Abstract
Natural transformation has a large impact on lateral gene flow and has contributed significantly to the ecological diversification and adaptation of bacterial species. Thermus thermophilus HB27 has emerged as the leading model organism for studies of DNA transporters in thermophilic bacteria. Recently, we identified a zinc-binding polymerization nucleoside triphosphatase (NTPase), PilF, which is essential for the transport of DNA through the outer membrane. Here, we present genetic evidence that PilF is also essential for the biogenesis of pili. One of the most challenging questions was whether T. thermophilus has any depolymerization NTPase acting as a counterplayer of PilF. We identified two depolymerization NTPases, PilT1 (TTC1621) and PilT2 (TTC1415), both of which are required for type IV pilus (T4P)-mediated twitching motility and adhesion but dispensable for natural transformation. This suggests that T4P dynamics are not required for natural transformation. The latter finding is consistent with our suggestion that in T. thermophilus, T4P and natural transformation are linked but distinct systems.
INTRODUCTION
DNA transport machineries of natural transformation systems are related to type IV pili (T4P) and type II protein secretion systems (T2SSs) and sometimes even share components (1–5). The components of natural transformation systems in Gram-negative bacteria are hypothesized to form distinct subcomplexes: a DNA translocator rod comprising pilin-like subunits that extends from the cytoplasmic membrane through the periplasm and the outer membrane; a shaft, made up of multiple copies of the secretin protein, that guides the rod through the outer membrane; a cytoplasmic motor complex that powers assembly of the rod; and a DNA translocator subcomplex mediating the transport of the DNA through the cytoplasmic membrane (3, 6–9).
The finding that DNA translocators and T4P share components led to the following two hypotheses: one explanation for the dual function of components in both systems is that DNA is bound to the T4P and retraction of the dynamic T4P leads to the transport of DNA across the outer membrane. However, several findings are not consistent with this hypothesis, such as the fact that the expression of pilus and natural transformation do not always correlate (10–12). A second hypothesis is that the transformation machinery represents an alternative system: DNA binds to the tip of a pilus-like DNA translocator rod. Then, the translocator, guided through a secretin pore through the outer membrane, retracts, thus transporting DNA into the periplasm, where it is bound by DNA binding proteins and subsequently transferred to an inner membrane DNA transporter subcomplex (3).
To elucidate structural and functional aspects of DNA transporters, we have chosen the natural transformation system of the thermophilic bacterium Thermus thermophilus, which exhibits the highest natural transformation frequencies known to date (13, 14). A genetic screen led to the identification of 16 competence genes essential for natural transformation of T. thermophilus (3). Some of them, such as the secretin PilQ, the pilin PilA4, and the inner and outer membrane proteins PilM, PilN, PilO, and PilW, were found to play a dual role in piliation and transformation, whereas others, such as the traffic nucleoside triphosphatase (NTPase) PilF and the pilins PilA1 to PilA3, were found to be essential only for natural transformation (15, 16).
Extension and retraction of pili and the pilus-like DNA translocator rods obviously require energy, which is supplied by ATP hydrolysis and catalyzed by traffic NTPases belonging to the PilF/PilB (pilus extension/assembly) and PilT/PilU (pilus retraction) subfamilies (17, 18). Members of these two subfamilies are known to power T4P and/or DNA translocators in many different Gram-negative bacteria, such as Pseudomonas aeruginosa, Neisseria meningitidis, Neisseria gonorrhoeae, Myxococcus xanthus, Francisella tularensis, and Pseudomonas stutzeri (19–28). Despite their broad distribution in T4P and DNA transporter systems, analysis of their structure and function is still in its infancy.
In T. thermophilus, so far only one traffic ATPase, PilF, was identified to be essential for natural transformation (29). PilF is similar to traffic NTPases of the PilF/PilB subfamily, which catalyze pilus extension (17, 21, 30–33). However, PilF differs from all other members of this subfamily by its bipartite structure with an unusually long N terminus comprising three GSPII domains (Fig. 1A). The N termini of traffic ATPases are suggested to mediate stable interactions with other proteins of T4P (34, 35), whereas the C termini harbor the ATP-binding sites and a conserved tetracysteine motif (33, 36, 37). The latter was found to be essential for Zn2+ binding of PilF (33, 36).
FIG 1.
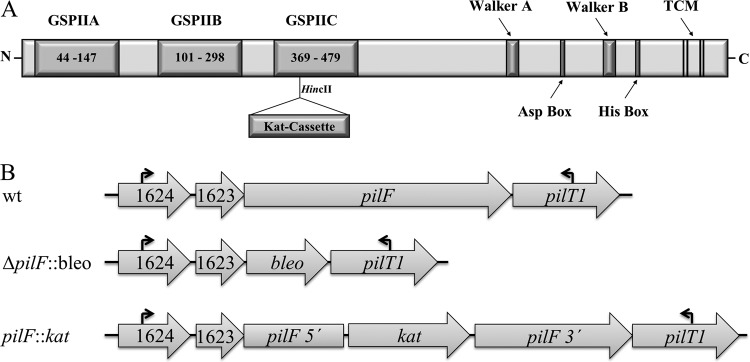
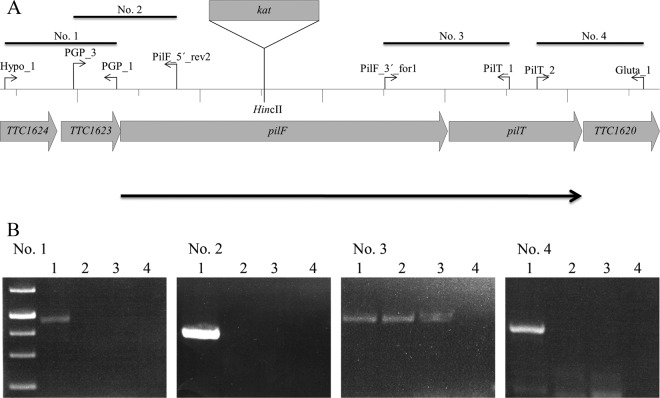
(A) Domain organization and kanamycin cassette insertion site in PilF; (B) organization of the pilF wild-type (wt) and mutant loci.
Interestingly, a pilF insertion mutant was unaffected in piliation but lost its transformation phenotype (29). This finding was puzzling, since members of the PilF/PilB subfamily often exhibit a dual function in both natural transformation and piliation (2) and are thought to power T4P rod extension and polymerization of the DNA translocator shaft.
