified AHP CS1 hydrolysate by GLBRCY87 into more than 40 g/liter ethanol, highlighting the potential of both the pretreatment and strain for industrial processes. In contrast, hydrolysates produced from other pretreatments commonly require chemical, physical, or biological detoxification prior to hydrolysis (e.g., overliming, activated carbon treatment, or washing) for suitable sugar conversion (32).
Large chromosomal changes were found in the GLBRCY87 strain with enhanced viability and faster xylose consumption in the presence of p-coumaric and ferulic acids.
When the GLRCY73 and -87 strains were cultured on glucose, we observed significant effects on their viabilities in the presence of pCA and FA (Fig. 5A), suggesting that the impact of pCA and FA was not specific to xylose. While p-hydroxycinnamic acids significantly impacted the viability of GLBRCY73 when xylose was the sole available sugar source, GLBRCY87 interestingly appeared to recover from pCA and FA exposure after 44 h, with significantly greater cell viability than that of GLBRCY73. This response is particularly intriguing since it may provide an advantage for the evolved GLBRCY87 strain in xylose fermentation, which usually occurs at later times with pretreated hydrolysates after glucose has been mostly consumed. This may ultimately shorten the residence time during fermentation of feedstocks containing high concentrations p-hydroxycinnamic acids compared to other strains. In addition, we found that 9.0 mM FA had significant impacts at later stages of growth and xylose consumption by GLBRCY73 and -87, while equimolar concentrations of pCA had little effect (Fig. 5B to D). Moreover, we found a significant improvement in xylose consumption with FA by the evolved GLBRCY87 strain over the GLBRCY73 parent after 68 h of growth. Consistent with this, a previous study determined that FA significantly reduced the S. cerevisiae biomass yield on glucose, while pCA had relatively little impact (22). This suggests that the adaptation of the GLBRCY87 strain primarily occurred through improved FA tolerance that enhanced cell viability during xylose consumption.
To identify any major genetic alterations incurred during directed evolution on xylose in the presence of pCA and FA, we compared the genome sequences of the evolved GLBRCY87 and parental GLBRCY73 strains. Our sequencing analysis did not reveal new spontaneous mutations that emerged in GLBRCY87 from GLBRCY73. Instead, we uncovered two major chromosomal differences between these two diploid strains: (i) LOH in the left arm of both ChrVII copies of the GLBRCY87 strain relative to the heterozygous GLBRCY73 strain (Fig. 6A) and (ii) duplication of ChrXIV from two copies in GLBRCY73 to three copies in GLBRCY87 (Fig. 6B). Loss of heterozygosity, which can occur during mitotic recombination (86), can influence strain phenotypes by unmasking recessive traits hidden in heterozygous strains (87). LOH identified in the left arm of ChrVII in GLBRCY87 relative to GLBRCY73 suggests that this event occurred during the adaptation on xylose and pCA or FA and may play an important role in determining the pCA and FA tolerance in GLBRCY87. Of note, this region contains PDR1, which encodes one of two zinc finger transcription factors that regulate the pleiotropic drug response and subsequent expression of multidrug resistance transporters (88–91), including Pdr5p, which has been shown to be involved in tolerance to gallic acid (92), another phenolic acid. The LOH event caused a difference in amino acid 94 of Pdr1p from alanine/threonine in the heterozygous GLBRCY73 strain to threonine/threonine in GLBRCY87, potentially contributing to an altered transcriptional response in downstream targets involved in xenobiotic resistance.
In addition to LOH, GLBRCY87 acquired an additional copy of ChrXIV during the evolution from GLBRCY73. Chromosomal aneuploidy has been previously found in strains evolved under nutrient limitation (93, 94) and shown in some cases to directly impact yeast phenotypes (95). Thus, one possible explanation for the GLBRCY87 phenotype is that increased dosage of one or more genes on ChrXIV may be important for p-coumaric and ferulic acid tolerance. One group of interesting genes on this chromosome includes PDR18, which encodes another ABC transporter that is important for tolerance to herbicides, metal ions (96), and high concentrations of ethanol (97), as well as PDR16 and PDR17, which encode homologous regulators of lipid biosynthesis and membrane composition (98–100). Interestingly, both PDR16 and PDR17 play important roles in xenobiotic resistance for yeast, and expression of PDR16 is known to be controlled by Pdr1p (101, 102). Thus, it is possible that the adaptative changes in GLBRCY87, which include LOH in PDR1 and increased copy numbers of PDR16 to PDR18, led to an enhanced PDR gene response that subsequently allowed for enhanced cell survival in high concentrations of pCA and FA and a greater number of functional cells able to ferment xylose.
