Abstract
Pyrosequencing of the bacterial community associated with a cosmopolitan marine diatom during enrichment with crude oil revealed several Arenibacter phylotypes, of which one (OTU-202) had become significantly enriched by the oil. Since members of the genus Arenibacter have not been previously shown to degrade hydrocarbons, we attempted to isolate a representative strain of this genus in order to directly investigate its hydrocarbon-degrading potential. Based on 16S rRNA sequencing, one isolate (designated strain TG409T) exhibited >99% sequence identity to three type strains of this genus. On the basis of phenotypic and genotypic characteristics, strain TG409T represents a novel species in the genus Arenibacter, for which the name Arenibacter algicola sp. nov. is proposed. We reveal for the first time that polycyclic aromatic hydrocarbon (PAH) degradation is a shared phenotype among members of this genus, indicating that it could be used as a taxonomic marker for this genus. Kinetic data for PAH mineralization rates showed that naphthalene was preferred to phenanthrene, and its mineralization was significantly enhanced in the presence of glass wool (a surrogate for diatom cell surfaces). During enrichment on hydrocarbons, strain TG409T emulsified n-tetradecane and crude oil, and cells were found to be preferentially attached to oil droplets, indicating an ability by the strain to express cell surface amphiphilic substances (biosurfactants or bioemulsifiers) as a possible strategy to increase the bioavailability of hydrocarbons. This work adds to our growing knowledge on the diversity of bacterial genera in the ocean contributing to the degradation of oil contaminants and of hydrocarbon-degrading bacteria found living in association with marine eukaryotic phytoplankton.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are a particularly important class of pollutants. Based on their poor water solubility, toxicity, persistence, and potential to bioaccumulate, these compounds are recognized as high-priority pollutants and are of significant concern for human health (1). Entry of PAHs into the marine environment involves a variety of sources including biological sources (marine and terrestrial plants), anthropogenic sources (fluvial input, atmospheric fallout, surface runoff, and oil spills), and emanation at active tectonic zones (hydrothermal plumes, natural oil seeps, and volcanic eruption and fallout). Since PAHs can comprise as much as 25 to 35% of total hydrocarbon content in crude oils (2), oil spills are an important source for the release of these chemicals. For example, the recent Deepwater Horizon blowout of April 2010 released an estimated 4.4 million barrels (0.7 million tons) of crude oil into the Gulf of Mexico over a period of 84 days (3). Following this release, high concentrations of low-molecular-weight PAHs (189 μg/liter) were measured in subsurface waters (1,000 to 1,400 m), whereas higher-molecular-weight PAHs predominated in surface waters (85 μg/liter) (4, 5). Upon their entry, these chemicals become subjected to abiotic (e.g., photolysis, chemical oxidation) and biotic (e.g., microbial degradation) influences that will determine their fate. Although a proportion of these chemicals will reach the sea floor and become buried within the underlying sediment, most will be degraded and mineralized by microorganisms. Essentially, microorganisms play a critical role in the ultimate removal of these compounds from contaminated sites.
The introduction of PAHs into the marine environment often leads to the enrichment of indigenous populations of marine bacteria that can break down and utilize these compounds. These organisms can be strongly selected for in oil-impacted environments, where they successively increase in numbers from nearly undetectable levels to 70 to 90% of the total bacterial community (2). Several bacterial taxa that can degrade and utilize PAH compounds have been identified, many of which have been isolated or identified by molecular techniques in samples taken from oil-impacted sites or during enrichment experiments with crude oil or petrochemical derivatives. Recent work by our group has revealed eukaryotic phytoplankton to be a new and previously unexplored source of novel PAH-degrading bacteria. Three novel PAH degraders, including one new family, were isolated from laboratory cultures of marine phytoplankton: Algiphilus aromaticivorans strain DG1253T (6) and Porticoccus hydrocarbonoclasticus strain MCTG13dT (7) were both isolated from the marine dinoflagellate Lingulodinium polyedrum CCAP1121/2, whereas Polycyclovorans algicola strain TG408T was isolated from the marine diatom Skeletonema costatum CCAP1077/1C (8). These organisms all represent novel “specialist” hydrocarbon degraders; i.e., they exhibit an almost exclusive requirement for hydrocarbons as their preferred carbon and energy source. Interestingly, these organisms are not well represented in clone libraries from oil-impacted marine environments, possibly because they occupy a specific and relatively unexplored niche, i.e., the cell surface of eukaryotic phytoplankton.
The association of PAH-degrading bacteria with phytoplankton cells is thought to stem from the ability of the latter to adsorb and accumulate PAH molecules from the surrounding seawater (9, 10). This, we believe, would create a PAH-enriched zone around the phytoplankton cell surface, where PAH-degrading bacteria can thrive. The rationale for this association is further evidenced in a few studies that present data correlating the influence of eukaryotic phytoplankton, particularly during periods of bloom, with the removal of PAHs and other hydrocarbons from the marine water column (9–11). It has been inferred that this process can lead to the subsequent transport of these chemicals to the sea floor by sedimentation (9, 10). There is also evidence that phytoplankton produce PAHs (12, 13), which may also explain their close association with PAH-degrading bacteria. In the present study, we identify for the first time members of the genus Arenibacter in a laboratory culture of the marine diatom Skeletonema costatum and show the ability of this genus to degrade PAHs. Moreover, a novel species was isolated from this marine eukaryotic phytoplankton, which exhibited the capacity to utilize aromatic hydrocarbons as a sole source of carbon and energy. The capacity to mineralize naphthalene was enhanced by the presence of glass wool, presumably as a surrogate for the pellicle of the phytoplankton. Production of amphiphilic substances, attachment to oil droplets, and efficient emulsification of oil were additional properties possibly associated to contributing an ecological role in marine hydrocarbon degradation.
MATERIALS AND METHODS
Strains used.
A nonaxenic laboratory culture of the marine diatom Skeletonema costatum CCAP 1077/1C (origin, North Sea) was obtained from the Culture Collection of Algae and Protozoa (CCAP; Oban, Scotland) and maintained in algal medium under conditions recommended by the CCAP. Arenibacter certesii KMM 3941T, Arenibacter latericius KCTC 12957T, Arenibacter troitsensis KCTC 12362T, and Arenibacter echinorum KCTC 22013T were obtained from the Korean Collection of Type Cultures (KCTC). Arenibacter palladensis LMG 21972T was obtained from the BCCM/LMG. Arenibacter nanhaiticus NH36AT was kindly provided by Zongze Shao.
Enrichment experiments and isolation.
Enrichments of Skeletonema costatum CCAP 1077/1C with Alaska North Slope (ANS) crude oil were prepared in order to assess the dynamics of the diatom-associated bacteria in response to challenge with the oil. For this, four 1-liter Erlenmeyer flasks were prepared, each containing ca. 500 ml of f/2 algal medium amended with Na2SiO3 (14), and inoculated (to 1%, vol/vol) using a growing culture of the diatom. All four flasks were then incubated (corresponding to day zero) at 16°C in a temperature-controlled illuminated incubator with a 12:12 light/dark cycle and at a photon flux density of 25 μmol s−1 m−2. At day 7, filter-sterilized (0.2 μm) ANS oil (to 1%, vol/vol) was added to one pair of flasks (labeled flasks 1 and 2); no oil was added to the other pair of flasks (labeled flasks 4 and 5) to act as the oil-untreated controls. Samples of 5 ml from each flask were taken at days 0, 7, 10, 35, and 64. Each sample was combined with ca. 50 ml of filter-sterilized seawater (0.2 μm) and passed through a 0.2-μm filter (Millipore). The filters were stored at −20°C for extraction of DNA and subsequent molecular analysis (see below).
To isolate hydrocarbon-degrading bacteria associated with S. costatum CCAP 1077/1C, enrichment cultures were prepared by inoculating 2 ml of a growing culture of the diatom in 48 ml of a marine broth (ZM/10) containing the following mixture of PAHs (final concentrations, in mg/liter): phenanthrene (500), anthracene (50), fluorene (50) and dibenzothiophene (50). ZM/10 broth is composed of three-quarters-strength naturally aged seawater, peptone (0.05%), and yeast extract (0.01%) and was supplemented after autoclaving with filter-sterilized (0.2 μm) trace elements and vitamins to final concentrations as previously described (15). After an incubation period of 3 weeks with shaking in the dark (120 rpm; 28°C), samples of the culture were spread onto plates of ZM/10 agar. Several colonies displaying different colonial morphologies formed, one of which was abundant and distinctly orange. It was purified, labeled strain TG409T, and stored in 20% (vol/vol) glycerol at −80°C for further study.
Measurement of PAH degradation.
