Abstract
We describe here the construction of a series of 71 vectors to silence central carbon metabolism genes in Escherichia coli. The vectors inducibly express antisense RNAs called paired-terminus antisense RNAs, which have a higher silencing efficacy than ordinary antisense RNAs. By measuring mRNA amounts, measuring activities of target proteins, or observing specific phenotypes, it was confirmed that all the vectors were able to silence the expression of target genes efficiently. Using this vector set, each of the central carbon metabolism genes was silenced individually, and the accumulation of metabolites was investigated. We were able to obtain accurate information on ways to increase the production of pyruvate, an industrially valuable compound, from the silencing results. Furthermore, the experimental results of pyruvate accumulation were compared to in silico predictions, and both sets of results were consistent. Compared to the gene disruption approach, the silencing approach has an advantage in that any E. coli strain can be used and multiple gene silencing is easily possible in any combination.
INTRODUCTION
Recent advances in biotechnology have allowed genome-wide studies of metabolic pathways (1). In Escherichia coli, a single-gene disruption library of all nonessential genes (the Keio collection) has been constructed and distributed all over the world (2, 3). In addition, predicting metabolic flux change after gene disruption is becoming possible with in silico simulation (4, 5). Information obtained by these studies is used for understanding the metabolic pathway as a system and for rationally designing the metabolic pathway to produce valuable compounds. However, the Keio collection is not applicable to all E. coli strains and does not include growth essential genes.
We previously reported the construction of vectors that express antisense RNAs (asRNAs) to silence the function of any gene in E. coli (6, 7). The vectors are isopropyl-β-d-thiogalactoside (IPTG)-inducible, allowing for the conditional silencing of target genes. Moreover, the asRNAs have novel structures called paired-terminus (PT) asRNAs, consisting of 38-bp inverted repeats that create paired double-stranded RNA termini for any antisense sequence. The PTasRNAs have much higher silencing efficacies than ordinary asRNAs because of their improved stability, which increases the abundance of asRNAs in cells. The advantages of the gene silencing approach over the gene disruption approach include their inducibility, portability, and high throughput.
The final goal of our research is the construction of PTasRNA expression vectors for all E. coli genes (ca. 4,000 to 5,000) in order to understand the whole cell as a system. As a first step, we focused on central carbon metabolism, since it is a good example of a miniaturized cellular system. Similarly, in silico simulation study initially focused on 14 central carbon-metabolism genes and 4 additional genes for the metabolic precursors needed for cell growth (the first core model) (8), whereas the current simulation model contains over 1,000 genes.
In the present study, we report the construction of 71 PTasRNA expression vectors targeted to central carbon metabolism genes. The targeted genes encode proteins involved in glycolysis and fermentation, the tricarboxylic acid (TCA) cycle, the respiratory chain, and global transcriptional regulation. We also show that proper information for metabolic engineering can be obtained via this method and that comparing experimental and in silico simulation data is possible. The vectors facilitate comprehensive analyses of central carbon metabolism, and they can be used for the rational design of metabolic pathways as desired.
MATERIALS AND METHODS
E. coli strains, plasmids, and general techniques.
The wild-type E. coli MG1655 strain (The Coli Genetic Stock Center; CGSG6300) was used as a host for expressing PTasRNAs throughout the study. If not otherwise stated, E. coli cells were cultured in Luria broth (LB) at 37°C. The cydA-disrupted cells were always cultured at 30°C because they grew poorly at 37°C. If necessary, IPTG (1 mM for both liquid and solid media) and/or appropriate antibiotics were added to express PTasRNAs and to maintain vectors. Other general techniques used are described elsewhere (6, 7, 9).
The pHN1257 vector is an empty vector for expressing PTasRNAs in an IPTG-inducible manner and carries an expression element consisting of the trc promoter (Ptrc; IPTG-inducible), a 5′-end paired terminus, a multiple cloning site, and a 3′-end paired terminus (6). It also carries a kanamycin resistance gene, the pSC101H replication origin (ori), and the lactose repressor gene (lacIq; to ensure IPTG-inducible expression). The pHN1464 vector, another empty vector, contains the same expression element as pHN1257, a chloramphenicol-resistance gene, and the pACYC ori. This vector was constructed by digesting pHN678 (7) with PstI and self-ligating to remove the unnecessary lacIq gene. The PTasRNA expression vectors were constructed by cloning PCR fragments of antisense sequences into the NcoI and XhoI sites of pHN1257 and pHN1464 in an antisense orientation with respect to Ptrc (9). Oligonucleotides used to amplify antisense sequences from the MG1655 genome and the resultant PTasRNA expression vectors are listed in Tables S1 and S2 in the supplemental material.
To express PTasRNAs in liquid culture, transformants containing PTasRNA expression vectors were pregrown overnight in the absence of IPTG, diluted 1:400 with fresh media containing IPTG, and cultured up to the mid- to late-logarithmic phase. Silencing efficacies were calculated by comparing the protein activity or mRNA amount in a silenced strain with that in a control strain (that harbors pHN1257); both strains were cultured in the presence of IPTG.
Gene disruption vector construction and gene disruption method.
The 5′- and 3′-flanking regions for cra were PCR-amplified from genomic DNA from the MG1655 strain using specific primer sets (see Table S3 in the supplemental material; the cra1/cra2 set and the cra3/cra4 set, respectively). The termini of the fragments were treated with NsiI and XhoI or with XhoI and NcoI, respectively, and both fragments were cloned into the PstI and NcoI sites in pHN1234 (10). The vector pHN1234 contains the pSC101ts (temperature-sensitive) ori, a chloramphenicol resistance gene, and the sacB gene; the vector is used to disrupt genes according to the method developed by Hamilton and others (11–13). The resulting plasmid was named pHN2051.
For construction of disruption vectors for pflB, ppsA, sucC, pykF, and cydA, the primer sets shown in Table S3 in the supplemental material were used similarly. In all cases, the fragments were cloned into the PstI and NcoI sites of pHN1234. The resulting vectors were named pHN1458 (pflB), pHN2038 (ppsA), pHN2040 (sucC), pHN2039 (pykF), and pHN2050 (cydA).
Real-time quantitative RT-PCR.
Real-time quantitative reverse transcription-PCR (RT-PCR) was performed with the One-Step SYBR PrimeScript RT-PCR kit II (TaKaRa Bio Co., Japan) and the Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA) as described previously (6). Oligonucleotides for amplifying target mRNAs were designed so as not to amplify the corresponding PTasRNAs, and the sequences are listed in Table S3 in the supplemental material. 16S rRNA was chosen as a reference for normalization.
Protein assays.
Enzymatic activities for Pta (6), AckA (7), Zwf (14), Mdh (14), LdhA (14), TpiA (14), Gnd (14), AceE (14), GltA (14), Pgi (14), Pgk (14), IcdA (15), AceA (16), and Eno (17) were determined as described previously. Cells were disrupted in phosphate-buffered saline (8 g of NaCl, 0.2 g of KCl, 1.2 g of Na2HPO4, and 0.2 g of KH2PO4 liter−1) using glass beads and a Multi-Beads shocker (Yasui Kikai, Japan) at 4°C; however, since the glass bead method did not work for the AceE assay, for this assay, cells were disrupted with sonication. Cell extracts were prepared by centrifugation at 20,000 × g for 15 min at 4°C, and the total protein concentration was determined using the Bio-Rad Bradford assay kit (Bio-Rad, Hercules, CA). Each reaction mixture (180 μl) was prepared in a microplate well (Nunc, Thermo Fisher, NY; product 269620). The absorbance was read per minute using the Safire microplate reader (Tecan Group, Ltd., Switzerland) and the LS-PLATEmanager 2004 data analysis program (Wako Pure Chemicals Co., Japan).
