Abstract
One of the functions of the mammalian large intestinal microbiota is the fermentation of plant cell wall components. In ruminant animals, the majority of their nutrients are obtained via pregastric fermentation; however, up to 20% can be recovered from microbial fermentation in the large intestine. Eight-week continuous culture enrichments of cattle feces with cellulose and xylan-pectin were used to isolate bacteria from this community. A total of 459 bacterial isolates were classified phylogenetically using 16S rRNA gene sequencing. Six phyla were represented: Firmicutes (51.9%), Bacteroidetes (30.9%), Proteobacteria (11.1%), Actinobacteria (3.5%), Synergistetes (1.5%), and Fusobacteria (1.1%). The majority of bacterial isolates had <98.5% identity to cultured bacteria with sequences in the Ribosomal Database Project and thus represent new species and/or genera. Within the Firmicutes isolates, most were classified in the families Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, and Clostridiaceae I. The majority of the Bacteroidetes were most closely related to Bacteroides thetaiotaomicron, B. ovatus, and B. xylanisolvens and members of the Porphyromonadaceae family. Many of the Firmicutes and Bacteroidetes isolates were related to species demonstrated to possess enzymes which ferment plant cell wall components; the others were hypothesized to cross-feed these bacteria. The microbial communities that arose in these enrichment cultures had broad bacterial diversity. With over 98% of the isolates not represented as previously cultured, there are new opportunities to study the genomic and metabolic capacities of these members of the complex intestinal microbiota.
INTRODUCTION
A function of the gut microbiota in mammalian herbivores and omnivores is the fermentation of plant cell wall components; mammals lack the enzymes required to breakdown β-(1,4) bonds and other linkages between the monosaccharides that make up plant cell walls (1, 2). The plant cell wall is made up of cellulose and hemicelluloses, the primary structural components of plants, and contain smaller and variable amounts of pectins, β-glucans, oligosaccharides, lignins, and glycoproteins (3).
The physiology of ruminant animals, such as cattle, goats, and sheep, includes a foregut, where fermentation occurs and which allows the animal to directly utilize the short-chain fatty acids (SCFAs) produced by the anaerobic microbial fermentation of plant cell walls for energy (2). Thus, ruminal fermentation provides between 60% and 80% of the animal's energy requirements (4, 5). Ruminants obtain energy from the SCFAs produced in the large intestine, which provides from 0 to 20% of their dietary requirements, similar to the levels reported for other mammals (4, 5).
A number of intestinal microbial metagenomes, including those of cattle (6, 7), human (8, 9), pig (10), and chicken (11), have been demonstrated to be enriched in carbohydrate transport and metabolism genes. Ley et al. (1) found that the type of diet (herbivorous, omnivorous, or carnivorous) strongly predicted the fecal microbial composition across a broad sampling of mammals. The microbiotas in the rumen and feces of cows are distinct from each other (12, 13), with an ∼60% difference between the bacterial profiles in the rumen and feces being reported (14). Most metagenomic studies in ruminants have focused on the rumen, and while a number of studies have determined the phylogenetic structure of the microbial communities in cattle feces (14–16), only one (17) reported the functional genes in the feces of beef cattle, of which 12.8% were carbohydrate-related genes.
While a great deal of information is currently being gathered using metagenomic techniques, it is still useful to have isolated microorganisms available for study. Few cultured bacteria, other than pathogens, from the feces of cattle are included in phylogenetic databases. In order to determine the bacteria associated with cellulose and hemicellulose fermentation in the large intestine of the cow, continuous culture fermentors were used to enrich for those microorganisms. The aim of the present study was to isolate a broad range of bacteria from these enrichments and further characterize them by 16S rRNA gene phylogeny. This work makes available a large collection of bacterial isolates for further study.
MATERIALS AND METHODS
Animals and fermentor operation.