The ability of strain TG409T and other closely related Arenibacter type strains to degrade PAHs was determined in acid-washed (0.1 N HCl) steam-sterilized glass test tubes (13 by 100 mm) fitted with screw caps lined with Teflon-lined silicone septa (Chromacol). Stock solutions of phenanthrene (ca. 3,000 mg/liter) and naphthalene (ca. 9,000 mg/liter) dissolved in acetone were prepared. For each strain, two sets of 3 test tubes were prepared, each containing 2.9 ml of ONR7a, which is a defined synthetic seawater medium (16). To one set of tubes phenanthrene was added, and naphthalene was added to the second set. All 6 test tubes were then inoculated with 100 μl of a washed cell suspension of the test strain. Uninoculated controls, acid-killed controls, and tubes that were inoculated but without any added PAH were also prepared. The initial amounts of phenanthrene and naphthalene were 0.25 ± 0.01 mg and 0.71 ± 0.02 mg, respectively. All test tubes were incubated in the dark with gentle shaking (100 rpm) at 21°C. PAH degradation was determined by high-pressure liquid chromatography (HPLC). For this, the test tubes from each PAH incubation were sacrificed after 14 days for extraction with ethyl acetate (HPLC grade). This was performed by adding 2 ml of ethyl acetate to each tube and then vortexing for 30 s. Aliquots of the nonaqueous top layer were stored at −20°C in hermetically sealed HPLC vials for subsequent analysis. A significant decrease (P < 0.05) in the PAH concentration measured in the inoculated test tubes, relative to the uninoculated controls, was indicative of degradation.
14C mineralization experiments.
The ability of strain TG409T to mineralize decane, n-hexadecane, phenanthrene, anthracene, and naphthalene was evaluated in 40-ml amber EPA vials containing 5 ml of ONR7a medium and one of the radiolabeled compounds to about 20,000 dpm. Triplicate incubations were prepared for each compound. A test tube containing a piece of filter paper saturated with 2 N KOH was then placed inside each vial to act as the CO2 trap. Each incubation was inoculated with washed cells that had been grown in ZM/10 broth. The vials were hermetically sealed with alumina-lined screw-top lids and incubated on a rotary shaker (100 rpm) at 21°C. At 3-day intervals over the course of 12 days, the filter paper was removed and the amount of 14CO2 captured was measured with a Packard (Meriden, CT) Tri-Carb Liquid Scintillation Analyzer, model 1900 TR. The KOH-saturated filter paper from each vial was replaced at each sampling point for the course of the experiment.
To compare the rate and extent at which strain TG409T could mineralize 14C-labeled phenanthrene and naphthalene, a highly sensitive 14C-respirometer system (17) was used. This is a flask-based system designed to monitor substrate mineralization for up to 10 weeks with multiple respirometer-opening events. About 20,000 dpm of each 14C-labeled compound was diluted with its unlabeled counterpart and added to a final concentration of 10 mg/liter. In replicate incubations run in parallel, a piece of sterile glass wool was added to the medium to test if it could serve as a substratum for attachment of the cells (as a surrogate for the diatom surface) and enhance the mineralization of the compounds. The percentage of added 14C mineralized was calculated as 100 × [(14CO2 captured from each of the triplicate experimental measurements) − (mean of the values of 14CO2 captured from sterile controls)]/(total activity [dpm] of 14C added).
Morphological, physiological, and biochemical characteristics.
Phenotypic characterization typically used marine medium 2216 (Difco) or ONR7a medium (16). Unless indicated otherwise, all biochemical tests and growth assessments were performed at 30°C and monitored over a period of 1 to 2 weeks. Uninoculated medium and medium without an added carbon source were used as controls. All incubations were performed in at least triplicate. The cell morphology of strain TG409T was examined by transmission electron microscopy from cells that had been grown in marine broth (18). The Gram reaction, oxidase, catalase, lipase (Tween 80), and gelatinase tests and production of H2S and poly-β-hydroxybutyrate (PHB) granules were performed using standard procedures (19). Total phosphatase activity and motility were evaluated as previously described (20). Gliding motility was evaluated on marine agar medium 2216 and by direct observation under the light microscope. The Schaeffer-Fulton staining method was used for the determination of endospore formation (19). To determine nitrate reduction, the organism was grown in ONR7a medium amended with 0.1% KNO3, 0.1% Bacto agar, and glucose as the sole carbon source, and the result was observed after 1 week of incubation. Growth was evaluated at 4, 10, 15, 20, 30, 37, and 40°C, in the presence of increasing concentrations of chloramphenicol, ampicillin, kanamycin, and streptomycin, and under anaerobic conditions. The pH range for growth was determined in marine broth adjusted to different pH values with the following buffers: 25 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.5 to 6.5; 25 mM 3-[N-Tris(hydroxymethyl)methylamino]-2-hydroxypropanesulfonic acid (TAPSO), pH 7.5; and 25 mM Tris base/Tris HCl, pH 8.5 to 9.5. Oxidative-fermentation tests were performed in ONR7a broth (16) amended with bromocresol purple as the indicator and with one of the following at 0.5% (wt/vol) final concentration: glucose, mannitol, fructose, xylose, and arabinose. The strain's requirement and tolerance to NaCl was determined with ONR7a broth containing increasing concentrations of NaCl: 0.0, 1.0, 3.0, 6.0, 9.0, 12.0, 15.0, 20.0, and 25.0% (wt/vol). Tolerance to KCl was performed in the same way, but this time in the absence of added NaCl. Carbon source utilization was examined using the Biolog GN2 Microplate system according to the manufacturer's instructions. To determine whether various aromatic and aliphatic hydrocarbons could be used as sole sources of carbon, they were added to ONR7a liquid medium. n-Hexadecane, n-tetradecane, and British Petroleum (BP) light crude oil were added at 0.2%, vol/vol. The volatile substrates benzene, toluene, p-xylene, biphenyl, naphthalene, and phenol were supplied via the vapor phase as previously described (21). The cultures were incubated at 30°C under aerobic conditions, and growth was monitored spectrophotometrically over 2 to 3 weeks. Growth was assessed on chocolate blood agar, MacConkey's agar, sheep blood agar, and horse blood agar. Growth on methane was tested by inoculating ONR7a agar plates and placing them in a desiccator, after which 30% of the air atmosphere was replaced with methane.
Fatty acid, respiratory quinone, and polar lipid analysis.
Whole-cell fatty acid profiles, obtained following extraction of biomass of strain TG409T and methylation of the total lipid fraction, were analyzed (22) from cells that had been grown under identical conditions reported for other type species of Arenibacter. Respiratory quinones were extracted from lyophilized cells, and the samples were purified and analyzed by the DSMZ Identification Service (23, 24). Polar lipid analysis was performed using an Iatroscan MK V TH10 thin-layer chromatography-flame ionization detector (TLC-FID) analyzer (Iatron Laboratories, Tokyo, Japan) as previously described (25).
16S rRNA gene sequencing and phylogenetic analysis.
The partial 16S rRNA gene sequence of TG409T was determined as previously described (6). The sequence was submitted to GenBank and checked for close relatives and phylogenetic affiliation using the BLASTn search program (26) and RDP-II (27). Clustal_X (28) was used to align the sequences and construct neighbor-joining trees with Treeview (WIN32) version 1.5.2 (29). The trees were bootstrapped 1,000 times, and gaps in the alignment were ignored. To evaluate the dendrogram topology, sequences were imported and automatically aligned in the ARB-SILVA SSURef 94 database (30) and manually refined, taking into account the secondary structural information of the rRNA molecule. Tree reconstruction was performed using the neighbor-joining (ARB), maximum parsimony (DNAPars), and maximum likelihood (RAxML) (31, 32) methods with and without the application of 30%, 40%, and 50% positional conservation filters.
DNA-DNA hybridization and DNA G+C content.
DNA-DNA hybridization experiments were conducted by the DSMZ, Germany, between strain TG409T and the type species with 16S rRNA gene sequence similarities of >97.0%: Arenibacter certesii KMM 3941T; Arenibacter latericius KCTC 12957T; Arenibacter nanhaiticus NH36AT; Arenibacter palladensis LMG 21972T; Arenibacter troitsensis KCTC 12362T; and Arenibacter echinorum KCTC 22013T. Cells were disrupted by using a French pressure cell (Thermo Spectronic), and the DNA in the crude lysate was purified by chromatography on hydroxyapatite as described by Cashion et al. (33). DNA-DNA hybridization was carried out as described by De Ley et al. (34) under consideration of the modifications described by Huss et al. (35) using a Cary model 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with in situ temperature probe (Varian). Two independent determinations were carried out for each experiment, and the results reported are mean values.
The DNA G+C content of TG409T was determined by HPLC, as previously described (36).
Growth experiments and emulsification assays.
