Abstract
A betaproteobacterium, shown by molecular techniques to have widespread global distribution in extremely acidic (pH 2 to 4) ferruginous mine waters and also to be a major component of “acid streamer” growths in mine-impacted water bodies, has proven to be recalcitrant to enrichment and isolation. A modified “overlay” solid medium was devised and used to isolate this bacterium from a number of mine water samples. The physiological and phylogenetic characteristics of a pure culture of an isolate from an abandoned copper mine (“Ferrovum myxofaciens” strain P3G) have been elucidated. “F. myxofaciens” is an extremely acidophilic, psychrotolerant obligate autotroph that appears to use only ferrous iron as an electron donor and oxygen as an electron acceptor. It appears to use the Calvin-Benson-Bassham pathway to fix CO2 and is diazotrophic. It also produces copious amounts of extracellular polymeric materials that cause cells to attach to each other (and to form small streamer-like growth in vitro) and to different solid surfaces. “F. myxofaciens” can catalyze the oxidative dissolution of pyrite and, like many other acidophiles, is tolerant of many (cationic) transition metals. “F. myxofaciens” and related clone sequences form a monophyletic group within the Betaproteobacteria distantly related to classified orders, with genera of the family Nitrosomonadaceae (lithoautotrophic, ammonium-oxidizing neutrophiles) as the closest relatives. On the basis of the phylogenetic and phenotypic differences of “F. myxofaciens” and other Betaproteobacteria, a new family, “Ferrovaceae,” and order, “Ferrovales,” within the class Betaproteobacteria are proposed. “F. myxofaciens” is the first extreme acidophile to be described in the class Betaproteobacteria.
INTRODUCTION
Bacteria are microscopic entities, and individual cells, with the notable exception of the sulfur-depositing marine bacterium Thiomargarita namibiensis, are not visible to the naked eye. However, by producing extracellular polymeric substances (EPS) and forming biofilms and, in some cases, larger macroscopic structures such as “streamers,” “snottites,” and “microbial stalactites,” bacterial communities can be very evident in some situations. Streamer-like growths are particularly common in some extreme environments, such as geothermal sites and extremely acidic streams (1). While this might reflect the absence or limited abundance of higher life forms in these environments, grazing of bacteria by protozoa and Rotifera in extremely acidic water bodies has been documented (e.g., see reference 2).
Mine-impacted water bodies (MIWs) associated with active and abandoned metal and coal mines and mine spoils are often acidic (sometimes extremely so) and usually contain elevated concentrations of soluble sulfate, iron and other metals, and metalloids such as arsenic (3). Acidity derives from the microbiological oxidation of reduced forms of sulfur and iron (and the subsequent hydrolysis of ferric iron), both of which occur in the most ubiquitous of all sulfide minerals, pyrite (FeS2). The most extreme MIWs that have been described are located within the Richmond mine in Iron Mountain, California, where pools of negative pH have been recorded (4). Microbial slimes and snottites have been observed in the warm (∼40°C) and extremely acidic (pH ∼1) waters within the mine, and the dominant indigenous microorganisms have been identified as belonging to known genera of acidophilic bacteria (Leptospirillum spp.) and archaea (“Ferroplasma acidarmanus”) (5, 6). Waters within the Richmond mine are, however, atypical of most reported MIWs, where temperatures tend to be lower and pH values somewhat higher (>2).
Examination of acid streamer growths at a chalybeate spa and two abandoned mines (a copper mine and a sulfur/pyrite mine) in north Wales using a variety of molecular techniques revealed that the numerically dominant indigenous organism in all three situations was an unknown betaproteobacterium (7) related to ammonium-oxidizing, autotrophic, neutrophilic bacteria and also to the currently nonvalidated iron-oxidizing neutrophile “Ferritrophicum radicicola” (8). Like most MIWs, the streams in which these macroscopic growths were found contained almost negligible amounts of ammonium (<1 mg liter−1) and dissolved organic carbon (<5 mg liter−1) but large concentrations of ferrous iron (14 to 725 mg liter−1), and it therefore seemed unlikely, given the scale of the streamer growths (∼100 m3 within one of the mines), that the unknown betaproteobacterium was either an ammonium-oxidizing autotroph or an obligate heterotroph. However, all attempts at enriching and isolating this bacterium, using a range of liquid and solid media, met initially with no success. Known acidophiles (iron-oxidizing Acidithiobacillus spp. and Leptospirillum ferrooxidans and heterotrophic Acidiphilium spp.) dominated enrichment cultures and were the sole bacteria isolated on solid media (9). A bacterium that was the dominant prokaryote in some of the most massive microbial accumulations that had been described was seemingly “unculturable.”
“Overlay” solid media, in which a heterotrophic acidophile is included in the lower layer of a 2-layer gel in order to remove trace amounts of toxic pyruvic acid and other organic materials that arise from acid hydrolysis of the gelling material, have long been used to isolate acidophilic prokaryotes from natural and human-made environments (e.g., see references 10 and 11). The chance isolation of a bacterium that grew as a ferric iron-encrusted colony from water draining a coal mine, and whose 16S rRNA gene had 99% similarity to that of the unknown streamer-forming betaproteobacterium (K. B. Hallberg and D. B. Johnson, unpublished data), indicated that the lack of growth of the latter was not due to the absence of a necessary growth factor in the standard overlay plate formulation. This single isolate, which could not be transferred to a liquid medium, was noted to grow in very close proximity to colonies of heterotrophic acidophiles, suggesting that the iron-oxidizing isolate could be even more sensitive to some soluble carbon compounds than the most fastidious chemolithotrophic acidophile known at that time (Leptospirillum ferrooxidans). This observation led to a strategy being devised that has resulted in the isolation of the previously unknown betaproteobacterium from a number of acidic mine waters. An axenic culture of an isolate (“Ferrovum myxofaciens”) obtained from acid streamer growths in one of the first sites of this prokaryote to be reported has been characterized and confirmed to be an obligately chemolithotrophic iron-oxidizing acidophile. This is the first, and to date the only, representative of a proposed new bacterial family, “Ferrovaceae,” and order, “Ferrovales.”
MATERIALS AND METHODS
Isolation of “Ferrovum myxofaciens” P3G and cultivation in ferrous iron medium.
Strain P3G was isolated on a modified overlay solid medium (“iFeo”) (12). This is essentially an “inorganic” medium, which contains only ferrous sulfate (25 mM) as an energy source. Acidiphilium cryptum (strain SJH) was included in the lower gel layer to remove traces of organic materials, as with most other overlay media. Plates were inoculated with disrupted acid streamers growing in a stream draining the abandoned Mynydd Parys copper mine in north Wales (Fig. 1A) (7, 13) and incubated at 20 or 30°C for up to 4 weeks. Other overlay media (Feo, FeSo, FeTo, and YE3o) (12) were also used to isolate acidophiles from the disrupted streamers.
