Abstract
Gulls are prevalent in beach environments and can be a major source of fecal contamination. Gulls have been shown to harbor a high abundance of fecal indicator bacteria (FIB), such as Escherichia coli and enterococci, which can be readily detected as part of routine beach monitoring. Despite the ubiquitous presence of gull fecal material in beach environments, the associated microbial community is relatively poorly characterized. We generated comprehensive microbial community profiles of gull fecal samples using Roche 454 and Illumina MiSeq platforms to investigate the composition and variability of the gull fecal microbial community and to measure the proportion of FIB. Enterococcaceae and Enterobacteriaceae were the two most abundant families in our gull samples. Sequence comparisons between short-read data and nearly full-length 16S rRNA gene clones generated from the same samples revealed Catellicoccus marimammalium as the most numerous taxon among all samples. The identification of bacteria from gull fecal pellets cultured on membrane-Enterococcus indoxyl-β-d-glucoside (mEI) plates showed that the dominant sequences recovered in our sequence libraries did not represent organisms culturable on mEI. Based on 16S rRNA gene sequencing of gull fecal isolates cultured on mEI plates, 98.8% were identified as Enterococcus spp., 1.2% were identified as Streptococcus spp., and none were identified as C. marimammalium. Illumina deep sequencing indicated that gull fecal samples harbor significantly higher proportions of C. marimammalium 16S rRNA gene sequences (>50-fold) relative to typical mEI culturable Enterococcus spp. C. marimammalium therefore can be confidently utilized as a genetic marker to identify gull fecal pollution in the beach environment.
INTRODUCTION
Gulls and other shorebirds and waterfowl are prevalent in beach environments, and their feces are considered a major source of fecal indicator bacteria (FIB) in coastal and lake waters worldwide (1–5). Escherichia coli and enterococci are commonly used to monitor the presence of pathogenic organisms in beach environments and assess the relative human health risks associated with recreational water use (6); however, these FIB are ubiquitous, as they are found in the feces of humans and animals alike, limiting their utility to accurately predict human health risks (7). Although there is evidence to support the shedding of human pathogens (8–11), the human health risks associated with shorebird and waterfowl feces are inherently lower than those from human fecal inputs (12, 13). Furthermore, bird droppings are a more concentrated point source of contamination on beaches, while sewage is more readily dispersed, presenting a larger radius of contamination risk. Beach advisories are implemented to protect public health, but many unnecessary beach closings result from FIB seeding by gulls (2, 3). These closings can have huge economic implications, especially in coastal areas (14); therefore, the ability to discriminate the source of fecal contamination is critical for both human health risk assessments and mitigation of economic losses.
Currently, most health-related surface water monitoring programs use selective media to culture FIB, usually E. coli and enterococci. Gulls often shed high densities (105 to 109 CFU of E. coli and 104 to 108 CFU of Enterococcus spp. g−1 feces) of these organisms via their droppings to surface waters and beach environments (3, 15). Thus, gull contamination can confound the monitoring efforts of recreational water sources proposed by the U.S. Environmental Protection Agency (16). Gull populations have steadily increased in urban areas over the past 30 years because of more readily abundant food supplies, long life spans, and few natural predators (17–19). Recreational water quality monitoring efforts are, in turn, impacted by these growing gull populations.
Traditional culture-based monitoring methods have been successful at establishing human health risk thresholds; however, these methods generally lack the ability to identify contamination sources (7). The inability to distinguish sources of fecal contamination—human sewage, avian species, domestic pets, or other urban animals—hinders management efforts and conveys an inexact measure of human health risks associated with recreational water use. Discriminating between contamination sources in beach and coastal environments can improve risk assessment and mitigation strategies and help limit unnecessary beach advisories and closings caused by sources that do not carry pathogenic organisms.
Despite the ubiquitous presence of gulls and their impact on water quality monitoring efforts, analysis of the organisms harbored in gull feces has been limited to traditional cloning methods (20–22) and a comprehensive microbial community profile has yet to be investigated. Next generation sequencing platforms have gained popularity in recent years as a cost-effective way to deeply explore the microbial community compositions of many samples, while providing taxonomic resolution nearly equivalent to full-length 16S rRNA gene sequences (23, 24). Deeper resolution of microbial communities from gull feces can aid in discerning the gulls' contribution to the total fecal load in beach environments. Furthermore, an increased understanding of the gull fecal bacterial composition could assist in the development and validation of source-specific assays to enhance beach monitoring efforts.
Previous attempts to monitor gull pollution have resulted in the development of assays targeting specific bacteria as markers of fecal pollution. The Gull-2 PCR assay and the gull2, gull3, gull4, and LeeSeaGull real-time PCR assays target the 16S rRNA gene of Catellicoccus marimammalium (Gull-2, gull2, and gull4) and the genus Streptococcus (gull3) (21, 22, 25). The assays targeting C. marimammalium seem the most promising, as this organism is commonly found in gull fecal samples, is host specific compared to nonavian animals, and is detectable in water sources with suspected gull contamination (21). Geographic variability can factor into the efficiency and breadth of microbial source tracking (MST) marker use, so the regional utility of a marker must be well tested and validated. A recent nationwide, multilaboratory assessment of PCR methods targeting C. marimammalium discovered that MST methods designated for gull detection were cross-reactive with pigeon feces from California (26). The prevalence, geographic scope, and ecology of C. marimammalium in host birds warrants further investigation, and future assessments should encompass environmental samples from diverse geographic regions (26). Given these findings, it will be important to note the abundance of C. marimammalium and general composition of gull fecal samples from Lake Michigan.
In this study, we used the Roche 454 and Illumina MiSeq deep sequencing platforms to examine the composition and relative abundance of bacteria in gull fecal samples. We obtained 16S rRNA gene sequences for bacterial species cultured on mEI plates using Sanger sequencing and determined their proportion in the overall community. We explored the potential for gull fecal contamination to confound current water quality monitoring efforts through their contribution of commonly cultured FIB. Last, we evaluated the regional utility of the Gull-2 assay for beaches along Lake Michigan using gull fecal samples and environmental samples with elevated mEI plate counts.
MATERIALS AND METHODS
Study area, sample collection, and DNA extraction of gull fecal samples.