Our work highlights the importance of selecting the appropriate starting strain for directed engineering and evolution for fermentation of the pretreated lignocellulosic hydrolysate of choice. By first identifying and quantifying the composition of inhibitors in AHP-pretreated lignocellulosic hydrolysates, followed by genetic engineering and directed evolution of a wild S. cerevisiae strain specifically selected for tolerance to AHP hydrolysate inhibitors, we were able to improve xylose fermentation in industrially relevant lignocellulosic hydrolysates within relatively few generations of selection. We also showed that natural isolates of S. cerevisiae, which are increasingly more available to scientific researchers from various culture collections and have a number of published genomes, are viable genetic backgrounds to study tolerance to industrial stresses. These results also highlight the unique types of mutations and evolutionary paths that can emerge from wild heterozygous diploid strains, which can rapidly evolve desired traits through LOH and chromosomal aneuploidy events. Finally, our work provides evidence that AHP pretreatment coupled with the appropriate ethanologen is a promising pretreatment for lignocellulosic ethanol production.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We thank Jeff Lewis, Clete Kurtzmann, and Justin Fay for yeast strains, Ragothaman Avanasi for technical support, Yury Bukhman for GCAT assistance, Bob Landick for scientific advice, Wendy Schackwitz and Joel Martin for advice on sequencing analysis, and Christa Pennachio for coordination of Illumina sequencing.
Footnotes
Published ahead of print 8 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01885-13.
REFERENCES
- 1.Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund M. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147–177. 10.1007/10_2007_062 [DOI] [PubMed] [Google Scholar]
- 2.Saha BC, Nichols NN, Cotta MA. 2011. Ethanol production from wheat straw by recombinant Escherichia coli strain FBR5 at high solid loading. Bioresour. Technol. 102:10892–10897. 10.1016/j.biortech.2011.09.041 [DOI] [PubMed] [Google Scholar]
- 3.Hodge DB, Karim MN. 2002. Modeling and advanced control of recombinant Zymomonas mobilis fed-batch fermentation. Biotechnol. Prog. 18:572–579. 10.1021/bp0155181 [DOI] [PubMed] [Google Scholar]
- 4.Mohagheghi A, Evans K, Chou Y-C, Zhang M. 2002. Cofermentation of glucose, xylose, and arabinose by genomic DNA-integrated xylose/arabinose fermenting strain of Zymomonas mobilis AX101. Appl. Biochem. Biotechnol. 98-100:885–898 [PubMed] [Google Scholar]
- 5.Almeida JR, Runquist D, Sanchez i Nogue V, Liden G, Gorwa-Grauslund MF. 2011. Stress-related challenges in pentose fermentation to ethanol by the yeast Saccharomyces cerevisiae. Biotechnol. J. 6:286–299. 10.1002/biot.201000301 [DOI] [PubMed] [Google Scholar]
- 6.Van Vleet JH, Jeffries TW. 2009. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol. 20:300–306. 10.1016/j.copbio.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 7.Almeida JRM, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82:340–349. 10.1002/jctb.1676 [DOI] [Google Scholar]
- 8.Attfield PV, Bell PJL. 2006. Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEMS Yeast Res. 6:862–868. 10.1111/j.1567-1364.2006.00098.x [DOI] [PubMed] [Google Scholar]
- 9.Jeffries TW. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320–326. 10.1016/j.copbio.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 10.Brat D, Boles E, Wiedemann B. 2009. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75:2304–2311. 10.1128/AEM.02522-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Ho NW. 1993. Cloning and improving the expression of Pichia stipitis xylose reductase gene in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 39-40:135–147 [DOI] [PubMed] [Google Scholar]