The objective of this study was to investigate the role of the competence protein PilF in T4P biogenesis and function and to search for potential depolymerization NTPases acting as counterplayers of the polymerization ATPase PilF. We discovered that full-length PilF is essential for T4P biogenesis. Two depolymerization ATPases, PilT1 (TTC1621) and PilT2 (TTC1415), were identified to be essential for T4P-mediated twitching motility and adhesion. Together with the hyperpiliation phenotype of pilT1 and pilT2 mutants, this provides evidence that PilT1 and PilT2, essential for T4P dynamics, are the counterplayers of PilF. Interestingly, the pilT mutants and the pilT double mutants had no defect in transformation, indicating that T4P depolymerization is not required for natural transformation.
MATERIALS AND METHODS
Organisms and cultivation.
Escherichia coli cells were grown under standard conditions at 37°C in LB complex medium. Antibiotics were added when appropriate (100 μg/ml hygromycin B and/or 100 μg/ml ampicillin). T. thermophilus HB27 was grown at 68°C on TM+ complex medium (containing 8 g/liter tryptone, 4 g/liter yeast extract, 3 g/liter NaCl, 0.6 mM MgCl2, and 0.17 mM CaCl2) or at 60°C when hygromycin was added. Antibiotics were added when appropriate (bleomycin at 5 μg/ml or 15 μg/ml, hygromycin at 100 μg/ml, or kanamycin at 20 μg/ml or 40 μg/ml) in liquid and solid medium.
Generation of a ΔTTC1415 (pilT2)::hygro deletion mutant.
For the construction of a ΔTTC1415::hygro mutant, the 723-bp upstream region and the 806-bp downstream region of TTC1415 were amplified using the primers panto_for1_EcoRI (GGATTAGAATTCCTTCGTGGTGGTCTCCGTCTTCGTCAAC), panto_rev1_XbaI (ATTATCTAGACTCATGGGTAGACCTCCAGGTTGTCTATAAG), fructo_for1_PstI (TAATCTGCAGCGCCCTTTCCCTTGCCCGCCTGGCCC), and fructo_rev1_HindIII (ATTGAAGCTTGGCCCGGAAGCGCTCCACGAGCTCTTG). The hygromycin cassette was amplified from the vector pH118 (38) using the primers Hyg_XbaI (AGCCGGAGTCTAGACCCGGGAGTATAAC) and Hyg_rev2_PstI (AGGCGGCTGCAGACGTTCAAAATGGTATGCGTTTTGACAC). The DNA fragments were cloned into the vector pUC19, resulting in pUC19-ΔTTC1415_Hyg. To generate a ΔTTC1415 mutant, pUC19-ΔTTC1415_Hyg was linearized with PciI and transformed into T. thermophilus HB27. Transformants were selected at 60°C on TM+ medium with 100 µg hygromycin/ml. The mutants were verified by PCR, using the primers Ü_1415_for1 (TAGCTTCTCCTGCCGCTTCTTCTTGCCCAC) and Ü_1415_rev1 (ATGGCCTGGGTGAACTCCATGTTGTTGGTG).
Generation of a ΔpilF::bleo deletion mutant.
To construct a ΔpilF::bleo deletion mutant, the 685-bp upstream region of pilF was amplified from genomic DNA using the primers Put_for1 (EcoRI) (ATACGAATTCTAGGGAGGAGCGGATCTAC) and Put_rev1 (AAGCATAAGCGGCCGCAAATTTAAACCATGGTCATCTCGTCACACCC) and the 545-bp downstream region was amplified using the primers PilT_for1 (PstI) (AATACTGCAGAGGAGGAAAGCGATGCCCAAAG) and PilT_rev (HindIII) (CCGTAAGCTTCTTGTGGAAGAACTCTATGGG). The bleomycin cassette was amplified from the vector pWUR112 (39) using the primers Bleo-for-EcoRV (AGATATCGGCGGCGCAGGCCTTCCTGG) and Bleo-rev-PstI (AAACTGCAGCTTCCGGCTCGTATGTTGTGTGG). The three fragments were digested with the corresponding restriction enzymes and inserted into the vector pUC19, resulting in pUC19_putative_Bleo_pilT. The vector was linearized using HindIII and transformed into T. thermophilus HB27. Transformants were selected on TM+ medium with 15 µg bleomycin/ml. The deletion mutant was verified by PCR analysis with the primers DeltaF_for2 (GGGCGTGCTACCAGGTGGGCGAGGAGGTGGTG) and DeltaF_rev2 (CGGCGGCGATGGTCTCGTAGTCCCGCATCTCCC), binding 785 bp upstream and 664 bp downstream of pilF, respectively.
Generation of a TTC1621::kat (pilT1) ΔTTC1415::hygro (pilT2) double mutant.
To generate a pilT1 pilT2 double mutant, the pilT1 mutant (33) was transformed with linearized (PciI) plasmid pUC19-ΔTTC1415_Hyg. Transformants were selected at 60°C on TM+ medium with 100 µg hygromycin and 40 µg kanamycin per ml. The mutants were verified by PCR, using the primers Ü_1415_for1 (TAGCTTCTCCTGCCGCTTCTTCTTGCCCAC) and Ü_1415_rev1 (ATGGCCTGGGTGAACTCCATGTTGTTGGTG).
Complementation of the ΔpilF::bleo mutant with pilF.
The pilF gene was amplified from the T. thermophilus HB27 genome using the primers pilF_pDM12_for1 (ATTACATATGCACCATCATCACCACCATCATAGCGTGCTCACCATAGGGGACAAAAGG) and pilF_rev (ACTTAGATGCGGCCGCTTACTCAATGGTACGCGCCAG), which contain NdeI and NotI restriction sites, respectively. The resulting 2,714-bp DNA fragment was restricted with NdeI and NotI and inserted into the E. coli/Thermus shuttle vector pDM12, which carried a bc1 promoter from T. thermophilus. Electroporation of the ΔpilF mutant led to the ΔpilF::bleo (pilF) mutant strain.
Electroporation.
Exponentially grown Thermus cells were harvested and washed 2 times with 10% glycerol. One hundred-microliter stocks were incubated for 5 min with DNA and then electroporated. A voltage of 12.5 V and a pulse length of 5 ms were used. The cells were regenerated at 68°C in prewarmed TM+ medium for 2 h and then plated. Electroporation of T. thermophilus was performed as described by de Grado et al. (40).
Western blot analyses.