Production of pyruvate.
For batch flask culture, cells were pregrown overnight and diluted 1:20 with 8 ml of N2G medium in a 100-ml Sakaguchi flask. The N2G medium contained 40 g of glucose, 5 g of (NH4)2HPO4, 1 g of K2HPO4, 2 g of NaCl, 0.24 g of MgSO4·7H2O, 0.011 g of CaCl2·2H2O, and 20.9 g of 3-morpholino propanesulfonic acid (MOPS)-NaOH (pH 7.5) liter−1. The flask was shaken vigorously to ensure aerobic growth at 37°C. After 24 and 48 h, the broth supernatant was sampled. At 24 h, 5 M NaOH was added to raise the pH to near 6.8, if necessary. The N5G media used for jar fermenter cultures contained 60 g of glucose, 10 g of (NH4)2SO4, 1 g of K2HPO4, 2 g of NaCl, 0.24 g of MgSO4·7H2O, and 0.011 g of CaCl2·2H2O liter−1. Pregrown cells were diluted 1:20 with 160 ml of N5G medium in a 250-ml vessel. The temperature was maintained at 37C, and the pH was regulated with a pH controller (DJ-1023P; Able & Biott Co., Ltd., Tokyo, Japan) using 4 M NaOH and maintained at pH 6.2 to 6.7. The flow rate of sparged air was fixed at 0.63 volumes of air per volume of medium per minute (vvm).
High-performance liquid chromatography (HPLC) was performed using a cationic exchange column, Aminex HPX-87H, and a Micro-Guard Column, Aminex 85H (Bio-Rad Labs, Richmond, CA). The chromatographic conditions were as follows: mobile phase, 4 mM H2SO4; flow rate, 0.5 ml min−1; and column oven temperature, 45°C. Organic acids were detected with an UV indicator at 210 nm.
In silico metabolic simulation.
We used the original genome-scale metabolic model (GSM) of E. coli named iAF1260 (18), which contains 1,260 open reading frames (ORFs), 2,077 metabolic and transport reactions, and 1,038 unique metabolites. The metabolic flux distribution on the GSM was simulated using flux balance analysis (FBA) (4, 5). Briefly, a pseudosteady state of the metabolite profile was assumed, and the maximum and minimum ranges of the flux values for each reaction were defined. These constraints provided a feasible space for the flux distribution in the metabolic model. To obtain the optimal flux distribution in the feasible space, an objective function was introduced and a linear programming technique was applied. This problem is represented by the following equation: to maximize cT·v and subject to S·v = 0, vmin ≤ v ≤ vmax, where S represents the stoichiometric matrix of metabolites in the metabolic reactions, and v indicates the vector of flux for each metabolic reaction. The values vmin and vmax represent the minimum and maximum constraint flux values for each reaction, and c is a vector that represents the objective function to be maximized or minimized. In the present study, biomass production was used as the objective function to be maximized, with the assumption that the cell evolved its metabolic network to maximize the growth rate. All simulations were performed using Matlab (Mathworks, Inc., Natick, MA) with glpk (http://glpkmex.sourceforge.net/), which is a solver for linear programming.
For all simulations, glucose was used as the sole carbon source, and its uptake rate was set to 10 mmol g (dry cell weight)−1 h−1. The oxygen uptake rate was set to 15 and 20 mmol g (dry cell weight)−1 h−1. Other external metabolites, such as CO2 and NH3, were allowed to transport freely through the cell membrane (19). After the maximal biomass production was obtained, the variability of pyruvate production was checked by maximization and minimization of pyruvate production under fixed biomass production on the maximal value.
Since the metabolic model was constructed using linear equations, the metabolic fluxes changed in linear proportion to the glucose uptake rate. Thus, we simulated all mutants using the same rates of glucose uptake and oxygen uptake. We used the pyruvate yields from glucose, which were not affected by the glucose uptake rate in the simulation, for comparison with the experimental results.
RESULTS
Genes chosen and construction of PTasRNA expression vectors.
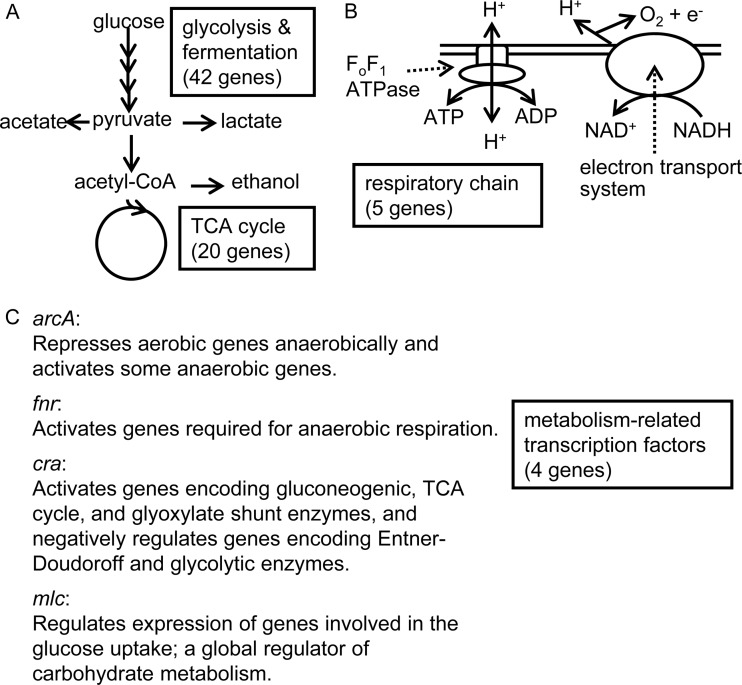
The 71 genes whose products are involved in central carbon metabolism were chosen as targets (Fig. 1 and Table 1) (20). In cases where one metabolic step is catalyzed by more than one gene product redundantly, all of the genes were included as targets, and in cases where the metabolic steps are catalyzed by a complex of multigene products, only one of the genes was chosen.
FIG 1.
Schematic map of central carbon metabolism and the 71 target genes. A more detailed map is shown in Fig. S1 in the supplemental material.
TABLE 1.