- 12.Jeppsson M, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604–1609. 10.1128/AEM.68.4.1604-1609.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925–934. 10.1016/j.femsyr.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 14.Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, LaButti KM, Sun H, Clum A, Pangilinan JL, Lindquist EA, Lucas S, Lapidus A, Jin M, Gunawan C, Balan V, Dale BE, Jeffries TW, Zinkel R, Barry KW, Grigoriev IV, Gasch AP. 2011. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc. Natl. Acad. Sci. U. S. A. 108:13212–13217. 10.1073/pnas.1103039108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laluce C, Schenberg ACG, Gallardo JCM, Coradello LFC, Pombeiro-Sponchiado SR. 2012. Advances and developments in strategies to improve strains of Saccharomyces cerevisiae and processes to obtain the lignocellulosic ethanol—a review. Appl. Biochem. Biotechnol. 166:1908–1926. 10.1007/s12010-012-9619-6 [DOI] [PubMed] [Google Scholar]
- 16.Young E, Lee S-M, Alper H. 2010. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol. Biofuels 3:24. 10.1186/1754-6834-3-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonderegger M, Sauer U. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 69:1990–1998. 10.1128/AEM.69.4.1990-1998.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scalcinati G, Otero JM, Van Vleet JRH, Jeffries TW, Olsson L, Nielsen J. 2012. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res. 12:582–597. 10.1111/j.1567-1364.2012.00808.x [DOI] [PubMed] [Google Scholar]
- 19.Wahlbom CF, van Zyl WH, Jönsson LJ, Hahn-Hägerdal B, Otero RRC. 2003. Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res. 3:319–326. 10.1016/S1567-1356(02)00206-4 [DOI] [PubMed] [Google Scholar]
- 20.Hendriks ATWM, Zeeman G. 2009. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour. Technol. 100:10–18. 10.1016/j.biortech.2008.05.027 [DOI] [PubMed] [Google Scholar]
- 21.Hodge DB, Andersson C, Berglund KA, Rova U. 2009. Detoxification requirements for bioconversion of softwood dilute acid hydrolyzates to succinic acid. Enzyme Microb. Technol. 44:309–316. 10.1016/j.enzmictec.2008.11.007 [DOI] [Google Scholar]
- 22.Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant N-O, Jönsson L. 2000. Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 84-86:617–632 [DOI] [PubMed] [Google Scholar]
- 23.Klinke HB, Olsson L, Thomsen AB, Ahring BK. 2003. Potential inhibitors from wet oxidation of wheat straw and their effect on ethanol production of Saccharomyces cerevisiae: Wet oxidation and fermentation by yeast. Biotechnol. Bioeng. 81:10. 10.1002/bit.10523 [DOI] [PubMed] [Google Scholar]
- 24.Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant N-O. 1999. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb. Technol. 24:9 [Google Scholar]
- 25.Qureshi N, Saha BC, Hector RE, Cotta MA. 2008. Removal of fermentation inhibitors from alkaline peroxide pretreated and enzymatically hydrolyzed wheat straw: production of butanol from hydrolysate using Clostridium beijerinckii in batch reactors. Biomass Bioenergy 32:1353–1358. 10.1016/j.biombioe.2008.04.009 [DOI] [Google Scholar]
- 26.Persson P, Andersson J, Gorton L, Larsson S, Nilvebrant N-O, Jönsson LJ. 2002. Effect of different forms of alkali treatment on specific fermentation inhibitors and on the fermentability of lignocellulose hydrolysates for production of fuel ethanol. J. Agric. Food Chem. 50:5318–5325. 10.1021/jf025565o [DOI] [PubMed] [Google Scholar]
- 27.Chundawat SPS, Vismeh R, Sharma LN, Humpula JF, da Costa Sousa L, Chambliss CK, Jones AD, Balan V, Dale BE. 2010. Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Bioresour. Technol. 101:8429–8438. 10.1016/j.biortech.2010.06.027 [DOI] [PubMed] [Google Scholar]
