Abstract
The development and implementation of effective antimicrobial interventions by the beef processing industry in the United States have dramatically reduced the incidence of beef trim contamination by Escherichia coli O157:H7. However, individual processing plants still experience sporadic peaks in contamination rates where multiple E. coli O157:H7-positive lots are clustered in a short time frame. These peaks have been referred to as “high event periods” (HEP) of contamination. The results reported here detail the characterization of E. coli O157:H7 isolates from 21 HEP across multiple companies and processing plants to gain insight regarding the mechanisms causing these incidents. Strain genotypes were determined by pulsed-field gel electrophoresis, and isolates were investigated for characteristics linking them to human illness. Through these analyses, it was determined that individual HEP show little to no diversity in strain genotypes. Hence, each HEP has one strain type that makes up most, if not all, of the contamination. This is shown to differ from the genotypic diversity of E. coli O157:H7 found on the hides of cattle entering processing plants. In addition, it was found that a large proportion (81%) of HEP are caused by strain types associated with human illness. These results pose a potential challenge to the current model for finished product contamination during beef processing.
INTRODUCTION
The development and implementation of effective antimicrobial interventions by the beef processing industry in the United States have reduced the incidence of beef trim contamination by Escherichia coli O157:H7. These improvements have resulted in decreased contamination rates of raw beef trim by the bacterial pathogen E. coli O157:H7, to an estimated national prevalence of 0.39% (1). However, individual processing plants experience sporadic peaks in contamination rates where multiple positive lots are clustered in a short time frame. These peaks have been referred to as “high event periods” (HEP) of contamination. The Food Safety and Inspection Service (FSIS) of the USDA has defined HEP as production intervals during which slaughter establishments experience high rates of positive results for E. coli O157:H7 (or Shiga-toxigenic E. coli [STEC] or virulence markers) in trim samples (2). Typically, a cause/source for a HEP is not identified, and the contamination event will be resolved before notable correction of the process can be performed.
The current model of finished product contamination during beef processing starts with the pathogen load on the hides of cattle entering the processing plant. Several studies (3–5) have identified the hide as the major source of E. coli O157:H7 contamination of carcasses during processing. Once contamination has been transferred from the hide to the carcass during dehiding, it must be removed or destroyed through antimicrobial interventions to prevent finished product contamination. However, research has indicated that interventions or even systems of multiple interventions can be overwhelmed by high concentrations of bacteria and fail to prevent finished product contamination (6). In addition to exceeding the threshold of properly functioning interventions, the model assumes that finished product contamination will occur when interventions are not functioning at optimal levels or processing personnel are not working within the guidelines of the industry's best practices.
It has been assumed that HEP would follow the basic premise of this contamination model and be a function of incoming pathogen load. However, there is a large knowledge gap regarding the mechanism of HEP. Due to the intricacies of the beef harvest process, most studies of beef processing can follow contamination only from the incoming animal through the kill floor to the point where the carcasses are chilled after all interventions have been applied. Following the chilling process, carcasses are graded and sorted into similar weight/grade categories to facilitate marketing prior to further processing of the carcass into primal and subprimal cuts and the production of beef trim. Due to the sorting of carcasses into groups that were harvested at different times, combined with the typically low levels of E. coli O157 contamination, sample numbers too high to be feasible are required to track contamination beyond the chilled carcass to the finished product.
To gain insight into the cause of HEP contamination events, we employed molecular typing of E. coli O157:H7 isolates collected from beef trim produced during HEP. Organisms from multiple trim lots and time points within a HEP and across multiple HEP were typed to gain information regarding the source of contamination, specifically whether HEP contamination is derived from a single point source or from multiple sources. The latter would be expected if the incoming load were exceeding the capacity of in-plant interventions. Genetic typing of HEP strains would also provide information regarding where in the process (slaughter floor versus fabrication) HEP contamination may be occurring and if particular strains are more commonly associated with events.
The objectives of this work were (i) to describe the diversity of strains within and among individual HEP, (ii) to determine if HEP occurring in the same processing plant are caused by the same strains, and (iii) to characterize HEP strains for attributes related to human disease.
MATERIALS AND METHODS
Experimental design.
Beef trim enrichment samples (n = 639; isolates were recovered from 566) representing 21 HEP (referred to as HEP-A through HEP-U) (Table 1) were received from nine beef processing plants operated by multiple companies and management systems. The processing plants were located in Beef Industry Food Safety Council (BIFSCo) regions 1, northwest (WA, OR, and ID); 3, southwest (AZ, NM, and TX); 5, upper Midwest (NE, ND, SD, MN, and WI); and 8, northeast (IL, IN, KY, MA, ME, MD, MI, NJ, NY, NH, CN, RI, OH, WV, VA, VT, PA, and DE). The number of HEP sample sets received from individual plants ranged from one to seven. All processing plants participating in this study harvest over 200 head per hour.
TABLE 1.