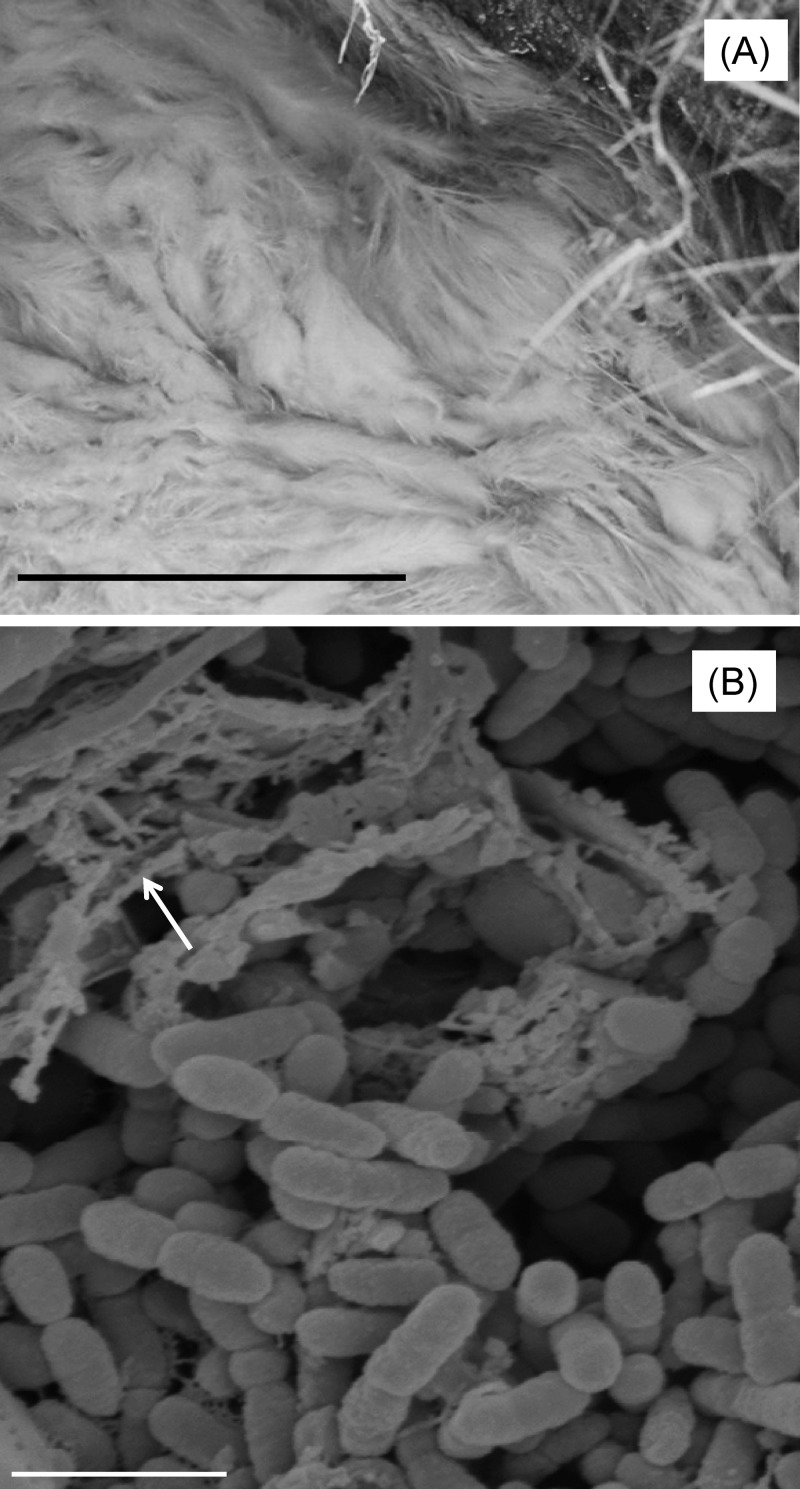
FIG 1.
(A) Acid streamers at Mynydd Parys, which served as the source of “F. myxofaciens” P3G (bar, 50 cm). (B) Scanning electron micrograph of “F. myxofaciens” P3G, showing aggregates of cells and dehydrated EPS (shown with an arrow) (bar, 2 μm). The sample was fixed in glutaraldehyde, critical point dried in liquid CO2, and viewed in an Hitachi S-120 scanning electron microscope.
Over 20 ferric iron-encrusted colonies, varying in size from <1 mm to ∼10 mm in diameter, which grew on iFeo medium were transferred into 200 μl of liquid medium (in 1-ml Eppendorf tubes) containing 5 mM ferrous sulfate, basal salts, and trace elements, pH 2.5 (14). Cultures in which the ferrous iron oxidized (indicated by a change in color to orange/brown and deposition of ferric iron) were transferred into small volumes (5 ml) of the same medium of progressively lower pH (to pH 2.1) to avoid hydrolysis and precipitation of ferric iron. In the absence of precipitates in the oxidized medium, small streamer-like growths were evident in some cases. Amplification of 16S rRNA genes using primers that were specific to “Ferrovum”-like isolates (see below) confirmed that the novel proteobacterium was present in many of the latter cultures, though not in those where small streamer growths were not evident. Culture purity was confirmed by terminal restriction fragment length polymorphism (T-RFLP) analysis, using restriction enzymes that separate “Ferrovum”-like bacteria from other acidophiles (13), and also by plating 50 μl of oxidized medium onto Feo medium (which supports the growth of other acidophilic iron oxidizers) and a solid medium containing 5 mM fructose and 0.02% (wt/vol) yeast extract (to test for the presence of heterotrophs, such as Acidiphilium and Acidocella spp.). One culture confirmed as pure (isolate P3G) was characterized, as described below.
Oxidation of ferrous iron by “F. myxofaciens” P3G and tests for growth on alternative electron donors.
Oxidation of ferrous iron by “F. myxofaciens” P3G in shake flask cultures containing 100 ml of 5 mM ferrous sulfate liquid medium (pH 2.1) and incubated at 30°C was monitored by measuring time-related changes in concentrations of ferrous iron, using the ferrozine assay (15). The ability of isolate P3G to use alternative inorganic electron donors was tested by harvesting ferrous iron-grown biomass and inoculating it into liquid medium (pH 3.0) containing elemental sulfur (0.1%, wt/vol), potassium tetrathionate (2.5 mM), or ammonium sulfate (10 mM). The higher initial pH used in these tests was because oxidation of these substrates generates acidity, in contrast to ferrous iron oxidation, which is a proton-consuming reaction. As well as monitoring any changes in pH, concentrations of sulfate (sulfur and tetrathionate cultures) and nitrite and nitrate (ammonium cultures) were measured at regular intervals, using ion chromatography. Autotrophic growth via oxidation of hydrogen was tested using a protocol described elsewhere (16). In brief, this entailed incubating “F. myxofaciens” P3G in liquid (5 mM ferrous sulfate) and solid (iFeo plates) media in a sealed jar with an atmosphere containing nitrogen, hydrogen, oxygen, and carbon dioxide. Control cultures containing only nitrogen, oxygen, and carbon dioxide were set up at the same time. Cultures were inspected after 2 and 4 weeks to look for differences in colony morphologies (switching to hydrogen causes colonies of iron oxidizers to become larger and cream/gray colored) and to determine whether there was enhanced growth in the presence of hydrogen. Finally, oxidative dissolution of pyrite (FeS2) was tested by inoculating iron-grown “F. myxofaciens” P3G in a pH 2.5 liquid medium containing 1% (wt/vol) finely ground (<100-μm) pyrite (Strem Chemicals, Inc., MA) and monitoring changes in total soluble iron concentrations (by ion chromatography). All of the above-described tests were carried out in duplicate, and Acidithiobacillus ferrooxidans (ATCC 23270T), which oxidizes ferrous iron, sulfur, tetrathionate, hydrogen, and pyrite, though not ammonium, was used as a positive control.
Carbon assimilation and nitrogen fixation by “F. myxofaciens” P3G.
Liquid media containing either 100 μM or 5 mM ferrous sulfate, basal salts, and trace elements, adjusted to pH 2.1, were amended with the following potential organic substrates: glucose, fructose, citric acid, glutamic acid, glycerol (at 5 mM), and peptone or yeast extract (at 0.02%, wt/vol). Cultures were incubated at 30°C for 10 days, biomass was assessed visually (by comparing sizes of streamer growths), and yields were determined by measuring the protein contents of harvested cultures (17). Additionally, genes encoding the large subunit of type I (cbbL) and type II (cbbM) RubisCO were amplified and sequenced, as described below.