Fresh gull fecal samples were collected aseptically with sterile spatulas from parking lot surfaces at Bradford Beach (Milwaukee, WI) and Grant Park (South Milwaukee, WI). Samples were collected within 2 to 3 min after deposition to prevent overgrowth with nonfecal bacteria. In total, 57 gull fecal samples were collected at Bradford Beach (n = 42) and Grant Park (n = 13), and Point Beach (n = 2) in Two Rivers, WI, between December 2011 and October 2012. The collected samples were stored in sterile 2-ml tubes, transported to the laboratory within 2 h, and stored at −80°C. DNA extractions were performed on 200 mg of the fecal pellets using the Qiagen stool kit (Qiagen, Valencia, CA) according to the manufacturer's protocol with minor modifications (doubling the amount of proteinase K, transferring 2× supernatant volumes, and eluting with both DES [ultrapure water; MP Biomedicals, Solon, OH] and AE buffer). Extracted DNA was stored at −20°C until further analysis. Four samples from Grant Park (2012) and four from Bradford Beach (two each from 2011 and 2012) were further purified using the Mo Bio PowerClean DNA cleanup kit for sequencing (Mo Bio Laboratories, Carlsbad, CA). DNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Pittsburg, PA).
Next-generation sequencing and 16S rRNA gene data set analysis.
454 pyrosequencing targeting the V4-V6 hypervariable regions of the 16S rRNA gene generated ∼7,000 quality reads per gull fecal sample. Forward primer 518F (5′-CCAGCAGCYGCGGTAAN-3′) and reverse primer 1064R (5′-CGACRRCCATGCANCACCT-3′) amplified DNA, and primer 565F (5′-TGGGCGTAAAG-3′) allowed for bioinformatic trimming (27) of raw sequence reads from the Roche genome sequencer GS-FLX. Illumina sequencing was also performed for six of eight gull samples (gull 1 and gulls 4 to 8). The V4-V5 hypervariable regions were amplified in these samples according to protocols developed at the Josephine Bay Paul Center at the Marine Biological Laboratory, Woods Hole, MA (28). Sequences from both data sets were trimmed, quality controlled, and aligned; data were stored in the Visualization and Analysis of Microbial Population Structures (VAMPS) database (http://vamps.mbl.edu). Taxonomic assignments were made for all sequences using Global Alignment for Sequence Taxonomy (GAST) (23). VAMPS taxonomic count tables were normalized to maximum for microbial community comparisons.
Enterococcus columbae and Catellicoccus marimammalium reference sequence comparisons and Gull-2 alignment.
Similarity entropy plots comparing full-length sequences of E. columbae (GAST annotation of the most abundant species of bacteria in gull samples) and C. marimammalium (identified by Lu et al. [21] as the dominant species in gulls and target for the Gull-2 assay) were made with Vector NTI software (Life Technologies, Grand Island, NY). NCBI reference sequences (E. columbae [AF061006] and C. marimammalium [AJ854484]) were used for comparison.
16S rRNA clone library generation from gull fecal samples.
Three of the eight gull fecal samples (gulls 1, 2, and 5) were used to generate 16S rRNA gene clone libraries to enable a more definitive classification of the most abundant taxa in the gull microbial communities. PCR was performed with universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). Products were purified and cloned as described previously (20); sequencing was carried out with both the M13F and M13R primers using the ABI BigDye terminator kit (Applied Biosystems, Foster City, CA) on an ABI Prism 3700xi genetic analyzer (Applied Biosystems, Foster City, CA). Nearly full-length (∼1,400-bp) contigs were constructed from forward and reverse sequence reads with sufficient overlap (CLC Genomics Workbench, Cambridge, MA). A total of 223 clones were analyzed; incomplete and partial sequences were removed from the data set. Quality sequences were aligned with mother software program (29) using the SILVA alignment as a reference. Chimeras were identified using DECIPHER (30) and removed. Quality sequences were uploaded into Ribosomal Database Project (RDP) for classification (31). BLAST (32) was used to obtain species-level identification of sequences identified by RDP as Catellicoccus. Sequence alignment comparisons were done using MEGA5 software program (33) to evaluate the number of times the Gull-2 assay would match our nearly full-length C. marimammalium contigs (n = 176).
Sequencing and phylogenetic analysis of gull fecal bacterial isolates cultured on mEI plates.
Fresh fecal samples were collected aseptically, as described above, from Grant Park, South Milwaukee, WI, on 18 October 2012. Fecal pellets were resuspended in a 1:1 (wt/vol) of phosphate-buffered saline (PBS); further dilutions (∼1:10 and 1:100) were made using sterile water. The diluted fecal solutions were filtered onto 47-mm-diameter, 0.45-μm-pore-size nitrocellulose filters (Millipore, Billerica, MA) and placed on membrane-Enterococcus indoxyl-β-d-glucoside (mEI) agar plates (34) and incubated for 24 h at 41°C. Plates that generated enough colonies, without being too numerous to count (TNTC), were selected for colony PCR with universal primers 8F and 1492R to amplify the 16S rRNA gene. PCR products were cleaned up using ExoSAP-IT (Affymetrix, Santa Clara, CA), and sequencing was carried out as described above, but with only the 8F primer.
Phylogenetic analyses of high-quality forward reads from the mEI isolates, ∼874 bp long, were conducted using MEGA5 (33). Clones representing the 111 unique sequences were selected out of a total of 342. Four isolates that were identified by BLAST (32) as Streptococcus lutetiensis were removed from the final phylogenetic tree. A neighbor-joining phylogenetic tree based on evolutionary distances estimated using the p-distance method was generated with the unique sequence representatives and reference sequences in MEGA5 (33); distances are given as the number of base differences per site. A bootstrap test was performed with 1,000 replicates. Isolates that clustered around a known reference sequence, with 99% sequence identity or greater, were grouped together and annotated as such. Isolates that clustered with two or more reference sequences were left in the phylogenetic tree without classification.
An alignment and cluster analysis was performed using mothur (29) to verify the proportion of mEI culturable organisms within our data set. Sequence reads from the Illumina data set and Enterococcus species sequences obtained from the mEI isolates (albeit not the same gull samples as were sequenced, but from the same study area) were compared; only exact matches were counted.
Sequences within the Illumina data set identified as C. marimammalium (n = 224,082) were compared to sequences identified as an exact match to our cultured mEI isolates to generate the proportional relationship between C. marimammalium and culturable Enterococcus spp. Proportions of culturable Enterococcus identified to the species level (e.g., E. faecium, E. faecalis, E. hirae, and E. durans; n = 142) and sequences that may represent organisms that might be cultured on mEI but were not identified to the species level (i.e., Enterococcus species not available [species N/A]; n = 4,132), were also compared to C. marimammalium sequences.