Western blot analyses were performed as described by Rose et al. (33). Polyclonal PilF rabbit antibodies were used at a dilution of 1:7,500.
Sub-agar-surface adhesion studies.
Sub-agar-surface adhesion studies were performed on minimal medium containing 1% bovine serum albumin (BSA) and 0.01% yeast extract (41). Plates were incubated at 64°C under humid conditions in a plastic bag for 3 days. Cells were stab inoculated through the solid medium down to the petri dish. To visualize adhering cells, the solid medium was removed. Cells were stained with Coomassie blue. The petri dish bottom was washed 4 times for 30 min each time with 10 ml 25 mM Tris HCl and 100 mM NaCl. Adhering cells retained the blue color.
Twitching motility studies.
Cells were grown on BSA containing minimal medium. The agar was stained with Coomassie blue (42), and after removal of the solution, the agar was washed for 1 min with 10 ml water. Then, the cells were detached from the agar surface by suspending them in 10 ml of water. After decantation of the cell suspension, a colorless region which corresponded to the attachment zones of the cells was visible. This zone was defined as the twitching zone, which was visible as a dark zone due to the inversion of the picture.
Electron microscopy.
Electron microscopy was performed to analyze the piliation phenotype of Thermus cells. The cells were transferred to a copper grid (400 mesh). The cells were shadowed with a pressure of 3 × 10−5 to 4 × 10−5 Pa at 28°C and at a unidirectional angle of 25°, and a 1.5-nm thickness of platinum-carbon was used, as reported recently (11).
Transformation studies.
Transformation studies were performed on TM+ agar medium using 5 μg of genomic DNA of a spontaneously streptomycin-resistant HB27 mutant (15). The transformation frequencies were calculated as the number of transformants per the number of live cells (i.e., the living count). All transformation assays were performed in triplicate.
RT-PCR.
Briefly, for reverse transcription-PCR (RT-PCR), total Thermus RNA was isolated from cells harvested in the exponential growth phase by use of a NucleoSpin RNA cleanup kit and treated with RQ1 RNase-free DNase (Promega, Mannheim, Germany) to remove genomic DNA. The final RNA preparation was used to generate cDNA. This cDNA was used as the template in a PCR using primers (listed in Table 1) that bridge the intergenic regions. DNA and RNA were used as controls.
TABLE 1.
Primers used in this study
| Primer | Sequence | Usea |
|---|---|---|
| Hypo_1 | GCTGAAGGAAGAGCCTGGCCTCCTCGG | TTC1624 and TTC1623 |
| PGP_1 | GGAGCTTCCAGCATGCGTATGAAACGGGTG | |
| PGP_3 | CCGAGCGCTTCGCCCGCGTGCAGCGCCTCC | TTC1623 and pilF (TTC1622) |
| PilF_5′_rev2 | CGGGAAGGCCGAGCTCCGGGTAGTGTTTGG | |
| PilF_3′_for1 | GGGACTCCGAGACGGCCAAGATCGCCACC | pilF (TTC1622) and pilT1 (TTC1621) |
| PilT_1 | GGTCCTCAATGGTGACGATGTGGCAGGCCTTGCGTTC | |
| PilT_2 | CCATAGACCGCATCGTGGACGTCTTCCC | pilT1 (TTC1621) and TTC1620 |
| Gluta_1 | GCTGAAGGAAGAGCCTGGCCTCCTCGG |
Use as a bridge between the indicated two genes.
RESULTS
Generation of a pilF deletion mutant.
In a previously characterized pilF insertion mutant, a kanamycin cassette was inserted close to the HincII restriction site near the 5′end encoding the third GSPII domain of pilF (29). This mutant lost its transformation phenotype but was unaffected in piliation. Since the possibility of the production of a C-terminal domain of PilF in this mutant that was still able to function in piliation could not be excluded, a ΔpilF deletion mutant was generated by marker exchange mutagenesis using a bleomycin cassette. Transformants which had acquired the bleomycin cassette by allelic replacement of the pilF gene (Fig. 1B) were selected on TM+ medium containing bleomycin. The genotype of the ΔpilF mutant was analyzed by PCR. Wild-type cells and the pilF::kat insertion mutant carrying a kanamycin cassette in the HincII site (Fig. 1A) were used as controls. A ≈4-kb fragment was obtained with wild-type DNA, a ≈5-kb fragment was obtained with the DNA from the pilF::kat insertion mutant, and a ≈2.3-kb fragment was obtained with DNA from the ΔpilF mutant; these correspond to the calculated sizes of 4.0 kb for the wild type, 5.3 kb for the pilF::kat mutant, and 2.2 kb for the ΔpilF mutant. These data demonstrate a single insertion of the bleomycin cassette into the target locus by a double recombination event, thereby leading to a deletion of the pilF gene.
pilF is essential for both natural transformation and piliation.
The ΔpilF mutant was found to be completely impaired in natural transformation (≤108 transformants per living count). This corresponds to our former results that showed that the pilF::kat insertion mutant was completely defective in natural transformation and confirms that PilF is essential (29, 41). Next, we analyzed the piliation phenotype of the ΔpilF mutant by electron microscopy. T. thermophilus wild-type cells and the pilF::kat mutant were used as controls. The piliation phenotypes of both control strains were comparable. Sixty-eight percent of the pilF::kat mutant cells (Fig. 2C) and 78% of the wild-type cells exhibited 6.5 ± 5.5 pili at one of the cell poles (Fig. 2A). In contrast, the ΔpilF deletion mutant did not exhibit any pilus structures at all (Fig. 2B).
FIG 2.
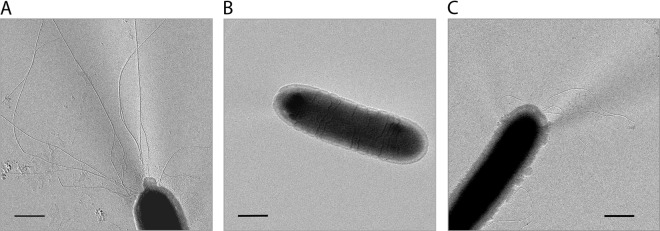
Electron micrographs of a representative sample of the T. thermophilus HB27 wild type (A), the ΔpilF deletion mutant (B), and the pilF::kat insertion mutant (C). Electron microscopic investigations were conducted by shadowing the cells with platinum-carbon. A total of 250 to 300 cells of each strain were analyzed. Bars = 500 nm.