To assess whether isolate P3G could fix dinitrogen, a liquid medium containing 20 mM ferrous iron and nitrogen-free basal salts/trace elements was prepared and duplicate flasks were inoculated with an active culture of the iron oxidizer. The flasks were placed in a desiccator containing sulfuric acid to absorb any ammonia present in the atmosphere and incubated at 30°C. The oxidized cultures were subcultured (twice) into liquid medium of the same composition. Parallel cultures using standard inorganic N-containing basal salts were also set up. Genetic evidence for the capacity of P3G to fix nitrogen was obtained by amplifying a portion of the nifH gene, as described below.
Growth and specific rates of ferrous iron oxidation by “F. myxofaciens” P3G at different temperatures and pH values.
“F. myxofaciens” P3G was grown in batch culture in a pH- and temperature-controlled 2-liter bioreactor (Electrolab, United Kingdom) with 20 mM ferrous sulfate. The reactor was aerated (1 liter min−1) and stirred at 100 rpm. Concentrations of ferrous iron were determined at regular intervals, and growth rates of strain P3G at each preset condition were determined from the exponential phases identified in semilogarithmic plots of iron oxidized against time. As is the case with other acidophilic chemolithotrophic iron oxidizers such as Acidithiobacillus and Leptospirillum spp., ferrous iron oxidation was coupled to bacterial growth in actively growing cultures. To determine optimum growth temperature, the bioreactor was maintained at pH 2.4, and to determine optimum pH, the temperature was set at 25°C.
Specific rates of ferrous iron oxidation by “F. myxofaciens” P3G at different temperatures were determined using a protocol described elsewhere (18). In brief, the isolate was grown in 5-liter shake flasks on ferrous iron, and the macroscopic growths were harvested, washed, and homogenized in pH 2.2 basal salts. Aliquots of dispersed streamers were then inoculated into 10 ml of basal salts containing 1 mM ferrous sulfate in triplicate 100-ml flasks, which were incubated at 5 to 35°C. Changes in ferrous iron concentrations with time were recorded (using the ferrozine assay), and the protein concentration in the homogenized inoculum was determined using the Bradford reagent (17).
Production of EPS.
An approximate value of the water and ash contents and the relative amounts of protein and EPS produced by “F. myxofaciens” P3G was obtained using an acid streamer sample from Mynydd Parys where this bacterium accounted for >90% of the cells present (from T-RFLP analysis). One hundred grams of wet streamers was first dried at 105°C and then ignited at 450°C, to determine water and ash contents, respectively. Duplicate 1-g samples of the same acid streamer sample were dissolved in 20 ml of 0.5 M NaOH, and the protein and carbohydrate contents (as glucose equivalents) were determined using the Bradford and phenol (19) colorimetric assays, respectively. Parallel analysis of proteins and carbohydrates was carried out using harvested biomass of “F. myxofaciens” P3G. To identify the sugars present in EPS from both native acid streamers and a pure culture of “F. myxofaciens” P3G, EPS were first extracted and hydrolyzed (20) and monosaccharides were identified using ion chromatography (21).
Transition metal tolerance.
“F. myxofaciens” P3G was grown in 5 mM ferrous sulfate liquid medium (pH 2.1) containing 0 to 100 mM ferric iron, copper, zinc, nickel, manganese(II), and aluminum or 0 to 1 mM sodium molybdate. Media containing up to 100 mM ferrous iron and no other transition metal were also prepared. Duplicate cultures were set up in 25-ml universal bottles and incubated at 30°C for up to 10 days, at which time they were tested for concentrations of residual ferrous iron.
Biomolecular analyses.
“F. myxofaciens” P3G was grown in shake flasks by decanting oxidized medium, retaining the streamer-like growths, and replacing the medium with fresh medium containing 5 mM ferrous iron (pH 2.1). When the streamer biomass had reached an estimated 500 mg (wet weight), it was removed and washed in pH 2 basal salts solution, and DNA was extracted (22). This served as the template for amplification PCRs in which portions of the 16S rRNA gene (11) and the genes coding for the large subunit of type I (cbbL) and type II (cbbM) RubisCO (23) as well as the nifH genes (24) were amplified; all of the PCR products obtained were sequenced. Individual colonies on overlay plates were confirmed as strains of “Ferrovum” via a PCR that used a reverse primer (NitrosoR) that was specific to this iron-oxidizing acidophile (5′-TCC AGG TTA TTC GCC TGA AC-3′) in combination with the generic 27F primer (11).
Phylogenetic analysis.
The 16S rRNA gene sequence of “F. myxofaciens” P3G was analyzed, using BLAST, at the NCBI database (http://ncbi.nlm.nih.gov/BLAST) and added to a database of over 200,000 prokaryotic 16S rRNA gene sequences. The sequence was aligned together with other related sequences using ClustalX (25); gene sequence alignments were corrected manually, and alignment uncertainties were omitted in the phylogenetic analysis. Phylogenetic trees were generated with a subset of 45 nearly full-length sequences (>1,000 bp) by DNA parsimony, neighbor-joining, and maximum-likelihood analyses. In all cases, general tree topology and clusters were stable, and the reliability of the tree topologies was confirmed by bootstrap analysis using 1,000 replicate alignments. As the topologies of trees generated by all three methods were nearly identical, only the neighbor-joining tree is presented.
Nucleotide sequence accession numbers.
Newly determined sequences were deposited in GenBank under accession numbers HQ322122, HQ322123, and HM044161.
RESULTS
Isolation, purification, and maintenance of “F. myxofaciens.”
Numbers of ferric iron-encrusted colonies that grew on iFeo plates inoculated with disrupted Mynydd Parys acid streamers that appeared (from T-RFLP analysis) to be dominated by the unknown betaproteobacterium were about 1 to 3 orders of magnitude greater than those found on other overlay media. Most of these colonies were very small (<1 mm) and visible only after 2 to 3 weeks of incubation at both 20°C and 30°C. The larger (and less numerous) ferric iron-stained colonies were identified as mostly Acidithiobacillus ferrivorans (22). The specific 16S rRNA gene primers designed to identify “Ferrovum”-like bacteria were successful at differentiating between colonies of this acidophile and those of other iron-oxidizing bacteria. Many of the “Ferrovum”-like colonies that were transferred to liquid medium failed to grow, and most of those that did were subsequently found to be mixed cultures of “F. myxofaciens” and Acidiphilium spp. The latter were mostly A. cryptum SJH, which had presumably migrated from the gel underlayers, which is a common occurrence when plate incubation is protracted, as was necessary to obtain colonies of “Ferrovum.” Obtaining pure cultures of “F. myxofaciens” from liquid cultures that also contained Acidiphilium spp. was found to be extremely difficult, as numbers of planktonic-phase heterotrophic bacteria were usually far greater than those of the iron oxidizer, even when grown in ferrous sulfate medium, since most of the “F. myxofaciens” P3G cells were present in the small streamers. This meant that culture purification using serial dilution was a nonviable option. Alternative methods used to obtain pure cultures of other iron-oxidizing acidophiles, e.g., using the greater tolerance of acidithiobacilli to copper or of the leptospirilli to extreme acidity to eliminate heterotrophic contaminants (12), were also not applicable in the case of “F. myxofaciens.” Ultimately, the only approach that produced axenic cultures of “Ferrovum” was to transfer colonies from iFeo plates that were obtained directly from environmental samples into liquid medium. Due to the problems described above, very few colonies were found to be both viable and axenic, and therefore, large numbers (>50) of colonies needed to be individually processed. A pure culture of “F. myxofaciens” (strain P3G) was obtained from Mynydd Parys acid streamers, but only mixed cultures (with Acidiphilium-like heterotrophs) were obtained from other sites, including the Cae Coch mine, Wales (26); a water treatment plant at Nochten, Germany (27); and the Pyhäsalmi copper/zinc mine, Finland (C. M. Kay and D. B. Johnson, unpublished data). It was also necessary to subculture axenic cultures on a regular basis (every 5 to 7 days for active [30°C] cultures or monthly for those maintained at 4°C) to maintain their viability. Attempts to revive “F. myxofaciens” P3G from stored freeze-dried cultures, or from cultures frozen (at −70°C) in glycerol, glycine betaine, or dimethyl sulfoxide, were not successful.