Detection and prevalence of C. marimammalium gull fecal and environmental samples from the Milwaukee region.
Gull-2 assay (primers Gull-2F [5′-TGCATCGACCTAAAGTTTTGAG-3′] and Gull-2R [5′-GTCAAAGAGCGAGCAGTTACTA-3′]) PCR amplifications were carried out as described by Lu et al. (21) on fecal samples collected between December 2011 and October 2012 (n = 57). Preliminary tests revealed that the Gull-2 assay amplified with better efficiency when bovine serum albumin (BSA) was added to the reaction mixture. Sensitivity tests were also conducted on the four gull fecal samples with the highest concentrations of DNA to test the detection limit of the Gull-2 assay. PCR was done on serial dilutions (100 to 106) from a 1:1 (wt/vol) mixture of fecal matter and PBS.
Environmental samples collected during the 2010 and 2011 field seasons from Bradford, Atwater, Point, and McKinley beaches in and near Milwaukee, WI, with elevated plate counts of both E. coli and enterococci were selected for Gull-2 screening. Sand and water samples were previously extracted using the Mo Bio soil isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA) and stored at −80°C. PCR amplifications of the Gull-2 assay were carried out as described above using 5 μl of DNA in a final reaction volume of 25 μl.
Nucleotide sequence accession numbers.
Isolate and clone library 16S rRNA gene sequences were deposited into GenBank under the following accession numbers: KF250762 to KF250872 (mEI isolate sequences) and KF250873 to KF251018 (clone library sequences). The 454 pyrosequences and Illumina sequences are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) under project SRP033277.
RESULTS
Microbial community structure in gull fecal samples.
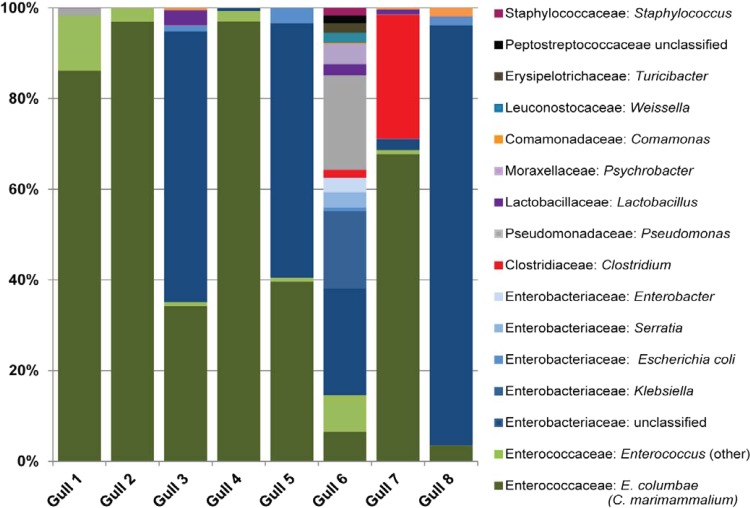
A total of 56,428 pyrosequences were generated from eight gull fecal samples. Enterococcaceae and Enterobacteriaceae were the two most abundant families, comprising 58% (Enterococcaceae) and 29% (Enterobacteriaceae) of the samples (Fig. 1). Nearly all samples contained ≥50% Enterococcaceae or Enterobacteriaceae, with the exception of the fecal sample from gull 6, which was also composed of Pseudomonadaceae, Erysipelotrichaceae, Moraxellaceae, Lactobacillaceae, Leuconostocaceae, Clostridiaceae, and Staphylococcaceae and had the highest diversity by far of any of the fecal samples.
FIG 1.
Microbial community populations of eight gull fecal samples. The V4-V6 hypervariable regions of the 16S rRNA gene from fecal genomic DNA were amplified and pyrosequenced. Taxonomy was assigned to sequences using GAST, and taxonomic counts were normalized to the maximum number of sequences. Only the most abundant genera (≥1% of at least one sample) are presented. Fecal samples from gulls 1 to 4 were collected from Bradford Beach, Milwaukee, WI, on 20 December 2011 (gulls 1 and 2) and 5 January 2012 (gulls 3 and 4), and samples from gulls 5 to 9 were collected from Grant Park, South Milwaukee, WI, on 1 January 2012.
Catellicoccus was the most abundant genus on average, accounting for 55% of the total pyrosequences recovered from the eight gull fecal samples (Table 1). Enterococcus, Escherichia/Shigella, Clostridium, Klebsiella, and Pseudomonas were also common genera, each accounting for >10% of the sequences obtained in at least one fecal sample. Catellicoccus and Enterococcus were the only genera present in all samples. The fecal sample from gull 6 had a more heterogeneous distribution of genera than the others, with relatively high proportions of Pseudomonas, Klebsiella, and Psychrobacter. A few typically nonfecal genera were also relatively common in our samples, including Serratia, Staphylococcus, and Comamonas.
TABLE 1.
Comparison of taxonomic assignments for gull fecal samples
| Sequence and taxonomic assignment | Proportion (%) of bacteria from fecal sample from: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gull 1 | Gull 2 | Gull 3 | Gull 4 | Gull 5 | Gull 6 | Gull 7 | Gull 8 | |
| Pyrosequences | ||||||||
| Enterococcus | 12.3 | 3.4 | 1.1 | 2.5 | 1.0 | 7.5 | 1.1 | 0.1 |
| Staphylococcus | 1.8 | |||||||
| Catellicoccus | 85.4 | 96.4 | 33.6 | 96.7 | 39.3 | 5.9 | 67.0 | 3.4 |
| Lactobacillus | 0.1 | 3.2 | 2.5 | 1.1 | 0.1 | |||
| Leuconostoc | 0.4 | |||||||
| Weissella | 2.1 | |||||||
| Clostridium | 1.6 | 27.0 | 0.1 | |||||
| Turicibacter | 1.9 | |||||||
| Comamonas | 0.5 | 0.3 | 0.3 | 1.7 | ||||
| Enterobacter | 3.0 | |||||||
| Escherichia | 1.2 | 3.3 | 0.8 | 0.1 | 1.8 | |||
| Escherichia/Shigella | 58.6 | 0.6 | 55.7 | 21.1 | 2.3 | 90.7 | ||
| Klebsiella | 15.4 | |||||||
| Serratia | 3.1 | |||||||
| Pasteurella | 0.7 | 0.3 | 0.1 | |||||
| Psychrobacter | 4.0 | |||||||
| Pseudomonas | 1.6 | 0.1 | 0.1 | 18.8 | 0.2 | 0.2 | ||
| Clone library sequences | ||||||||
| Catellicoccusa | 98.6 | 100.0 | N/Ab | N/A | 83.8 | N/A | N/A | N/A |
| Escherichia/Shigella | N/A | N/A | 9.4 | N/A | N/A | N/A | ||
| Salmonella | 1.4 | N/A | N/A | 5.4 | N/A | N/A | N/A | |
| Streptococcus | N/A | N/A | 1.3 | N/A | N/A | N/A | ||
Catellicoccus sequences (n = 210) uploaded to BLAST were a 99% (n = 208) and 96% (n = 2) match to C. marimammalium reference sequence NR042357.