Distributions of PFGE types, lineages, and tir alleles of strains isolated from HEPa
| HEP | No. of positive enrichments received | No. of enrichments from which an isolate was obtained | No. (%) of isolates identical to predominant RDP | No. (%) of isolates closely related to predominant RDP | LSPA lineage | tir allele |
|---|---|---|---|---|---|---|
| A | 8 | 8 | 8 (100) | 8 (100) | I/II | T |
| B | 16 | 9 | 9 (100) | 9 (100) | I | T |
| C | 11 | 10 | 9 (90) | 9 (90) | I/II | T |
| D | 9 | 9 | 9 (100) | 9 (100) | I/II | T |
| E | 7 | 7 | 6 (86) | 6 (86) | I, II | T, A |
| F | 12 | 8 | 7 (88) | 8 (100) | I | T |
| G | 7 | 6 | 6 (100) | 6 (100) | I/II | T |
| H | 21 | 18 | 13 (72) | 18(100) | I/II | T |
| I | 20 | 20 | 15 (75) | 20 (100) | I | T |
| J | 20 | 17 | 16 (94) | 16 (94) | I | T |
| Kb | 32 | 10 | 10 (100) | 10 (100) | I/II | T |
| L | 9 | 9 | 9 (100) | 9 (100) | I | T |
| M | 13 | 12 | 11 (92) | 11 (92) | I/II | T |
| N | 18 | 18 | 9 (50) | 16 (89) | I/II | T |
| O | 44 | 44 | 43 (98) | 44 (100) | I | T |
| P | 65 | 61 | 61 (100) | 61 (100) | I | T |
| Q | 50 | 50 | 50 (100) | 50 (100) | II | A |
| R | 50 | 35 | 33 (94) | 35 (100) | II | A |
| S | 44 | 43 | 42 (98) | 42 (98) | I/II | T |
| T | 17 | 15 | 15(100) | 15 (100) | II | A |
| U | 166 | 157 | 157 (100) | 157 (100) | I/II | T |
Abbreviations: PFGE, pulsed-field gel electrophoresis; HEP, high event period; RDP, restriction digest pattern.
Low recovery of isolates attributed to enrichments received after 10 mo of storage at 4°C.
All samples had been determined previously to harbor E. coli O157:H7, and product represented by each sample was either diverted to a cooking process or destroyed. Upon arrival at the lab, enrichments were cultured to recover E. coli O157:H7. Pure strains recovered from each culture were analyzed by a novel, non-PulseNet pulsed-field gel electrophoresis (PFGE) method. In addition, strain lineages and tir alleles were determined to identify commonalities between strains causing contamination events. For HEP-A, -B, and -C, two E. coli O157:H7 isolates per sample were selected for PFGE analysis, while four isolates per sample were analyzed for HEP-Q. It was determined that multiple isolates from the same enrichment yielded the same PFGE pattern. For the remaining HEP, when E. coli O157:H7 was recovered from an enrichment, a single isolate was used to represent that sample for characterization.
In order to determine the diversity of E. coli O157:H7 isolates on incoming cattle hides for comparison to HEP, PFGE analyses conducted for previous studies (3, 7) were utilized. Incoming load diversity for E. coli O157:H7 hide isolates was evaluated from two sampling designs: consecutive animal sampling within a lot and sampling across an 8-h shift. Hide samples collected to represent an 8-h shift were thought to simulate the total incoming load that would contribute to the widespread contamination issues observed in HEP. Incoming hide isolates were obtained from 100 head per day for 3 days each at three different processing plants.
Alternatively, consecutive sampling of individual cattle within a lot was used to determine the incoming diversity associated with single source animals. For consecutive sampling, the number of cattle sampled per trip ranged from 56 to 149 for six different lots (see Table 3). All processing plants from which hide samples were collected operated in excess of 200 head per hour. Hide samples were not associated with HEP. Hide samples were processed as described previously (3). When a sample was positive, a single isolate was used to represent each sample for PFGE.
TABLE 3.
E. coli O157:H7 PFGE types from consecutive cattle hide samples
| Lot | No. of head sampled | No. of isolates | No. of unique RDP |
|---|---|---|---|
| 1 | 81 | 63 | 6 |
| 2 | 149 | 134 | 15 |
| 3 | 56 | 56 | 23 |
| 4 | 87 | 81 | 19 |
| 5 | 88 | 34 | 11 |
| 6 | 127 | 98 | 29 |
Isolation of E. coli O157:H7 from HEP samples.
Beef trim samples were collected by processing plant personnel and analyzed in accordance with each plant's routine trim testing program. Aliquots of each enrichment were typically sent to the U.S. Meat Animal Research Center (USMARC) within 1 week following the determination of a HEP having occurred; however, one set of samples was stored at 4°C for 10 months following the HEP. Upon arrival at the lab, the enriched HEP sample aliquots were vortexed vigorously for 30 s and allowed to set for 1 min, and then 10 μl was removed to streak for isolated colonies onto ntCHROMagar (CHROMagar-O157 [DRG International, Mountainside, NJ] supplemented with novobiocin [5 mg/liter; Sigma, St. Louis, MO] and potassium tellurite [2.5 mg/liter; Sigma]). Simultaneously, the samples were processed by immunomagnetic separation, in which 1 ml from each enrichment was subjected to immunomagnetic bead cell concentration using 20 μl of anti-E. coli O157 beads (Invitrogen, Carlsbad, CA). The beads were extracted from enrichment samples and washed two times in phosphate-buffered saline–Tween 20 (PBS-Tween; Sigma) by using an automated magnetic particle processor (KingFisher 96; Thermo Fisher Scientific, Inc., Waltham, MA). The beads were resuspended in 100 μl of PBS-Tween. Fifty microliters of the final bead-bacterium complexes were spread plated onto ntCHROMagar. All plates were incubated at 37°C for 18 to 20 h. After the plates were incubated, up to three presumptive positive colonies were picked for confirmation. Multiplex PCR (8) was used to confirm that each E. coli isolate harbored genes for the O157 antigen, H7 flagella, gamma intimin, and at least one of the Shiga toxins. All isolates were maintained as frozen stocks in 15% glycerol (Sigma) for later use in PFGE.
PFGE.
In order to obtain E. coli O157:H7 isolates from commercial processors, an agreement was reached that HEP isolates would not be analyzed by XbaI-PFGE and therefore would not be connected inappropriately to human disease isolates simply by inference from similar PFGE patterns. To satisfy this requirement, a novel PFGE technique was developed. Isolates from HEP (n = 743) were analyzed by PFGE using separation of SpeI-digested genomic DNA. To validate the resolution of SpeI-PFGE, a comparison was performed between SpeI-PFGE and XbaI-PFGE. The PFGE comparison utilized 77 E. coli O157:H7 isolates previously collected from cattle hides (7) that represented the breadth of XbaI-PFGE diversity in the USMARC strain collection. The indices of discrimination for the resulting dendrograms were calculated as described by Hunter and Gaston (9).