Seventy-one vectors that have sufficient silencing efficacy
| Gene | Mean silencing efficacy (% reduction) ± SD and observed phenotype(s)a |
|---|---|
| accA | 76 ± 1 (mRNA), severe growth |
| aceA | 87 ± 2 (mRNA), 85 ± 15 (protein) |
| aceB | 98 ± 0.2 (mRNA) |
| aceE | 71 ± 1 (mRNA), >99 (protein) |
| ackA | 51 ± 1 (mRNA), 75 ± 4 (protein) |
| acnA | 91 ± 6 (mRNA) |
| acnB | 96 ± 2 (mRNA) |
| acs | 78 ± 3 (mRNA) |
| adhE | 62 ± 1 (mRNA) |
| arcA | 79 ± 1 (mRNA) |
| atpF | 84 ± 13 (mRNA) |
| cra | 61 ± 3 (mRNA) |
| cydA | 83 ± 3 (mRNA) |
| cyoA | 83 ± 6 (mRNA) |
| eda | 91 ± 3 (mRNA) |
| edd | 61 ± 2 (mRNA) |
| eno | 92 ± 0.02 (mRNA), 82 ± 5 (protein) |
| fbaA | 81 ± 1 (mRNA) |
| fbaB | 69 ± 5 (mRNA) |
| fbp | 77 ± 8 (mRNA) |
| fnr | 62 ± 16 (mRNA) |
| frdA | 70 ± 4 (mRNA) |
| fumA | 72 ± 9 (mRNA) |
| fumB | 77 ± 15 (mRNA) |
| fumC | 80 ± 4 (mRNA) |
| gapA | 93 ± 1 (mRNA), severe growth |
| gcd | 60 ± 11 (mRNA) |
| glcB | 75 ± 5 (mRNA) |
| glkA | 78 ± 5 (mRNA) |
| gltA | 95 ± 0.05 (mRNA), 92 ± 0.4 (protein) |
| gnd | 77 ± 17 (mRNA), 60 ± 11 (protein) |
| gntK | 67 ± 5 (mRNA) |
| gpmA | 72 ± 2 (mRNA) |
| gpmB | 64 ± 5 (mRNA) |
| gpmI | 60 ± 2 (mRNA) |
| icdA | 64 ± 9 (mRNA), 93 ± 2 (protein) |
| idnK | 83 ± 0.07 (mRNA) |
| ldhA | 52 ± 6 (mRNA), 77 ± 4 (protein) |
| maeA | 80 ± 6 (mRNA) |
| maeB | 86 ± 6 (mRNA) |
| mdh | 95 ± 3 (mRNA), 84 ± 10 (protein) |
| mlc | 64 ± 4 (mRNA) |
| mqo | 87 ± 7 (mRNA) |
| ndh | 63 ± 6 (mRNA) |
| nuoA | 95 ± 2 (mRNA) |
| pckA | 86 ± 3 (mRNA) |
| pfkA | 87 ± 3 (mRNA)b |
| pfkB | 87 ± 0.3 (mRNA) |
| pflB | 70 ± 2 (mRNA) |
| pgi | 90 ± 1 (mRNA), 95 ± 3 (protein)c |
| pgk | 79 ± 4 (mRNA), 75 ± 4 (protein) |
| pgl | 92 ± 2 (mRNA) |
| poxB | 97 ± 1 (mRNA) |
| ppc | 63 ± 13 (mRNA) |
| ppsA | 86 ± 10 (mRNA) |
| pta | 77 ± 1 (mRNA), 80 ± 8 (protein) |
| ptsG | 98 ± 0.3 (mRNA)d |
| pykA | 88 ± 2 (mRNA) |
| pykF | 82 ± 8 (mRNA) |
| rpe | 61 ± 8 (mRNA) |
| rpiA | 82 ± 2 (mRNA) |
| rpiB | 61 ± 9 (mRNA) |
| sdhC | 64 ± 6 (mRNA) |
| sucA | 87 ± 2 (mRNA) |
| sucC | 74 ± 2 (mRNA) |
| talA | 76 ± 13 (mRNA) |
| talB | 95 ± 1 (mRNA) |
| tktA | 84 ± 3 (mRNA) |
| tktB | 98 ± 0.1 (mRNA) |
| tpiA | 54 ± 7 (mRNA), 68 ± 5 (protein) |
| zwf | 76 ± 3 (mRNA), 88 ± 3 (protein) |
Numbers indicate the reductions (%) of mRNA amounts (mRNA) or the enzymatic activities (protein) compared to control cells that carry an empty vector (pHN1257). Data are given as means from triplicate experiments.
Defect in carbon catabolite repression, i.e., (6.2 ± 2.7)-fold higher activity of LacZ in the presence of IPTG and glucose than for the control.
Defect in carbon catabolite repression, i.e., (41 ± 4.6)-fold higher activity of LacZ in the presence of IPTG and glucose than for the control.
Defect in carbon catabolite repression, i.e., (8.6 ± 1.0)-fold higher activity of LacZ in the presence of IPTG and glucose than for the control.
The empty vector used to construct PTasRNA expression vectors was pHN1257 (see Materials and Methods). For target sequences, the ribosome-binding site (RBS) and start codon regions (typically 70 to 160 bp) were always chosen, because these regions are thought to be the most sensitive to silencing (21).
Validation of vectors.
To determine whether the constructed vectors could silence target genes with sufficient efficacy, the mRNA and protein levels of the target genes were measured. When a PTasRNA expression vector was shown to have inefficient silencing efficacy, the target sequence was changed by a few nucleotides, and then the vector was constructed again.
For all 71 genes, mRNA amounts remaining after silencing were determined by using real-time quantitative RT-PCR. It was possible to use this method for evaluation because, based on our previous results, target mRNAs are degraded upon silencing (9, 22). For all of the genes except ackA, ldhA, and tpiA, the silencing vectors reduced mRNA levels by at least 60% (Table 1).
The enzymatic activities of 14 gene products were measured directly in cell extracts (Table 1), and for all genes tested, activity decreased by at least 60%. For ackA, ldhA, and tpiA, mRNA levels decrease poorly, but the protein levels decrease efficiently, indicating the presence of silencing mechanisms that do not involve mRNA degradation. Probably, these PTasRNAs prevent ribosomes from recognizing the RBS and thus inhibit translation without mRNA degradation. Probably, these PTasRNAs prevent ribosomes from recognizing the RBS and thus inhibit translation without mRNA degradation. The precise silencing mechanisms used by artificial asRNAs and native small regulatory RNAs are still controversial (21).
Taken together, the results indicate that efficient silencing vectors were constructed for all 71 genes. During the study, however, it was necessary to remake the vectors for 18 of the genes. The PTasRNA expression vectors that had insufficient silencing efficacy are listed in Table S2 in the supplemental material, along with the validation results. Surprisingly, in the case of sucC silencing, an antisense sequence that was shorter than the original by only 5 nucleotides had a higher silencing efficacy than the original sequence (74% ± 2% versus 39% ± 16% reduction in mRNA, respectively; Table 1 and see Table S2 in the supplemental material). The secondary structures of both the sucC PTasRNAs were predicted using the STAR program (see Fig. S2 in the supplemental material) (23). The result indicated that the PT structures for both sucC PTasRNAs are conserved, but that the structure of the antisense sequence portion (the loop portion) is different. The secondary structures of efficient and inefficient PTasRNAs for acs, arcA, and cyoA were also compared, and showed similar results (see Fig. S3, S4, and S5 in the supplemental material). However, it was not possible to find uniform structural features among the efficient PTasRNAs; therefore, predicting antisense sequences with high silencing efficacy remains a challenge.
Phenotypes resulting from gene silencing.
To determine whether gene silencing could cause the same phenotypes as gene disruption, some known phenotypes were investigated after silencing with the constructed vectors.
The accA and gapA genes are annotated as “growth essential” (2, 20, 24). As expected, cells containing a PTasRNA expression vector targeting one of these genes showed severe growth inhibition on LB media containing IPTG (see Fig. S6 in the supplemental material). The eno, fbaA, and pgk genes are also annotated as growth essential, but silencing of these genes did not cause severe growth inhibition (data not shown); the silencing efficacies were 79 to 92% at the mRNA level (Table 1). Residual activity following silencing may be the reason for the difference between the silencing results and the disruption results, since it has been shown that the expression level required to support cell growth differs from gene to gene for the essential genes (22).
Disruption mutants of pfkA, pgi, or ptsG are known to express the lacZ gene in the presence of both glucose and lactose (or its analog, IPTG), although wild-type strains do not (25). This phenomenon in wild-type strains is known as carbon catabolite repression. Therefore, PTasRNA expression vectors for these genes were tested by observing whether they could relieve carbon catabolite repression in the presence of both glucose and IPTG. The results (Table 1) showed that carbon catabolite repression is in fact relieved upon expression of any one of these PTasRNAs. These results indicate that silencing with PTasRNAs can replicate the phenotypes seen in the corresponding disruptants.