N/A, not analyzed by the clone library method.
Not only was the general taxonomic diversity in V4-V6 reads from gull fecal samples low, but sequences within the Catellicoccus group shared high similarity. A single sequence accounted for over 16% of the Catellicoccus sequence reads. Furthermore, ∼33% of the pyrosequences shared ≥99% sequence identity over a >400-bp read length.
Clone libraries constructed from a subset of the same DNA samples verified the pyrosequencing results. Over 94% of the 223 clone library-based sequences were classified as Catellicoccus. Catellicoccus accounted for >98% of clones in the fecal sample from gull 1 (1.4% as Salmonella), 100% in the fecal sample from gull 2, and 84% in the fecal sample from gull 5 (9% Escherichia/Shigella, 5% Salmonella, and 1% Streptococcus; Table 1). Nearly all sequence reads identified as Catellicoccus (208/210) were ≥99% match to the C. marimammalium reference sequence NR042357. The two remaining clones were a 96% match to the same reference sequence. A comparison between the clone libraries and 454 pyrosequences revealed that more than 90% of the Catellicoccus sequences shared ≥99% sequence identity in the V5-V6 regions.
Taxonomic resolution of Catellicoccus marimammalium, the dominant taxonomic group in gull feces.
Enterococcus columbae was the initial taxonomic assignment of the most numerous sequence group in the 454 pyrosequence data set. However, comparison to the RDP and NCBI databases showed that this sequence group matched most closely to Catellicoccus marimammalium (Table 1). The majority of nearly full-length (∼1,400-bp) 16S rRNA gene sequences of these libraries exactly matched the pyrosequences in the V4-V6 region, confirming that C. marimammalium was the dominant species in our gull fecal samples.
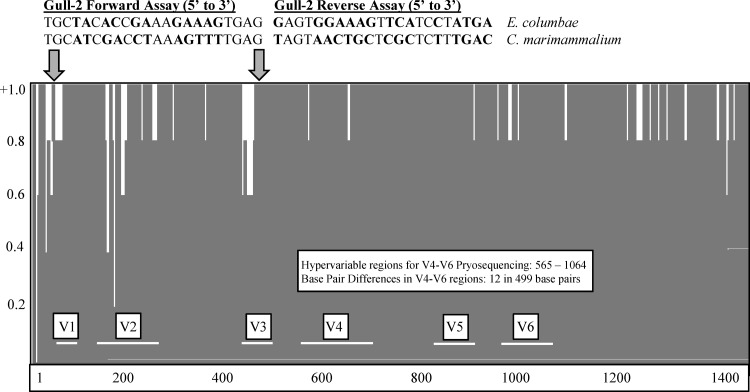
E. columbae and C. marimammalium reference sequences shared the highest identity (>97%) at the end of the 16S rRNA gene within the 499-bp hypervariable region targeted for pyrosequencing (V4-V6). The highest variability occurred within the V1-V3 regions, which are targeted by the Gull-2 PCR assay (21) (Fig. 2). The high sequence identity in the V4-V6 regions of the two species explained the incorrect annotation of E. columbae within our 454 pyrosequence data set.
FIG 2.
Entropy plot comparison of E. columbae and C. marimammalium reference sequences and Gull-2 PCR assay alignment. The entropy plot illustrates base pair agreement and dissimilarity in hypervariable regions (V1-V6). The entropy plot was generated using Vector NTI; identical base pairs have a value of +1.0, similar base pairs have a value of 0.5, and weakly similar base pairs have a value of 0.2, as indicated on the y axis. Variable regions, shown on the x axis, were identified using the E. coli system of nomenclature noted in Chakravorty et al. (37). Variable regions targeted by the Gull-2 PCR assay (21) include V1 and V3, and hypervariable regions selected for 454 pyrosequencing span the V4-V6 regions. Within the V1 and V3 regions targeted by the Gull-2 assay, there was 41% sequence identity within 44 bp, and 98% sequence identity with the V4-V6 regions, which spanned 499 bp. The GenBank accession numbers of the compared reference sequences are as follows: AF061006 for E. columbae and AJ854484 for C. marimammalium. Nucleotides in boldface signify mismatches between E. columbae and C. marimammalium.
Identification and comparison of bacteria isolated on mEI plates.
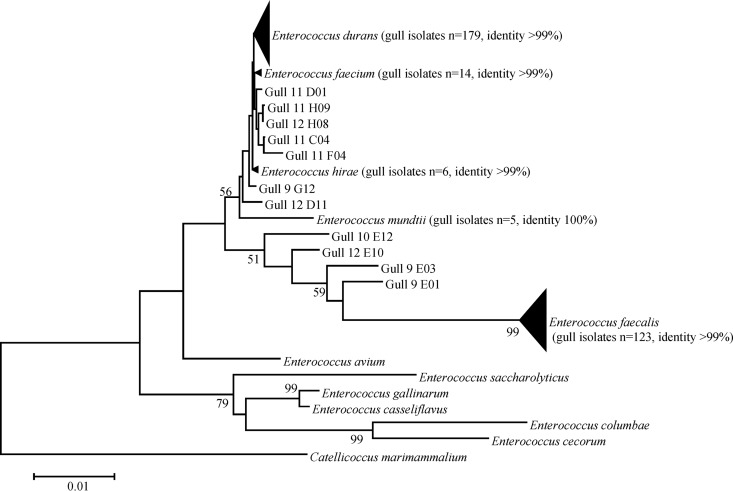
The quantity of enterococci cultured on mEI from four gull fecal samples ranged from 1.08 × 104 to 4.80 × 104 CFU g−1 (Table 2). The majority (338/342) of these isolates were identified as Enterococcus spp., and four were identified as Streptococcus spp. Both E. durans and E. faecalis were the most abundant species identified in each gull sample, but the distribution and proportions varied slightly between individuals. The overall Enterococcus species distribution was as follows: E. durans (52%), E. faecalis (36%), E. faecium (4%), E. mundtii (1%), and E. hirae (2%) (Fig. 3). All isolates shared ≥99% sequence identity to a reference sequence, with the exception of 11 Enterococcus isolates whose identity could not be confidently resolved to the species level. No isolates were closely related to C. marimammalium, the most abundant species from our sequencing approaches.