E. coli O157:H7 XbaI fingerprints were generated for cattle hide isolates to describe the incoming diversity. This analysis utilized the PFGE separation of XbaI-digested genomic DNA, as currently used by members of PulseNet (10). Briefly, pulsed-field gel certified agarose (SeaKem Gold agarose) was obtained from Cambrex Bio Science Rockland Inc. (Rockland, ME), and Tris-borate-EDTA (TBE) running buffer and proteinase K were purchased from Sigma. XbaI was purchased from New England BioLabs (Beverly, MA). Salmonella enterica serotype Braenderup strain H9812 was used as a control and for standardization of gels (11). Banding patterns were analyzed and comparisons made using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium), employing the Dice similarity coefficient in conjunction with the unweighted-pair group method using average linkages for clustering. Position tolerance settings used 1.5% optimization and 1.5% band tolerance.
SpeI-PFGE analysis was carried out as for XbaI-PFGE, with the following modifications. Genomic DNA was digested with SpeI (Promega, Madison, WI). The SpeI electrophoresis conditions utilized an initial switch time of 1.79 s and a final switch time of 18.66 s, with a gradient of 6 V/cm and an included angle of 120°. The run time was 17.5 h in 0.5× TBE (Sigma).
LSPA.
The lineage-specific polymorphism assay (LSPA) was carried out as previously reported (12), with the modifications described by Hartzell et al. (13). Reference strains for lineage I (FRIK 523) and lineage II (FRIK 920) were generously provided by Andrew Benson at the University of Nebraska-Lincoln.
A set of 75 E. coli O157:H7 isolates obtained from routine ground beef and beef trim testing was kindly provided by the FSIS. The strain set consisted of a random collection of isolates collected between 2009 and 2012. These isolates were analyzed by LSPA for comparison to HEP isolates.
tir single nucleotide polymorphism (SNP) genotyping.
E. coli O157:H7 HEP isolates were genotyped for either the tir 255 T or A allele by real-time PCR genotyping as described previously (14). Each reaction mixture consisted of TaqMan Universal PCR master mix (2×) (Applied Biosystems), 0.5 ng of genomic DNA, 1× assay mix (0.9 μM [each] primers and 0.2 μM [each] fluorescent probes), and molecular-grade water to a final volume of 25 μl. Amplification and detection were carried out in optical-grade 96-well plates, which were sealed with optical film in a Chromo4 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). The reaction mixtures were cycled at 50°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, with optical reading of the fluorophore taken after the extension step. Opticon 3.0 application software (Bio-Rad Laboratories) was used to determine the tir allele for each strain.
RESULTS
Comparison of SpeI- and XbaI-PFGE.
The 77-strain E. coli O157:H7 diversity panel was analyzed by SpeI- and XbaI-PFGE. Panel isolates produced 51 unique restriction digest patterns (RDP) by SpeI and 54 unique RDP by XbaI (Fig. 1). The diversity indices were calculated for both resulting dendrograms. The diversity index for the SpeI-digested panel was 0.967, and that for the XbaI-digested panel was 0.972 (Fig. 1).
FIG 1.
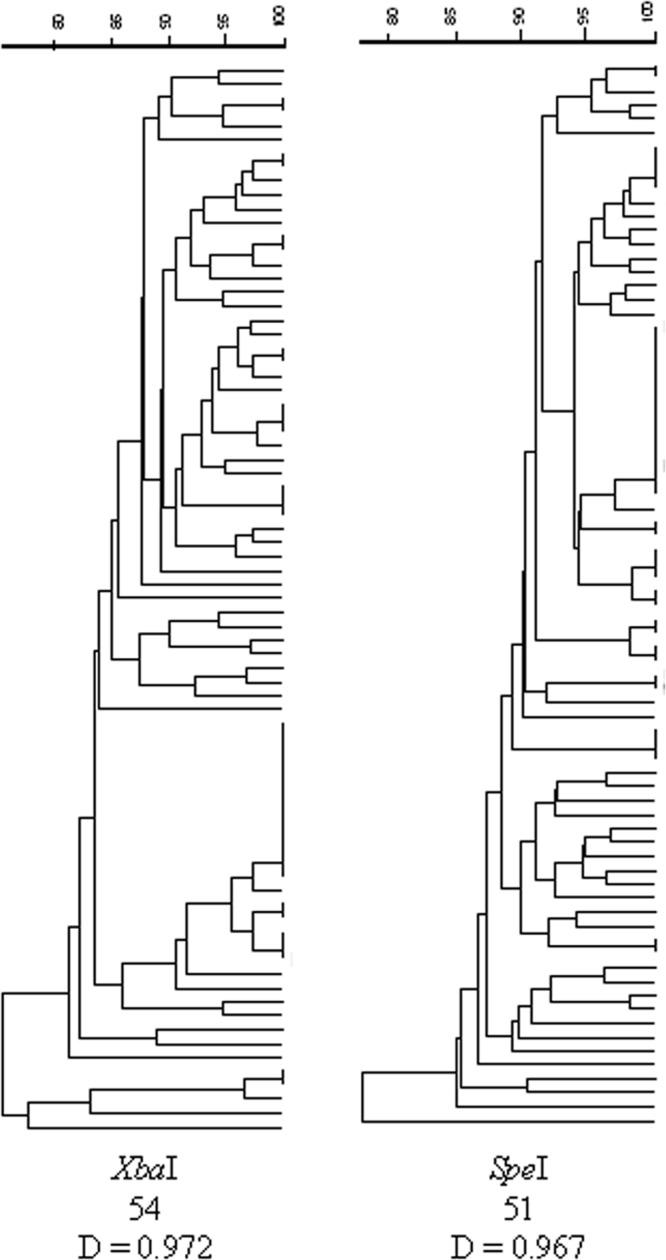
Comparison of the discriminatory powers of SpeI- and XbaI-PFGE analyses. Dendrograms for each enzyme digest are shown. The number of indistinguishable groups is provided at the bottom, along with the calculated discriminatory power (D) for each method.