To exclude the polar effects of the bleomycin marker in the ΔpilF mutant, complementation studies were performed with pDM12-pilF. Electroporation of the ΔpilF deletion mutant with this plasmid gave rise to mutants which had transformation frequencies indistinguishable from those of the wild-type strain (2.8 × 10−3) (Fig. 3).
FIG 3.
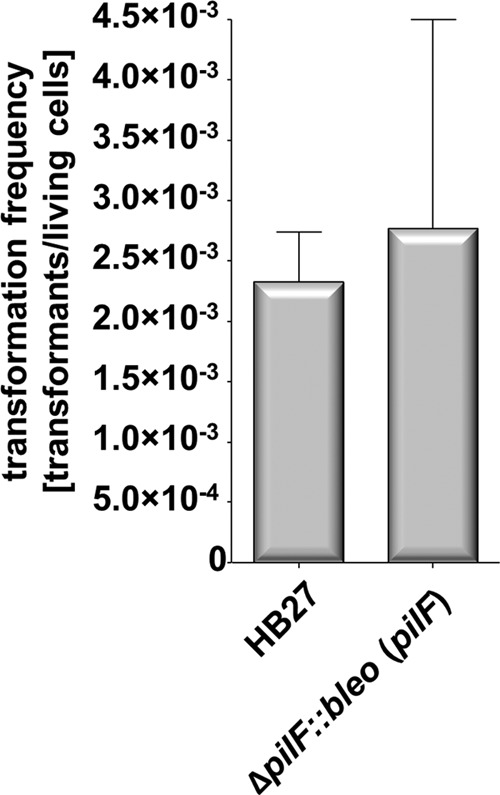
Natural competence of T. thermophilus HB27 and the complemented ΔpilF::bleo (pilF) mutant.
PilF is essential for T4P-mediated twitching motility and adhesion.
Recently, we showed that T. thermophilus pili are essential for twitching motility and adhesion (11). The finding that the pilF::kat mutant still exhibited wild-type piliation led to the question of whether the pili of the pilF::kat mutant are still functional with respect to twitching motility and adhesion. Therefore, the twitching phenotype of the pilF::kat mutant was analyzed using an assay in which we inoculated cells on an agar surface. The HB27 wild type and the nonpiliated ΔpilF mutant were used as controls. Cells exhibiting twitching motility moved away from the inoculation point, forming a spreading zone. To document the spreading zones, plates were flushed with Coomassie blue, which led to staining of the agar but not the spreading zone. Cells were then removed, resulting in a clear zone that was documented by photography, and then the pictures were inverted. Wild-type cells formed large, uniform motility zones of 1 to 2 cm in diameter after 3 days at 64°C (Fig. 4A). In contrast, the nonpiliated ΔpilF deletion mutant (Fig. 4C) was completely defective in twitching motility. Interestingly, the same was observed for the pilF::kat insertion mutant (Fig. 4B). This led to the conclusion that PilF is essential for pilus-mediated twitching motility and that T4P dynamics are impaired in the pilF::kat mutant. This suggests that the truncated PilF protein in the pilF::kat mutant is still sufficient for piliation but not for the pilus dynamics essential for twitching motility.
FIG 4.
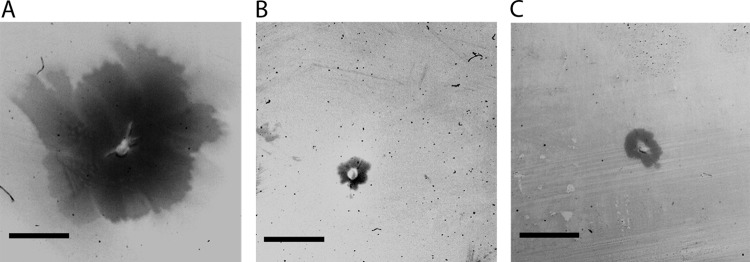
Twitching motility of cells of the T. thermophilus HB27 wild type (A), the pilF::kat insertion mutant (B), and the ΔpilF deletion mutant (C). The cells were inoculated on minimal medium containing 1% BSA and cultivated for 72 h at 64°C under a water-saturated atmosphere. Agar and cells were stained with Coomassie blue. The cells and the Coomassie dye were removed, and the twitching zone corresponding to the noncolored agar region was determined. The pictures were inverted. Experiments were done in triplicate and gave identical results. Bars = 0.5 cm.
Next, we examined the role of PilF in adhesion by a sub-agar-adhesion assay, based on growth of the cells at the interphase between the solid medium and the bottom of the petri dish. Wild-type cells adhered to the petri dish (Fig. 5A). In contrast, both the nonpiliated ΔpilF mutant and the piliated pilF::kat mutant were completely defective in adhesion (Fig. 5B and C). This corroborates our previous finding that full-length PilF is essential for pilus function.
FIG 5.
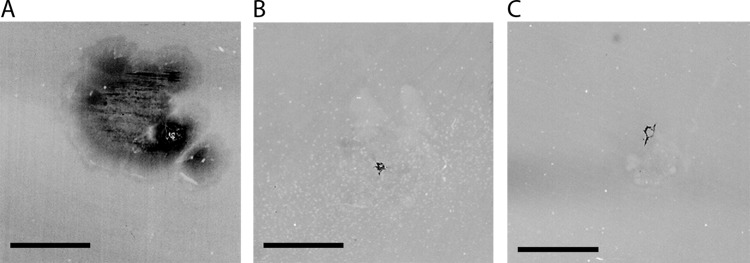
Adhesion of cells of the T. thermophilus wild type (A), the pilF::kat insertion mutant (B), and the ΔpilF deletion mutant (C). Cells were stab inoculated through the solid medium down to the petri dish and cultivated on minimal medium containing 1% BSA for 72 h at 64°C. After removal of the agar, the petri dish was washed four times and adhering cells were stained with Coomassie blue. Experiments were done in triplicate and gave identical results. Bars = 0.5 cm.
pilF and TTC1621 form a transcriptional unit.