Changes in metabolite accumulation and pyruvate production.
Next, we wanted to determine whether the vector set would be useful for obtaining information on metabolic engineering. Here, pyruvate was chosen as a suitable example to study because it is a valuable compound that is commonly used as the starting material for the production of pharmaceutical compounds, is a food additive, and is used in agriculture (26).
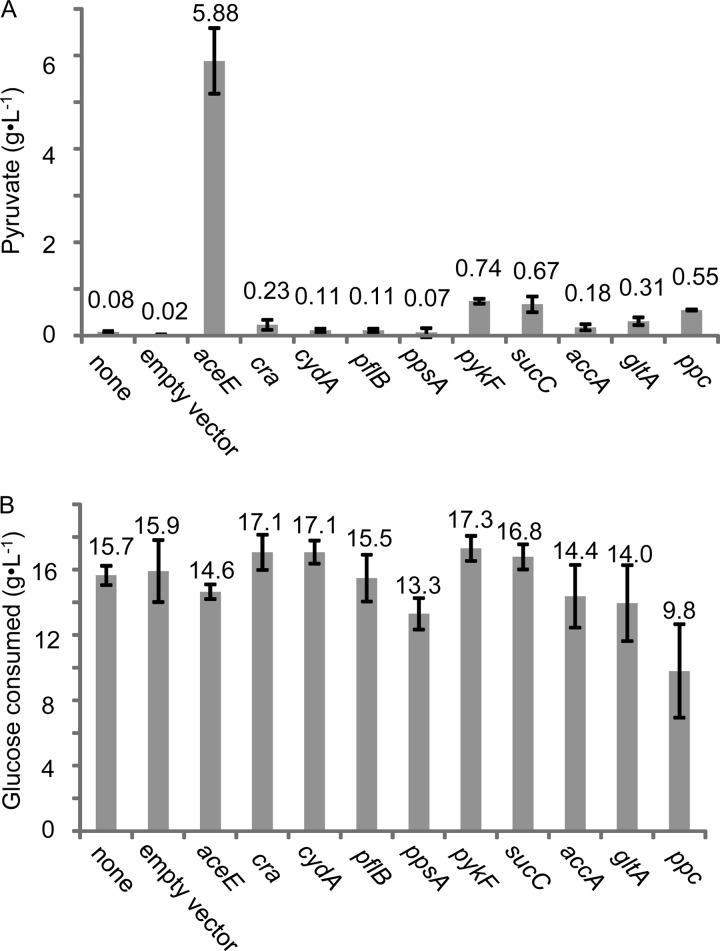
Each transformant was cultured for 24 h in N2G medium (see Materials and Methods), which is optimized for flask and aerobic production of pyruvate in the presence of IPTG. Metabolites accumulated in the medium were analyzed by HPLC. The first screening indicated that silencing of the following 10 genes increased pyruvate accumulation by more than ∼10-fold: accA (10-fold), aceE (427-fold), cra (10-fold), cydA (36-fold), gltA (15-fold), pflB (9.9-fold), ppc (39-fold), ppsA (13-fold), pykF (48-fold), and sucC (35-fold) (the full data set is available in Table S4 in the supplemental material). The data for these genes were taken from 3 separate experiments (Fig. 2), and each time pyruvate accumulation was confirmed, particularly for aceE silencing.
FIG 2.
Effect of gene silencing on pyruvate production. (A) Productivity of pyruvate in a 24-h culture is shown. A wild-type strain was used as a host, and silenced genes are shown under the graph. The terms “none” and “empty vector” indicate the results obtained with a nontransformant and a pHN1257 transformant, respectively. The data are given as means ± the standard deviations from triplicate experiments. (B) Glucose consumption in the same experiments as in panel A is shown.
The aceE gene product is a component of the pyruvate dehydrogenase (PDH) complex and mediates conversion of pyruvate to acetyl coenzyme A (acetyl-CoA) under aerobic conditions (27). A mutation in the PDH complex genes is known to cause the accumulation of pyruvate (28, 29). Therefore, in the next experiment, aceE silencing was fixed, and double silencing and silencing in combination with gene disruption were tested.
Of the 10 genes discussed above, ppc, gltA, and aceE are critical for growth in media containing glucose as a sole carbon source (20), and accA is growth essential under any conditions (2, 24); consequently, it would be desirable for metabolic engineering purposes for these 4 genes to be silenced rather than disrupted. Therefore, a new aceE PTasRNA expression vector for double silencing was constructed from a vector containing a chloramphenicol-resistance gene and the pACYC ori (pHN1464) (see Materials and Methods). This vector is compatible and cotransformable with pHN1257, which contains a kanamycin-resistance gene and the pSC101H ori. Plasmids carrying pACYC ori have a lower plasmid copy number than plasmids carrying pSC101H ori and therefore show lower silencing efficacy in general (6). However, in the case of aceE silencing, the efficacy was 71% ± 1% and 64% ± 9% at the mRNA level for the pHN1257- and pHN1464-based vectors, respectively, and the efficacy was >99% at the protein level for both vectors, indicating that both vectors worked efficiently. Note that, prior to this experiment, it had been confirmed that the silencing efficacy of a silencing vector is not affected by it being cotransformed with another silencing vector (6). In addition, in the double silencing experiments, 24 h after inoculation, 5 M NaOH was added to raise the pH to 6.8, after which the cells were cultured for an additional 24 h.
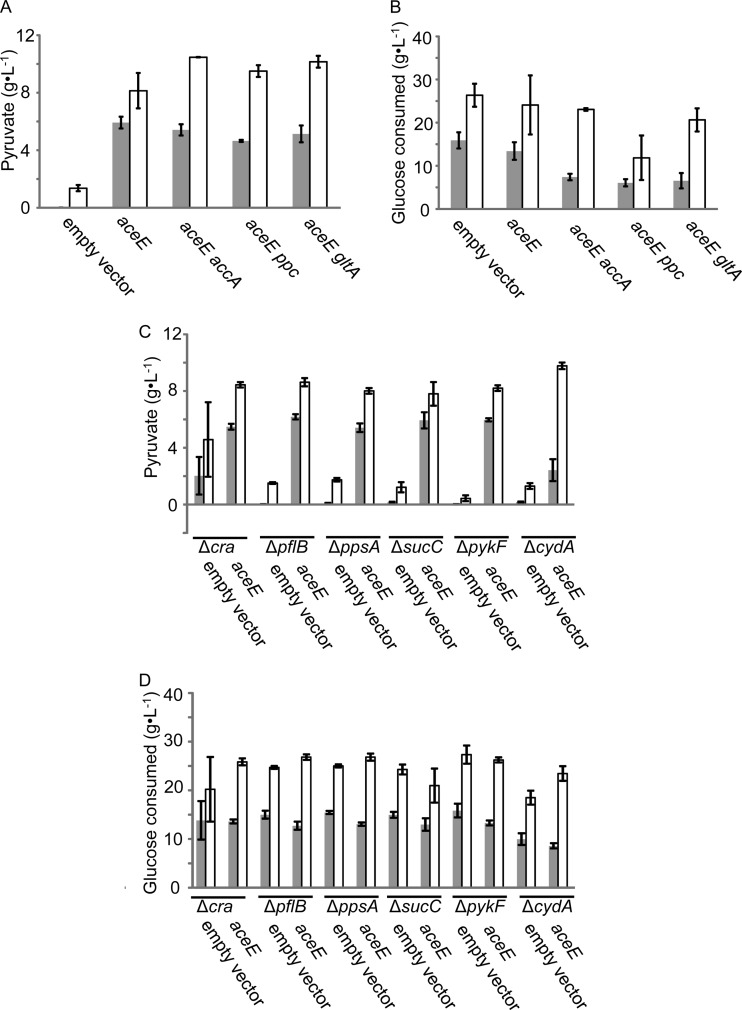
The results showed that double silencing of aceE with either accA, ppc, or gltA resulted in higher productivity of pyruvate than did aceE silencing alone (Fig. 3A) and that double silencing of aceE and accA resulted in the highest productivity (10.5 ± 0.02 g·liter−1). Glucose consumption was low in the aceE and ppc double-silenced strain compared to the other double-silenced strains (Fig. 3B), leading to a high theoretical yield.