PFGE analysis of individual HEP.
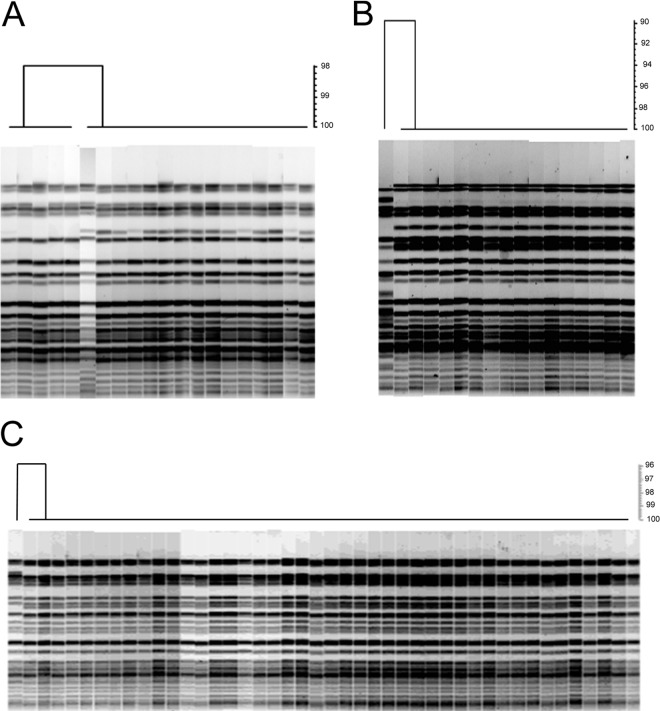
Isolates from 21 HEP were analyzed by SpeI-PFGE. Typical PFGE results are shown in Fig. 2. In all cases but one, HEP were found to consist of a predominant strain. That is not to say that for all HEP the same strain was isolated, but within each HEP there was little to no strain diversity. For nine HEP, all isolates analyzed within a HEP were indistinguishable by PFGE (Table 1). An additional six HEP would be considered to have essentially the same strain throughout the HEP by using the definition of “closely related” strains put forward by Tenover et al. (15). Overall, with the exception of HEP-N, the predominant indistinguishable strain within each HEP represented ≥72% of the samples, while closely related strains represented ≥86% of the isolates within a HEP (Table 1).
FIG 2.
Typical HEP PFGE profiles. Cluster analyses and dendrograms are shown for HEP-I (A), HEP-J (B), and HEP-O (C). Each cluster analysis and dendrogram are the results of SpeI-PFGE analysis.
Diversity of incoming E. coli O157:H7 isolates.
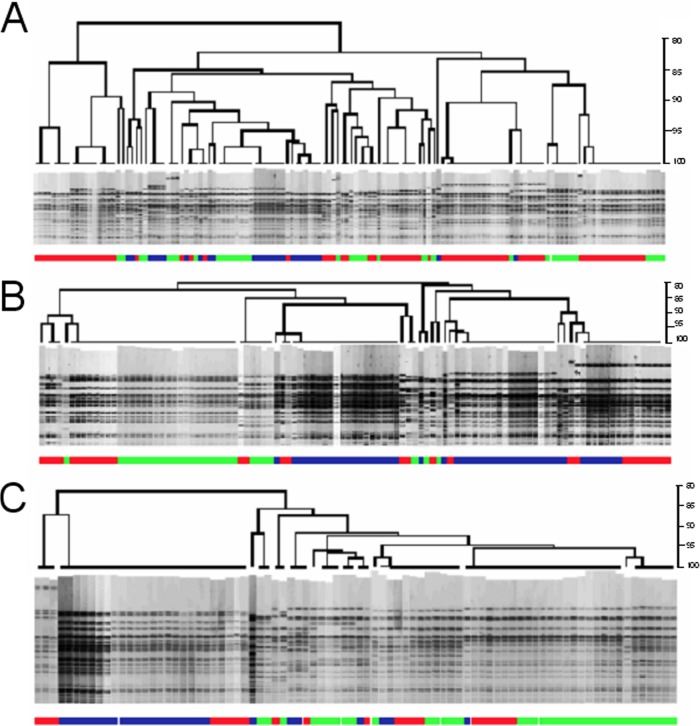
PFGE analysis of cattle hide isolates collected in previous sampling projects (3, 7) was utilized to determine the typical diversity of E. coli O157:H7 isolates associated with incoming cattle. Sponge samples for cattle hides, analyzed by individual trip and overall, showed much more diversity in isolate genotypes on incoming cattle than that observed for HEP.
Hide samples characterizing an 8-h shift were analyzed, and the results are presented in Table 2 and Fig. 3. For 100 head per day sampled for 3 days at each of three processing plants, the number of E. coli O157:H7 isolates obtained per day ranged from 22 to 76. The number of unique RDP obtained per day ranged from 6 to 24.
TABLE 2.
E. coli O157:H7 PFGE types from 100 cattle hide samples collected each day for 3 days
| Processing plant | Day | No. of isolates | No. of unique RDP |
|---|---|---|---|
| 1 | 1 | 36 | 18 |
| 2 | 76 | 24 | |
| 3 | 26 | 12 | |
| 2 | 1 | 29 | 6 |
| 2 | 30 | 12 | |
| 3 | 48 | 9 | |
| 3 | 1 | 38 | 10 |
| 2 | 22 | 7 | |
| 3 | 26 | 7 |
FIG 3.
Diversity of incoming load on cattle throughout production shift. Dendrograms, produced by XbaI restriction digests, represent the genotypic diversity of E. coli O157:H7 strains during an 8-h production shift each day for 3 days. Three separate processing plants are represented: plant 1 (A), plant 2 (B), and plant 3 (C). Each dendrogram combines isolates collected on three separate days: green, day 1; red, day 2; blue, day 3.