So far, a rod depolymerization traffic NTPase, a counterplayer of PilF, has not been identified in T. thermophilus. Inspection of the genome led to the identification of two PilT-like traffic NTPase genes, TTC1415 and TTC1621, but neither of them was found to be essential for pilus biogenesis and natural transformation (29, 33). However, one of the two genes, TTC1621, flanked pilF downstream, which indicated a functional relationship of the two genes. The open reading frame upstream of pilF, TTC1623, encodes a protein with 62% similarity to a haloacid dehalogenase, whereas the open reading frame downstream of TTC1621, TTC1620, encodes a protein with 72% similarity to an aspartyl/glutamyl-tRNA amidotransferase (Fig. 6A). Therefore, the open reading frames upstream of pilF and downstream of TTC1621 are apparently not in a functional relationship with pilF and TTC1621. To analyze the transcriptional organization of the pilF locus, mRNA was isolated from T. thermophilus cells and transcribed into cDNA. This cDNA was then used as a template in a PCR using primers that bridge the intergenic regions between TTC1623 and pilF, pilF and TTC1621, and TTC1621 and TTC1620. No PCR product was obtained with primers for TTC1623 and pilF and primers for TTC1621 and TTC1620, indicating that these genes are not cotranscribed. In contrast, a 611-bp DNA fragment was generated when primers for pilF and TTC1621 were used, indicating that pilF and TTC1621 form a transcriptional unit (Fig. 6B).
FIG 6.
Transcriptional organization of the pilF locus. To examine the transcriptional organization, RNA was isolated and subjected to RT-PCR analyses. (A) The primers and expected fragments are indicated and labeled 1 to 4. RNA was used as a negative control; chromosomal DNA was used as a positive control. (B) The resulting DNA fragments were separated on a 0.8% agarose gel. Lanes 1, chromosomal DNA from T. thermophilus; lanes 2, cDNA from T. thermophilus HB27; lanes 3, cDNA from T. thermophilus pilF::kat; lanes 4, RNA control.
TTC1621 encodes a motor ATPase essential for twitching motility and adhesion but dispensable for natural transformation.
TTC1621 encodes a 39.9-kDa protein which is closely related to members of the T4P retraction ATPase, PilT/PilU. TTC1621 is 54, 53, and 50% identical to PilT proteins from P. aeruginosa, P. stutzeri, and gonococci, respectively. Therefore, TTC1621 was designated PilT1. PilT proteins from P. stutzeri and gonococci are essential for both twitching motility and natural transformation, whereas PilT from P. aeruginosa is implicated only in twitching motility, since P. aeruginosa is not able to take up free DNA (43–45). Unlike P. stutzeri and gonococcal mutants, the piliated T. thermophilus pilT1 mutants were unaffected in natural transformation (2 × 10−3; see Fig. 8) (33). To address the challenging question of whether PilT1 plays a role in pilus retraction, 250 pilT1 mutant cells were analyzed by electron microscopy. A total of 99.7% of the mutant cells were piliated, whereas only 78% of the wild-type cells were piliated. A total of 22.5 ± 12.5 pili were detected on the surface of the pilT1 mutant and were assembled in a unipolar pattern (Fig. 7B), whereas the wild-type strain maximally exhibited 13 pilus structures (Fig. 7A). Seventeen percent of the pilT1 mutant cells exhibited less than 10 pili. This finding indicates a hyperpiliation phenotype of the pilT1 mutant and suggests that PilT1 is essential for T4P depolymerization.
FIG 8.
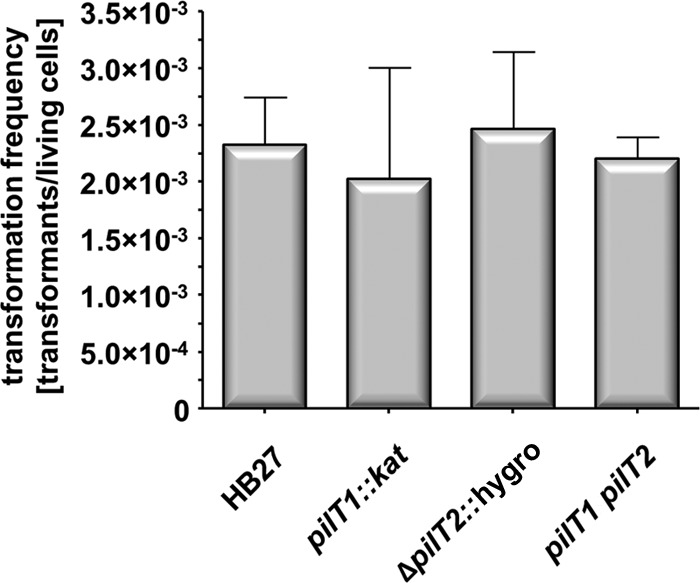
Transformation frequencies of T. thermophilus HB27, the pilT1::kat (TTC1621) and ΔpilT2::hygro (TTC1415) mutants, and the pilT1 pilT2 double mutant.
FIG 7.
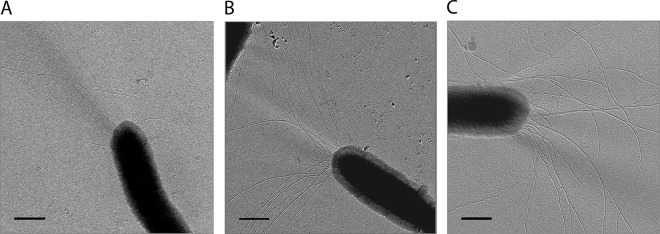
Piliation of T. thermophilus and hyperpiliation of the pilT1 and pilT2 mutants. Electron microscopic investigations were conducted by shadowing the cells with platinum-carbon. A total of 250 to 300 cells were analyzed, and the results for one representative sample of cells of the wild type (A), the pilT1 mutant (B), and the pilT2 mutant (C) are shown. Bars = 500 nm.
To elucidate the role of PilT1 in pilus functions, the twitching motility and adhesion phenotype of pilT1 mutants were analyzed as described above. These experiments revealed that the pilT1 mutant was defective in both twitching motility and adhesion to petri dishes (data not shown). The finding that the cells were defective in adhesion, despite their hyperpiliation, might be due to the twitching defect, leading to rigid pilus structures and, therefore, reduced interactions of the cells with the petri dish. Taken together, these data suggest that PilT1 is required for T4P-mediated twitching motility and adhesion and is indeed the counterplayer of the T4P extension NTPase PilF. Moreover, the wild-type transformation phenotype of the twitching-defective pilT1 mutant suggests that pilus dynamics are not required for natural transformation.
TTC1415 is also required for pilus functions but dispensable for natural transformation.