FIG 3.
Effect of gene silencing and disruption, together with aceE silencing on pyruvate production. (A and C) Productivity of pyruvate in a 24-h ( ) and 48-h (□) culture. A wild-type strain was used as a host in panel A, and disruptants of indicated genes (with “Δ”) were used in panel C. Silenced genes are indicated at the bottom of the graphs. The data are given as means ± the standard deviations from triplicate experiments. (B and D) Glucose consumption in the same experiments as in panels A and C is shown.
) and 48-h (□) culture. A wild-type strain was used as a host in panel A, and disruptants of indicated genes (with “Δ”) were used in panel C. Silenced genes are indicated at the bottom of the graphs. The data are given as means ± the standard deviations from triplicate experiments. (B and D) Glucose consumption in the same experiments as in panels A and C is shown.
Since the remaining six genes were not critical for growth, they were disrupted and aceE was silenced, and pyruvate productivity was investigated as described above (Fig. 3C and D). Surprisingly, pyruvate did not accumulate significantly in pflB-, ppsA-, sucC-, pykF-, and cydA-disrupted strains carrying the empty vector (Fig. 3C). This result is contradictory to the silencing result shown in Fig. 2. The reason for this is unclear, but residual activity after silencing might affect.
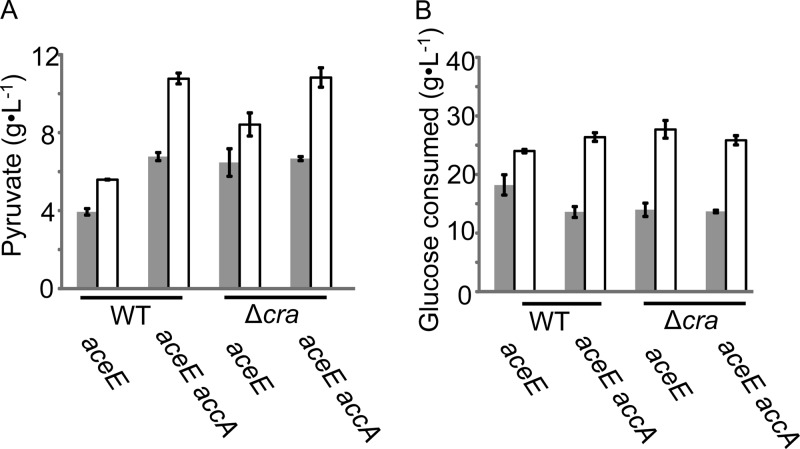
We also silenced both the aceE and accA genes in cra-disrupted strains. As hosts, the wild-type and Δcra strains were chosen here, because disruption of cra led to a more rapid rate of pyruvate production (although the final productivity was unchanged) (Fig. 3). However, no increase in pyruvate productivity was observed (Fig. 4). The ΔcydA strain showed fine productivity of pyruvate (Fig. 3A) but was not used here, because it was temperature sensitive (see Materials and Methods).
FIG 4.
Effect of aceE and accA silencing on pyruvate production in a wild-type (WT) or Δcra strain. (A) Pyruvate yields after 24 h ( ) and 48 h (□). Silenced genes are indicated at the bottom of the graphs. The data are given as means ± the standard deviations from triplicate experiments. (B) Glucose consumption in the same experiments as in panel A.
) and 48 h (□). Silenced genes are indicated at the bottom of the graphs. The data are given as means ± the standard deviations from triplicate experiments. (B) Glucose consumption in the same experiments as in panel A.
Pyruvate production with a jar fermentor.
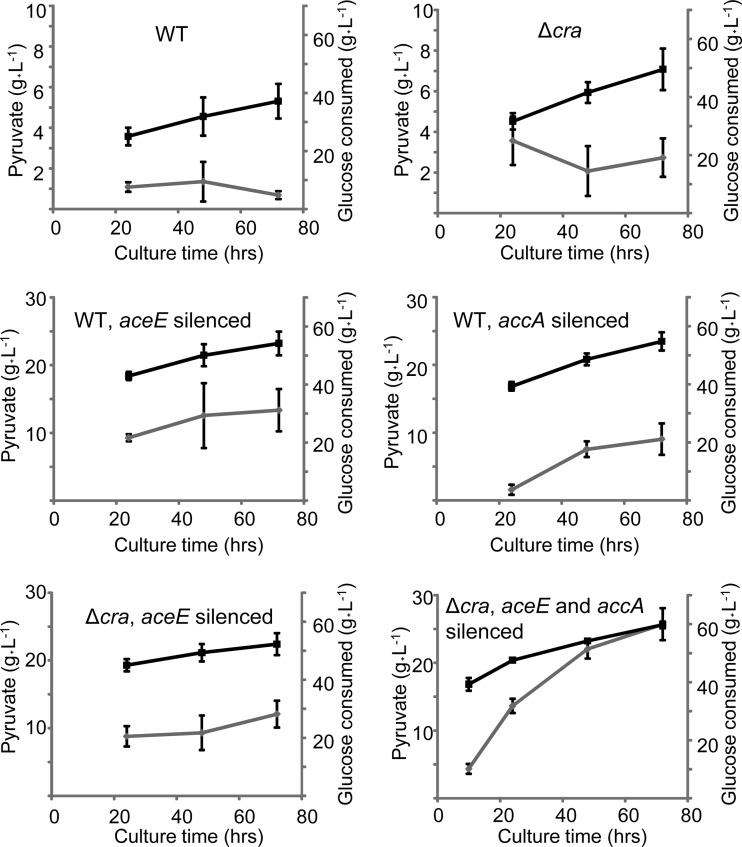
We attempted to produce pyruvate aerobically in a jar fermentor in which high glucose was used as the carbon source and pH was maintained at 6.3 to 6.8 throughout the culture. After 72 h, the largest amount of pyruvate (25.7 ± 2.4 g liter−1) was produced by a strain in which cra was disrupted and both aceE and accA were silenced (Fig. 5). In contrast to the results seen with flask cultures (Fig. 4), cra disruption, aceE silencing, and accA silencing were all found to be necessary to achieve the highest production of pyruvate. A possible explanation for this result is that in the flask cultures, glucose consumption is low due to a limited oxygen supply; therefore, pyruvate production reaches a plateau even though the cells have further production capacity.
FIG 5.
Pyruvate production in a jar fermentor. The amounts of pyruvate produced and glucose consumed are indicated by gray and black lines, respectively. As a host, a wild-type (WT) or Δcra strain was used. Silenced genes (if any) are indicated in each panel.
In silico simulation of pyruvate production and its comparison to the silencing result.