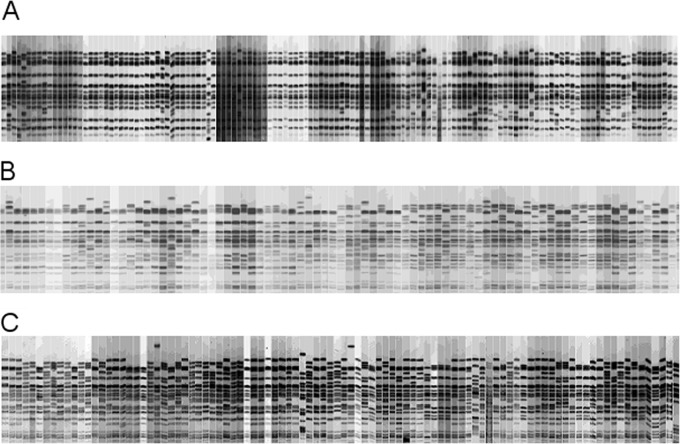
For consecutive sampling across individual lots of cattle, the number of isolates obtained from each trip ranged from 34 to 134 per lot (Table 3 and Fig. 4). Lot 1 produced the fewest unique RDP, with 63 isolates being categorized by six unique RDP. Lot 6 had the most unique RDP (n = 29), from 98 isolates.
FIG 4.
Diversity of incoming E. coli O157:H7 isolates on cattle hides by individual lots. Each image depicts the XbaI restriction digest patterns for E. coli O157:H7 isolates in sequential order for each animal in a lot. The number of unique genotypes for each lot can be found in Table 3. (A) Lot 2; (B) lot 4; (C) lot 6.
Indistinguishable isolates across multiple HEP.
In analyzing the HEP isolates as a whole, one indistinguishable strain type was found to be the predominant strain in five different HEP (HEP-A, -C, -G, -K, and -M). In addition, this strain type was indistinguishable from the minority strain in HEP-H, which was closely related to the predominant strain in that HEP. These HEP were from three different plants, operated by two different companies, but they were located within the same BIFSCo region. HEP-A and -C occurred in the same plant, and HEP-G, -H, and -M occurred in another. Both of these plants had additional HEP associated with unrelated strains. Aside from this strain type, there were no other HEP that shared a common strain.
Lineage and tir alleles for HEP isolates.
Seventeen of the 21 (81%) HEP consisted of strain lineages typically associated with human disease, i.e., lineages I and I/II (Table 1). Of those 17 HEP, 7 HEP had only lineage I strains and 10 HEP contained only lineage I/II strains. Only, HEP-E, -Q, -R, and -T yielded strains of lineage II. While HEP-Q, -R, and -T were populated by lineage II strains in every sample, HEP-E consisted of indistinguishable lineage II strains for 6 of 7 samples and a lineage I strain in the remaining sample (Table 1).
The tir allele results for HEP strains were similar to the lineage determinations. All lineage I and I/II strains harbored the human illness-associated tir T allele, while the lineage II strains carried the tir A allele. Hence, tir T allele-containing strains were found to be the predominant constituents for the vast majority of HEP (81%) (Table 1). The predominant strains in HEP-E, -Q, -R, and -T were the only strains found to harbor the tir A allele. HEP-E was the only HEP that consisted of strains differing in lineage or tir allele. For all other HEP, even when different PFGE patterns were identified within a HEP, all strains within the HEP were of the same lineage and tir type.
Lineage determination for non-HEP beef trim and ground beef isolates.
Lineages I and I/II had 31 and 30 isolates, respectively, among the 75 total beef trim and ground beef isolates provided by FSIS. This resulted in a combined prevalence of 81.3% human-biased lineages (data not shown). The remaining 14 (18.7%) isolates were lineage II.
DISCUSSION
The findings of this study indicate that most HEP from large commercial beef processing plants consist of a singular dominant E. coli O157:H7 strain type within each HEP (Table 1). In these cases, the dominant strains were found across multiple product types (trim from multiple lines originating from different sections of the carcass) and spread over substantial spans of time (occasionally more than one 8-h shift) and product (tens of thousands of pounds or greater). These findings would appear to be in disagreement with the current model of beef contamination, which states that finished product contamination originates on the kill floor and occurs when interventions malfunction, dressing practices are improper, or incoming load (hide carriage of the pathogen inadvertently transferred to the carcass surface) exceeds the capacity of the in-plant interventions to remove carcass contamination (3–5). In this model, one would expect to observe diverse E. coli O157:H7 isolates in the finished product, similar to the case for the hides of incoming cattle. The results obtained herein do not appear to support that hypothesis.
It should be noted that the hide samples discussed here provide a snapshot of the typical diversity in E. coli O157:H7 isolates entering beef processing plants and were not linked to HEP. The determination that a HEP has taken place occurs at least 48 h after the cattle have been harvested. Therefore, it is not possible to collect hide samples for a HEP, whose occurrence cannot be determined a priori.
The conversion of live animal to finished product for human consumption is a complicated process and should not be thought of as a linear progression through a system but rather as a complex network of pathways and branch points based on the assignment of product grades and the sorting of carcasses into like marketing groups to facilitate production and packaging of final products. The tracking of E. coli O157:H7 through this network is further complicated due to numerous sources inputting multiple pathogen types throughout the system. A group of cattle exit a production setting such as a feedlot and enter the processing plant as a lot. Typically, this lot will have a shared diet and management regimen, and previous reports indicate that as a lot, cattle may share a predominant E. coli O157:H7 strain (16, 17) in the feedlot environment. Our group and others (3, 18–20) have shown that upon arrival at the beef processing plant, the lairage environment can result in significant pathogen contamination of the cattle hide. This additional contamination adds many new strain types to the hide microflora, which may subsequently be transferred to the dehided carcass (3, 7).