- 28.Du B, Sharma LN, Becker C, Chen S-F, Mowery RA, van Walsum GP, Chambliss CK. 2010. Effect of varying feedstock-pretreatment chemistry combinations on the formation and accumulation of potentially inhibitory degradation products in biomass hydrolysates. Biotechnol. Bioeng. 107:430–440. 10.1002/bit.22829 [DOI] [PubMed] [Google Scholar]
- 29.Helm RF, Jervis J, Ray WK, Willoughby N, Irvin B, Hastie J, Schell DJ, Nagle N. 2010. Mass spectral analyses of corn stover prehydrolysates to assess conditioning processes. J. Agric. Food Chem. 58:12642–12649. 10.1021/jf1031197 [DOI] [PubMed] [Google Scholar]
- 30.Klinke HB, Thomsen AB, Ahring BK. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 66:10–26. 10.1007/s00253-004-1642-2 [DOI] [PubMed] [Google Scholar]
- 31.Liu Z. 2011. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl. Microbiol. Biotechnol. 90:809–825. 10.1007/s00253-011-3167-9 [DOI] [PubMed] [Google Scholar]
- 32.McMillan JD. 1994. Conversion of hemicellulose hydrolyzates to ethanol. ACS Symp. Ser. Am. Chem. Soc. 566:411–437 [Google Scholar]
- 33.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour. Technol. 74:25–33 http://dx.doi.org/10.1016/S0960-8524(99)00161–3 [Google Scholar]
- 34.Richardson TL, Harner NK, Bajwa PK, Trevors JT, Lee H. 2011. Approaches to deal with toxic inhibitors during fermentation of lignocellulosic substrates. ACS Symp. Ser. Am. Chem. Soc. 1067:171–202. 10.1021/bk-2011-1067.ch007 [DOI] [Google Scholar]
- 35.Banerjee G, Car S, Liu T, Williams DL, Meza SL, Walton JD, Hodge DB. 2012. Scale-up and integration of alkaline hydrogen peroxide pretreatment, enzymatic hydrolysis, and ethanolic fermentation. Biotechnol. Bioeng. 109:922–931. 10.1002/bit.24385 [DOI] [PubMed] [Google Scholar]
- 36.Saha BC, Cotta MA. 2006. Ethanol production from alkaline peroxide pretreated enzymatically saccharified wheat straw. Biotechnol. Prog. 22:449–453. 10.1021/bp050310r [DOI] [PubMed] [Google Scholar]
- 37.Saha BC, Cotta MA. 2007. Enzymatic saccharification and fermentation of alkaline peroxide pretreated rice hulls to ethanol. Enzyme Microb. Technol. 41:528–532. 10.1016/j.enzmictec.2007.04.006 [DOI] [Google Scholar]
- 38.Harris PJ, Stone BA. 2009. Chemistry and molecular organization of plant cell walls, p 61–93 In Himmel ME. (ed), Biomass recalcitrance. Blackwell Publishing Ltd, Oxford, United Kingdom [Google Scholar]
- 39.Heer D, Heine D, Sauer U. 2009. Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl. Environ. Microbiol. 75:7631–7638. 10.1128/AEM.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW. 2004. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 31:345–352 [DOI] [PubMed] [Google Scholar]
- 41.Peña PV, Glasker S, Srienc F. 2013. Genome-wide overexpression screen for sodium acetate resistance in Saccharomyces cerevisiae. J. Biotechnol. 164:26–33. 10.1016/j.jbiotec.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 42.Yang S, Land ML, Klingeman DM, Pelletier DA, Lu T-YS, Martin SL, Guo H-B, Smith JC, Brown SD. 2010. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 107:10395–10400. 10.1073/pnas.0914506107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson S, Nilvebrant NO, Jönsson L. 2001. Effect of overexpression of Saccharomyces cerevisiae Pad1p on the resistance to phenylacrylic acids and lignocellulose hydrolysates under aerobic and oxygen-limited conditions. Appl. Microbiol. Biotechnol. 57:167–174 [DOI] [PubMed] [Google Scholar]
- 44.Mukai N, Masaki K, Fujii T, Kawamukai M, Iefuji H. 2010. PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109:564–569. 10.1016/j.jbiosc.2009.11.011 [DOI] [PubMed] [Google Scholar]
- 45.Dhar R, Sägesser R, Weikert C, Yuan J, Wagner A. 2011. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol. 24:1135–1153. 10.1111/j.1420-9101.2011.02249.x [DOI] [PubMed] [Google Scholar]