Cells of strain TG409T were grown (120 rpm; 21°C) in preautoclaved 250-ml Erlenmeyer flasks containing 50 ml of ONR7a liquid medium amended with glucose, n-tetradecane, or crude oil. Glucose was added at 1% (wt/vol) and each of the oils at 4% (vol/vol). The crude oil used in these experiments was from the Gulf of Mexico (British Petroleum). During incubation of these cultures, samples were periodically taken over several days for emulsification assays and to monitor growth spectrophotometrically at 600 nm. For emulsification assays, both cell-free supernatants (13,000 × g; 10 min) and cells washed twice with 0.1 M phosphate-buffered saline were assayed to determine the production of extracellularly released and cell-bound emulsifying agents, respectively. Emulsification assays were performed by mixing samples (1 ml) with an equal volume of n-tetradecane or n-hexadecane in acid-washed (0.1 N HCl) screw-cap glass tubes (100 by 13 mm). The tubes were manually shaken (15 s) and vortexed (15 s) to homogeneity, left to stand for 10 min, and shaken as before, and the height of the emulsion layer, denoted as emulsification index (EI24), was measured after allowing the mixture to stand for 24 h at room temperature. In addition, subsamples from the n-tetradecane and crude oil incubations were stained with acridine orange (AO) (37) and imaged with a fluorescein isothiocyanate (FITC) filter on a Zeiss Axioskop (Carl Zeiss, Germany). Light microscopy was also used to observe the development of emulsion formation in the n-tetradecane and crude oil incubations.
Barcoded amplicon pyrosequencing.
DNA was extracted from samples taken during the 64-day crude oil enrichment with S. costatum CCAP 1077/1C. For this, one quarter of each frozen filter was cut into small pieces, and the DNA was extracted using the FastDNA Spin kit for Soil (MP Biomedicals, Solon, OH, USA) as per the manufacturer's instructions, with the exception that the DNA was eluted in Tris-EDTA (TE; 10 mM Tris-HCl, 1 mM EDTA, pH 8.0). All DNA extracts were then purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA, USA), eluted into TE buffer, and stored at −20°C until used for barcoded 16S rRNA gene pyrosequencing. For pyrosequencing, a two-step PCR method was used as previously described (38), employing primer pairs 27f and 338r, which were modified to incorporate an identical 8-bp barcode sequence that was unique for each sample and a 2-bp spacer on the 5′ end of the primer sequence (39). DNA concentrations of PCR amplicons were measured using a NanoDrop ND-3300 fluorospectrometer (Thermo) and the Quant-iT Picogreen double-stranded DNA (dsDNA) kit (Invitrogen) prior to combining into a single sample at a concentration suitable for pyrosequencing. The sample tube was then submitted to the High-Throughput Sequencing Facility at the University of North Carolina—Chapel Hill for sequencing using the 454 Life Sciences Titanium platform (Roche Diagnostics, Branford, CT, USA). Pyrosequencing reads were trimmed, filtered, and demultiplexed as previously described (40). To form operational taxonomic units (OTUs), the reads were clustered at 97% sequence identity with UCLUST (41), and the most abundant unique read within each cluster was used as its representative sequence. Representative sequences were identified using BLASTn (26).
Statistical analysis.
Student's t test was used to test for significant differences (P < 0.05) in the degradation and mineralization of hydrocarbons between the different treatments.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain TG409T was deposited with GenBank under accession number FJ176555.
RESULTS
Enrichment of S. costatum with crude oil and strain isolation.
Barcoded 16S rRNA gene pyrosequencing was used to analyze the bacterial community response associated with S. costatum CCAP 1077/1C after exposure to crude oil. Here, we report on the response of phylotypes identified belonging to the genus Arenibacter, whereas a more complete presentation of the bacterial community structure and dynamics from the pyrosequencing analysis is reported elsewhere (S. Mishamandani, T. Gutierrez, D. Berry, M. Aitken, submitted for publication). The total number of high-quality partial gene sequences obtained for each of the duplicate oil enrichments (flasks 1 and 2) and duplicate untreated controls (flasks 3 and 4) is presented in Table S1 in the supplemental material. From these libraries, 13 phylotypes were identified with ≥95% (approximately genus level) sequence identity to strain TG409T. The highest sequence similarity (>96%) to strain TG409T occurred with OTUs 103, 157, and 202. OTU-202 in particular was found to increase from near-undetectable levels in all four flasks from day 7 (before the addition of oil) to 1.2% ± 0.1% of the total bacterial community by day 64 in the oil enrichments (flasks 1 and 2). In contrast, its abundance was lower (0.7% ± 0.03%) in the untreated controls (flasks 4 and 5) (see Table S1 in the supplemental material).
Agar plates that were streaked from the enrichment of Skeletonema costatum CCAP 1077/1C with PAHs produced a number of colonial morphotypes that were picked and subcultured to obtain pure cultures. One isolate, designated strain TG409T, developed orange colonies that were predominant on ZM/10 agar plates and was selected for further study.
Degradation and mineralization of PAHs.
Enrichments were performed with phenanthrene or naphthalene in ONR7a liquid medium, and degradation of each compound by strain TG409T and closely related Arenibacter type strains was measured. All of the strains tested degraded phenanthrene, and except for Arenibacter latericius strain KCTC 12957T, all strains degraded naphthalene. The amounts of these compounds that were degraded after 2 weeks by each of the strains are shown in Table 1. Uninoculated controls showed no significant loss of these PAHs (data not shown).
TABLE 1.
Degradation of phenanthrene and naphthalene by strain TG409T and all type strains of the genus Arenibacter in ONR7a liquid medium
| Organism | Avg % (± SD) of degradeda: |
|
|---|---|---|
| Phenanthrene | Naphthalene | |
| Strain TG409T | 3.0 ± 0.4 | 10.8 ± 2.9 |
| Arenibacter palladensis strain LMG 21972T | 12.8 ± 0.1 | 40.1 ± 6.2 |
| Arenibacter echinorum strain KCTC 22013T | 4.2 ± 0.2 | 19.8 ± 7.3 |
| Arenibacter troitsensis strain KCTC 12362T | 16.1 ± 0.5 | 17.1 ± 2.9 |
| Arenibacter nanhaiticus strain NH36AT | 10.9 ± 0.6 | 18.1 ± 5.8 |
| Arenibacter certesii strain KMM 3941T | 9.1 ± 0.1 | 48.9 ± 9.2 |
| Arenibacter latericius strain KCTC 12957T | 6.1 ± 0.5 | 0.0 ± 0.0 |
Values are from triplicate HPLC measurements and expressed as a percentage of compound degraded after 2 weeks relative to uninoculated controls.
Using 14C-labeled substrates, strain TG409T was found to be capable of mineralizing phenanthrene and naphthalene. Decane, n-hexadecane, and anthracene were not mineralized. Using the validated and highly sensitive flask-based 14C-respirometer system of Reid et al. (17), detectable mineralization of phenanthrene and naphthalene occurred within 2 days and continued up until day 40, at which time these incubations were terminated (Fig. 1). The rate at which phenanthrene was mineralized was similar for incubations amended with and without glass wool. Mineralization rates for naphthalene were higher than that for phenanthrene, and glass wool enhanced its mineralization. A “packed button” of orange cells also formed on the surface of the glass wool over the 40-day incubations.
FIG 1.
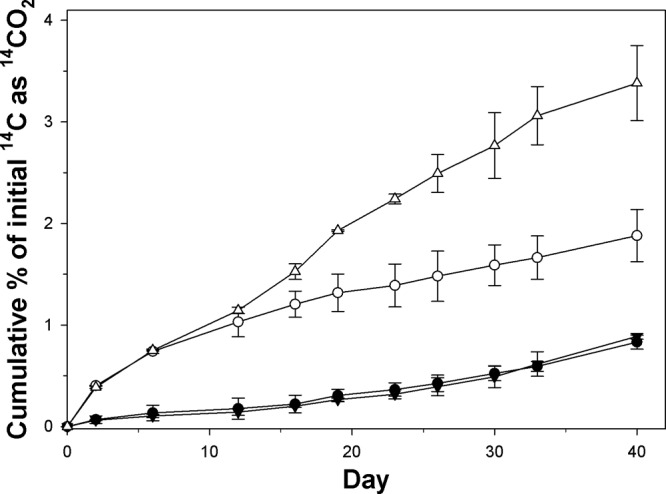
Cumulative 14CO2 recovered from incubations with [14C]phenanthrene and naphthalene by strain TG409T in ONR7a liquid medium amended with (upright and inverted triangles) and without (solid and open circles) sterile glass wool. Each data point represents the mean of results from triplicate incubations ± standard deviations. Filled symbols, phenanthrene; open symbols, naphthalene. Some error bars are smaller than the symbols.
Phylogenetic analysis of strain TG409T.