The carcasses are maintained as a lot as they progress through the abattoir kill floor, where multiple antimicrobial interventions are applied, followed by entry into the cooler. Following the 24- to 48-h carcass chilling period, carcasses are graded and sorted such that lots are no longer maintained together. Sorting carcasses by grades results in carcasses from multiple sources being intermingled before further processing. During further processing, called fabrication, the carcasses are broken down into primal and subprimal cuts, with individual carcass sections being routed to specific cutting lines to achieve the multitude of final products from each carcass.
At essentially every step in the fabrication process, small portions of meat are trimmed away from the main product. These trim pieces, consisting of lean and fat, are collected in 2,000-lb lots, referred to as beef trim combos, and are ultimately used in the production of ground beef. With a typical feedlot-produced steer or heifer, one would estimate that ≈140 lb of beef trim would be produced per carcass, which would be distributed among several combos, depending on a variety of factors (original primal and subprimal source, desired fat/lean ratios, etc.). The filled combo is the endpoint in this process and is the point where most beef processors conduct pathogen testing prior to release of the trim material for ground beef production.
A detailed understanding of the breakdown of carcasses into final products is necessary to give context to the results of the study described herein. It is easy to see through this description why the hypothesis of this study was that HEP would contain a diverse array of strain types originating from the hides of incoming cattle. As shown in Fig. 3 and Table 2, many different strain types can be found on incoming cattle over a time frame consistent with many HEP. Most plants of the capacity sampled herein will process in excess of 1,500 cattle in separate lots originating from multiple sources over an 8-h shift. Aside from the E. coli O157:H7 diversity presented by multiple incoming lots, there is also a continuous deposition of E. coli O157:H7-laden feces in the lairage environment (3) that will contribute to the within-lot diversity of hide contamination as shown in Fig. 4 and Table 3. In light of the incoming diversity and the intermingling of carcasses as well as carcass products, it was surprising to observe such a high degree of homogeneity in E. coli O157:H7 strain types when HEP occurred.
The most striking example comes from HEP-U. This HEP had the largest number of positive samples for any HEP studied herein, and all E. coli O157:H7 isolates were of the same PFGE type. The 157 positive samples all came from 2,000-lb combos totaling 314,000 lb of beef trim. Given that the typical carcass yield of trim is ≈140 lb, the minimum number of carcasses represented by this HEP would be estimated to be 2,243. The actual number of carcasses contributing to this HEP was likely much higher, because the trimmings from individual carcasses are not contained as discrete units within a combo but are dispersed into multiple combos. It is difficult to imagine a mechanism of contamination for such an event. The scenario would require a source containing a single E. coli O157:H7 genotype and being of sufficient concentration and volume to be spread over such a large amount of product.
While there has been research showing that various E. coli O157:H7 strains will emerge as predominant over time within a group of cattle in a production setting, the exclusivity is not nearly to the degree seen for HEP. LeJeune et al. (16) used PFGE to show that 230 isolates obtained from eight feedlot pens consisted of 56 unique genotypes. Isolates belonging to a group of four closely related genetic subtypes made up 60% of all isolates collected over the sampling period. Carlson et al. (17) collected 132 E. coli O157:H7 isolates representing 32 different PFGE subtypes from 788 feedlot cattle in five pens. A single, predominant PFGE subtype accounted for 53% of the 132 isolates. In addition, Rice et al. (21) found up to 11 PFGE subtypes per farm, with up to 7 subtypes/farm identified from a single date.
Upon exiting the production environment, cattle are exposed to additional E. coli O157:H7 contamination during transportation to the processing plant (18, 19, 22). Arthur et al. (18) found that up to 10% of the E. coli O157:H7 isolates obtained from carcasses within a lot during processing matched genotypes found in the trucks they were transported on, which were different from the genotypes found in the feedlot the cattle originated from.
As cattle are placed in lairage at the processing plant, further contamination of the hide by E. coli O157:H7 occurs, which results in further increased strain diversity in the incoming load (3, 19, 20). This diversity can be observed in the hide sampling results presented in Table 3. As many as 23 unique E. coli O157:H7 genotypes could be identified within as few as 56 head from the same lot sampled consecutively. Hide contamination has been shown to be the source of carcass contamination, and as such, the diverse isolates observed on hides are subsequently transferred to the carcass. Arthur et al. (18) reported that 80% (67 of 80 isolates representing 10 genotypes) of the isolates recovered from carcasses sampled prior to evisceration did not come from the feedlot of origin for those cattle but were attributed to hide contamination acquired in the lairage environment. Similarly, Dodd et al. (23) also reported high levels of diversity (17 subtypes from 39 positive carcasses among 1,503 total carcass samples) among E. coli O157:H7 isolates from preevisceration carcasses.
While the homogeneity in genotypes within HEP appears to differ with respect to the diversity of the incoming load and what is found on the carcass during processing, there does seem to be agreement with genotypic profiles obtained from beef recalls and disease outbreaks. Investigations into beef-related outbreaks of disease due to E. coli O157:H7 have found a similar high degree of strain homogeneity. Most of the isolates (16 of 18 isolates) from a 1997 outbreak and associated recall were determined to have indistinguishable PFGE patterns, while the remaining two isolates differed from the predominant pattern by one band (24). In a 2002 outbreak/recall, 354,200 lb of ground beef were implicated, and illnesses spanned seven states. The genotypes of all isolates (19 of 19 isolates) collected from human illness cases (n = 18) and one ground beef sample were determined to be indistinguishable by PFGE analysis (25).