To test whether the experimental data on pyruvate production agrees with in silico metabolic simulation data, the GSM E. coli K-12 MG1655 iAF1260 (18) was used, and FBA was performed. FBA predicts metabolic flux distributions under the assumption that the metabolism of a cell is at steady state and is regulated to maximize growth rate (4, 5). When pyruvate production is predicted after a single-gene disruption, pyruvate production is expected to lead to ATP generation and/or adjustment of the NAD+/NADH ratio, thereby maximizing the growth rate.
For a wild-type strain and all single-gene disruptants, no pyruvate production was predicted under aerobic conditions [limit of oxygen uptake rate, 20 mmol g (dry cell weight)−1 h−1]. Among all combinations of double-gene disruptants, only the aceE pflB double disruptant was predicted to produce pyruvate at 0.11 g of pyruvate g of glucose−1. Both AceE and PflB catalyze the conversion of pyruvate to acetyl-CoA, but PflB is known to work very poorly under aerobic conditions (30, 31). Given that PflB was largely inactive in our experimental conditions, the experimental and simulation data are consistent. For further metabolic simulations, the aceE pflB double disruptant was treated as identical to the aceE disruptant.
Next, using the genes shown in Fig. 2, the pyruvate yield was further simulated in two situations. In one situation, all of the genes were disrupted, and in the other, ppc, gltA, and aceE were silenced with the remaining six genes disrupted. For simulation of the gene disruptants, maximum and minimum flux values for the corresponding reaction were set to zero. For simulation of the gene silenced strains, taking into consideration silencing efficiencies (Table 1), the maximum flux of the corresponding reaction was set to the following values (compared to the flux calculated by FBA in a wild-type strain): aceE, 3%; accA, 3%; ppc, 37%; and gltA, 5%. The simulated pyruvate yields are shown in Table 2, along with the experimental pyruvate yields calculated from the result shown in Fig. 3. All simulation results used in Table 2 are summarized in Data Set S1 in the supplemental material. The results indicated that pyruvate production was predicted in all strains in which aceE was silenced or disrupted. When the limit of oxygen uptake rate was set to 20 or 15 mmol g (dry cell weight)−1 h−1, the simulation data showed good agreement with the experimental data (Table 2). In addition, pyruvate yields were predicted to increase in the aceE accA, aceE ppc, and aceE gltA double-silenced strains, which is consistent with the results shown in Fig. 3. Disruption of ppsA, sucC, pykF, and cydA were predicted not to affect pyruvate yield, which is also consistent with the experimental data.
TABLE 2.
Comparison of experimental and simulation dataa
| Experimental data |
Simulation datab |
||||
|---|---|---|---|---|---|
| Gene silenced or disrupted | Pyruvate yield (g of pyruvate g of glucose−1) | Gene disruption | Pyruvate yield (g of pyruvate g of glucose−1) | Gene silenced or disrupted | Pyruvate yield (g of pyruvate g of glucose−1) |
| None | 0.05 | 0 | 0 | ||
| aceE | 0.34 | ΔaceE | 0.34 | aceE | 0.33 |
| aceE accA | 0.45 | ΔaceE ΔaccA | Lethal | aceE accA | 0–1.02 |
| aceE ppc | 0.80 | ΔaceE Δppc | 0.37 | aceE ppc | 0.35 |
| aceE gltA | 0.49 | ΔaceE ΔgltA | Lethal | aceE gltA | 0–0.93 |
| aceE Δcra | 0.33 | ΔaceE Δcra | – | aceE Δcra | – |
| aceE ΔpflB | 0.32 | ΔaceE ΔpflB | 0.34 | aceE ΔpflB | 0.33 |
| aceE ΔppsA | 0.30 | ΔaceE ΔppsA | 0.34 | aceE ΔppsA | 0.33 |
| aceE ΔsucC | 0.37 | ΔaceE ΔsucC | 0.34 | aceE ΔsucC | 0.33 |
| aceE ΔpykF | 0.31 | ΔaceE ΔpykF* | 0.34 | aceE ΔpykF* | 0.33 |
| aceE ΔcydA | 0.42 | ΔaceE ΔcydA* | 0.34 | aceE ΔcydA* | 0.33 |
Disrupted genes are indicated by a “Δ” symbol, and silenced genes are shown without a “Δ” symbol. The pyruvate yield was calculated from the data shown in Fig. 3. “ΔaceE” represents the disruption of both aceE and pflB in these columns (see the text).
The data with the limit of oxygen uptake rate set to 15 mmol·g dry cell weight−1·h−1 is shown. Lethal, the accA and gltA genes were predicted as growth essential. –, the cra gene is not included in the iAF1260. *, for pykF and cydA, isozyme genes are known, and the pyruvate yields in the simulation were calculated as though all isozyme genes were disrupted.
DISCUSSION
The antisense silencing approach shown here is advantageous over the gene disruption approach for large-scale screening and manipulating growth-critical genes. However, the silencing approach is not always advantageous. We believe that gene disruption is much more suitable for manipulating small numbers of genes and genes that are not critical for growth. The weakest point of antisense silencing is that complete removal of gene function is impossible, and therefore, residual function may lead to misinterpretation of the results. Furthermore, it is known that the presence of multicopy plasmids places a “metabolic burden” on cells caused by the increased demand for nucleotides for plasmid replication and plasmid gene translation (32). Indeed, in the pyruvate production experiment described here, we used the silencing approach for narrowing down the number of genes; however, after this, the small number of genes that were not growth critical was disrupted but not silenced (Fig. 3, 4, and 5). We believe that, depending on the purpose of experiments, combining the antisense silencing and gene disruption approaches is very important.
When a gene forms an operon with one or more other genes, silencing of the gene may affect the expression of adjacent genes undesirably. In our previous study on an operon containing ackA and pta (in that order), silencing of the first gene, ackA, caused 68 and 41% reductions in AckA and Pta activities, respectively, and silencing of the second gene, pta, caused 79 and 7% reductions in Pta and AckA activities, respectively (6). A similar polar effect by antisense silencing has been reported elsewhere (33, 34). Of the 71 genes targeted here, 31 genes clearly form operons. Among them, aceE (35), atpF (36), cydA (37), cyoA (37), frdA (38), and nuoA (39) encode one component of multisubunit enzymes and form operons with other components. Therefore, the operon problem is unlikely to affect the interpretation of our results for these genes. The sucA, sucC, and sdhC genes are located in close proximity to each other and form several types of polycistronic mRNA species, such as sdhCDAB-sucABCD, sdhCDAB, sucABCD, sucAB, and sucCD (40). Similarly, the eno gene is transcribed as both a monocistronic mRNA and a polycistronic mRNA with pyrG (encoding a subunit of CTP synthase) (41). Therefore, when targeting a gene within an operon, data should be interpreted carefully depending on the nature of the operon. However, the production of similar undesirable effects on adjacent genes is a serious issue in gene disruption as well. As an example, disruption of the ackA gene caused a 31% reduction of Pta activity even though the pta gene was kept intact, and disruption of the pta gene caused a 38% reduction of AckA activity (6).