Morphological characteristics and EPS production.
In liquid medium containing ferrous iron as an electron donor, “F. myxofaciens” P3G produced small streamer-like growths in shake flask cultures, and planktonic-phase cells were also evident. Cells were nonmotile and oval in shape (1.0 ± 0.1 μm long by 0.6 ± 0.04 μm wide), and no endospores were observed. When grown in continuous culture in bioreactors, the streamers became attached to metal supports and other structures within the growth vessel and formed long filaments (28). When grown at pH >2.3, macroscopic growths of “F. myxofaciens” P3G became heavily encrusted with ferric iron precipitates as oxidation progressed, while in media containing ferrous iron poised initially at 2.1 (where ferric iron remained in solution), the gelatinous bacterial growths were free of such precipitates and off-white in color.
Electron microscopy revealed that large amounts of EPS were associated with bacterial cells in the small, streamer-like growths of “F. myxofaciens” P3G that grew in liquid media (Fig. 1B). Insufficient biomass of pure culture was available to determine accurately the water and ash contents of the small streamer-like growths produced in vitro. However, analysis of acid streamers from Mynydd Parys that were dominated by “Ferrovum”-like bacteria found that they consisted of 99% water and that the dry streamers had an ash content of 53%. Of the organic fraction of the streamer biomass, 31% was polysaccharide (glucose equivalent) and 17% was protein. Analysis of hydrolyzed EPS of both iron-grown “F. myxofaciens” P3G and Mynydd Parys acid streamers found the same monosaccharides to be present, with glucose identified as the major sugar and ribose, rhamnose, galactose and mannose present in smaller relative amounts. No acidic or basic sugar monomers were identified in hydrolyzed EPS of either native acid streamers or “F. myxofaciens” P3G streamers.
Electron donors and acceptors.
“F. myxofaciens” strain P3G was shown to be a highly specialized bacterium that grew only by using ferrous iron as an electron donor and oxygen as an electron acceptor. Repeated tests for growth on reduced sulfur, ammonium, hydrogen, and various organic carbon compounds all proved negative. The isolate was, however, able to catalyze the oxidative dissolution of pyrite, which was anticipated in view of its ability to generate ferric iron (the main oxidant of pyrite in acidic liquors). The rate of pyrite dissolution by “F. myxofaciens” P3G was much lower than that by the A. ferrooxidans type strain, and culture pH values did not fall so much (Fig. 2), probably because A. ferrooxidans (unlike “F. myxofaciens”) can oxidize reduced sulfur as well as ferrous iron. Only oxygen was used by “F. myxofaciens” as a terminal electron acceptor.
FIG 2.
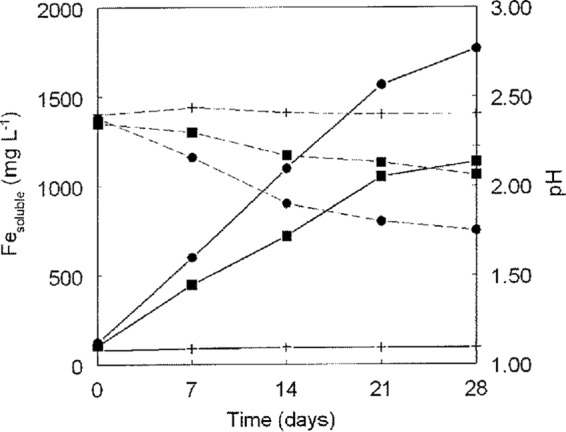
Comparison of pyrite oxidation by “F. myxofaciens” P3G and that by the A. ferrooxidans type strain, showing changes in soluble iron concentrations (solid lines) and pH (broken lines). Symbols: ■, “F. myxofaciens” P3G; ●, A. ferrooxidans type strain; +, abiotic controls. Data points are mean values of duplicate cultures, and error bars (where visible) depict data ranges.
Carbon and nitrogen assimilation.
“F. myxofaciens” P3G could be maintained by subculturing it in liquid media containing only ferrous iron and basal salts; amending liquid media with different organic materials did not result in increased cell numbers and often inhibited iron oxidation, indicating that this acidophile, like many others, is an obligate autotroph. Successful amplification and sequencing of a portion of the large subunit type I (cbbL) RubisCO gene (GenBank accession number HQ322122) implied that isolate P3G fixes carbon dioxide using the Calvin-Benson-Bassham cycle, but despite varying conditions of PCR (including variations in Mg2+ and annealing temperature), no PCR product was obtained using primers that targeted the type II cbbM gene. “F. myxofaciens” P3G could be successfully transferred in liquid medium devoid of inorganic (and organic) nitrogen and under an ammonium-free atmosphere. This, together with the successful amplification of the nifH gene (GenBank accession number HQ322123) from DNA extracted from “F. myxofaciens” P3G, provided evidence that this novel iron-oxidizing acidophile is, like Acidithiobacillus and Leptospirillum spp., a diazotroph.
Effect of temperature and pH on growth and iron oxidation by “F. myxofaciens” P3G.
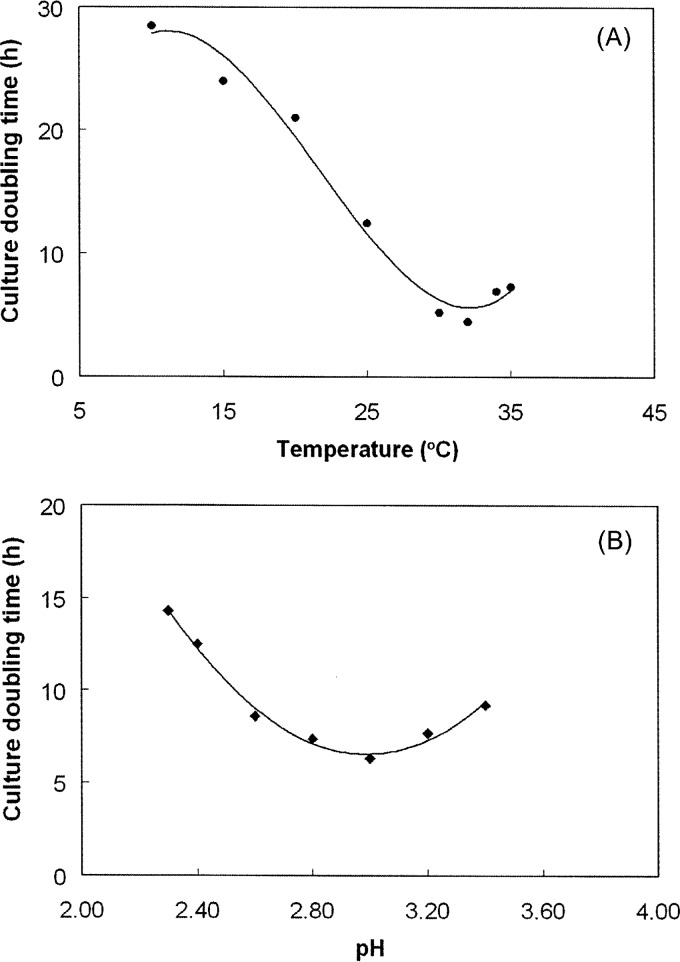
The optimum pH and temperature for growth of “F. myxofaciens” P3G were 3.0 and 32°C, respectively (Fig. 3), with lower limits of pH 2.0 and 4°C; no growth was observed at 37°C. On the basis of these data, “F. myxofaciens” P3G can be categorized as a psychrotolerant extreme acidophile. When grown under optimum conditions of temperature and pH, the culture doubling time of isolate P3G was 4 h (equivalent to a growth rate of 0.17 h−1).