At this time, it is difficult to resolve the dichotomy that E. coli O157:H7 contamination on cattle hides and carcasses consists of a high degree of diversity, while HEP show little to no strain diversity. One argument states that there is no dichotomy and that the current model of incoming load overwhelming antimicrobial interventions remains applicable through one of three possible scenarios. The first of these scenarios would focus on animals shedding E. coli O157:H7 at extremely high levels, i.e., supershedders. It is plausible that a lot containing multiple supershedders not only would contaminate themselves and their cohorts but also deposit large amounts of a particular strain type in the lairage environment to contaminate subsequent cattle lots. It can be speculated that this would provide a large concentration and volume of strain-specific contamination that would need to be reduced through proper dressing and functional interventions. However, this scenario seems unlikely, because supershedders make up approximately 2% of the cattle population (26), and multiple supershedders likely enter processing plants on a daily basis during high shedding season. As shown in Tables 2 and 3, there is little evidence of incoming cattle hides being contaminated with predominantly one strain type. Even acknowledging the lack of data in this regard, it is unreasonable to conclude that HEP occur only when a singular genotype dominates the incoming load.
The second scenario also pertains to supershedder-derived contamination. The basis for this scenario would be the gross contamination of a small group of carcasses with very high concentrations of E. coli O157:H7. Cross-contamination of workers and contact surfaces would occur to transmit the contamination to multiple lots of finished product. This scenario relies on poor dressing practices and the inability of antimicrobial interventions to reduce the contamination load. There are two main concerns with this model. First, it is difficult to imagine contamination of a few carcasses providing enough material to be spread across large HEP such as HEP-U. Second, it seems just as likely to achieve gross contamination of carcasses with a mixed strain population, leading to HEP with multiple genotypes. If scenario 1 or 2 were occurring, it seems likely that one would observe HEP with one dominant strain and HEP with multiple strains, which was not the case in this study.
In the third scenario, the diversity seen in carcass contamination is reduced by multihurdle intervention schemes employed by the processing plants, but through this reduction a selection of robust strains is facilitated. This would seem unlikely for a variety of reasons. First, while there was one strain found in multiple HEP, most of the HEP were caused by unique strains, indicating that there are multiple strain types that can survive this selection, which should be manifest in more diverse HEP. Second, there are limited data from previous studies comparing the effects of antimicrobial interventions on multiple E. coli O157:H7 genotypes, and no significant differences in their survival were observed (27, 28). However, many more strain types need to be evaluated to validate this point.
An opposing argument suggests that HEP contamination occurs post-kill floor. While it is unknown at this time what the mechanism for such contamination would be, this would explain why beef trim from carcasses harvested several hours apart would share a contaminant genotype. Currently, there are few to no additional data to support or refute this model, but it is difficult to imagine a source of widespread contamination post-kill floor. It does not appear to be plant-specific endemic contamination, as several plants had multiple HEP caused by differing strains of E. coli O157:H7.
Another significant finding of this work is the bias toward human illness-related E. coli O157:H7 strains among those isolated from HEP. Seventeen of the 21 (81%) HEP consisted exclusively of strains associated with human illness (tir allele T). This was significant, as the tir alleles were previously found in cattle populations at rates of 55% T and 44% A but were heavily biased toward the T allele (99% T versus 1% A) among E. coli O157:H7 strains isolated from human illness cases (14). To further investigate the potential bias toward the tir T allele in HEP strains, a set of E. coli O157:H7 isolates was obtained from the raw beef sampling program conducted by FSIS for tir analysis. The FSIS isolates had a similar high rate (81.3%) of human illness-associated strain types, indicating that the tir T allele may not be associated specifically with HEP but rather with beef trim in general. It should be noted that the tir T allele was recently found to have a prevalence of 71% among E. coli O157:H7 strains isolated from supershedding cattle (26). More data will be needed to determine if human illness-associated strains are associated with beef trim and if supershedding plays a role in such an association.
In conclusion, much more work needs to be done to determine the mechanism responsible for HEP. The difficulties in such work are that there is no way to know when HEP are going to occur and that HEP are not detected until approximately 24 to 48 h after the contamination has taken place. It may be and is quite likely that both models are correct and contamination events can occur from both kill floor and post-kill floor contamination. The data reported herein suggest that whatever the mechanism, HEP occurring at large beef processing plants typically show little to no diversity in E. coli O157:H7 genotypes, and the majority consist of human illness-related strains.
ACKNOWLEDGMENTS
We thank Nicole Burns, Mallory Suhr, Sandy Fryda-Bradley, and Frank Reno for technical support. We thank Andy Benson and the Food Safety Inspection Service for kindly providing strains.