A near-full-length sequence of the 16S rRNA gene (1,492 nucleotides [nt]) of TG409T was obtained and used for phylogenetic analysis. From a BLASTn analysis, the highest level of sequence identity (99.8%) for TG409T was to Flexibacter aggregans strain IFO 15975 (originally named Microscilla aggregans strain Q-3), which had been isolated from marine sand in Split, Yugoslavia (42). The highest levels of sequence identities to type strains occurred with Arenibacter palladensis strain LMG 21972T (99.5%), Arenibacter troitsensis strain KCTC 12362T (99.4%), and Arenibacter echinorum strain KCTC 22013T (99.3%). All other known type strains represented within the Arenibacter genus exhibited <96% sequence identity for TG409T. These and other related sequences were used to construct the neighbor-joining phylogenetic tree shown in Fig. 2.
FIG 2.
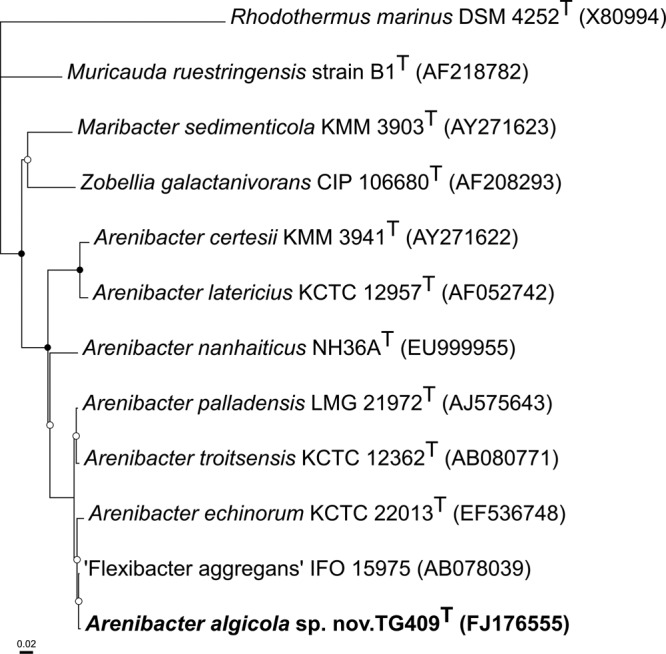
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences (>1,200 bp), showing the relationships between strain TG409T and closely related representative strains within the family Flavobacteriaceae. Rhodothermus marinus in the class Betaproteobacteria was selected as the outgroup. Nodes marked with an open or a closed circle indicate bootstrap support values (from 1,000 replications) of >50% or >95%, respectively. GenBank accession numbers are shown in parentheses. Bar, 2 substitutions per 100 nucleotide positions. The maximum parsimony and maximum likelihood trees showed essentially the same topology (data not shown).
The DNA-DNA relatedness of the new isolate to related type strains was below 70% (Table 2). Since this is below the threshold value of 70% DNA-DNA for inclusion of the strain in one of the established type species (43), strain TG409T appears therefore to represent a novel species.
TABLE 2.
Percentage of DNA-DNA similarity between the reference strain TG409T and all type strains of the genus Arenibacter
| Arenibacter type species | DNA-DNA similarity to strain TG409T (%) |
|---|---|
| A. palladensis strain LMG 21972T | 44 |
| A. echinorum strain KCTC 22013T | 37 |
| A. troitsensis strain KCTC 12362T | 34 |
| A. nanhaiticus strain NH36AT | 24 |
| A. certesii strain KMM 3941T | 11 |
| A. latericius strain KCTC 12957T | 11 |
Phenotypic and biochemical characterization of strain TG409T.
On marine agar plates, colonies were orange, slightly raised, and with entire margins and diameters of 2 to 3 mm after 7 days. The cells were also pigmented orange when grown in liquid media, and no diffusible pigments were observed. Various clinical media, including chocolate blood agar, MacConkey's agar, sheep blood agar, and horse blood agar (19), did not support growth. Strain TG409T was aerobic, catalase positive, and oxidase negative. Cells were long, non-spore-forming rods, 2.5 to 3.0 by 0.5 μm in average size, occurring singly or in pairs (Fig. 3). They stained Gram negative and were nonmotile. Staining with Nile blue did not reveal the presence of PHB granules, which was confirmed by spectral analysis. Nitrate was reduced to nitrite.
FIG 3.
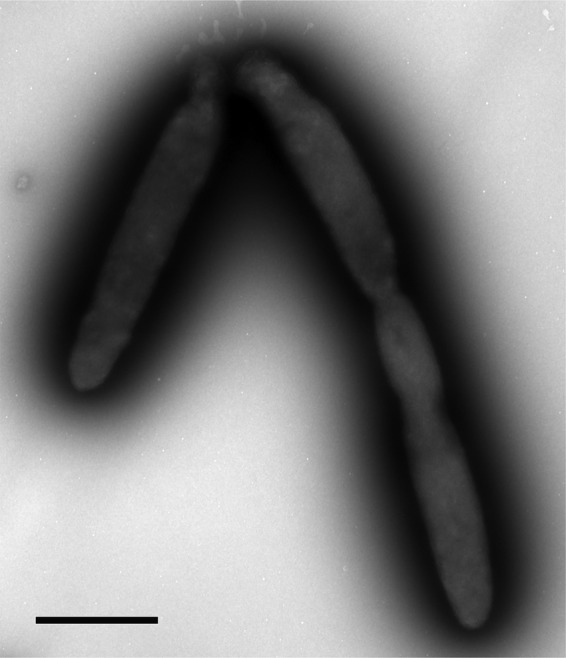
Transmission electron micrograph showing negatively stained cells of strain TG409T. Bar, 1 μm.
The strain displayed a wide nutritional spectrum for utilizing various organic carbon substrates. Using the Biolog GN system, strain TG409T was positive for the oxidation of dextrin, Tween 20, Tween 40, Tween 80, N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, l-arabinose, d-cellobiose, d-fructose, l-fucose, d-galactose, gentiobiose, α-d-glucose, m-inositol, α-d-lactose, lactulose, maltose, d-mannitol, d-mannose, d-melibiose, β-methyl-d-glucoside, d-psicose, d-raffinose, l-rhamnose, sucrose, d-trehalose, turanose, pyruvic acid methyl ester, succinic acid mono-methyl ester, acetic acid, d-galactonic acid lactone, d-galactonic acid, d-gluconic acid, d-glucuronic acid, α-ketobutyric acid, α-ketovaleric acid, d,l-lactic acid, succinic acid, glucuronamide, l-alaninamide, d-alanine, l-alanine, l-alanyl-glycine, l-asparagine, l-aspartic acid, l-glutamic acid, glycyl-l-aspartic acid, glycyl-l-glutamic acid, hydroxyl-l-proline, l-ornithine, l-proline, l-serine, l-threonine, d,l-carnitine, urocanic acid, uridine, phenylethylamine, glycerol, d,l-α-glycerol phosphate, α-d-glucose-1-phosphate, and d-glucose-6-phosphate. Growth on glycogen, d-arabitol, d-sorbitol, citric acid, d-glucosaminic acid, α-hydroxybutyric acid, α-ketoglutaric acid, propionic acid, quinic acid, d-saccharic acid, l-histidine, l-leucine, γ-aminobutyric acid, inosine, thymidine, putrescine, 2-aminoethanol, and 2,3-butanediol was weak. No growth was observed on α-cyclodextran, adonitol, i-erythritol, xylitol, cis-aconitic acid, formic acid, β-hydroxybutyric acid, γ-hydroxybutyric acid, p-hydroxyphenylacetic acid, itaconic acid, malonic acid, sebacic acid, bromosuccinic acid, succinamic acid, l-phenylalanine, l-pyroglutamic acid, and d-serine. The strain grew on benzene, p-xylene, n-tetradecane, n-hexadecane, and crude oil, whereas sodium benzoate, sodium salicylate, phenol, biphenyl, phenanthrene, anthracene, naphthalene, pyruvate, fluorene, and toluene did not support growth. The strain grew on methanol but not on methane.
Strain TG409T grew at temperatures ranging from 10 to 30°C (optimal temperature, 30°C) and at pH values ranging from 6.5 to 8.5 (optimal pH, 8.0). The strain was negative for lipase (Tween 80) and gelatinase. The strain was positive for phosphatase activity and for the fermentation of d-glucose, d-mannitol, d-fructose, d-xylose, and l-arabinose. The strain exhibited slight halotolerance, since it grew in medium containing NaCl concentrations of up to 6%. Higher NaCl concentrations completely inhibited growth. The strain grew optimally in the absence of any added NaCl. Strain TG409T is therefore slightly halotolerant and can be considered a marine strain (44). The strain did not, however, exhibit a requirement for the Na+ cation since good growth was observed in medium without Na+ added. Addition of Na+ did not stimulate growth. Substituting Na+ for K+ in the growth medium did not inhibit the growth of strain TG409T for KCl concentrations of up to 1%. Growth was markedly inhibited at a KCl concentration of 3% and completely inhibited at concentrations of ≥6%. The strain was resistant to ampicillin (up to 166 μg/ml), kanamycin (166 μg/ml), and streptomycin (8.3 μg/ml) but susceptible to chloramphenicol. Table 3 lists the differential characteristics for strain TG409T relative to the type strains of Arenibacter with the highest 16S rRNA gene sequence similarity (see below).