TABLE 2.
Enterococcal counts from gull fecal samples cultured on mEI
| Gull | Fecal mass (g) | Enterococcal count (CFU g−1 feces)a |
|---|---|---|
| Gull 9 | 0.45 | 2.06 × 104 |
| Gull 10 | 0.72 | 1.08 × 104 |
| Gull 11 | 0.76 | 4.80 × 104 |
| Gull 12 | 0.64 | 1.25 × 104 |
CFU counts were obtained from 1:10 or 1:100 dilutions plated on mEI (see Materials and Methods). Isolates for 16S rRNA sequencing were harvested from plates with well-dispersed colonies.
FIG 3.
Phylogenetic tree of gull fecal samples cultured on mEI. The unrooted consensus phylogram from neighbor-joining phylogenetic analysis shows evolutionary relationships among bacterial isolates cultured on mEI from fecal pellets collected on 18 October 2012 from Grant Park, South Milwaukee, WI. Enterococcus species reference sequences (GenBank accession numbers AF061006, AB012212, AJ301830, AF039900, AF133535, AF061009, Y17302, AF061013, AF039903, AJ276354, and AF061004) were used for isolate clustering and classification. Catellicoccus reference sequences (NR042357 and AJ854484), which clustered together, were used as an outgroup. Enterococcus species reference sequences are listed next to the isolate name or cluster, along with the isolate's percent identity to the reference sequence. Gull fecal isolates were classified to the species level if they were >99% identical to GenBank reference sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Only nodes with ≥50% confidence during bootstrapping are labeled. The scale bar indicates 1% sequence divergence.
Use of Illumina deep sequencing to explore the rare occurrence of mEI-culturable bacteria in the gull fecal microbial community.
MiSeq Illumina sequencing provided greater sequencing depth (∼65,000 reads per sample) and further confirmed C. marimammalium as the dominant taxonomic group within our gull fecal samples. The identity and abundance of microbial taxa were generally in good agreement with the distributions seen in the pyrosequence data set. This deeper sequencing method highlighted the disparity between C. marimammalium and the low-abundance mEI-culturable Enterococcus spp.: E. faecium, E. faecalis, E. durans, and E. hirae. Of the 389,838 bacterial sequences generated by MiSeq Illumina sequencing, 57% were identified as C. marimammalium, <2.0% of sequences represented organisms commonly cultured on mEI, and 0.01% were an exact match to mEI-cultured isolates. Comparison of the mEI isolate sequences to the Illumina data set revealed only four unique matches, representing 46 total sequences (45 Enterococcus species N/A sequences and 1 E. durans sequence). Escherichia coli made up 0.05% of the Illumina data set, and sequences that could not be distinguished between Escherichia and Shigella made up 24% (Table 3).
TABLE 3.
Relative abundance comparisons of C. marimammalium and commonly cultured FIBa
| FIB type | No. of sequences from gull fecal specimen |
Total no. of sequencesb | |||||
|---|---|---|---|---|---|---|---|
| Gull 1 | Gull 4 | Gull 5 | Gull 6 | Gull 7 | Gull 8 | ||
| Enterobacteriaceae | |||||||
| Escherichia/Shigellac | 28 | 80 | 23,921 | 20,996 | 333 | 48,545 | 93,903 |
| Escherichia coli | 0 | 0 | 41 | 45 | 1 | 121 | 208 |
| mEI-culturable organismsd | |||||||
| E. faecium | 1 | 0 | 0 | 105 | 0 | 0 | 105 |
| E. faecalis | 0 | 0 | 0 | 28 | 0 | 0 | 28 |
| E. hirae | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. durans | 0 | 0 | 0 | 9 | 0 | 0 | 9 |
| Enterococcus species N/Ae | 74 | 90 | 43 | 3854 | 59 | 12 | 4,132 |
| Identical to mEI isolatesf | |||||||
| E. durans | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Enterococcus species N/A | 0 | 0 | 0 | 45 | 0 | 0 | 45 |
| C. marimammalium | 64,376 | 64,731 | 40,366 | 5,561 | 41,965 | 7,083 | 224,082 |
| Total Illumina sequences from gulls | 64,974 | 64,983 | 64,980 | 64,938 | 64,971 | 64,992 | 389,838 |
Totals are shown in boldface type.
Gull samples 2 and 3 were not analyzed by Illumina sequencing.
Sequences not distinguishable between Escherichia and Shigella.
Species previously observed to grow on mEI.
Sequences that may represent organisms that are typically cultured on mEI but could not be assigned.
Subset of Enterococcaceae whose sequences exactly matched our mEI isolate sequences.
Validation of the Gull-2 PCR assay with clone library contigs, gull fecal samples, and environmental samples.
The primers for the Gull-2 PCR assay were an exact match to 170 of the 176 nearly full-length C. marimammalium sequences generated from gull fecal samples. Four of the contigs had a single base pair mismatch in either the forward or reverse primer regions, and two had base pair mismatches in both the forward and reverse primer regions. Fifty-seven gull fecal samples were tested with the Gull-2 PCR assay; 54 (95%) tested positive. Sensitivity tests on the four samples with the highest DNA concentrations revealed that the Gull-2 assay was detected down to the 1:1 × 105 dilution. However, only 6 (27%) of 22 environmental samples (sand and water) tested with the Gull-2 assay were positive (Table 4).
TABLE 4.