- 46.Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sá-Correia I. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 337:95–103. 10.1016/j.bbrc.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 47.Keating JD, Panganiban C, Mansfield SD. 2006. Tolerance and adaptation of ethanologenic yeasts to lignocellulosic inhibitory compounds. Biotechnol. Bioeng. 93:1196–1206. 10.1002/bit.20838 [DOI] [PubMed] [Google Scholar]
- 48.Fujitomi K, Sanda T, Hasunuma T, Kondo A. 2012. Deletion of the PHO13 gene in Saccharomyces cerevisiae improves ethanol production from lignocellulosic hydrolysate in the presence of acetic and formic acids, and furfural. Bioresour. Technol. 111:161–166. 10.1016/j.biortech.2012.01.161 [DOI] [PubMed] [Google Scholar]
- 49.Sanda T, Hasunuma T, Matsuda F, Kondo A. 2011. Repeated-batch fermentation of lignocellulosic hydrolysate to ethanol using a hybrid Saccharomyces cerevisiae strain metabolically engineered for tolerance to acetic and formic acids. Bioresour. Technol. 102:7917–7924. 10.1016/j.biortech.2011.06.028 [DOI] [PubMed] [Google Scholar]
- 50.Wright J, Bellissimi E, de Hulster E, Wagner A, Pronk JT, van Maris AJA. 2011. Batch and continuous culture-based selection strategies for acetic acid tolerance in xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. 11:299–306. 10.1111/j.1567-1364.2011.00719.x [DOI] [PubMed] [Google Scholar]
- 51.Hector RE, Dien BS, Cotta MA, Qureshi N. 2011. Engineering industrial Saccharomyces cerevisiae strains for xylose fermentation and comparison for switchgrass conversion. J. Ind. Microbiol. Biotechnol. 38:1193–1202. 10.1007/s10295-010-0896-1 [DOI] [PubMed] [Google Scholar]
- 52.Ismail KSK, Sakamoto T, Hatanaka H, Hasunuma T, Kondo A. 2013. Gene expression cross-profiling in genetically modified industrial Saccharomyces cerevisiae strains during high-temperature ethanol production from xylose. J. Biotechnol. 163:50–60. 10.1016/j.jbiotec.2012.10.017 [DOI] [PubMed] [Google Scholar]
- 53.Koppram R, Albers E, Olsson L. 2012. Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol. Biofuels 5:32. 10.1186/1754-6834-5-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demeke M, Dietz H, Li Y, Foulquie-Moreno M, Mutturi S, Deprez S, Den Abt T, Bonini B, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein J. 2013. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol. Biofuels 6:89. 10.1186/1754-6834-6-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fay JC, Benavides JA. 2005. Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics 170:1575–1587. 10.1534/genetics.105.042283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP. 2010. Exploiting natural variation in Saccharomyces cerevisiae to identify fenes for increased ethanol resistance. Genetics 186:1197–1205. 10.1534/genetics.110.121871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, Liti G, Moses A, Blomberg A. 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7:e1002111. 10.1371/journal.pgen.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, Foster C, Pu Y, Holmes D, Saffron C, Ragauskas A, Hodge DB. 2012. Structural characterization of alkaline hydrogen peroxide pretreated grasses exhibiting diverse lignin phenotypes. Biotechnol. Biofuels 5:38. 10.1186/1754-6834-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin M, Sarks C, Gunawan C, Bice BD, Simonett SP, Avanasi Narasimhan R, Willis LB, Dale BE, Balan V, Sato TK. 2013. Phenotypic selection of a wild Saccharomyces cerevisiae strain for simultaneous saccharification and co-fermentation of AFEXTM pretreated corn stover. Biotechnol. Biofuels 6:108. 10.1186/1754-6834-6-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richards FJ. 1959. A flexible growth function for empirical use. J. Exp. Bot. 10:290–301. 10.1093/jxb/10.2.290 [DOI] [Google Scholar]