TABLE 3.
Key phenotypic characteristics that differentiate strain TG409T from related type strainsa
| Characteristic | Strainb |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Gliding motility | + | + | − | + |
| Na+ requirement | − | − | + | − |
| Nitrate reduction | + | − | + | − |
| Oxidase | − | + | + | + |
| H2S production | + | − | + | − |
| Degradation of: | ||||
| Gelatin | + | − | + | − |
| Urea | − | − | − | − |
| Tween 20 | + | − | − | + |
| Tween 40 | + | − | + | + |
| Tween 80 | + | − | − | − |
| Growth conditions, range (optimal) | ||||
| Temp (°C) | 10–30 (30) | 4–38 (24) | 10–42 (30) | 4–35 (25) |
| pH range | 6.5–8.5 (8.0) | ND | 5.5–10.0 (8.0) | ND |
| Salt concn (%) | 0–6 (0) | 1–6 | 0–8 | |
| Utilization of: | ||||
| d-Galactose | + | + | − | − |
| Maltose | + | + | − | + |
| l-Arabinose | + | + | + | + |
| Glycerol | + | − | − | − |
| N-Acetyl-β-glucosamine | + | + | − | − |
| Mannitol | + | − | − | + |
| Malate | w | − | − | + |
| Citrate | w | − | − | + |
| Acid production from: | ||||
| d-Glucose | + | + | − | + |
| d-Xylose | + | + | − | + |
| Susceptibility to streptomycin | − | − | − | + |
| DNA G+C (mol%) | 41.9 | 40.0 | 40.0 | 39–40 |
Symbols and abbreviations: ND, not determined; +, positive; −, negative; w, weakly positive. Data are from Nedashkovskaya et al. (45, 56, 57), Sun et al. (59), and this study.
Strains: 1, strain TG409T; 2, Arenibacter palladensis LMG 21972T; 3, Arenibacter troitsensis KCTC 12362T; 4, Arenibacter echinorum KCTC 22013T.
The fatty acid profile of strain TG409T was similar to those of reference strains that were grown under identical conditions (Table 4). The dominant fatty acids for strain TG409T were iso-C15:0 (21.7%), iso-C17:0 3-OH (17.6%), iso-C17:1 (11.2%), iso-C15:1 (10.2%), C15:0 (9.1%), C16:1ω7c (7.2%), iso-C15:0 3-OH (5.8%), C16:0 (5.2%), and iso-C17:0 (3.6%). Fatty acids detected as minor components (<2%) were iso-C14:0 (1.7%) and C17:0 2-OH (1.1%). The fatty acid composition of strain TG409T is in accordance with those reported for the species of the genus Arenibacter (45). Strain TG409T contained 3 main phospholipids, with phosphatidyl ethanolamine (32%) as the major class present, followed by phosphatidic acid (28%) and phosphatidyl glycerol (24%). A higher-eluting and therefore less polar unidentified class (16%) was also present as a minor component. The predominant respiratory quinone was MK-6 (92.4%), with MK-5 (7.6%) as a minor component. The DNA G+C content of strain TG409T was 41.9 mol%, which was slightly higher than that found in all other type strains of the Arenibacter genus and in most members of the Flavobacteriaceae family.
TABLE 4.
Cellular fatty acid composition of strain TG409T and closely related type strains
| Fatty acid | % of total fatty acids in straina: |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Iso-C14:0 | 1.7 | |||
| Iso-C15:0 | 21.7 | 8.7 | 6.8 | 6.4 |
| Anteiso-C15:0 | 3.3 | 3.2 | 1.8 | |
| Iso-C15:1 | 10.2 | 12.7 | 12.2 | 13.1 |
| C15:0 | 9.1 | 15.0 | 13.6 | 22.0 |
| C15:1 ω6c | 2.6 | 1.2 | 4.2 | |
| Iso-C16:0 | 0.2 | 0.3 | 1.0 | |
| C16:0 | 5.2 | |||
| Iso-C17:0 | 3.6 | |||
| Iso-C17:1 ω9cb | 11.2 | 4.0 | 5.3 | 3.9 |
| C17:1 ω8c | 0.5 | 0.9 | 1.1 | |
| C17:1 ω6c | 1.4 | 1.1 | 2.9 | |
| Iso-C15:0 3-OH | 5.8c | 5.3 | 5.1 | 3.5 |
| C15:0 3-OH | 2.2 | 1.6 | 2.0 | |
| Iso-C16:0 3-OH | 1.6 | 2.2 v | 1.0 | |
| C16:0 3-OH | 2.0 | 2.2 | 1.0 | |
| Iso-C17:0 3-OH | 17.6 | 17.4 | 21.9 | 10.5 |
| C17:0 2-OH | 1.1 | 1.0 | 1.7 | 0.3 |
| Summed feature 1d | 7.2 | 11.1 | 9.6 | 10.6 |
Strains: 1, strain TG409T; 2, Arenibacter palladensis LMG 21972T; 3, Arenibacter troitsensis KCTC 12362T; 4, Arenibacter echinorum KCTC 22013T. Identities of strains and references are as in Table 3. Components that represented <0.1% in all strains are not included.
In strain 1, the double-bond position of iso-C17:1 was not determined; in strains 2 to 4, this fatty acid was reported as iso-C17:1 ω9c.
Fatty acid composition contributed also by iso-C15:0 2-OH.
Summed feature 1 values represent fatty acids that could not be separated by gas-liquid chromatography with the MIDI system for strains LMG 21972T, KCTC 12362T, and KCTC 22013T. This feature consists of iso-C15:0 2-OH, C16:1 ω7c, and C16:1 ω7t.
Emulsifier production and oil emulsion formation.
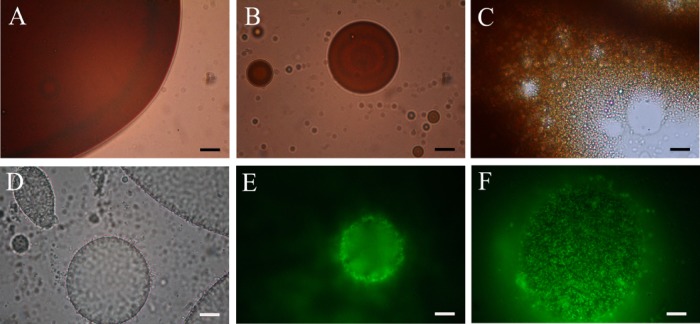
The capacity of strain TG409T to produce cell-bound and/or extracellularly released emulsifying agents during growth on glucose is shown in Fig. 4. Emulsification activities were assessed against both n-hexadecane and n-tetradecane—substrates that serve as a sole carbon and energy source for the strain—and were found to be highly similar. Hence, only the emulsification results against n-tetradecane are reported. Emulsification activity was associated only with the cells, and no activity was detected in the cell-free culture supernatant fractions. Activity was highest during the early stationary phase of growth, falling dramatically thereafter. Lower emulsification activities (EI24 values, <20%) were measured for cells grown on n-tetradecane or crude oil (results not shown) compared to growth on glucose. However, visual inspection of the overlying n-tetradecane and crude oil phases in these cultures indicated that they became increasingly emulsified during these incubations. Microscopic examination of samples taken from the crude oil incubation confirmed that the oil was breaking up into smaller droplets than in the original culture and uninoculated controls (Fig. 5A to C). Similarly, microscopic examination of samples taken from the n-tetradecane enrichments showed the development of small oil droplets that appeared to be stabilized by the attachment of strain TG409T cells onto the surface of the droplets (Fig. 5D to F). AO-stained samples from the crude oil incubation could not be imaged due to significant autofluorescence from the crude oil.
FIG 4.