Through comprehensive silencing of 71 genes, we identified 10 genes to disrupt or silence for enhanced pyruvate production (Fig. 2). It has been shown that cra disruption causes pyruvate accumulation, which is caused by the increased transcription of glycolysis genes, since transcription from such genes is negatively regulated by Cra (42). The cydA gene encodes cytochrome bd-I terminal oxidase subunit I (43). Silencing of cydA probably causes an increase in NADH, which is a competitive inhibitor of the PDH complex (43), leading to pyruvate accumulation in the same way that aceE silencing does. Also, silencing of cyoA which encodes the other cytochrome oxidase bo led to pyruvate accumulation, although the accumulation amount was one fourth compared to silencing of cydA (see Table S4 in the supplemental material). Regarding pykF, this gene encodes pyruvate kinase and seems to be necessary for pyruvate production, but pykF silencing was actually found to increase pyruvate production (Fig. 2). A possible explanation for this result is that pykF silencing increases carbon flux into pyruvate through the pentose phosphate pathway (PPP). Consistent with this possibility, in the pykF− mutant, the flux through PPP has been shown to increase (44). Alternatively, because the metabolic flux analysis showed the acceleration of carbon flux from phosphoenolpyruvate to pyruvate via Ppc, Mdh, and MaeAB in the pykF− mutant, this pathway may be used in the pykF-silenced strain to accumulate pyruvate (44).
Silencing experiments with accA, ppc, and gltA demonstrated the advantages of using this technology, since these genes are growth critical or critical in media containing glucose as a sole carbon source. In the metabolic simulation experiment, the pyruvate yield is predicted to increase in the aceE accA, aceE ppc, and aceE gltA double-silenced strains (Table 2), which is consistent with the experimental result (Fig. 3). The accA gene product catalyzes the first step of fatty acid synthesis from acetyl-CoA, which is essential for cell growth (45). In the aceE accA double-silenced strain, a metabolic state was predicted in which excessive carbon would be secreted as pyruvate due to impaired fatty acid synthesis. Similarly, in the aceE gltA double-silenced strain, pyruvate production was predicted to be a consequence of the emission of excessive carbon. This was because gltA silencing inhibited the production of metabolites within the TCA cycle (GltA catalyzes the first step of the TCA cycle), as well as the production of further metabolites such as glutamate and asparagine (46). As for the aceE ppc double-silenced strain, the mechanism of increase in pyruvate production by FBA was considered to be as follows: aceE silencing increased the flux of the reactions catalyzed by pyruvate oxidase (PoxB) and acetyl-CoA synthetase (Acs) to produce acetyl-CoA; since Acs consumes ATP as a cofactor (47), this reaction was disadvantageous for growth, and flux into the TCA cycle and NADH production decreased; instead, NADH required for the respiratory chain to produce ATP was supplied by the transhydrogenase reaction from NADPH, which is produced in the PPP. Since NADH oxidation by the respiratory chain is dependent on oxygen supply, the limitation of oxygen uptake decreased the flux of PPP to balance the NAD+/NADH ratio. This resulted in an increase of the flux into glycolysis and pyruvate production in the aceE-silenced strain, as shown in Table 2. The silencing of ppc in the aceE-silenced strain increased the flux of the glyoxylate pathway and the reaction of malate dehydrogenase (Mdh) to provide oxaloacetate required for biomass synthesis. Increase of NADH production by Mdh decreased the flux of PPP to balance the NAD+/NADH ratio. Consequently, pyruvate production was predicted to increase in the aceE ppc double-silenced strain.
The final pyruvate productivity reported here (25.7 ± 2.4 g liter−1) is similar to that obtained from a previous study (25.5 g liter−1) using a lipoic acid auxotroph mutant (lipoic acid is a cofactor for the PDH) (29) and is less than the productivity obtained using a PDH mutant (35 g liter−1) (28). However, in these preceding results, expensive compounds such as yeast extract, polypeptone, lipoic acid (for lipoic acid auxotroph strains), acetate (PDH mutants require acetate), and/or succinate must be included in the media for maximum productivities. In our study, only inexpensive inorganic compounds (except for glucose) were used. Causey et al. reported the production of 52 g of pyruvate liter−1 using a pflB, frdBC, ldhA, atpFH, adhE, sucA, poxB, and ackA mutant strain in a medium containing only inexpensive inorganic compounds and glucose (26). In that study, the fermentation process was regulated intricately; the dissolved oxygen level was strictly controlled depending on growth phase using an O2-air mixture. We expect that our strain can be engineered further and production processes optimized to increase productivity.
We also simulated the production of fumarate, which is an intermediate metabolite in the TCA cycle. In all single-gene disruptants, no fumarate production was predicted. In strains in which accA, gapA, acnB, gltA, icdA, or acnA was silenced, a high fumarate yield was predicted, but the yields varied from 0 to 1.11 g of pyruvate g of glucose−1 (indefinite results). From the experimental results (see Table S4 in the supplemental material), silencing of adhE (0.00026 g of pyruvate g of glucose−1), mqo (0.00022 g of pyruvate g of glucose−1), pykF (0.00020 g of pyruvate g of glucose−1), fumC (0.00020 g of pyruvate g of glucose−1), aceA (0.00019 g of pyruvate g of glucose−1), and talB (0.00019 g of pyruvate g of glucose−1) increased fumarate yield (wild type; 0.00013 g of pyruvate g of glucose−1). However, the experimental yields of all strains were very low in consistent with the simulation result. According to the simulation of double disruption, increase of fumarate yield was maximally predicted in frdA fumA, frdA fumB, and frdA fumC disrupted strains as definite results, 0.06 g of pyruvate g of glucose−1. Therefore, in this case, silencing tests with the 71 vectors might have to be carried out in a strain in which frdA, fumA, fumB, and fumC had been disrupted beforehand so as to raise fumarate productivity. This point should be confirmed in further studies.
Recently, the in silico metabolic network of the MG1655 strain was updated to iJO1366, which contains 1366 genes (48). We are continuing to construct PTasRNA expression vectors to experimentally verify the number of genes in iJO1366. The expanded vector set should allow integration of information obtained in silico and in vivo, thereby accelerating metabolic engineering for bioproduction. In the future, we aim to construct PTasRNA expression vectors for all E. coli genes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Yuya Murakami and Kazuhiro Takahashi for technical assistance and to Liam Good and Shan Goh for valuable discussions.
This study was supported in part by KAKENHI (23780096 and 24246134) and by the Kato Memorial Bioscience Foundation.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 8 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02376-13.