Detection of Gull-2 PCR assay in gull fecal and environmental samples
| Source | No. of samples |
|
|---|---|---|
| Tested | Positive | |
| Gull feces | ||
| Grant Park | 13 | 12 |
| Bradford Beach | 42 | 40 |
| Point Beach | 2 | 2 |
| Lake Michigana | ||
| Beach water | 9 | 4 |
| Sand | 13 | 2 |
Bradford Beach, Atwater Beach, Point Beach, and McKinley Beach.
DISCUSSION
Enterococcaceae and Enterobacteriaceae families dominant within gull fecal samples.
Enterococcaceae and/or Enterobacteriaceae dominated the 454 pyrosequencing libraries, with most Enterococcaceae sequences matching the C. marimammalium reference strain. With the exception of a few sequences, Enterobacteriaceae sequences could not be classified beyond the family level. There appeared to be a dichotomy between gull populations at the two sampling locations, Bradford Beach (Milwaukee, WI) (gulls 1 to 4) and Grant Park (South Milwaukee, WI) (gulls 5 to 8). Samples collected at Bradford Beach consisted almost entirely of Enterococcaceae, while those collected at Grant Park were generally more diverse and were dominated by Enterobacteriaceae species. Variations in gull gut microflora can occur even within small geographic regions of a city (20) and highlight the need for an assay with wide geographic applicability. While both Bradford Beach and Grant Park are beaches on Lake Michigan, differences between them may account for the observed differences in gull fecal communities. Grant Park is relatively isolated and minimally impacted by human activities. Bradford Beach, within the Milwaukee city limits, is a well-utilized and highly impacted tourist beach. Human activities affect the abundance and diversity of food sources available to gulls. Furthermore, when gulls inhabit areas where contact with human and domestic pet excrement may occur, host-enteric bacterial interactions and cosmopolitan population structures can develop (35).
Catellicoccus marimammalium is numerically dominant, but not culturable on mEI.
C. marimammalium has been identified in multiple studies (21, 22) as the dominant species in gull fecal microbial communities. Our study confirms those findings with a comprehensive analysis using three separate sequencing platforms, each targeting a different region and length of sequence. Results were generally in good agreement but did show that taxonomic classification was influenced by amplification region and assignment method. Proper taxonomic classification is limited by the length of the sequence read, the region targeted, and reference sequences available in the taxonomic databases. Targeting different hypervariable regions within the same sequence can result in different taxonomic annotations (the V4-V6 regions compared to the V1-V3 regions of the same sequence) (36, 37). We were able to demonstrate the benefits of pairing smaller, full-length sequence libraries with high-abundance, shorter sequence read libraries to elucidate the abundance and proper classification of C. marimammalium. Sequence comparisons illustrated that the 5′ end of the 16S rRNA gene sequences, notably the V1-V3 regions, appear to be more discriminatory for taxonomic classification than the central portion of the gene (V4-V6 region).
We identified near clonal populations of C. marimammalium species in our gull fecal samples as the most highly abundant sequences. Lu et al. (21) also found predominance (26%) of high-identity (≥99%) C. marimammalium sequences in gulls from West Virginia. Collectively, these findings suggest that C. marimammalium has relatively low sequence diversity in gull fecal samples across large temporal and spatial gradients. High abundance of C. marimammalium in our gull fecal samples collected from Milwaukee beaches, combined with near clonal populations within the species, further supports the selection of C. marimammalium as the target for the microbial source tracking of gulls in this region.
The abundance of C. marimammalium in DNA samples suggests that the gull gut is good habitat for this organism, yet we failed to detect it on any of the mEI plates that we screened, despite the fact that C. marimammalium is within the Enterococcaceae family. Identification of gull fecal bacteria cultured on mEI plates revealed that the intestinal tracts of gulls harbor several other, lower abundance, Enterococcaceae spp. that are readily culturable. The mEI medium selects for the growth of Enterococcus spp., notably E. faecalis, E. casseliflavus, and E. faecium, but other enterococci (E. mundtii, E. hirae, E. gallinarum, E. avium, and E. durans) have also been isolated on mEI (38, 39). The gull fecal isolates cultured on our mEI plates were similar to those identified in a previous study by Layton et al. (40) in which C. marimammalium was notably absent as well, suggesting that it is not mEI culturable.
Use of deep sequencing to explore the rare occurrence of culturable Enterococcus spp. in relation to high-abundance C. marimammalium.
A noteworthy finding from our study is the rare occurrence of mEI-culturable Enterococcus spp. in relation to the high abundance of C. marimammalium within the Illumina data set. Only a small proportion (46/180,888) of Illumina sequences assigned to the Enterococcus genus were an exact match to our mEI isolates. Using the relative abundances of taxonomic counts from our six Illumina samples, we can extrapolate the proportional relationship between C. marimammalium and culturable Enterococcus spp. to infer that for each colony of enterococci observed on mEI plates, there are ∼4,900 C. marimammalium present in gull fecal samples. If we take into account all Enterococcus spp., and not just our exact sequenced isolate matches, the relative fold abundance is ∼50 C. marimammalium per Enterococcus CFU. Enterococcus spp. typically have four to six rrnA operons, whereas C. marimammalium have only one, so these are minimum estimates of the relative fold enrichment (41, 42). Comparing the quantity of enterococci cultured from our four gull fecal samples, which averaged 2.3 × 104 CFU g−1, we can estimate that there would be over 3 orders of magnitude greater density (∼1.1 × 108 cells g−1) of C. marimammalium also present. Our results may help to better understand the relationship between organisms identified by traditional culture methods versus culture-independent molecular techniques.
Implications for screening assays, source tracking, and human health.
The prevalence of C. marimammalium in fecal samples (55/57; 95%) was higher than previously reported (71%) (21), based on screening with the Gull-2 PCR assay. Detection of C. marimammalium in 27% of environmental samples was considerably lower than previously observed (21). Lu et al. (21) reported that 96% of 48 freshwater samples with presumed gull contamination, including eight samples from Grant Park Beach in South Milwaukee, WI, were positive by the Gull-2 PCR assay (21); however, our environmental samples were not necessarily presumed to have gull contamination. Factors affecting detection can include different fecal loads at the time of sampling, seasonal effects on bacterial survival, lower proportions of gull fecal pollution, or dilution of C. marimammalium DNA sequences to below the limit of detection of the assay. Our serial dilutions showed an average detection limit of 0.3 pg of gull fecal DNA per reaction mixture, which is 20 times lower than the value reported by Lu et al. (21), indicating that the gulls we sampled contained a higher concentration of C. marimammalium, a possible explanation for the increased Gull-2 assay efficiency on our gull fecal samples. Although the Gull-2 assay is sensitive to gull fecal contamination (21, 22), the high volume of water in beach environments, particularly Lake Michigan beaches, may dilute C. marimammalium DNA to below detectable limits. Sand and water sources may additionally impact the persistence and survival of C. marimammalium, which would also affect assay detection.