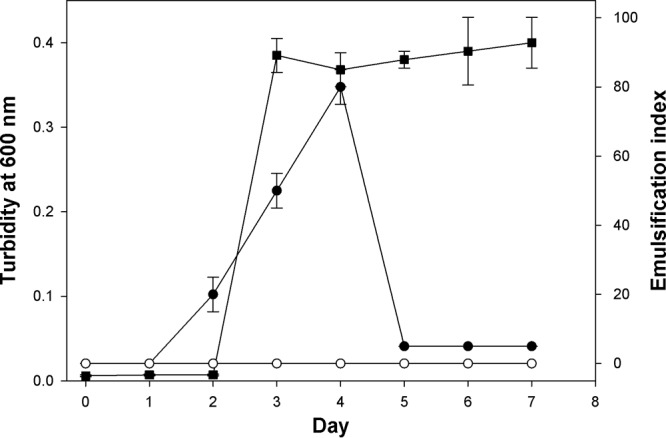
Growth and emulsifier production by strain TG409T in ONR7a liquid medium amended with glucose (1%, wt/vol). Emulsification assays were conducted against n-tetradecane. Squares, cell culture turbidity at 600 nm; solid circles, emulsifying activity of washed cells; open circles, emulsifying activity of cell-free culture medium after removal of the cells by centrifugation. Error bars represent the standard deviations of at least three independent measurements from different culture flasks. Some error bars are smaller than the symbols.
FIG 5.
Microscopic images of oil emulsion formation by strain TG409T during incubation in ONR7a medium with crude oil or n-tetradecane. (A to C) Light microscopic images showing the development of a crude oil emulsion from day 0 (A) to day 10 (B) and day 14 (C). (D) Light microscopic image of emulsified oil droplets from an enrichment with n-tetradecane showing the higher contrast around the circumference of the droplets as produced by the concentrated attachment of cells. (E and F) Epifluorescence images of a sample from the n-tetradecane enrichment stained with acridine orange and showing cells attached to the periphery (E) and entire surface (F) of an emulsified droplet as captured using a differential depth of field. Bars, 10 μm.
TAXONOMY
Description of Arenibacter algicola sp. nov.
Arenibacter algicola sp. nov. (al.gi′co.la. M. L. n. alga, alga or seaweed; L. suff. -cola from L. n. incola, an inhabitant or dweller; N. L. fem. n. algicola, alga dweller).
Cells are Gram-negative, strictly aerobic, and nonmotile rods (2.5 to 3.0 by 0.5 μm). Colonies on 2216 marine medium are orange, slightly raised, with entire margins, and 2 to 3 mm in diameter after 7 to 10 days of incubation at 30°C. Growth occurs at 10 to 30°C and pH 6.5 to 8.5. The optimal temperature for growth is 30°C. Optimal pH for growth is 8.0. NaCl is not essential for growth, since growth is detected in NaCl concentrations of 0 to 6% (wt/vol). Optimal growth occurred in the absence of added NaCl. Substituting Na+ for K+ inhibited growth at KCl concentrations of ≥3%. Oxidase negative and catalase positive. Positive for nitrate reduction and phosphatase activity. Gelatin liquefaction and lipase (Tween 80) activity are negative. In Biolog GN2 microplates, positive for oxidation of dextrin, Tween 40, Tween 80, N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, l-arabinose, d-cellobiose, d-fructose, l-fucose, d-galactose, gentiobiose, α-d-glucose, m-inositol, α-d-lactose, lactulose, maltose, d-mannitol, d-mannose, d-melibiose, β-methyl-d-glucoside, d-psicose, d-raffinose, l-rhamnose, sucrose, d-trehalose, turanose, pyruvic acid methyl ester, succinic acid mono-methyl ester, acetic acid, d-galactonic acid lactone, d-galactonic acid, d-gluconic acid, d-glucuronic acid, α-ketobutyric acid, α-ketovaleric acid, d,l-lactic acid, succinic acid, glucuronamide, l-alaninamide, d-alanine, l-alanine, l-alanyl-glycine, l-asparagine, l-aspartic acid, l-glutamic acid, glycyl-l-aspartic acid, glycyl-l-glutamic acid, hydroxyl-l-proline, l-ornithine, l-proline, l-serine, l-threonine, d,l-carnitine, urocanic acid, uridine, phenylethylamine, glycerol, d,l,α-glycerol phosphate, α-d-glucose-1-phosphate, and d-glucose-6-phosphate. Glycogen, d-arabitol, d-sorbitol, citric acid, d-glucosaminic acid, α-hydroxybutyric acid, α-ketoglutaric acid, propionic acid, quinic acid, d-saccharic acid, l-histidine, l-leucine, γ-aminobutyric acid, inosine, thymidine, putrescine, 2-aminoethanol, and 2,3-butanediol were weakly utilized. Susceptible to chloramphenicol but resistant to ampicillin, kanamycin, and streptomycin. The cells can mineralize phenanthrene and naphthalene as sole or principal sources of carbon and energy. Growth on methanol but not methane as a sole carbon source. The major fatty acids are iso-C15:0, iso-C17:0 3-OH, iso-C17:1, and iso-C15:1. The major polar lipid is phosphatidyl ethanolamine (32%), followed by phosphatidic acid (28%) and phosphatidyl glycerol (24%). The DNA G+C content of the type strain of the type species is 41.9 mol%. Isolated from a laboratory culture of the marine diatom Skeletonema costatum. The type and only species is Arenibacter algicola. The type strain is TG409T (ATCC BAA-2265, DSM 24761).
DISCUSSION
Physiology and ecology.
To our knowledge, strain TG409T represents the first Arenibacter strain isolated from a marine eukaryotic phytoplankton. This strain grows in medium containing NaCl concentrations of up to 6% (wt/vol), indicating that it is slightly halotolerant (44). Strain TG409T displayed a wide nutritional spectrum, which is typical of members of the Arenibacter genus. The strain was also capable of degrading various aromatic hydrocarbons. Its ability to mineralize naphthalene, phenanthrene, and n-hexadecane, albeit partially, and inability to grow on these substrates are not uncommon with other species of bacteria. We had initially hypothesized that the ability to degrade PAHs was unique to strain TG409T within the Arenibacter genus. However, subsequent testing revealed that all Arenibacter type species are capable of degrading phenanthrene and, with the exception of A. latericius strain KCTC 12957T, naphthalene. Given the apparent conservation of this phenotype within the Arenibacter genus, it should be considered quite useful as a taxonomic marker in future systematic descriptions of members belonging to this genus.
Interestingly, the rate of naphthalene but not phenanthrene mineralization by strain TG409T was enhanced by the presence of glass wool. Naphthalene adsorbs to glass wool more readily than phenanthrene (46), which may explain this difference. After 1 week of growth on naphthalene, an orange button of TG409T cells developed on the glass wool surface, which coincided with an increase in the rate of naphthalene mineralization. Hence, the glass wool may have acted as a hot spot of concentrated naphthalene or heightened cell activity that accelerated the mineralization. Silica (SiO2) is a major structural component in the cell walls of diatoms (47), which glass wool might mimic. These results suggest that the eukaryotic partner S. costatum of strain TG409T could stimulate mineralization rates of PAHs by passively concentrating these pollutants in the marine water column.
Eukaryotic phytoplankton, in particular diatoms, produce copious quantities of extracellular polymeric substances (EPS), which they may either express on their cell surface or release into the surrounding seawater (48, 49). By nature of their stickiness, these polymers participate in the aggregation and subsequent sedimentation of diatoms (48, 49). Although S. costatum generally produces lower quantities of EPS (50) than other species of diatoms, microscopic observation of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells of strain CCAP1077/1C showed the presence of an abundant community of bacteria localized within a polymer matrix surrounding the diatom cells (results not shown). This zone, referred to as the phycosphere, can be summarily described as the immediate layer around each microalgal cell. Whether through biogenic synthesis or adsorption from the surrounding seawater, the phycosphere of marine eukaryotic phytoplankton may be regarded as a hot spot to which hydrocarbon-degrading bacteria are associated. This concept is supported by our recent discovery of several novel hydrocarbon-degrading bacteria in laboratory cultures of marine phytoplankton (6–8). Notably, these bacteria conferred the ability to utilize PAHs as sole sources of carbon and energy and exhibited a nutritional preference for hydrocarbons over other carbon substrates. We hypothesize that the underlying rationale to explain these alga-bacterium associations is the potential of phytoplankton to adsorb and accumulate PAHs from the surrounding seawater (9, 10) or their capacity to synthesize these compounds (12, 13) and translocate them into the algal cell wall (51–53). Hence, the microalgal phycosphere could, under certain circumstances, become enriched with hydrocarbons (e.g., PAHs) and thereby sustain hydrocarbon-degrading bacteria, such as strain TG409T.