REFERENCES
- 1.Keasling JD. 2012. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 14:189–195. 10.1016/j.ymben.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 2.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006–2008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, Mori H. 2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335. 10.1038/msb.2009.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis NE, Nagarajan H, Palsson BØ. 2012. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 10:291–305. 10.1038/nrmicro2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zomorrodi AR, Suthers PF, Ranganathan S, Maranas CD. 2012. Mathematical optimization applications in metabolic networks. Metab. Eng. 14:672–686. 10.1016/j.ymben.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 6.Nakashima N, Tamura T. 2009. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res. 37:e103. 10.1093/nar/gkp498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakashima N, Tamura T, Good L. 2006. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 34:e138. 10.1093/nar/gkl697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majewski RA, Domach MM. 1990. Simple constrained-optimization view of acetate overflow in Escherichia coli. Biotechnol. Bioeng. 35:732–738. 10.1002/bit.260350711 [DOI] [PubMed] [Google Scholar]
- 9.Nakashima N, Goh S, Good L, Tamura T. 2012. Multiple-gene silencing using antisense RNAs in Escherichia coli. Methods Mol. Biol. 815:307–319. 10.1007/978-1-61779-424-7_23 [DOI] [PubMed] [Google Scholar]
- 10.Nakashima N, Tamura T. 2012. A new carbon catabolite repression mutation of Escherichia coli, mlc*, and its use for producing isobutanol. J. Biosci. Bioeng. 114:38–44. 10.1016/j.jbiosc.2012.02.029 [DOI] [PubMed] [Google Scholar]
- 11.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447–1457. 10.1111/j.1365-2958.1991.tb00791.x [DOI] [PubMed] [Google Scholar]
- 13.Emmerson JR, Gally DL, Roe AJ. 2006. Generation of gene deletions and gene replacements in Escherichia coli O157:H7 using a temperature sensitive allelic exchange system. Biol. Proc. Online 8:153–162. 10.1251/bpo123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng L, Shimizu K. 2003. Global metabolic regulation analysis for Escherichia coli K-12 based on protein expression by 2-dimensional electrophoresis and enzyme activity measurement. Appl. Microbiol. Biotechnol. 61:163–178. 10.1007/s00253-002-1202-6 [DOI] [PubMed] [Google Scholar]
- 15.Fatania HR, al-Nassar KE, Thomas N. 1993. Chemical modification of rat liver cytosolic NADP(+)-linked isocitrate dehydrogenase by N-ethylmaleimide: evidence for essential sulphydryl groups. FEBS Lett. 322:245–248 [DOI] [PubMed] [Google Scholar]
- 16.Serrano JA, Camacho M, Bonete MJ. 1998. Operation of glyoxylate cycle in halophilic archaea: presence of malate synthase and isocitrate lyase in Haloferax volcanii. FEBS Lett. 434:13–16. 10.1016/S0014-5793(98)00911-9 [DOI] [PubMed] [Google Scholar]
- 17.Lal SK, Johnson S, Conway T, Kelley PM. 1991. Characterization of a maize cDNA that complements an enolase-deficient mutant of Escherichia coli. Plant Mol. Biol. 16:787–795. 10.1007/BF00015071 [DOI] [PubMed] [Google Scholar]
- 18.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3:121. 10.1038/msb4100155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinfuku Y, Sorpitiporn N, Sono M, Furusawa C, Hirasawa T, Shimizu H. 2009. Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb. Cell Fact. 8:43. 10.1186/1475-2859-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Copley SD. 2007. Why metabolic enzymes are essential or nonessential for growth of Escherichia coli K-12 on glucose. Biochemistry 46:12501–12511. 10.1021/bi7014629 [DOI] [PubMed] [Google Scholar]
- 21.Stefan A, Schwarz F, Bressanin D, Hochkoeppler A. 2010. Shine-Dalgarno sequence enhances the efficiency of lacZ repression by artificial anti-lac antisense RNAs in Escherichia coli. J. Biosci. Bioeng. 110:523–528. 10.1016/j.jbiosc.2010.05.012 [DOI] [PubMed] [Google Scholar]
- 22.Goh S, Boberek JM, Nakashima N, Stach J, Good L. 2009. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One 4:e6061. 10.1371/journal.pone.0006061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrahams JP, Vandenberg M, Vanbatenburg E, Pleij C. 1990. Prediction of RNA secondary structure, including pseudoknotting, by computer-simulation. Nucleic Acids Res. 18:3035–3044. 10.1093/nar/18.10.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN, Osterman AL. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673–5684. 10.1128/JB.185.19.5673-5684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimata K, Tanaka Y, Inada T, Aiba H. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20:3587–3595. 10.1093/emboj/20.13.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Causey TB, Shanmugam KT, Yomano LP, Ingram LO. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc. Natl. Acad. Sci. U. S. A. 101:2235–2240. 10.1073/pnas.0308171100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quail MA, Haydon DJ, Guest JR. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95–104. 10.1111/j.1365-2958.1994.tb00998.x [DOI] [PubMed] [Google Scholar]
- 28.Tomar A, Eiteman MA, Altman E. 2003. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 62:76–82. 10.1007/s00253-003-1234-6 [DOI] [PubMed] [Google Scholar]
- 29.Yokota A, Terasawa Y, Takaoka N, Shimizu H, Tomita F. 1994. Pyruvic acid production by an F1-ATPase-defective mutant of Escherichia coli W1485lip2. Biosci. Biotechnol. Biochem. 58:2164–2167. 10.1271/bbb.58.2164 [DOI] [PubMed] [Google Scholar]
- 30.Hasona A, Kim Y, Healy FG, Ingram LO, Shanmugam KT. 2004. Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J. Bacteriol. 186:7593–7600. 10.1128/JB.186.22.7593-7600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knappe J, Blaschkowski HP, Gröbner P, Schmitt T. 1974. Pyruvate formate-lyase of Escherichia coli: acetyl-enzyme intermediate. Eur. J. Biochem. 50:253–263. 10.1111/j.1432-1033.1974.tb03894.x [DOI] [PubMed] [Google Scholar]
- 32.Seo JH, Bailey JE. 1985. Effects of recombinant plasmid content on growth properties and cloned gene product formation in Escherichia coli. Biotechnol. Bioeng. 27:1668–1674. 10.1002/bit.260271207 [DOI] [PubMed] [Google Scholar]
- 33.Dryselius R, Nikravesh A, Kulyté A, Goh S, Good L. 2006. Variable coordination of cotranscribed genes in Escherichia coli following antisense repression. BMC Microbiol. 6:97. 10.1186/1471-2180-6-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestka S, Daugherty BL, Jung V, Hotta K, Pestka RK. 1984. Anti-mRNA: specific inhibition of translation of single mRNA molecules. Proc. Natl. Acad. Sci. U. S. A. 81:7525–7528. 10.1073/pnas.81.23.7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer ME, Guest JR. 1985. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol. Gen. Genet. 200:145–154. 10.1007/BF00383328 [DOI] [PubMed] [Google Scholar]
- 36.Kasimoglu E, Park SJ, Malek J, Tseng CP, Gunsalus RP. 1996. Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J. Bacteriol. 178:5563–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cotter PA, Chepuri V, Gennis RB, Gunsalus RP. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng CP, Albrecht J, Gunsalus RP. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bongaerts J, Zoske S, Weidner U, Unden G. 1995. Transcriptional regulation of the proton-translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol. 16:521–534. 10.1111/j.1365-2958.1995.tb02416.x [DOI] [PubMed] [Google Scholar]
- 40.Cunningham L, Guest JR. 1998. Transcription and transcript processing in the sdhCDAB-sucABCD operon of Escherichia coli. Microbiology 144:2113–2123. 10.1099/00221287-144-8-2113 [DOI] [PubMed] [Google Scholar]
- 41.Weng M, Makaroff CA, Zalkin H. 1986. Nucleotide sequence of Escherichia coli pyrG encoding CTP synthetase. J. Biol. Chem. 261:5568–5574 [PubMed] [Google Scholar]
- 42.Sakai T, Nakamura N, Umitsuki G, Nagai K, Wachi M. 2007. Increased production of pyruvic acid by Escherichia coli RNase G mutants in combination with cra mutations. Appl. Microbiol. Biotechnol. 76:183–192. 10.1007/s00253-007-1006-9 [DOI] [PubMed] [Google Scholar]
- 43.Kim Y, Ingram LO, Shanmugam KT. 2008. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol. 190:3851–3858. 10.1128/JB.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. 2004. Effect of a pyruvate kinase (pykF− gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 235:25–33. 10.1111/j.1574-6968.2004.tb09563.x [DOI] [PubMed] [Google Scholar]
- 45.Li SJ, Cronan JE. 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 175:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilde RJ, Guest JR. 1986. Transcript analysis of the citrate synthase and succinate dehydrogenase genes of Escherichia coli K-12. J. Gen. Microbiol. 132:3239–3251 [DOI] [PubMed] [Google Scholar]
- 47.Kumari S, Beatty CM, Browning DF, Busby SJW, Simel EJ, Hovel-Miner G, Wolfe AJ. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173–4179. 10.1128/JB.182.15.4173-4179.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ. 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol. Syst. Biol. 7:535. 10.1038/msb.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.