Freshly voided feces (at least 500 g) were collected from 4 multiparous prepartum dairy cows fed a high-forage diet at the Iowa State University Dairy Farm and returned to the lab for processing within 30 min. A fecal slurry (1:10 wt/vol) was made using anaerobic phosphate-buffered saline (PBS) and mixing for 3 min in a lab blender (Seward Stomacher, model 400; Fisher Scientific) in an anaerobic chamber (Coy, Grass Lake, MI). Two 300-ml slurries from each fecal (n = 4) sample were prepared for inoculation into fermentors. One fermentor was enriched with cellulose (CAS no. 9004-34-6; Alphacel; ICN Biomedicals, Aurora, OH), and the other was enriched with xylan-pectin (2:1 oat spelt xylan [CAS no. 9014-63-5; Sigma, St. Louis, MO] and pectin [CAS no. 9000-69-5, ICN Biomedicals]). The fermentors were fed 3 g carbohydrate twice daily at 0600 and 1700 h.
BioFlo 110 fermentors (New Brunswick Scientific, Edison, NJ) to which 350 ml anoxic nutrient medium with no carbohydrate (Table 1) was previously added received the fecal slurry. Each fermentor was fed 3 g of polysaccharide, and the organisms were allowed to grow in batch culture without nutrient medium inflow until the next day (∼18 h). The fermentors were then allowed to reach the working volume of 700 ml (∼2.3 h) by addition of nutrient medium at a dilution rate of 0.03 h−1. The operating conditions of the fermentors were set to model conditions in the large intestine (cecum and colon) of cattle. The contents of the vessels were continuously mixed and sparged with nitrogen to maintain anaerobic conditions. The working volume was maintained by removal of contents with an outflow pump. Throughout the 8 weeks of operation, a temperature of 38°C and a pH of 6.7 were maintained.
TABLE 1.
Fermentor nutrient medium
| Ingredienta | Concn |
|---|---|
| Peptone water | 2.0 g/liter |
| Yeast extract | 2.0 g/liter |
| NaCl | 0.1 g/liter |
| K2HPO4 | 0.04 g/liter |
| KH2PO4 | 0.04 g/liter |
| MgSO4 · 7H2O | 0.01 g/liter |
| CaCl2 · 6H2O | 0.01 g/liter |
| NaHCO3 | 2.0 g/liter |
| Cysteine-HCl | 0.5 g/liter |
| Bile salts | 0.5 g/liter |
| Heminb | 0.05 g/liter |
| Tween 80 | 2.0 ml/liter |
| Vitamin K1 | 0.01 ml/liter |
All chemicals were obtained from Sigma.
Hemin solution consisted of 0.5 g hemin in 25 ml 1 M NaOH.
Bacterial isolations.
After 8 weeks, 1 ml of the fermentor contents was anaerobically transferred into 9 ml anaerobic 1/2-strength peptone water (Fisher Scientific) and then serially diluted through a 10−10 dilution. Dilutions of 10−5 through 10−10 were plated onto Wilkins-Chalgren agar plates. Carbohydrate-specific agar plates (18) (Table 2) were replicate plated (19); the carbohydrates used were cellulose, carboxymethyl cellulose (CAS no. 9000-11-7; Sigma), beech wood xylan (CAS no. 9014-63-5; Sigma), and pectin. The plates were incubated anaerobically at 38°C for 3 to 5 days. In order to get the greatest range of bacteria associated with cellulose and xylan-pectin fermentation, the plates with the lowest dilutions with distinct colonies were chosen for bacterial isolations.
TABLE 2.
Composition of carbohydrate-specific agars
| Ingredienta | Concn |
|---|---|
| Carbohydrate | 5.0 g/liter |
| Trypticase | 4.5 g/liter |
| Yeast extract | 0.5 g/liter |
| Agar | 20.0 g/liter |
| Mineral 1b | 40.0 ml/liter |
| Mineral 2c | 40.0 ml/liter |
| Hemind | 10.0 ml/liter |
| VFA solutione | 10.0 ml/liter |
| Resazurin (0.1% solution) | 1.0 ml/liter |
| Na2S–l-cysteine HCl solutionf | 10.0 ml/liter |
| Incubated clarified rumen fluidg | 0.0 ml/liter |
| Distilled water | 870.0 ml/liter |
All chemicals were purchased from Sigma.