A second PilT-like traffic NTPase gene, TTC1415, not associated with the pilF locus, is present in the T. thermophilus genome (29). The deduced protein is 46% identical to PilT2 from N. gonorrhoeae (46). Therefore, TTC1415 was designated PilT2. To analyze the role of pilT2 in piliation, a ΔTTC1415::hygro deletion mutant was generated by marker exchange mutagenesis. Mutant studies revealed that the pilT2 deletion mutant was unaffected in natural transformation (2.5 × 10−3 transformants per living count; Fig. 8). Electron microscopic studies of the pilT2 mutant revealed that the mutant exhibited a hyperpiliated phenotype (Fig. 7C). A total of 99.2% of the cells were piliated, whereas only 78% of the wild-type cells were piliated. A total of 28.5 ± 18.5 pili were detected on the surface of the pilT1 mutant and were assembled in a unipolar pattern (Fig. 7C). These experiments demonstrate that PilT2 is essential for T4P depolymerization.
To elucidate the role of PilT2 in pilus functions, the twitching motility and adhesion phenotype of the pilT2 mutant were analyzed. These studies revealed that the mutant was defective in both twitching motility and adhesion to petri dishes (data not shown). These data demonstrate that PilT2 is required for T4P-mediated twitching motility and adhesion and that PilT2 is indeed a counterplayer of the T4P extension NTPase PilF. Moreover, the wild-type transformation phenotype of the twitching-defective pilT2 mutant (Fig. 8) is consistent with our conclusion that pilus dynamics are not required for natural transformation.
To provide clear evidence that both PilT1 and PilT2 are not essential for natural competence, complementation of the pilT1 or the pilT2 defect by the wild-type pilT2 gene and pilT1 gene, respectively, had to be excluded. Therefore, a pilT1 pilT2 double mutant was generated and analyzed with respect to natural transformation. The pilT1 pilT2 double mutant exhibited a wild-type transformation frequency of 2.2 × 10−3 (Fig. 8). This provides clear evidence that neither PilT1 nor PilT2 is essential for natural competence in T. thermophilus.
DISCUSSION
The pilF deletion mutant generated in this study has a phenotype that is quite different from the phenotype of the insertion mutant described before (29). The difference in phenotype between the ΔpilF deletion mutant and the pilF::kat insertion mutant may be caused by the presence of the N-terminal truncated PilF in the insertion mutant, which is supported by the detection of a truncated PilF protein by Western blotting analyses (data not shown). This, together with the finding that the insertion mutant is active in piliation but defective in twitching motility and adhesion, argues for a domain structure of PilF with different domains having different functions. Recently, Collins et al. (47) determined the structure of PilF by cryoelectron microscopy. It consists of two distinct N-terminal and C-terminal domains forming a disk and a ring structure. The two domains are connected by a short stem-like structure. Upon binding of the ATP analogue adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), the C-terminal ring domain undergoes structural changes which are transmitted to the N-terminal disk-like structure via the connecting stem (47). It was suggested that this structural change confers a piston-like mechanism, which would trigger assembly of pilus structures.
The pilF insertion mutant was completely impaired in twitching motility, despite the presence of pilus structures. How can this be explained? The ATP binding site and the Asp and His boxes of motor ATPases, which are absolutely essential for T4P biogenesis and function (21, 32, 48), are still present in the C-terminal domain of PilF. However, it is known that despite energization by ATP hydrolysis, energy is transmitted via the N-terminal domain to N-terminal cytoplasmic domains of cytoplasmic membrane proteins. For example, the N terminus of XpsE of Xanthomonas campestris is essential for interaction with the cytoplasmic domain of XpsL (34). Analogously, a direct interaction between the N terminus of the secretion ATPase OutE from Erwinia chrysanthemi with the cytoplasmic domain of OutL (49) and the N terminus of EpsE with the cytoplasmic domain of EpsL was found (35). Analyses of the functional role of the proteins interacting with the N termini of secretion ATPases suggest that these interactions stimulate the ATPase activity of the secretion ATPases (50–53) and target the ATPases to the membrane. The absence of twitching motility of the PilF insertion mutants is consistent with these findings, since the absence of the N terminus in the truncated PilF protein might result in the loss of PilF-mediated pilus dynamics due to abrogation of membrane targeting. This might lead to an uncoupling of the PilF ATPase activity from pilus dynamics. This is supported by the finding that an XpsE (R286A) mutant exhibits increased ATPase activities but is nonfunctional in protein secretion via the T2SS (50).
One of the most challenging questions was whether T. thermophilus HB27 T4P dynamics and DNA translocation require depolymerization ATPases. Here we demonstrate that the two traffic NTPases, PilT1 and PilT2, are essential for T4P-mediated twitching motility and adhesion but dispensable for natural transformation. The finding that pilT1 and pilT2 mutants are hyperpiliated is consistent with the hyperpiliation phenotype of Synechocystis sp. strain PCC 6803 pilT mutants (54, 55), P. stutzeri pilT mutants (43), and P. aeruginosa pilT and pilU mutants (26, 45). Therefore, we conclude that the two traffic NTPases, PilT1 and PilT2, are T4P depolymerization ATPases acting as counterplayers of the T4P polymerization ATPase PilF, thereby mediating retraction of T4P.
The finding that PilT1 and PilT2 both display a crucial role in T4P dynamics but are not essential for the T4P-linked natural transformation system is rather unique. In contrast, pilT mutants of N. gonorrhoeae and P. stutzeri were defective in both twitching motility and natural transformation (43, 44). However, it must be noted that expression of a pilin where the C-terminal amino acids were replaced by a His tag in a P. stutzeri pilT mutant restored natural transformation (43). This led to the hypothesis that pilin subunits or rudimentary pilus-like complexes might mimic depolymerized pilus structures. These complexes were suggested to interact with the outer membrane pores, leading to their conformational change and subsequent DNA uptake (43). This suggestion is also supported by the finding that a low level of expression of pilus structures in N. gonorrhoeae still leads to high natural transformation frequencies (10, 56).
A linkage of natural transformation systems and T4P has been observed in many Gram-negative bacteria (2, 3, 57, 58). It has been shown that the dynamics of T4P structures and DNA translocation require two types of traffic NTPases, polymerization and depolymerization ATPases. Our findings that PilT1 and PilT2 are both T4P depolymerization ATPases and that they are the only ATPases of the PilT subfamily present in T. thermophilus but dispensable for natural transformation lead to the conclusion that retraction of T4P is not required for DNA uptake. This suggests that the T. thermophilus DNA translocator is linked to T4P but is a distinct system. This is also supported by our recent finding that a pilQ mutant had a defect in piliation and pilus functions but had only a minor change in natural transformation (11). Moreover, the transformability of the pilT1 and pilT2 mutants indicates that either depolymerization of the DNA translocator is not required for DNA uptake or PilF can display both activities, polymerization and depolymerization of the DNA translocator. These important questions will be the subject of further investigations.