Product names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print 8 November 2013
REFERENCES
- 1.FSIS 2011. National prevalence estimate of pathogens in domestic beef manufacturing trimmings (trim). www.fsis.usda.gov/wps/wcm/connect/f07f5e1d-63f2-4ec8-a83a-e1661307b2c3/Baseline_Data_Domestic_Beef_Trimmings_Rev.pdf?MOD=AJPERES
- 2.FSIS 2012. Compliance guideline for establishments sampling beef trimmings for Shiga toxin-producing Escherichia coli (STEC) organisms or virulence markers. www.fsis.usda.gov/wps/wcm/connect/e0f06d97-9026-4e1e-a0c2-1ac60b836fa6/Compliance_Guide_Est_Sampling_STEC_0512.pdf?MOD=AJPERES
- 3.Arthur TM, Bosilevac JM, Brichta-Harhay DM, Kalchayanand N, King DA, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Source tracking of Escherichia coli O157:H7 and Salmonella contamination in the lairage environment at commercial U.S. beef processing plants and identification of an effective intervention. J. Food Prot. 71:1752–1760 [DOI] [PubMed] [Google Scholar]
- 4.Barkocy-Gallagher GA, Arthur TM, Siragusa GR, Keen JE, Elder RO, Laegreid WW, Koohmaraie M. 2001. Genotypic analyses of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the Midwestern states of the United States. Appl. Environ. Microbiol. 67:3810–3818. 10.1128/AEM.67.9.3810-3818.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nou X, Rivera-Betancourt M, Bosilevac JM, Wheeler TL, Shackelford SD, Gwartney BL, Reagan JO, Koohmaraie M. 2003. Effect of chemical dehairing on the prevalence of Escherichia coli O157:H7 and the levels of aerobic bacteria and Enterobacteriaceae on carcasses in a commercial beef processing plant. J. Food Prot. 66:2005–2009 [DOI] [PubMed] [Google Scholar]
- 6.Vosough Ahmadi B, Velthuis AG, Hogeveen H, Huirne RB. 2006. Simulating Escherichia coli O157:H7 transmission to assess effectiveness of interventions in Dutch dairy-beef slaughterhouses. Prev. Vet. Med. 77:15–30. 10.1016/j.prevetmed.2006.05.011 [DOI] [PubMed] [Google Scholar]
- 7.Arthur TM, Bosilevac JM, Nou X, Shackelford SD, Wheeler TL, Koohmaraie M. 2007. Comparison of the molecular genotypes of Escherichia coli O157:H7 from the hides of beef cattle in different regions of North America. J. Food Prot. 70:1622–1626 [DOI] [PubMed] [Google Scholar]
- 8.Hu Y, Zhang Q, Meitzler JC. 1999. Rapid and sensitive detection of Escherichia coli O157:H7 in bovine faeces by a multiplex PCR. J. Appl. Microbiol. 87:867–876. 10.1046/j.1365-2672.1999.00938.x [DOI] [PubMed] [Google Scholar]
- 9.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 11.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Kovar J, Kim J, Nietfeldt J, Smith DR, Moxley RA, Olson ME, Fey PD, Benson AK. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854. 10.1128/AEM.70.11.6846-6854.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartzell A, Chen C, Lewis C, Liu K, Reynolds S, Dudley EG. 2011. Escherichia coli O157:H7 of genotype lineage-specific polymorphism assay 211111 and clade 8 are common clinical isolates within Pennsylvania. Foodborne Pathog. Dis. 8:763–768. 10.1089/fpd.2010.0762 [DOI] [PubMed] [Google Scholar]
- 14.Bono JL, Keen JE, Clawson ML, Durso LM, Heaton MP, Laegreid WW. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. 10.1186/1471-2334-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeJeune JT, Besser TE, Rice DH, Berg JL, Stilborn RP, Hancock DD. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377–384. 10.1128/AEM.70.1.377-384.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson BA, Nightingale KK, Mason GL, Ruby JR, Choat WT, Loneragan GH, Smith GC, Sofos JN, Belk KE. 2009. Escherichia coli O157:H7 strains that persist in feedlot cattle are genetically related and demonstrate an enhanced ability to adhere to intestinal epithelial cells. Appl. Environ. Microbiol. 75:5927–5937. 10.1128/AEM.00972-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur TM, Bosilevac JM, Brichta-Harhay DM, Guerini MN, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2007. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 70:280–286 [DOI] [PubMed] [Google Scholar]
- 19.Childs KD, Simpson CA, Warren-Serna W, Bellenger G, Centrella B, Bowling RA, Ruby J, Stefanek J, Vote DJ, Choat T, Scanga JA, Sofos JN, Smith GC, Belk KE. 2006. Molecular characterization of Escherichia coli O157:H7 hide contamination routes: feedlot to harvest. J. Food Prot. 69:1240–1247 [DOI] [PubMed] [Google Scholar]
- 20.Dewell GA, Simpson CA, Dewell RD, Hyatt DR, Belk KE, Scanga JA, Morley PS, Grandin T, Smith GC, Dargatz DA, Wagner BA, Salman MD. 2008. Impact of transportation and lairage on hide contamination with Escherichia coli O157 in finished beef cattle. J. Food Prot. 71:1114–1118 [DOI] [PubMed] [Google Scholar]
- 21.Rice DH, McMenamin KM, Pritchett LC, Hancock DD, Besser TE. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479–484. 10.1017/S0950268899002496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barham AR, Barham BL, Johnson AK, Allen DM, Blanton JR, Jr, Miller MF. 2002. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J. Food Prot. 65:280–283 [DOI] [PubMed] [Google Scholar]
- 23.Dodd CC, Renter DG, Fox JT, Shi X, Sanderson MW, Nagaraja TG. 2010. Genetic relatedness of Escherichia coli O157 isolates from cattle feces and preintervention beef carcasses. Foodborne Pathog. Dis. 7:357–365. 10.1089/fpd.2009.0415 [DOI] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention 1997. Escherichia coli O157:H7 infections associated with eating a nationally distributed commercial brand of frozen ground beef patties and burgers—Colorado, 1997. JAMA 278:891. 10.1001/jama.1997.03550110029013 [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention 2002. Multistate outbreak of Escherichia coli O157:H7 infections associated with eating ground beef—United States, June-July. JAMA 288:690–691. 10.1001/jama.288.6.690 [DOI] [PubMed] [Google Scholar]
- 26.Arthur TM, Ahmed R, Chase-Topping M, Kalchayanand N, Schmidt JW, Bono JL. 2013. Characterization of Escherichia coli O157:H7 strains isolated from supershedding cattle. Appl. Environ. Microbiol. 79:4294–4303. 10.1128/AEM.00846-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthur TM, Kalchayanand N, Bosilevac JM, Brichta-Harhay DM, Shackelford SD, Bono JL, Wheeler TL, Koohmaraie M. 2008. Comparison of effects of antimicrobial interventions on multidrug-resistant Salmonella, susceptible Salmonella, and Escherichia coli O157:H7. J. Food Prot. 71:2177–2181 [DOI] [PubMed] [Google Scholar]
- 28.Berry ED, Barkocy-Gallagher GA, Siragusa GR. 2004. Stationary-phase acid resistance and injury of recent bovine Escherichia coli O157 and non-O157 biotype I Escherichia coli isolates. J. Food Prot. 67:583–590 [DOI] [PubMed] [Google Scholar]