Geographic variability and host specificity can factor into the applicability of microbial source tracking markers. The gull fecal samples used in the Lu et al. (21) study were from West Virginia, Georgia, Ohio, Florida, and Ontario, Canada; our gull fecal samples were collected on Lake Michigan in Wisconsin. A more recent study assessed the prevalence of gull markers in animal feces collected from California, Ohio, Arkansas, Georgia, Delaware, and South Africa and detected C. marimammalium in >85% of the gull fecal samples tested (22). C. marimammalium is specific to gulls, compared to other poultry, waterfowl, and nonavian species, including pigs, dogs, cows, and humans (21, 22, 25, 43). Aside from C. marimammalium in the feces of pigeons, which was detected with the same sensitivity and specificity as that of gulls, most nontarget hosts have been near or below the lower limit of quantification or nondetectable (26). The detection of C. marimammalium in our gull fecal samples, the amplification of C. marimammalium with the Gull-2 PCR assay in our environmental samples, and the alignment of the Gull-2 primers (21) with our nearly full-length clone library contigs demonstrates the regional utility of this microbial source tracking marker for gull contamination in the Great Lakes region.
Our research provides information on the diversity and variability of the gull fecal microbial community, including the dominance of Enterococcaceae or Enterobacteriaceae spp. in most samples analyzed. Using three separate sequencing platforms, we were able to demonstrate that although taxonomic assignments can be biased by primer choice, amplification region, and sequence length, all methods clearly show C. marimammalium to be the dominant taxon within our gull fecal samples.
Plate count assays such as mEI agar are the industry standard for detection of FIB in sand and water samples. However, as is often the case with environmental samples, the most abundant organisms are not readily culturable (44). Although the gull gut appears to be a good habitat for organisms in the Enterococcaceae family, Enterococcus spp. that are readily cultured on mEI accounted for 0.04% of our sequence reads. Establishing the relationship between C. marimammalium and culturable enterococci allows better quantification of the total gull fecal load to beaches and coastal waters, which may help in understanding the true impact gulls have on water quality monitoring efforts. These findings also highlight the advantages of using culture-independent detection techniques for environmental monitoring, as they are more encompassing, time efficient, and increasingly more cost-effective. Although gulls harbor traditional FIB that can be observed in plate assays, non-mEI-culturable C. marimammalium dominates the gull fecal microbial community and is a more suitable genetic marker for tracking gull fecal pollution.
ACKNOWLEDGMENTS
This work was supported by the National Oceanic and Atmospheric Administration, University of Wisconsin Sea Grant Institute grant NA100AR4170070, project R/HCE-1, Oceans and Human Health Traineeship grant to A.M.K. (NA06OAR4310119), and by NIH funding (grant R01AI091829-01A1).
We thank members of the McLellan lab for technical assistance and guidance on this research, with a special thank you to Patricia Bower and Jessica VandeWalle. We also acknowledge the contributions of Hilary Morrison, Sharon Grim, Joe Vineis, and Mitchell Sogin at the Josephine Bay Paul Center of the Marine Biological Laboratory for sequencing and bioinformatics support.
Footnotes
Published ahead of print 15 November 2013
REFERENCES
- 1.Alderisio KA, DeLuca N. 1999. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada geese (Branta canadensis). Appl. Environ. Microbiol. 65:5628–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Converse RR, Kinzelman JL, Sams EA, Hudgens E, Dufour AP, Ryu H, Santo-Domingo JW, Kelty CA, Shanks OC, Siefring SD, Haugland RA, Wade TJ. 2012. Dramatic improvements in beach water quality following gull removal. Environ. Sci. Technol. 46:10206–10213. 10.1021/es302306b [DOI] [PubMed] [Google Scholar]
- 3.Fogarty LR, Haack SK, Wolcott MJ, Whitman RL. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94:865–878. 10.1046/j.1365-2672.2003.01910.x [DOI] [PubMed] [Google Scholar]
- 4.Lévesque B, Brousseau P, Simard P, Dewailly E, Meisels M, Ramsay D, Joly J. 1993. Impact of the ring-billed gull (Larus delawarensis) on the microbiological quality of recreational water. Appl. Environ. Microbiol. 59:1228–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meerburg BG, Koene MG, Kleijn D. 2011. Escherichia coli concentrations in feces of geese, coots, and gulls residing on recreational water in The Netherlands. Vector Borne Zoonotic Dis. 11:601–603. 10.1089/vbz.2010.0218 [DOI] [PubMed] [Google Scholar]
- 6.US Environmental Protection Agency 1986. Ambient water quality criteria for bacteria. Office of Water, Regulation and Standards, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 7.McLellan SL, Boehm AB, Shanks OC. 2013. Marine and freshwater fecal indicators and source identification, p 199–236 In Kanki P, Grimes DJ. (ed), Infectious diseases: selected entries from the encyclopedia of sustainability science and technology. Springer Science+Business Media, New York, NY [Google Scholar]
- 8.Albarnaz JD, Toso J, Correa AA, Simoes CMO, Barardi CRM. 2007. Relationship between the contamination of gulls (Larus dominicanus) and oysters (Crassostrea gigas) with Salmonella serovar Typhimurium by PCR-RFLP. Int. J. Environ. Health Res. 17:133–140. 10.1080/09603120701219816 [DOI] [PubMed] [Google Scholar]
- 9.Kinzelman J, McLellan SL, Amick A, Preedit J, Scopel CO, Olapade O, Gradus S, Singh A, Sedmak G. 2008. Identification of human enteric pathogens in gull feces at Southwestern Lake Michigan bathing beaches. Can. J. Microbiol. 54:1006–1015. 10.1139/W08-096 [DOI] [PubMed] [Google Scholar]
- 10.Lu JR, Ryu H, Domingo JWS, Griffith JF, Ashbolt N. 2011. Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta. Appl. Environ. Microbiol. 77:5034–5039. 10.1128/AEM.00018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quessy S, Messier S. 1992. Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring-billed gulls (Larus delawarensis). J. Wildl. Dis. 28:526–531. 10.7589/0090-3558-28.4.526 [DOI] [PubMed] [Google Scholar]