FIG 3.
Effect of temperature (A) and pH (B) on the growth of “F. myxofaciens” P3G.
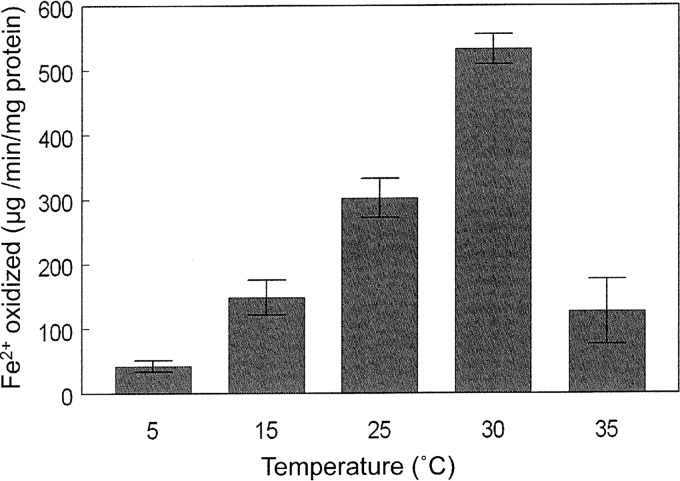
Specific rates of ferrous iron oxidation by “F. myxofaciens” P3G at different temperatures are shown in Fig. 4. These were slightly greater than those recorded for the iron-oxidizing acidithiobacilli at 30°C (18). At 5°C, specific rates of iron oxidation were 8% of that measured at the optimum temperature for this acidophile (30°C).
FIG 4.
Effect of temperature on the specific rates of ferrous iron oxidation by isolate P3G.
Transition metal tolerance.
As is the case with many extreme acidophiles, “F. myxofaciens” P3G displayed tolerance to many (cationic) transition metals frequently found at elevated concentrations in mine-impacted environments, including iron (both ferrous and ferric), aluminum, manganese, copper, and zinc (Table 1). However, this acidophile was far more sensitive to nickel than were iron-oxidizing Acidithiobacillus spp. and, in common with most other acidophiles, was highly intolerant of the oxyanion molybdate.
TABLE 1.
Tolerance of “F. myxofaciens” P3G to some transition metals and comparison with iron-oxidizing acidithiobacilli
| Metal | Concn (mM) of metala: |
|||
|---|---|---|---|---|
| “F. myxofaciens” P3G | Acidithiobacillus ferrooxidans type strain | Acidithiobacillus ferrivorans type strain | Acidithiobacillus ferridurans type strain | |
| Fe2+ | ≥100 | 400 (200) | 400 (200) | 600 (400) |
| Fe3+ | 100 (50) | 400 (200) | <100 | 300 (200) |
| Mn2+ | 30 (20) | ND | ND | ND |
| Al3+ | >100 | 400 (300) | 400 (300) | 400 (300) |
| Cu2+ | 40 (30) | 500 (400) | <50 | 300 (200) |
| Zn2+ | >100 | 1000 (800) | 300 (200) | 1000 (800) |
| Ni2+ | 5 (1) | 200 (100) | 300 (200) | 300 (200) |
| MoO42− | <0.5 | 0.25 (0.10) | <0.10 | 0.10 (0.04) |
Data show concentrations of metals that completely inhibited ferrous iron oxidation by the bacteria listed, and numbers in parentheses show the highest metal concentrations where iron oxidation was observed. ND, not determined.
Phylogenetic relationship.
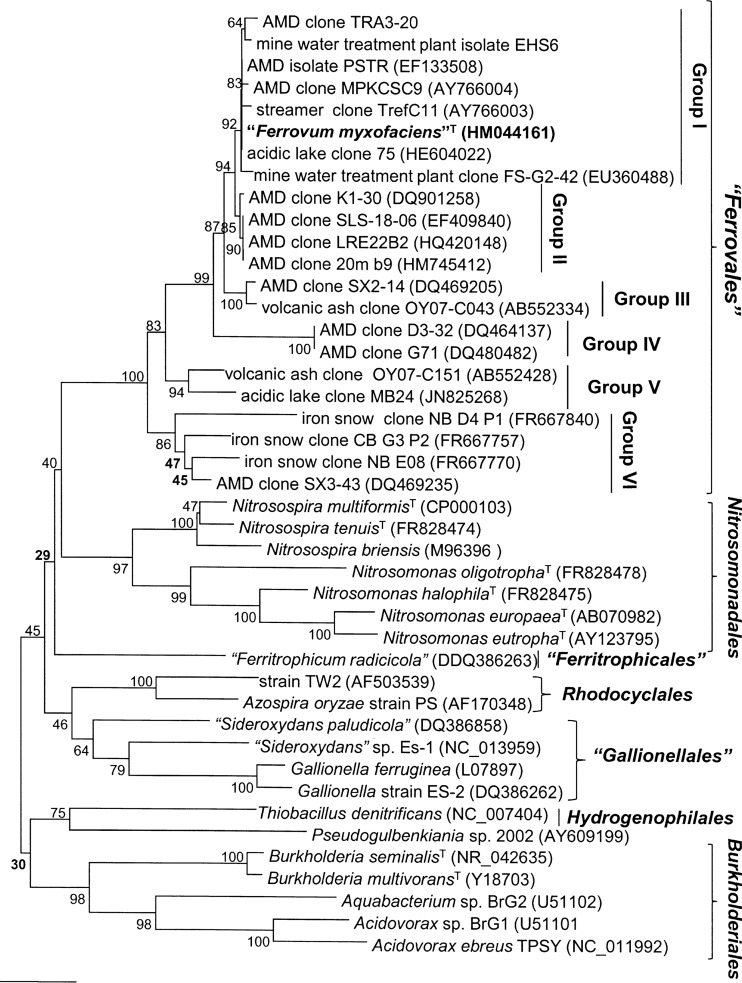
The 16S rRNA gene sequence of “F. myxofaciens” P3G (GenBank accession number HM044161) was nearly identical (>99%) to those cloned from a variety of acidic mine water-impacted environments (Fig. 5). Sequence analysis of the 16S rRNA gene firmly placed “F. myxofaciens” P3G in the Betaproteobacteria (100% confidence using the RDP Classifier), but confidence in the assignment of isolate P3G to any lower rankings of this group was low (e.g., 54% for inclusion in the order Nitrosomonadales). The closest cultivated relatives of “F. myxofaciens” in the database are strains belonging to the ammonium-oxidizing genus Nitrosospira (93% identity). As highlighted in Fig. 5, the phylogenetic relationship (based on the 16S rRNA gene) of “Ferritrophicum radicicola” to other members of the Betaproteobacteria varied when using different algorithms to construct phylogenetic trees. While dendrograms constructed with parsimony and neighbor-joining methods suggested that “Ferritrophicum radicicola” was more closely related to the proposed order “Ferrovales,” calculations based on the maximum-likelihood algorithm placed “Ferritrophicum radicicola” in a cluster with sequences belonging to the orders “Gallionellales” and Rhodocyclales.
FIG 5.
Neighbor-joining tree showing the phylogenetic relationship of the type strain (in bold) of the proposed species “Ferrovum myxofaciens” (belonging to the proposed family “Ferrovaceae” and order “Ferrovales”) to 16S rRNA gene sequences of some closely related clones and orders within the Betaproteobacteria. 16S rRNA gene sequences within the proposed order “Ferrovales” form at least six monophyletic groups. Numbers at the nodes represent bootstrap values out of 1,000 replicates, and GenBank accession numbers for sequences are given in parentheses. Bootstrap values in bold indicate branches which were not supported by the other analysis methods used. The scale bar represents 2% sequence divergence. The tree was rooted with the Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans type strains (not shown).