Mineral 1 is K2HPO4, 6 g liter−1.
Mineral 2 is KH2PO4, 6 g liter−1; (NH4)2SO4, 6 g liter−1; NaCl, 12 g liter−1; MgSO4, 2.45 g liter−1; and CaCl2 · 2H2O, 1.69 g liter−1.
Added as a solution of 0.5 g hemin in 25 ml 1 M NaOH.
Short chain fatty acid solution contained the following acids, in ml/liter: acetic, 6.85; propionic, 3.00; butyric, 1.85; isobutyric, 0.50; 2-methyl butyric, 0.55; N-valeric, 0.55; and isovaleric, 0.55.
Two hundred milliliters anaerobic solution at pH 10 with 2.5 g l-cysteine HCl · H2O and 2.5 g Na2S · 9H2O.
Incubated clarified rumen fluid modified from the work of Allison et al. (50) as follows: 400 ml rumen fluid with addition of 300 ml mineral 1, 300 ml mineral 2, and 4 g Na2CO3; addition of a second centrifugation after overnight storage at 4°C; and adjustment to pH 7.0 prior to autoclaving. The fluid was stored frozen at −20°C until it was used.
Colonies were selected from the carbohydrate-specific agar plates on the basis of differential colony morphology and plated onto anaerobic Wilkins-Chalgren agar to obtain individual isolates. The purity of the bacterial cultures was determined using Gram stain and microscopic observations. The isolated bacteria were cryopreserved using a Microbank bacterial preservation system (Pro-Lab Diagnostics, Austin, TX). DNA was isolated using a DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions for bacterial cells. PCR amplification of 16S rRNA genes was done with primers 27F (5′-AGAGGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (Invitrogen, Carlsbad, CA) (20) and a DYAD DNA Engine thermocycler (MJ Research, Watertown, MA). The PCR mixture (50 μl) contained 1× Qiagen PCR buffer, 1.25 U of Taq polymerase (Qiagen), 0.25 mM each deoxynucleoside triphosphate (Amresco, Solon, OH), 25 pmol of each primer, and 80 μg of template DNA. Amplified products were cleaned using QIAquick 96 PCR purification (Qiagen) and sequenced by the Iowa State University DNA Sequencing and Synthesis Facility (Ames, IA) using an ABI Prism 377 sequencer (Applied Biosystems, Foster City, CA).
Sequence analysis.
Sequences were made using VNTI (v.11.1) software (Invitrogen). Isolates with sequences ≥1,200 bp in length were included. The closest related sequences were identified using the Ribosomal Database Project (RDP) (21) 16S rRNA gene database for the nearly full-length sequences. Sequence similarity was analyzed using Bionumerics (v.6.5) software (Applied Maths, Austin, TX), and an unrooted dendrogram was created using standard pairwise alignment and unweighted pair group method with arithmetic mean (UPGMA) clustering. Bacteria isolated from the same fermentor with ≥99% similarity were removed from the final analysis in order to simplify the data set and reduce redundancy. Data are presented by phylum for all bacterial isolates except the Firmicutes and Bacteroidetes, which are divided into class and family groupings.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in GenBank under the accession numbers JQ607747 to JQ608320.
RESULTS
A total of 573 bacteria were isolated from the cellulose and xylan-pectin enrichments with the cow (n = 4) fecal inoculum. Exclusion of 114 bacteria because they had ≥99% similarity to bacteria isolated from the same fermentor and fecal donor resulted in a collection of 459 isolates. Six phyla were represented among the bacterial isolates: Firmicutes (n = 238), Bacteroidetes (n = 142), Proteobacteria (n = 51), Actinobacteria (n = 16), Synergistetes (n = 7), and Fusobacteria (n = 5). The majority of bacterial isolates had <98.5% identity to cultured bacteria with sequences in the RDP. The distribution of 259 bacterial isolates with <95% identity to previously cultured bacteria was 131 Firmicutes, 97 Bacteroidetes, 13 Proteobacteria, 9 Actinobacteria, 4 Synergistetes, and 5 Fusobacteria. A total of 191 isolates had identities to previously cultured bacteria of between 95 and 98.5% and were distributed across the phyla as follows: 99 Firmicutes, 46 Bacteroidetes, 30 Proteobacteria, 8 Actinobacteria, and 3 Synergistetes.