- 12.Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291. 10.1021/es903523q [DOI] [PubMed] [Google Scholar]
- 13.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44:4674–4691. 10.1016/j.watres.2010.06.049 [DOI] [PubMed] [Google Scholar]
- 14.Rabinovici SJM, Bernknopf R, Wein A. 2004. Economics and health risk trade-offs of swim closures at a Lake Michigan beach. Environ. Sci. Technol. 38:2737–2745. 10.1021/es034905z [DOI] [PubMed] [Google Scholar]
- 15.Edge TA, Hill S. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Res. 41:3585–3594. 10.1016/j.watres.2007.05.012 [DOI] [PubMed] [Google Scholar]
- 16.US Environmental Protection Agency 2002. National beach guidance and required performance criteria for grants EPA-823-B- 02–004 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 17.Engeman RM, Hartmann JW, Beckerman SF, Seamans TW, Abu-Absi S. 2012. Egg oiling to reduce hatch-year ring-billed gull numbers on Chicago's beaches during swim season and water quality test results. EcoHealth 9:195–204. 10.1007/s10393-012-0760-z [DOI] [PubMed] [Google Scholar]
- 18.Hartmann JW, Beckerman SF, Seamans TW, Engeman RM, Abu-Absi S. 8 February 2013. Report to the City of Chicago on Conflicts with Ring-Billed Gulls and the 2012 Integrated Ring-Billed Gull Damage Management Project. http://www.aphis.usda.gov/wildlife_damage/nwrc/publications/13pubs/engeman134.pdf
- 19.Sauer JR, Hines JE, Fallon J. 2008. The North American breeding bird survey, results and analysis 1966-2007. Version 5.15.2008. USGS Patuxent Wildlife Research Center, Laurel, MD: http://www.mbr-pwrc.usgs.gov/bbs/bbs2007.html [Google Scholar]
- 20.Jeter SN, McDermott CM, Bower PA, Kinzelman JL, Bootsma MJ, Goetz GW, McLellan SL. 2009. Bacteroidales diversity in ring-billed gulls (Laurus delawarensis) residing at Lake Michigan beaches. Appl. Environ. Microbiol. 75:1525–1533. 10.1128/AEM.02261-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 74:3969–3976. 10.1128/AEM.00019-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu H, Griffith JF, Khan IU, Hill S, Edge TA, Toledo-Hernandez C, Gonzalez-Nieves J, Santo Domingo JW. 2012. Comparison of gull feces-specific assays targeting the 16S rRNA genes of Catellicoccus marimammalium and Streptococcus spp. Appl. Environ. Microbiol. 78:1909–1916. 10.1128/AEM.07192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. 10.1371/journal.pgen.1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Nat. Acad. Sci. U. S. A. 103:12115–12120. 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, Marion J, Lee J. 2013. Development and application of a quantitative PCR assay targeting Catellicoccus marimammalium for assessing gull-associated fecal contamination at Lake Erie beaches. Sci. Total Environ. 454-455:1–8 [DOI] [PubMed] [Google Scholar]
- 26.Sinigalliano CD, Ervin JS, Van De Werfhorst LC, Badgley BD, Balleste E, Bartkowiak J, Boehm AB, Byappanahalli M, Goodwin KD, Gourmelon M, Griffith J, Holden PA, Jay J, Layton B, Lee C, Lee J, Meijer WG, Noble R, Raith M, Ryu H, Sadowsky MJ, Schriewer A, Wang D, Wanless D, Whitman R, Wuertz S, Santo Domingo JW. 5 July 2013. Multi-laboratory evaluation of the performance of Catellicoccus marimammalium PCR assays developed to target gull fecal sources. Water Res. 10.1016/j.watres.2013.02.059 [DOI] [PubMed] [Google Scholar]
- 27.Marteinsson VT, Runarsson A, Stefansson A, Thorsteinsson T, Johannesson T, Magnusson SH, Reynisson E, Einarsson B, Wade N, Morrison HG, Gaidos E. 2013. Microbial communities in the subglacial waters of the Vatnajokull ice cap, Iceland. ISME J. 7:427–437. 10.1038/ismej.2012.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison H, Grim S, Vineis J, Sogin M. 2013. 16S amplicon fusion primers and protocol for Illumina platform sequencing. figshare. http://dx.doi.org/10.6084/m9.figshare.833944.
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 78:717–725. 10.1128/AEM.06516-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.US Environmental Protection Agency 2002. Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-d-glucoside agar (mEI). EPA 821-R-02-022. Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 35.Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184–3191. 10.1128/AEM.71.6.3184-3191.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar PS, Brooker MR, Dowd SE, Camerlengo T. 2011. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS One 6:e20956. 10.1371/journal.pone.0020956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330–339. 10.1016/j.mimet.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonilla TD, Nowosielski K, Esiobu N, McCorquodale DS, Rogerson A. 2006. Species assemblages of Enterococcus indicate potential sources of fecal bacteria at a south Florida recreational beach. Mar. Pollut. Bull. 52:807–810. 10.1016/j.marpolbul.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 39.Mote BL, Turner JW, Lipp EK. 2012. Persistence and growth of the fecal indicator bacteria enterococci in detritus and natural estuarine plankton communities. Appl. Environ. Microbiol. 78:2569–2577. 10.1128/AEM.06902-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Layton BA, Walters SP, Lam LH, Boehm AB. 2010. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109:539–547 [DOI] [PubMed] [Google Scholar]
- 41.Lee ZM, Bussema C, Schmidt TM. 2009. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 37:D489–D493. 10.1093/nar/gkn689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigand MR, Ryu H, Bozcek L, Konstantinidis KT, Santo Domingo JW. 2013. Draft genome sequence of Catellicoccus marimammalium, a novel species commonly found in gull feces. Genome Announc. 1(1):e00019–00012. 10.1128/genomeA.00019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu H, Henson M, Elk M, Toledo-Hernandez C, Griffith J, Blackwood D, Noble R, Gourmelon M, Glassmeyer S, Santo Domingo JW. 2013. Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl. Environ. Microbiol. 79:196–204. 10.1128/AEM.02802-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]