Many species of marine bacteria are capable of producing EPS, such as for swarming behavior and in promoting their colonization or for biofilm formation (54). The capacity of strain TG409T cells to emulsify hydrocarbons appears to be related to production of cell-associated EPS. Although only very low levels of emulsifying activity were detected during growth on n-tetradecane or crude oil, these substrates were emulsified in the culture and cells became attached to the emulsified oil droplets. These results indicate that amphiphilic EPS were produced and expressed on the cell surface to mediate this attachment. Since n-tetradecane served as an excellent substrate for this strain, direct physical attachment of the cells to oil droplets can be inferred to be a mechanism to access this poorly soluble substrate. While the autofluorescence of crude oil hindered our ability to image cells attached to crude oil droplets, its emulsification and degradation probably also resulted from the production of amphiphilic EPS. This conclusion is supported by the ability of washed cells and cell-free fractions from the crude oil enrichment to emulsify fresh crude oil samples (results not shown).
The discovery of members of the genus Arenibacter with the ability to degrade aromatic hydrocarbons and be enriched during the crude oil enrichments with the diatom highlights the important role that these organisms could be playing in removing hydrocarbons in the marine environment, especially in surface waters enriched with diatoms. The potential of Arenibacter to degrade hydrocarbons has likely eluded discovery previously due to their low levels of enrichment in laboratory cultures and samples from oil-impacted environments. These organisms may be physiologically less adept at thriving under conditions of high petrochemical loading than other hydrocarbon degraders such as Alcanivorax, Marinobacter, and Cycloclasticus that are common at oil-impacted sites (2). Possibly, the phycosphere of marine eukaryotic phytoplankton may be a biotope that has not been sufficiently investigated for novel species of hydrocarbon-degrading bacteria like strain TG409T. From an ecological perspective, we posit that the close association between a hydrocarbon-degrading bacterium, such as strain TG409T, and a microalgal partner plays an important role in the removal of organic pollutants from the marine water column. While the hydrocarbon-degrading phenotype of these alga-derived bacteria remains active in the absence of the algal partner, as demonstrated by the capacity of these bacteria to grow on and degrade hydrocarbons in pure culture, the coupling of these organisms in their natural environment may facilitate their uptake of carbon energy sources in the marine oligotrophic environment. Further work will be needed to more fully comprehend the underlying mechanism(s) that defines these bacterium-alga associations and the role played by Arenibacter spp. in hydrocarbon degradation in the marine environment.
Phylogeny and taxonomy.
Phylogenetic analysis revealed that strain TG409T belongs to the genus Arenibacter. First proposed by Ivanova et al. (55), species within this genus are Gram negative, aerobic, heterotrophic, and dark-orange-pigmented marine bacteria. Currently, this genus is represented by six species, A. latericius, A. troitsensis, A. certesii, A. palladensis, A. echinorum, and A. nanhaiticus, that originated from various marine sources, including the edible holothurian Apostichopus japonicus, the brown alga Chorda filum, the green macroalga Ulva fenestrata, and the sea urchin Strongylocentrotus intermedius (45, 55–59). Strain TG409T is the first representative within the genus Arenibacter to be isolated from a marine eukaryotic phytoplankton. Since no data on the potential for any of the six type strains of Arenibacter to degrade aromatic hydrocarbons existed hitherto, we showed that all of these strains, including strain TG409T, are capable of degrading aromatic hydrocarbons as a sole source of carbon and energy. Based on this commonality among all type strains of this genus, we suggest that this phenotypic trait might serve as a genus level feature.
Based on 16S rRNA analysis, the highest levels (>97%) of sequence identity for strain TG409T to type strains were to Arenibacter palladensis strain LMG 21972T (99.5%), Arenibacter troitsensis strain KCTC 12362T (99.4%), and Arenibacter echinorum strain KCTC 22013T (99.3%). However, important phenotypic and biochemical properties distinguish strain TG409T from its closest relatives (Table 3). Notably, strain TG409T lacked oxidase, is capable of degrading Tween 80, and exhibited a narrower range of temperature tolerance than its closest relatives. Moreover, there are clear differences between the fatty acid profiles of strain TG409T and these reference strains. For example, one of the major fatty acids in strain TG409T was iso-C15:0 (21.7%), which was much less abundant (≤8.7%) in other closely related type strains (Table 4). Strain TG409T did not contain anteiso-C15:0, C15:1 ω6c, iso-C16:0, C17:1 ω8c, C17:1 ω6c, C15:0 3-OH, iso-C16:0 3-OH, and C16:0 3-OH, whereas these acids were identified in other closely related type strains. Conversely, iso-C14:0, C16:0, and iso-C17:0 were present in strain TG409T, whereas they were absent in these other type strains. Furthermore, the results of the DNA-DNA hybridization experiments showed that the strain is a novel Arenibacter species (43).
Based on this polyphasic analysis, strain TG409T may be classed as a distinct species within the genus Arenibacter, for which we propose the name Arenibacter algicola.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the 7th European Community Framework Programme. Partial support was also provided through the U.S. National Institute of Environmental Health Sciences, grant 5 P42ES005948.
We thank Victoria Madden (UNC's Microscopy Services Laboratory), who provided valuable assistance with the preparation of samples for electron microscopy and in the capture of electron micrographs, and Damien Shea and Robin Shields for providing crude oil samples. We also thank Danny Holdsworth, who managed the CSIRO GC and GC-MS facility, Chris Paynter for assistance with the 14C-respirometer experiments, the CCAP (Culture Collection for Algae & Protozoa, Oban, United Kingdom) for providing S. costatum, and Zongze Shao (State Oceanic Administration, Xiamen, China) for providing A. nanhaiticus NH36AT.
Footnotes
Published ahead of print 8 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03104-13.
REFERENCES
- 1.Schoeny R, Poirier K. 1993. EPA/600/R-93/089. U.S. Environmental Protection Agency, Office of Research and Development, Washington, DC [Google Scholar]
- 2.Head IM, Martin Jones D, Röling WFM. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173–182. 10.1038/nrmicro1348 [DOI] [PubMed] [Google Scholar]
- 3.Crone TJ, Tolstoy M. 2010. Magnitude of the 2010 Gulf of Mexico oil leak. Science 330:634. 10.1126/science.1195840 [DOI] [PubMed] [Google Scholar]
- 4.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill W, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakroborty R, Sonnenthal EL, D'haeseleer P, Holman H-YN, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. 10.1126/science.1195979 [DOI] [PubMed] [Google Scholar]
- 5.Wade TL, Sweet ST, Sericano JL, Guinasso NL, Dierks A-R, Highsmith RC, Asper VL, Joung DJ, Shiller AM, Lohrenz SE, Joye SB. 2011. Analyses of water samples from the Deepwater Horizon oil spill: documentation of the subsurface plume, p 77–82 In Liu Y, Macfadyen A, Ji Z-G, Weisberg RH. (ed), Monitoring and modeling the Deepwater Horizon oil spill: a record-breaking enterprise. Geophysical monograph series 195. American Geophysical Union, Washington, DC. 10.1029/2011GM001103 [DOI] [Google Scholar]
- 6.Gutierrez T, Green DH, Whitman WB, Nichols PD, Semple KT, Aitken MD. 2012. Algiphilus aromaticivorans gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from a culture of the marine dinoflagellate Lingulodinium polyedrum, and proposal of Algiphilaceae fam. nov. Int. J. Syst. Evol. Microbiol. 62:2743–2749. 10.1099/ijs.0.033324-0 [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez T, Nichols PD, Whitman WB, Aitken MD. 2012. Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 78:628–637. 10.1128/AEM.06398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez T, Green DH, Nichols PD, Whitman WB, Semple KT, Aitken MD. 2013. Polycyclovorans algicola gen. nov., sp. nov., an aromatic-hydrocarbon-degrading marine bacterium found associated with laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 79:205–214. 10.1128/AEM.02833-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binark N, Guven KC, Gezgin T, Unlu S. 2000. Oil pollution of marine algae. Bull. Environ. Contamin. Toxicol. 64:866–872. 10.1007/s001280000083 [DOI] [PubMed] [Google Scholar]