The number of isolates obtained from the enrichments with each carbohydrate differed, with 269 bacteria coming from the cellulose enrichments and 190 coming from the xylan-pectin enrichments. Furthermore, the distribution of isolates recovered from each polysaccharide differed across the phyla and the genera within a phylum (Table 3). The most striking differences in isolate numbers between the cellulose and xylan-pectin enrichments by phylum were for the Firmicutes (135 versus 105, respectively) and Proteobacteria (44 versus 7, respectively).
TABLE 3.
Distribution of bacteria isolated from cellulose or xylan-pectin enrichments of cow feces by phylum and genus
| Phylum | Genus | No. of isolates |
|
|---|---|---|---|
| Cellulose | Xylan-pectin | ||
| Firmicutes | Anaerofilum | 1 | 1 |
| Anaerosporobacter | 18 | 0 | |
| Anaerostipes | 0 | 1 | |
| Anaerovorax | 2 | 0 | |
| Blautia | 8 | 11 | |
| Butyricicoccus | 0 | 3 | |
| Clostridium | 21 | 4 | |
| Clostridium IV | 3 | 4 | |
| Clostridium XI | 0 | 1 | |
| Clostridium XIVa | 21 | 37 | |
| Clostridium XIVb | 2 | 0 | |
| Clostridium XVIII | 3 | 14 | |
| Enterococcus | 5 | 1 | |
| Erysipelotrichaceae incertae sedis | 3 | 15 | |
| Eubacterium | 2 | 0 | |
| Flavonifractor | 0 | 1 | |
| Lachnospiraceae incertae sedis | 3 | 0 | |
| Lachnospiraceae, unclassified | 5 | 1 | |
| Lactonifactor | 1 | 1 | |
| Megasphaera | 0 | 2 | |
| Paenibacillus | 1 | 0 | |
| Peptostreptococcus | 0 | 1 | |
| Oscillibacter | 10 | 5 | |
| Ruminococcus, unclassified | 5 | 1 | |
| Sedimentibacter | 1 | 0 | |
| Sporanaerobacter | 2 | 0 | |
| Staphylococcus | 1 | 0 | |
| Streptococcus | 1 | 1 | |
| Tissierella | 14 | 0 | |
| Bacteroidetes | Bacteroides | 39 | 55 |
| Dysgonomonas | 19 | 10 | |
| Parabacteroides | 9 | 2 | |
| Proteiniphilum | 8 | 0 | |
| Proteobacteria | Aeromonas | 6 | 1 |
| Citrobacter | 1 | 0 | |
| Desulfovibrio | 15 | 1 | |
| Escherichia | 0 | 1 | |
| Morganella | 1 | 0 | |
| Proteus | 21 | 4 | |
| Synergistetes | Cloacibacillus | 6 | 1 |
| Fusobacteria | Fusobacterium | 4 | 1 |
| Actinobacteria | Bifidobacterium | 0 | 4 |
| Olsenella | 0 | 1 | |
| Paraeggerthella | 1 | 0 | |
| Propionibacterium | 0 | 4 | |
| Micrococcineae, unclassified | 1 | 0 | |
| Actinomycetales, unclassified | 5 | 0 | |
While the total number of Bacteroidetes isolates between the polysaccharide enrichments was similar, the distribution of the isolates belong to different genera differed. Cellulose enrichments yielded greater numbers of species belong to the genera Dysgonomonas, Parabacteroides, and Proteiniphilum, while species belonging to the Bacteroides were more frequently isolated from xylan-pectin enrichments. The numbers of bacteria isolated from each fecal donor were comparable (88 from cow A, 114 from cow B, 154 from cow C, and 103 from cow D). There were differences in the distribution of bacterial isolates from the different fecal donors within the phyla, as follows. No Synergistetes or Bacilli (in the Firmicutes) were isolated from cow A, but all of the Fusobacteria isolated were from this fecal donor. In the Proteobacteria, all of isolates in the Aeromonadaceae were from cows A and C, while no Deltaproteobacteria were isolated from cow B.