In summary, the results of the present study provide compelling evidence that PilF plays an essential dual role in T4P polymerization and natural transformation. T4P dynamics are not required for DNA translocation in T. thermophilus, supporting our suggestion that T4P and the DNA translocator are distinct systems. The DNA translocator rod is suggested to extend from the inner membrane into the staggered ring structures of the secretin complex, where it might serve as a placeholder for the incoming DNA. This DNA translocator rod comprises the pilin-like subunits PilA1, PilA2, PilA3, and PilA4. The pilin-like proteins PilA1, PilA2, and PilA3 are highly specific for the DNA translocator and are not involved in T4P (59). This also supports the conclusion that T4P and the DNA translocator are distinct systems sharing components such as PilM, PilN, PilO, PilW, PilC, the pilin PilA4, the prepilin peptidase PilD, the motor ATPase PilF, and the secretin PilQ (3). As shown recently, the pilus structures are not essential for natural transformation (11). This, together with the absence of alternate secretins in T. thermophilus, suggests that some of the PilQ complexes guide the pili, whereas others guide the pilus-independent DNA transporter rods through the outer membrane and the periplasmic space.
Recently, it was found in Vibrio cholerae that the cells mostly contain a single pilus which colocalizes with the secretin PilQ but that PilQ forms several foci occasionally colocalized with the dynamic motor ATPase PilB (60). Analogously to the dynamic motor ATPase PilB in V. cholerae, PilF in T. thermophilus might be dynamic, thereby occasionally interacting with the PilQ complexes of T4P or the PilQ complexes guiding the DNA translocator rod. The interaction of the pilus-like rod with one of the ring structures of the PilQ complex might affect its conformation, thereby triggering the opening of the central channel of the complex. This would allow the binding of DNA at the cavity of the secretin pore. Then, the DNA might be pulled in a single step across the outer membrane and the periplasmic space through the secretin channel by the action of ComEA (15). ComEA might exert force on the bound DNA, thereby pulling the DNA through the secretin channel spanning the outer membrane and the periplasmic space (7). This is also supported by the finding that a T. thermophilus ComEA mutant exhibits a more than 50-fold increase in DNase-sensitive DNA bound to the cell surface (41). Taken together, the absence of a retraction ATPase in the natural transformation system and the dramatic effect of a comEA mutation on DNA import argue against pilus detraction/retraction pulling the DNA through to the outer membrane. Certainly, more studies are required to elucidate the mechanisms of DNA transport through the distinct barriers of the T. thermophilus cell periphery and to clarify the role of the motor ATPase PilF in DNA uptake.
ACKNOWLEDGMENTS
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (AV 9/6-1).
Furthermore, we thank Bernd Ludwig for providing plasmid pDM12.
Footnotes
Published ahead of print 8 November 2013
REFERENCES
- 1.Chen I, Dubnau D. 2003. DNA transport during transformation. Front. Biosci. 8:S544–S556. 10.2741/1047 [DOI] [PubMed] [Google Scholar]
- 2.Hobbs M, Mattick JS. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233–243. 10.1111/j.1365-2958.1993.tb01949.x [DOI] [PubMed] [Google Scholar]
- 3.Averhoff B. 2009. Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33:611–626. 10.1111/j.1574-6976.2008.00160.x [DOI] [PubMed] [Google Scholar]
- 4.Nunn D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402–408. 10.1016/S0962-8924(99)01634-7 [DOI] [PubMed] [Google Scholar]
- 5.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072. 10.1099/mic.0.26364-0 [DOI] [PubMed] [Google Scholar]
- 6.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249. 10.1038/nrmicro844 [DOI] [PubMed] [Google Scholar]
- 7.Krüger N-J, Stingl K. 2011. Two steps away from novelty—principles of bacterial DNA uptake. Mol. Microbiol. 80:860–867. 10.1111/j.1365-2958.2011.07647.x [DOI] [PubMed] [Google Scholar]
- 8.Hamilton HL, Dillard JP. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376–385. 10.1111/j.1365-2958.2005.04964.x [DOI] [PubMed] [Google Scholar]
- 9.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer TF. 1997. Transformation competence and type-IV pilus biogenesis in Neisseria gonorrhoeae. Gene 192:125–134. 10.1016/S0378-1119(97)00038-3 [DOI] [PubMed] [Google Scholar]
- 10.Long CD, Tobiason DM, Lazio MP, Kline KA, Seifert HS. 2003. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect. Immun. 71:6279–6291. 10.1128/IAI.71.11.6279-6291.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkhardt J, Vonck J, Langer JD, Salzer R, Averhoff B. 2012. Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation. J. Biol. Chem. 287:8484–8494. 10.1074/jbc.M111.334912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749–760. 10.1046/j.1365-2958.2002.03193.x [DOI] [PubMed] [Google Scholar]
- 13.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshima T, Imahori K. 1971. Isolation of an extreme thermophile and thermostability of its transfer ribonucleic acid and ribosomes. J. Gen. Appl. Microbiol. 17:513–517. 10.2323/jgam.17.513 [DOI] [Google Scholar]
- 15.Friedrich A, Hartsch T, Averhoff B. 2001. Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67:3140–3148. 10.1128/AEM.67.7.3140-3148.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rumszauer J, Schwarzenlander C, Averhoff B. 2006. Identification, subcellular localization, and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273:3261–3272. 10.1111/j.1742-4658.2006.05335.x [DOI] [PubMed] [Google Scholar]
- 17.Robien MA, Krumm BE, Sandkvist M, Hol WG. 2003. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333:657–674. 10.1016/j.jmb.2003.07.015 [DOI] [PubMed] [Google Scholar]
- 18.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U. S. A. 98:2503–2508. 10.1073/pnas.051436598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitag NE, Seifert HS, Koomey M. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16:575–586. 10.1111/j.1365-2958.1995.tb02420.x [DOI] [PubMed] [Google Scholar]
- 20.Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 95:14973–14978. 10.1073/pnas.95.25.14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakovljevic V, Leonardy S, Hoppert M, Sogaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421. 10.1128/JB.01793-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- 23.Kaiser D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:777–780. 10.1016/S0960-9822(00)00568-6 [DOI] [PubMed] [Google Scholar]