“F. myxofaciens” and related clone sequences form a monophyletic group within the Betaproteobacteria distantly related to classified orders (e.g., Nitrosomonadales) (Fig. 5). Based on their 16S rRNA gene sequences, environmental clones related to “F. myxofaciens” can further be subdivided into at least six monophyletic groups which might well represent further taxa within the class Betaproteobacteria. Signature nucleotides of the 16S rRNA gene sequences representing the proposed family “Ferrovaceae” and the order “Ferrovales” can be found in Escherichia coli nucleotide positions 258, 464, 559, 630, 721, 811 and 812, 817, 843, and 1109. Bootstrap values shown in Fig. 5 were relatively low for some of the nodes, indicating a lack of statistical significance of the respective branching points, which is probably due to minor differences in the primary structures of 16S rRNA genes of members of neighboring families. Also, most of the signature nucleotides are located in highly variable regions of the 16S rRNA genes, resulting in phylogenetic trees being based on regions which are subject to change. Despite that, similar branching of the orders and families in Fig. 5 occurs irrespective of the algorithm used, affirming their phylogenetic uniqueness (29).
DISCUSSION
The need to isolate prokaryotes that have been thought of as “unculturable” remains a major challenge in microbiology. Although the physiological capabilities of these “unknown” microorganisms may be inferred from construction and annotation of their genomes, (axenic) cultures are necessary to fully characterize novel prokaryotes, and isolation remains a prerequisite for using novel species in emerging biotechnologies. “F. myxofaciens,” which accounts for much of the biomass of some of the largest macroscopic prokaryotic growths that have been described, remained uncultivated for several years after it was detected by molecular methods (30) and decades after acid streamer growths, later shown to be dominated by this acidophile, had been reported (31).
The enigma of why such a visually obvious bacterium remained cryptic and difficult to isolate is due to a number of factors. First, its 16S rRNA gene sequence does not give major clues to its physiological potential, as its closest validated relatives (93% gene similarity) are ammonium-oxidizing neutrophiles of the genera Nitrosospira and Nitrosovibrio. Second, while its natural environment (acidic, ferrous iron-rich streams) might have suggested that the unknown proteobacterium was an iron oxidizer, it shares these habitats with other iron-oxidizing bacteria (notably Acidithiobacillus spp.) that outcompete “Ferrovum”-like bacteria (and other bacteria, such as Leptospirillum spp.) in enrichment cultures. Third, while the overlay media that had been developed prior to 2006 were adept at directly isolating iron-oxidizing and obligately heterotrophic bacteria from environmental samples, they were unsuitable for cultivating “Ferrovum”-like bacteria, due to the extremely fastidious nature of these betaproteobacteria. The introduction of “iFeo” solid medium circumvented these problems, though difficulties in obtaining pure cultures of “Ferrovum”-like bacteria highlighted previously and in maintaining cultures mean that these bacteria present more practical challenges than do other iron-oxidizing acidophilic bacteria. Although colonies of “Ferrovum”-like bacteria have been obtained from a number of mine waters, isolate “F. myxofaciens” P3G is the only strain known to exist at this time in pure culture.
“F. myxofaciens” P3G is a highly specialized bacterium that, like Leptospirillum spp., appears to be restricted to using a single electron donor (ferrous iron). Since it grows only at low pH, this defines “F. myxofaciens” P3G as an obligate aerobe, as oxygen is the only thermodynamically viable electron acceptor for ferrous iron oxidation in acidic liquors. In common with many other iron-oxidizing acidophiles, “F. myxofaciens” P3G is an obligate autotroph that also has the capacity to fix nitrogen.
Table 2 lists locations throughout the world where “Ferrovum”-like bacteria have been detected using molecular techniques. These are exclusively acidic, iron-rich waters, and many are flowing waters in which streamer growths have been reported. The pHs of these water bodies are generally between 2 and 4.8, reflecting the fact that “F. myxofaciens” P3G is less acidophilic than many other iron oxidizers that can grow at pHs of <2 (e.g., Acidithiobacillus, Sulfobacillus, and Ferrimicrobium spp.) or pHs of <1 (Leptospirillum and Ferroplasma spp.). Although its temperature optima for both growth and iron oxidation are ∼30°C, “F. myxofaciens” P3G is metabolically active at 4 to 5°C, confirming that it is psychrotolerant, a trait that it shares with A. ferrivorans (22).
TABLE 2.
Locations where “Ferrovum”-like bacteria have been detected, using molecular approaches
| Site and location (reference) | Physicochemical parameter |
|||
|---|---|---|---|---|
| pH | Temp (°C) | Fe2+ (g liter−1) | SO42− (g liter−1) | |
| Drainage stream, Iron Mountain, CA, USA (30) | 2.4 | 20 | NRc | NR |
| Stream draining a mineral tailing dam, New Zealand (33) | 3.5 | NR | NR | NR |
| Stream draining an abandoned copper mine, Spain (34) | 2.5–2.75 | 15–25 | 0.4–1.2 | 1.5 |
| Acid streamers; abandoned copper mine acid mine drainage, Wales (7) | 2.5 | 9 | 0.5 | 1.7 |
| Acid streamers: chalybeate spa, Wales (7) | 2.9 | 9.5 | 0.02 | NR |
| Water draining copper bioheaps, China (35) | 2.5 | 25 | 1.3b | 2 |
| Acid mine drainage, sulfide mine, China (36) | 2.5 | 25 | 4.0b | 4.3 |
| Water draining an abandoned uranium mine, Germany (37) | 2.6 | NR | NR | 0.8 |
| Water samples, copper mine, China (38) | 2.0 | NR | 1.9b | 3.6 |
| Water samples, pyrite mine, China (39) | 3.0 | 28 | 0.7b | 2.9 |
| Passive mine water remediation site, Australia (40) | 3.0–5.0 | NR | 0.6–1.2 | 0.5–1.7 |
| Water and solids, groundwater treatment plant, Germany (41) | 3.0 | 17 | 1.0 | 2.4 |
| Snottites, abandoned pyrite mine, Germany (42) | 2.6 | NR | 0.8 | 19.0 |
| Biofilms, abandoned uranium mine, Germany (43) | 2.5–2.9 | NR | 0.05–0.3 | 0.8–2.5 |
| Stream draining a surface coal mine; Pennsylvania, USA (44) | 2.4–3.5 | 2.4–3.5 | 0.5 | 3.0 |
| Acid streamers, underground pyrite mine, Wales (26) | 2.1–2.3 | 2.1–2.3 | 0.2–0.7 | 3.4–4.1 |
| Lignite mine pit lake, Germany (45) | 2.9 | 9–15 | 0.6–0.9 | 1.2–8.6 |
| Abandoned polymetallic sulfide mine, Spain (46) | 3.1 | 26 | 2.31 | 9.0 |
| Volcanic ash deposit, Japan (47) | 3.4 | 13 | NR | NR |
| Underground copper mine, Czech Republic (48) | 2.7–3.1 | 4.5–8.7 | 0.06–0.77b | 0.8–3.5 |
| Various acid mine drainage sites, southeastern China (49) | 1.9–4.1 | 13–39 | 0–4.8 | 2.7–5.9 |
| Rio Tinto, Spain (50) | 2.6–3.7 | NR | 0.31–0.75 | 0.8–1.6a |
| Acidic lake (Motykino), Russia (51) | 4.5–4.8 | NR | NR | NR |
| Metal mine pit lake, Spain (52) | 2.5–3.5 | 12–16 | 0–0.5 | 0.7–1.9 |
| Acid streamers, copper mine drainage stream, Wales (13) | 2.5 | 11 | 0.0.38 | 0.8 |
| “Iron snow”; lignite pit lake, Germany (53) | 4.0 | 9–15 | 0.39 g−1b | 0.17 g−1a |
| Water sample, Reiche Zeche mine, Germany (54) | 2.9 | NR | 0.26 | NR |
Determined as total sulfur.