- 62.Zwietering MH, Jongenburger I, Rombouts FM, van't Riet K. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Englezos P, Kalogerakis NN. 2001. Applied parameter estimation for chemical engineers. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 64.Cohen A. 1983. Comparing regression coefficients across subsamples: a study of the statistical test. Sociol. Methods Res. 12:77–94. 10.1177/0049124183012001003 [DOI] [Google Scholar]
- 65.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 89:19.10.1–19.10.21. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baranowski JD, Davidson PM, Nagel CW, Branen AL. 1980. Inhibition of Saccharomyces cerevisiae by naturally occurring hydroxycinnamates. J. Food Sci. 45:592–594. 10.1111/j.1365-2621.1980.tb04107.x [DOI] [Google Scholar]
- 69.Clausen M, Lamb CJ, Megnet R, Doerner PW. 1994. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene 142:107–112. 10.1016/0378-1119(94)90363-8 [DOI] [PubMed] [Google Scholar]
- 70.Bellissimi E, Van Dijken JP, Pronk JT, Van Maris AJA. 2009. Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose-isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Res. 9:358–364. 10.1111/j.1567-1364.2009.00487.x [DOI] [PubMed] [Google Scholar]
- 71.Hasunuma T, Sung K, Sanda T, Yoshimura K, Matsuda F, Kondo A. 2011. Efficient fermentation of xylose to ethanol at high formic acid concentrations by metabolically engineered Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 90:997–1004. 10.1007/s00253-011-3085-x [DOI] [PubMed] [Google Scholar]
- 72.Jordan DB, Braker JD, Bowman MJ, Vermillion KE, Moon J, Liu ZL. 2011. Kinetic mechanism of an aldehyde reductase of Saccharomyces cerevisiae that relieves toxicity of furfural and 5-hydroxymethylfurfural. Biochim. Biophys. Acta 1814:1686–1694. 10.1016/j.bbapap.2011.08.011 [DOI] [PubMed] [Google Scholar]
- 73.Liu ZL, Moon J. 2009. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10. 10.1016/j.gene.2009.06.018 [DOI] [PubMed] [Google Scholar]
- 74.Liu ZL, Moon J, Andersh B, Slininger P, Weber S. 2008. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:743–753. 10.1007/s00253-008-1702-0 [DOI] [PubMed] [Google Scholar]
- 75.Nilsson A, Gorwa-Grauslund MF, Hahn-Hägerdal B, Lidén G. 2005. Cofactor dependence in furan reduction by Saccharomyces cerevisiae in fermentation of acid-hydrolyzed lignocellulose. Appl. Environ. Microbiol. 71:7866–7871. 10.1128/AEM.71.12.7866-7871.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kinclova-Zimmermannova O, Gaskova D, Sychrova H. 2006. The Na+,K+/H+-antiporter Nha1 influences the plasma membrane potential of Saccharomyces cerevisiae. FEMS Yeast Res. 6:792–800. 10.1111/j.1567-1364.2006.00062.x [DOI] [PubMed] [Google Scholar]
- 77.Casey E, Sedlak M, Ho NWY, Mosier NS. 2010. Effect of acetic acid and pH on the cofermentation of glucose and xylose to ethanol by a genetically engineered strain of Saccharomyces cerevisiae. FEMS Yeast Res. 10:385–393. 10.1111/j.1567-1364.2010.00623.x [DOI] [PubMed] [Google Scholar]
- 78.Delgenes JP, Moletta R, Navarro JM. 1996. Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzyme Microb. Technol. 19:220–225. 10.1016/0141-0229(95)00237-5 [DOI] [Google Scholar]
- 79.Palmqvist E, Grage H, Meinander NQ, Hahn-Hägerdal B. 1999. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol. Bioeng. 63:46–55. [DOI] [PubMed] [Google Scholar]
- 80.Taherzadeh MJ, Lidén G, Gustafsson L, Niklasson C. 1996. The effects of pantothenate deficiency and acetate addition on anaerobic batch fermentation of glucose by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 46:176–182. 10.1007/s002530050801 [DOI] [PubMed] [Google Scholar]
- 81.Hawkins G, Doran-Peterson J. 2011. A strain of Saccharomyces cerevisiae evolved for fermentation of lignocellulosic biomass displays improved growth and fermentative ability in high solids concentrations and in the presence of inhibitory compounds. Biotechnol. Biofuels 4:49. 10.1186/1754-6834-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sundström L, Larsson S, Jönsson L. 2010. Identification of Saccharomyces cerevisiae genes involved in the resistance to phenolic fermentation inhibitors. Appl. Biochem. Biotechnol. 161:106–115. 10.1007/s12010-009-8811-9 [DOI] [PubMed] [Google Scholar]