- 10.Kowalewska G. 1999. Phytoplankton—the main factor responsible for transport of polynuclear aromatic hydrocarbons from water to sediments in the Southern Baltic ecosystem. ICES J. Mar. Sci. 56:219–222. 10.1006/jmsc.1999.0607 [DOI] [Google Scholar]
- 11.Witt G. 2002. Occurrence and transport of polycyclic aromatic hydrocarbons in the water bodies of the Baltic Sea. Mar. Chem. 79:49–66. 10.1016/S0304-4203(02)00035-X [DOI] [Google Scholar]
- 12.Andelman JB, Suess MJ. 1970. Polynuclear aromatic hydrocarbons in the water environment. Bull. World Health Organ. 43:479–508 [PMC free article] [PubMed] [Google Scholar]
- 13.Gunnison D, Alexander M. 1975. Basis for the resistance of several algae to microbial decomposition. Appl. Microbiol. 29:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillard RR. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60 In Smith WL, Chanley MH. (ed), Culture of marine invertebrate animals. Plenum Press, New York, NY [Google Scholar]
- 15.Blackburn SI, Hallegraeff GM, Bolch CJ. 1989. Vegetative reproduction and sexual life cycle of the toxic dinoflagellate Gymnodinium catenatum from Tasmania, Australia. J. Phycol. 25:577–590. 10.1111/j.1529-8817.1989.tb00264.x [DOI] [Google Scholar]
- 16.Dyksterhouse SE, Gray JP, Herwig RP, Cano Lara J, Staley JT. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116–123. 10.1099/00207713-45-1-116 [DOI] [PubMed] [Google Scholar]
- 17.Reid BJ, MacLeod CJA, Lee PH, Morriss AWJ, Stokes JD, Semple KT. 2001. A simple 14C-respirometric method for assessing microbial catabolic potential and contaminant bioavailability. FEMS Microbiol. Lett. 196:141–146. 10.1111/j.1574-6968.2001.tb10555.x [DOI] [PubMed] [Google Scholar]
- 18.Hayat MA, Miller SE. 1990. Negative staining. MacGraw-Hill Publishing Co., New York, NY [Google Scholar]
- 19.Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 20.MacFaddin JF. 2000. Biochemical tests for identification of medical bacteria, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21.Paje ML, Neilan B, Couperwhite I. 1997. A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiology 143:2975–2981. 10.1099/00221287-143-9-2975 [DOI] [PubMed] [Google Scholar]
- 22.Miller MR, Bridle AR, Nichols PD, Carter CG. 2008. Increased elongase and desaturase gene expression with stearidonic acid enriched diet does not enhance long-chain (n-3) content of seawater Atlantic salmon (Salmo salar L.). J. Nutr. 138:2179–2185. 10.3945/jn.108.091702 [DOI] [PubMed] [Google Scholar]
- 23.Tindall BJ. 1990. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13:128–130. 10.1016/S0723-2020(11)80158-X [DOI] [Google Scholar]
- 24.Tindall BJ. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66:199–202. 10.1111/j.1574-6968.1990.tb03996.x [DOI] [Google Scholar]
- 25.Volkman JK, Nichols PD. 1991. Applications of thin layer chromatography-flame ionization detection to the analysis of lipids and pollutants in marine and environmental samples. J. Planar Chromatogr. 4:19–26 [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 27.Maidak BL, Cole JR, Parker CT, Jr, Garrity GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen GJ, Overbreek R, Pramanik S, Schmidt TM, Tiedje JM, Woese CR. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171–173. 10.1093/nar/27.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL_X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 30.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.67. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 32.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 33.Cashion P, Hodler-Franklin MA, McCully J, Franklin M. 1977. A rapid method for base ratio determination of bacterial DNA. Anal. Biochem. 81:461–466. 10.1016/0003-2697(77)90720-5 [DOI] [PubMed] [Google Scholar]
- 34.De Ley J, Cattoir H, Reynaerts A. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133–142. 10.1111/j.1432-1033.1970.tb00830.x [DOI] [PubMed] [Google Scholar]
- 35.Huss VAR, Festl H, Schleifer KH. 1983. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4:184–192. 10.1016/S0723-2020(83)80048-4 [DOI] [PubMed] [Google Scholar]
- 36.Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167. 10.1099/00207713-39-2-159 [DOI] [Google Scholar]
- 37.Francisc DE, Mah RA, Rabin AC. 1973. Acridine orange-epifluorescence technique for counting bacteria in natural waters. Trans. Am. Microsc. Soc. 92:416–421. 10.2307/3225245 [DOI] [PubMed] [Google Scholar]
- 38.Berry D, Mahfoudh KB, Wagner M, Loy A. 2011. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 77:7846–7849. 10.1128/AEM.05220-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237. 10.1038/nmeth.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A. 2013. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 7:2091–2104. 10.1038/ismej.2013.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 42.Lewin RA, Lounsbery DM. 1969. Isolation, cultivation and characterization of Flexibacteria. J. Gen. Microbiol. 58:145–170. 10.1099/00221287-58-2-145 [DOI] [PubMed] [Google Scholar]
- 43.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463–464. 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- 44.Larsen H. 1986. Halophilic and halotolerant microorganisms—an overview and historical perspective. FEMS Microbiol. Rev. 39:3–7. 10.1111/j.1574-6968.1986.tb01835.x [DOI] [Google Scholar]
- 45.Nedashkovskaya OI, Vancanneyt M, Cleenwerck I, Snauwaert C, Kim SB, Lysenko AM, Shevchenko LS, Lee KH, Park MS, Frolova GM, Mikhailov VV, Bae KS, Swings J. 2006. Arenibacter palladensis sp. nov., a novel marine bacterium isolated from the green alga Ulva fenestrata, and emended description of the genus Arenibacter. Int. J. Syst. Evol. Microbiol. 56:155–160. 10.1099/ijs.0.63893-0 [DOI] [PubMed] [Google Scholar]
- 46.Gerde P, Scholander P. 1989. Adsorption of polycyclic aromatic hydrocarbons on to asbestos and man-made mineral fibres in the gas phase. IARC Sci. Publ. 90:140–148 [PubMed] [Google Scholar]
- 47.Martin-Jézéquel V, Hildebrand M, Brzezinski MA. 2000. Silicon metabolism in diatoms: implications for growth. J. Phycol. 36:821–840. 10.1046/j.1529-8817.2000.00019.x [DOI] [Google Scholar]
- 48.Passow U, Alldredge AL. 1994. Distribution, size and bacterial colonization of transparent exopolymer particles (TEP) in the ocean. Mar. Ecol. Progr. Ser. 113:185–198. 10.3354/meps113185 [DOI] [Google Scholar]
- 49.Passow U, Alldredge AL, Logan BE. 1994. The role of particulate carbohydrate exudates in the flocculation of diatom blooms. Deep Sea Res. I 4:335–357 http://www.engr.psu.edu/ce/enve/logan/publications/1994-Passow-etal.pdf [Google Scholar]
- 50.Myklestad SM. 1995. Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci. Total Environ. 165:155–164. 10.1016/0048-9697(95)04549-G [DOI] [Google Scholar]
- 51.Gol'man LP, Mikhaseva MF, Reznikov VM. 1973. Infrared spectra of lignin preparations of pteridophytes and seaweeds. Dokl. Akad. Nauk. BSSR. 17:1031–1033 [Google Scholar]
- 52.Pastuska G. 1961. Die Kieselgelschicht-Chromatographie von Phenolen und Phenolcarbensiuren. I. Z. Anal. Chem. 179:355–358. 10.1007/BF00462690 [DOI] [Google Scholar]
- 53.Zelibor JL, Romankiw L, Hatcher PG, Colwell RR. 1988. Comparative analysis of the chemical composition of mixed and pure cultures of green algae and their decomposed residues by 13C nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 54:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decho AW. 1990. Microbial exopolymer secretions in ocean environments: their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. Annu. Rev. 28:73–154 [Google Scholar]
- 55.Ivanova EP, Nedashkovskaya OI, Chun J, Lysenko AM, Frolova GM, Svetashev VI, Vysotskii MV, Mikhailov VV, Huq A, Colwell RR. 2001. Arenibacter gen. nov., new genus of the family Flavobacteriaceae and description of a new species, Arenibacter latericius sp. nov. Int. J. Syst. Evol. Microbiol. 51:1987–1995. 10.1099/00207713-51-6-1987 [DOI] [PubMed] [Google Scholar]
- 56.Nedashkovskaya OI, Kim SB, Han SK, Lysenko AM, Mikhailov VV, Bae KS. 2004. Arenibacter certesii sp. nov., a novel marine bacterium isolated from the green alga Ulva fenestrata. Int. J. Syst. Evol. Microbiol. 54:1173–1176. 10.1099/ijs.0.02872-0 [DOI] [PubMed] [Google Scholar]
- 57.Nedashkovskaya OI, Kim SB, Lysenko AM, Lee KH, Bae KS, Mikhailov VV. 2007. Arenibacter echinorum sp. nov., isolated from the sea urchin Strongylocentrotus intermedius. Int. J. Syst. Evol. Microbiol. 57:2655–2659. 10.1099/ijs.0.65251-0 [DOI] [PubMed] [Google Scholar]
- 58.Nedashkovskaya OI, Suzuki M, Vysotskii MV, Mikhailov VV. 2003. Arenibacter troitsensis sp. nov., isolated from marine bottom sediment. Int. J. Syst. Evol. Microbiol. 53:1287–1290. 10.1099/ijs.0.02384-0 [DOI] [PubMed] [Google Scholar]
- 59.Sun F, Wang B, Du Y, Liu X, Lai Q, Li G, Luo J, Shao Z. 2010. Arenibacter nanhaiticus sp. nov., isolated from marine sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 60:78–83. 10.1099/ijs.0.008573-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.