Total iron.
NR, not reported.
The other notable physiological characteristic of “F. myxofaciens” P3G is its propensity to generate copious amounts of EPS. This causes cells to adhere strongly to each other and also to metallic, glass, and plastic surfaces, limiting washout of cells in continuous flow systems, such as one described for remediating acidic, iron-rich mine waters (28). Because the cells attached to the sides of the reactor vessel and formed a mixed community (e.g., with Acidiphilium sp.) with filamentous growths that remained intact at high flow rates, the need to supply a growth support matrix (as in the case of other iron oxidizers) was avoided and the effective working volume of the reactor vessel was greatly enhanced. The psychrotolerant nature of “F. myxofaciens” P3G was also perceived to be an advantage in this system as many ferruginous mine waters have low temperatures.
The novel genus “Ferrovum” is distantly related to classified taxa within the class Betaproteobacteria, with its closest relatives being genera of the family Nitrosomonadaceae (order Nitrosomonadales), which are lithoautotrophic, ammonium-oxidizing bacteria often found in soil and freshwater environments. The closest related (92% 16S rRNA gene similarity) iron-oxidizing bacterium is the nonvalidated species “Ferritrophicum radicicola,” which is the only genus/species of the proposed family “Ferritrophicaceae” (order “Ferritrophicales”) and comprises neutrophilic, microaerophilic iron-oxidizing bacteria (8). Other bacteria distantly related to “F. myxofaciens” P3G (90 to 92% 16S rRNA gene similarity) include members of the family Gallionellaceae (Gram-negative microaerophiles that use ferrous iron as an energy source with CO2 as the sole carbon source). “Ferrovum myxofaciens” (denoting an iron-oxidizing, oval-shaped bacterium that generates copious amounts of slime) represents a new bacterial genus, and 16S rRNA data also suggest that it represents a new family (“Ferrovaceae”) and order (“Ferrovales”) within the class Betaproteobacteria. “F. myxofaciens” is the notably first extreme acidophile to be described in the class Betaproteobacteria.
Footnotes
Published ahead of print 15 November 2013
REFERENCES
- 1.Johnson DB. 2009. Extremophiles: acid environments, p 107–126 In Schaechter M. (ed), Encyclopaedia of microbiology. Elsevier, Oxford, United Kingdom [Google Scholar]
- 2.Johnson DB. 1998. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 27:307–317. 10.1111/j.1574-6941.1998.tb00547.x [DOI] [Google Scholar]
- 3.Nordstrom DK. 2000. Advances in the hydrogeochemistry and microbiology of acid mine waters. Int. Geol. Rev. 42:499–515. 10.1080/00206810009465095 [DOI] [Google Scholar]
- 4.Nordstrom DK, Alpers CN, Ptacek CJ, Blowes DW. 2000. Negative pH and extremely acidic minewaters from Iron Mountain, California. Environ. Sci. Technol. 34:254–258. 10.1021/es990646v [DOI] [Google Scholar]
- 5.Bond PL, Smriga SP, Banfield JF. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842–3849. 10.1128/AEM.66.9.3842-3849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond PL, Druschel GK, Banfield JF. 2000. Comparison of acid mine drainage communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962–4971. 10.1128/AEM.66.11.4962-4971.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallberg KB, Coupland K, Kimura S, Johnson DB. 2006. Macroscopic “acid streamer” growths in acidic, metal-rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022–2030. 10.1128/AEM.72.3.2022-2030.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss JV, Rentz JA, Plaia T, Neubauer SC, Merrill-Floyd M, Lilburn T, Bradburne C, Megonigal JP, Emerson D. 2007. Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol. J. 24:559–570. 10.1080/01490450701670152 [DOI] [Google Scholar]
- 9.Kimura S. 2005. Microbial communities and interactions in extremely acidic environments. Ph.D. thesis University of Wales, Cardiff, Wales, United Kingdom [Google Scholar]
- 10.Johnson DB, Rolfe S, Hallberg KB, Iversen E. 2001. Isolation and phylogenetic characterisation of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3:630–637. 10.1046/j.1462-2920.2001.00234.x [DOI] [PubMed] [Google Scholar]
- 11.Okibe N, Gericke M, Hallberg KB, Johnson DB. 2003. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred tank bioleaching operation. Appl. Environ. Microbiol. 69:1936–1943. 10.1128/AEM.69.4.1936-1943.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DB, Hallberg KB. 2007. Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms, p 237–261 In Rawlings DE, Johnson DB. (ed), Biomining. Springer-Verlag, Berlin, Germany [Google Scholar]
- 13.Kay CM, Rowe OF, Rocchetti L, Coupland K, Hallberg KB, Johnson DB. 2013. Evolution of microbial “streamer” growths in an acidic metal-contaminated stream draining an abandoned underground copper mine. Life 3:189–211. 10.3390/life3010189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osorio H, Mangold S, Denis Y, Ñancucheo I, Esparza M, Johnson DB, Bonnefoy V, Dopson M, Holmes DS. 2013. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79:2172–2181. 10.1128/AEM.03057-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stookey L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779–781. 10.1021/ac60289a016 [DOI] [Google Scholar]
- 16.Hedrich S, Johnson DB. 24 September 2013. Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 10.1111/1574-6968.12290 [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 18.Johnson DB, Kanao T, Hedrich S. 2012. Redox transformations of iron at extremely low pH: fundamental and applied aspects. Front. Microbiol. 3:96. 10.3389/fmicb.2012.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28:350–356. 10.1021/ac60111a017 [DOI] [PubMed] [Google Scholar]
- 20.Johnson DB, Kelso WI. 1981. Extracellular polymers of acid streamers from pyritic mines. Environ. Pollut. 24:291–301. 10.1016/0143-1471(81)90066-0 [DOI] [Google Scholar]
- 21.Ñancucheo I, Johnson DB. 2010. Production of glycolic acid by chemolithotrophic iron- and sulfur-oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral-oxidizing consortia. Appl. Environ. Microbiol. 76:461–467. 10.1128/AEM.01832-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hallberg KB, González-Toril E, Johnson DB. 2010. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14:9–19. 10.1007/s00792-009-0282-y [DOI] [PubMed] [Google Scholar]
- 23.Hallberg KB, Hedrich S, Johnson DB. 2011. Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 15:271–279. 10.1007/s00792-011-0359-2 [DOI] [PubMed] [Google Scholar]
- 24.Ueda T, Suga Y, Yahiro N, Matsuguchi T. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 26.Kimura S, Bryan CG, Hallberg KB, Johnson DB. 2011. Biodiversity and geochemistry of an extremely acidic, low temperature subterranean environment sustained by chemolithotrophy. Environ. Microbiol. 13:2092–2104. 10.1111/j.1462-2920.2011.02434.x [DOI] [PubMed] [Google Scholar]