- 83.Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54:519–546. 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- 84.Pan GX, Bolton JL, Leary GJ. 1998. Determination of ferulic and p-coumaric acids in wheat straw and the amounts released by mild acid and alkaline peroxide treatment. J. Agric. Food Chem. 46:5283–5288. 10.1021/jf980608f [DOI] [Google Scholar]
- 85.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioresour. Technol. 74:17–24 http://dx.doi.org/10.1016/S0960-8524(99)00160-1 [Google Scholar]
- 86.Llorente B, Smith CE, Symington LS. 2008. Break-induced replication: what is it and what is it for? Cell Cycle 7:859–864. 10.4161/cc.7.7.5613 [DOI] [PubMed] [Google Scholar]
- 87.Magwene PM, Kayikci O, Granek JA, Reininga JM, Scholl Z, Murray D. 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 108:1987–1992. 10.1073/pnas.1012544108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206–2214 [PubMed] [Google Scholar]
- 89.Carvajal E, van den Hazel H, Cybularz-Kolaczkowska A, Balzi E, Goffeau A. 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256:406–415. 10.1007/s004380050584 [DOI] [PubMed] [Google Scholar]
- 90.Katzmann DJ, Hallstrom TC, Mahé Y, Moye-Rowley WS. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271:23049–23054. 10.1074/jbc.271.38.23049 [DOI] [PubMed] [Google Scholar]
- 91.Mamnun YM, Pandjaitan R, Mahé Y, Delahodde A, Kuchler K. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46:1429–1440. 10.1046/j.1365-2958.2002.03262.x [DOI] [PubMed] [Google Scholar]
- 92.Pereira Rangel L, Fritzen M, Yunes RA, Leal PC, Creczynski-Pasa TB, Ferreira-Pereira A. 2010. Inhibitory effects of gallic acid ester derivatives on Saccharomyces cerevisiae multidrug resistance protein Pdr5p. FEMS Yeast Res. 10:244–251. 10.1111/j.1567-1364.2009.00603.x [DOI] [PubMed] [Google Scholar]
- 93.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 99:16144–16149. 10.1073/pnas.242624799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4:e1000303. 10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- 96.Cabrito TR, Teixeira MC, Singh A, Prasad R, Sá-Correia I. 2011. The yeast ABC transporter Pdr18 (ORF YNR070w) controls plasma membrane sterol composition, playing a role in multidrug resistance. Biochem. J. 440:195–202. 10.1042/BJ20110876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teixeira M, Godinho C, Cabrito T, Mira N, Sa-Correia I. 2012. Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation. Microb. Cell Fact. 11:98. 10.1186/1475-2859-11-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Šimová Z, Poloncová K, Tahotná D, Holič R, Hapala I, Smith AR, White TC, Griač P. 2013. The yeast Saccharomyces cerevisiae Pdr16p restricts changes in ergosterol biosynthesis caused by the presence of azole antifungals. Yeast 30:229–241. 10.1002/yea.2956 [DOI] [PubMed] [Google Scholar]
- 99.van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. 1999. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274:1934–1941. 10.1074/jbc.274.4.1934 [DOI] [PubMed] [Google Scholar]
- 100.Wu W-I, Routt S, Bankaitis VA, Voelker DR. 2000. A new gene involved in the transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, SEC14. J. Biol. Chem. 275:14446–14456. 10.1074/jbc.275.19.14446 [DOI] [PubMed] [Google Scholar]
- 101.Akache B, Turcotte B. 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277:21254–21260. 10.1074/jbc.M202566200 [DOI] [PubMed] [Google Scholar]
- 102.do Valle Matta MA, Jonniaux J-L, Balzi E, Goffeau A, van den Hazel B. 2001. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene 272:111–119. 10.1016/S0378-1119(01)00558-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.