- 24.Chakraborty S, Monfett M, Maier TM, Benach JL, Frank DW, Thanassi DG. 2008. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 76:2852–2861. 10.1128/IAI.01726-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U. S. A. 99:16012–16017. 10.1073/pnas.242523299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitchurch CB, Mattick JS. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079–1091. 10.1111/j.1365-2958.1994.tb00499.x [DOI] [PubMed] [Google Scholar]
- 27.Burrows LL. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888. 10.1111/j.1365-2958.2005.04703.x [DOI] [PubMed] [Google Scholar]
- 28.Clausen M, Jakovljevic V, Sogaard-Andersen L, Maier B. 2009. High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191:4633–4638. 10.1128/JB.00396-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedrich A, Prust C, Hartsch T, Henne A, Averhoff B. 2002. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745–755. 10.1128/AEM.68.2.745-755.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Possot OM, Pugsley AP. 1997. The conserved tetracysteine motif in the general secretory pathway component PulE is required for efficient pullulanase secretion. Gene 192:45–50. 10.1016/S0378-1119(97)00009-7 [DOI] [PubMed] [Google Scholar]
- 31.Chiang P, Habash M, Burrows LL. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187:829–839. 10.1128/JB.187.3.829-839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai D, Horiuchi T, Komano T. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 276:17968–17975. 10.1074/jbc.M010652200 [DOI] [PubMed] [Google Scholar]
- 33.Rose I, Biukovic G, Aderhold P, Müller V, Grüber G, Averhoff B. 2011. Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27. Extremophiles 15:191–202. 10.1007/s00792-010-0343-2 [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Shiue SJ, Huang CW, Chang JL, Chien YL, Hu NT, Chan NL. 2005. Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J. Biol. Chem. 280:42356–42363. 10.1074/jbc.M506843200 [DOI] [PubMed] [Google Scholar]
- 35.Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. 2005. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J. Mol. Biol. 348:845–855. 10.1016/j.jmb.2005.02.061 [DOI] [PubMed] [Google Scholar]
- 36.Salzer R, Herzberg M, Nies DH, Biukovic G, Gruber G, Muller V, Averhoff B. 2013. The DNA uptake ATPase PilF of Thermus thermophilus: a reexamination of the zinc content. Extremophiles 17:697–698. 10.1007/s00792-013-0544-6 [DOI] [PubMed] [Google Scholar]
- 37.Camberg JL, Sandkvist M. 2005. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J. Bacteriol. 187:249–256. 10.1128/JB.187.1.249-256.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cava F, Zafra O, Magalon A, Blasco F, Berenguer J. 2004. A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. J. Biol. Chem. 279:45369–45378. 10.1074/jbc.M404785200 [DOI] [PubMed] [Google Scholar]
- 39.Brouns SJ, Wu H, Akerboom J, Turnbull AP, de Vos WM, van der Oost J. 2005. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 280:11422–11431. 10.1074/jbc.M413623200 [DOI] [PubMed] [Google Scholar]
- 40.de Grado M, Castan P, Berenguer J. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241–245. 10.1006/plas.1999.1427 [DOI] [PubMed] [Google Scholar]
- 41.Schwarzenlander C, Haase W, Averhoff B. 2009. The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ. Microbiol. 11:801–808. 10.1111/j.1462-2920.2008.01801.x [DOI] [PubMed] [Google Scholar]
- 42.Weber K, Osborn M. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406–4412 [PubMed] [Google Scholar]
- 43.Graupner S, Weger N, Sohni M, Wackernagel W. 2001. Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J. Bacteriol. 183:4694–4701. 10.1128/JB.183.16.4694-4701.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfgang M, Lauer P, Park HS, Brossay L, Hébert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321–330. 10.1046/j.1365-2958.1998.00935.x [DOI] [PubMed] [Google Scholar]
- 45.Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. 2008. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT, and PilU. Microbiology 154:114–126. 10.1099/mic.0.2007/011320-0 [DOI] [PubMed] [Google Scholar]
- 46.Kurre R, Hone A, Clausen M, Meel C, Maier B. 2012. PilT2 enhances the speed of gonococcal type IV pilus retraction and of twitching motility. Mol. Microbiol. 86:857–865. 10.1111/mmi.12022 [DOI] [PubMed] [Google Scholar]
- 47.Collins RF, Hassan D, Karuppiah V, Thistlethwaite A, Derrick JP. 2013. Structure and mechanism of the PilF DNA transformation ATPase from Thermus thermophilus. Biochem. J. 450:417–425. 10.1042/BJ20121599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner LR, Lara JC, Nunn DN, Lory S. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175:4962–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Py B, Loiseau L, Barras F. 1999. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289:659–670. 10.1006/jmbi.1999.2803 [DOI] [PubMed] [Google Scholar]
- 50.Shiue SJ, Kao KM, Leu WM, Chen LY, Chan NL, Hu NT. 2006. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 25:1426–1435. 10.1038/sj.emboj.7601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. 2007. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 26:19–27. 10.1038/sj.emboj.7601481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ball G, Chapon-Herve V, Bleves S, Michel G, Bally M. 1999. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181:382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Possot OM, Vignon G, Bomchil N, Ebel F, Pugsley AP. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182:2142–2152. 10.1128/JB.182.8.2142-2152.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshihara S, Geng X, Okamoto S, Yura K, Murata T, Go M, Ohmori M, Ikeuchi M. 2001. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42:63–73. 10.1093/pcp/pce007 [DOI] [PubMed] [Google Scholar]
- 55.Okamoto S, Ohmori M. 2002. The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol. 43:1127–1136. 10.1093/pcp/pcf128 [DOI] [PubMed] [Google Scholar]
- 56.Aas FE, Winther-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JP, Koomey M. 2007. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 63:69–85. 10.1111/j.1365-2958.2006.05482.x [DOI] [PubMed] [Google Scholar]
- 57.Alm RA, Mattick JS. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89–98. 10.1016/S0378-1119(96)00805-0 [DOI] [PubMed] [Google Scholar]
- 58.Tønjum T, Koomey M. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships. Gene 192:155–163. 10.1016/S0378-1119(97)00018-8 [DOI] [PubMed] [Google Scholar]
- 59.Friedrich A, Rumszauer J, Henne A, Averhoff B. 2003. Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl. Environ. Microbiol. 69:3695–3700. 10.1128/AEM.69.7.3695-3700.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 110:17987–17992. 10.1073/pnas.1315647110 [DOI] [PMC free article] [PubMed] [Google Scholar]