- 27.Hedrich S, Heinzel E, Seifert J, Schlömann M. 2009. Isolation of novel iron-oxidizing bacteria from an acid mine water treatment plant. Adv. Mater. Res. 71-73:125–128. 10.4028/www.scientific.net/AMR.71-73.125 [DOI] [Google Scholar]
- 28.Hedrich S, Johnson DB. 2012. A modular continuous flow reactor system for the selective bio-oxidation and precipitation of iron in mine-impacted waters. Bioresour. Technol. 106:44–49. 10.1016/j.biortech.2011.11.130 [DOI] [PubMed] [Google Scholar]
- 29.Stackebrandt E, Rainey FA, Ward-Rainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria class nov. Int. J. Syst. Bacteriol. 47:479–491. 10.1099/00207713-47-2-479 [DOI] [Google Scholar]
- 30.Edwards KJ, Goebel BM, Rodgers TM, Schrenk MO, Gihring TM, Cardona MM, Hu B, McGuire MM, Hamers RJ, Pace NR, Banfield JF. 1999. Geomicrobiology of pyrite (FeS2) dissolution: a case study at Iron Mountain, California. Geomicrobiol. J. 16:155–179. 10.1080/014904599270668 [DOI] [Google Scholar]
- 31.Johnson DB, Kelso WI, Jenkins DA. 1979. Bacterial streamer growth in a disused pyrite mine. Environ. Pollut. 18:107–118. 10.1016/0013-9327(79)90086-7 [DOI] [Google Scholar]
- 32.Hedrich S, Johnson DB. 2013. Acidithiobacillus ferridurans, sp. nov.; an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic Gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 63:4018–4025. 10.1099/ijs.0.049759-0 [DOI] [PubMed] [Google Scholar]
- 33.Webster J, Lane V, Howarth R, Swedlund P, Saul D. 2000. Factors influencing the precipitation of sulphate-rich iron oxides, and their ability to adsorb trace metals. J. Conf. Abstr. 5(2):1073 http://goldschmidtabstracts.info/abstracts/abstractView?abstractId=2000001073 [Google Scholar]
- 34.Rowe OF, Sanchez-Espana J, Hallberg KB, Johnson DB. 2007. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ. Microbiol. 9:1761–1771. 10.1111/j.1462-2920.2007.01294.x [DOI] [PubMed] [Google Scholar]
- 35.Xie X, Xiao S, He Z, Liu J, Qiu G. 2007. Microbial populations in acid mineral bioleaching systems of Tong Shankou copper mine, China. J. Appl. Microbiol. 103:1227–1238. 10.1111/j.1365-2672.2007.03382.x [DOI] [PubMed] [Google Scholar]
- 36.He Z, Xie X, Xiao S, Liu J, Qiu G. 2007. Microbial diversity of mine water at Zhong Tiaoshan copper mine, China. J. Basic Microbiol. 47:485–495. 10.1002/jobm.200700219 [DOI] [PubMed] [Google Scholar]
- 37.Seifert J, Erler B, Seibt K, Rohrbach N, Arnold J, Schlömann M, Kassahun S, Jenk U. 2008. Characterization of the microbial diversity in the abandoned uranium mine Königstein, p 733–742 In Merkel B, Hasche-Berger A. (ed), Uranium, mining and hydrogeology. Springer, Berlin, Germany [Google Scholar]
- 38.Yin H, Cao L, Qiu G, Wang D, Kellogg L, Zhou J, Liu X, Dai Z, Ding J, Liu X. 2008. Molecular diversity of 16S rRNA and gyrB genes in copper mines. Arch. Microbiol. 189:101–110. 10.1007/s00203-007-0298-6 [DOI] [PubMed] [Google Scholar]
- 39.Tan GL, Shu WS, Zhou WH, Li XL, Lan CY, Huang LN. 2009. Seasonal and spatial variations in microbial community structure and diversity in the acid stream draining across an ongoing surface mining site. FEMS Microbiol. Ecol. 70:121–129. 10.1111/j.1574-6941.2009.00744.x [DOI] [PubMed] [Google Scholar]
- 40.Dann AL, Cooper RS, Bowman JP. 2009. Investigation and optimization of a passively operated compost-based system for remediation of acidic, highly iron- and sulfate-rich industrial waste water. Water Res. 43:2302–2316. 10.1016/j.watres.2009.02.030 [DOI] [PubMed] [Google Scholar]
- 41.Heinzel E, Hedrich S, Janneck E, Glombitza F, Seifert J, Schlömann M. 2009. Bacterial diversity in a mine water treatment plant. Appl. Environ. Microbiol. 75:858–861. 10.1128/AEM.01045-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegler S, Ackermann S, Majzlan J, Gescher J. 2009. Matrix composition and community structure analysis of a novel bacterial pyrite leaching community. Environ. Microbiol. 11:2329–2338. 10.1111/j.1462-2920.2009.01959.x [DOI] [PubMed] [Google Scholar]
- 43.Brockmann S, Arnold T, Schweder B, Bernhard G. 2010. Visualizing acidophilic microorganisms in biofilm communities using acid stable fluorescence dyes. J. Fluoresc. 20:943–951. 10.1007/s10895-010-0640-2 [DOI] [PubMed] [Google Scholar]
- 44.Brown JF, Jones DS, Mills DB, Macalady JL, Burgos WD. 2011. Application of a depositional facies model to an acid mine drainage site. Appl. Environ. Microbiol. 77:545–554. 10.1128/AEM.01550-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiche M, Lu S, Ciobota V, Neu TR, Nietzsche S, Rösch P, Popp J, Küsel K. 2011. Pelagic boundary conditions affect the biological formation of iron-rich particles (iron snow) and their microbial communities. Limnol. Oceanogr. 56:1386–1398. 10.4319/lo.2011.56.4.1386 [DOI] [Google Scholar]
- 46.González-Toril E, Águilera A, Souza-Egipsy V, Pamo EL, España JS, Amils R. 2011. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl. Environ. Microbiol. 77:2685–2694. 10.1128/AEM.02459-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujimura R, Sato Y, Nishizawa T, Nanba K, Oshima K, Hattori M, Kamijo T, Ohta H. 2012. Analysis of early bacterial communities on volcanic deposits on the island of Miyake (Miyake-jima), Japan: a 6-year study at a fixed site. Microbes Environ. 27:19–29. 10.1264/jsme2.ME11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falteisek L, Čepička I. 2012. Microbiology of diverse acidic and non-acidic microhabitats within a sulfidic ore mine. Extremophiles 16:911–922. 10.1007/s00792-012-0488-2 [DOI] [PubMed] [Google Scholar]
- 49.Kuang JL, Huang LN, Chen LX, Hua ZS, Li SJ, Hu M, Li JT, Shu WS. 2013. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J. 7:1038–1050. 10.1038/ismej.2012.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Moyano A, González-Toril E, Aguilera A, Amils R. 2012. Comparative microbial ecology study of the sediments and the water column of the Río Tinto, an extreme acidic environment. FEMS Microbiol. Ecol. 81:303–314. 10.1111/j.1574-6941.2012.01346.x [DOI] [PubMed] [Google Scholar]
- 51.Fedotova AV, Belova SE, Kulichevskaya IS, Dedysh SN. 2012. Molecular identification of filterable bacteria and archaea in the water of acidic lakes of Northern Russia. Microbiology 81:281–287. 10.1134/S002626171203006X [DOI] [Google Scholar]
- 52.Santofimia E, Gonzalez-Toril E, Lopez-Pamo E, Gomariz M, Amils R, Aquilera A. 2013. Microbial diversity and its relationship to physicochemical characteristics of the water in two extreme acidic pit lakes from the Iberian Pyrite Belt (SW Spain). PLoS One 8:e66746. 10.1371/journal.pone.0066746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu S, Chourey K, Reiche M, Nietzsche S, Shah MB, Neu TR, Hettich RL, Küsel K. 2013. Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (“iron snow”). Appl. Environ. Microbiol. 79:4272–4281. 10.1128/AEM.00467-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tischler JS, Jwair RJ, Gelhaar N, Drechsel A, Skirl A-M, Wiacek C, Janneck E, Schlömann M. 2013. New cultivation medium for “Ferrovum” and Gallionella-related strains. J. Microbiol. Methods 95:138–144. 10.1016/j.mimet.2013.07.027 [DOI] [PubMed] [Google Scholar]