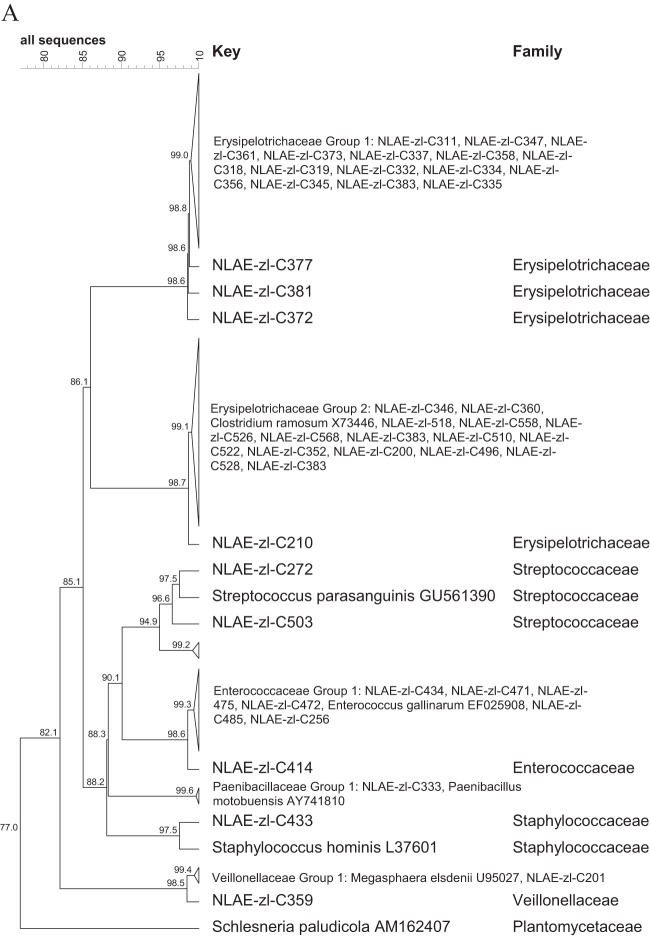
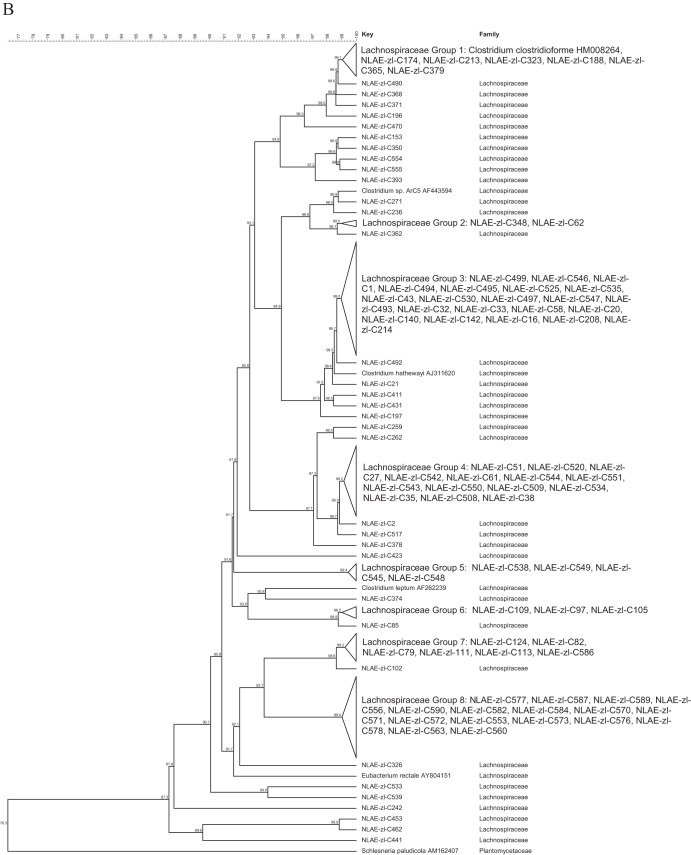
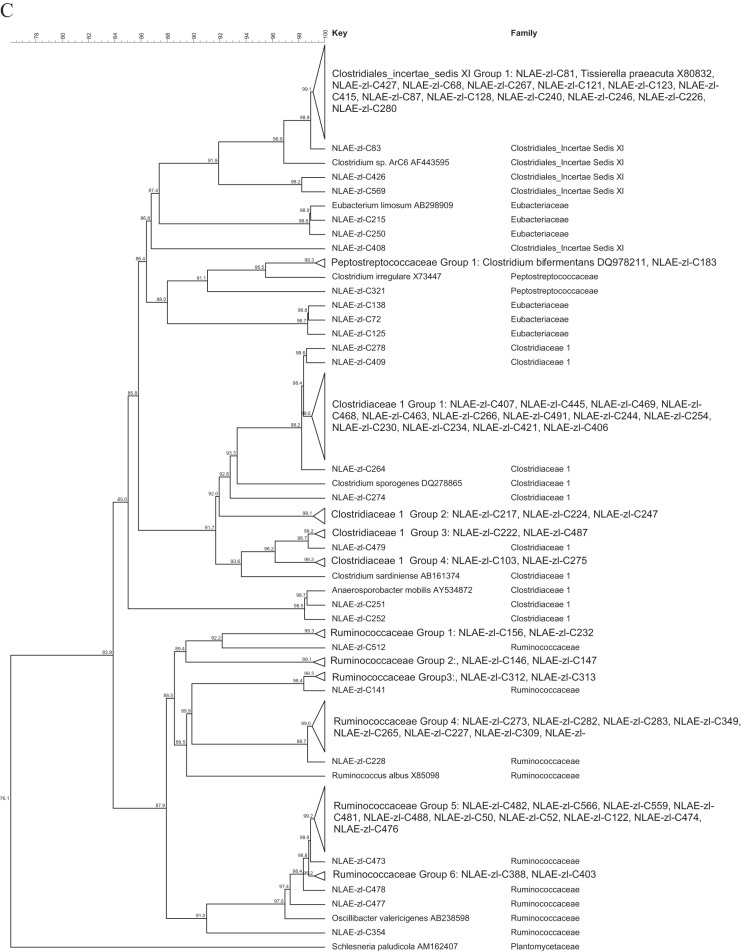
The largest proportion of the bacterial isolates were members of the Firmicutes (51.9%). For ease of discussion, the isolates in the Firmicutes were divided into the classes Bacilli (13 isolates), Clostridia (191 isolates), Erysipelotrichia (32 isolates), and Negativicutes (2 isolates), with further division based on family classification (Fig. 1). The Bacilli (Fig. 1A) were from 4 families: the Paenibacillaceae (1 isolate), Enterococcaceae (7 isolates), Staphylococcaceae (1 isolate), and Streptococcaceae (4 isolates). The Erysipelotrichia were all in the family Erysipelotrichaceae (32 isolates), and the Negativicutes were all in the family Veillonellaceae (2 isolates). The largest family grouping within the isolates was Lachnospiraceae (107 isolates) in the Clostridia class of the Firmicutes (Fig. 1B). Within the Lachnospiraceae, the majority of the bacteria fell into the RDP genus Clostridium XIVa (60 isolates), followed by the genera Blautia (19 isolates) and Anaerosporobacter (16 isolates). The other Clostridia were from the families Ruminococcaceae (33 isolates, 15 Oscillibacter isolates), Clostridiaceae I (27 isolates, 25 Clostridium sensu stricto), Clostridiales incertae sedis XI (17 isolates, 13 Tissierella sensu stricto), Eubacteriaceae (5 isolates), and Peptostreptococcaceae (2 isolates) (Fig. 1C).
FIG 1.
Cluster analysis with an unrooted dendrogram of bacterial isolates from cellulose and xylan-pectin enrichments of cow feces in the phylum Firmicutes. (A) Isolates in the Bacilli, Erysipelotrichia (family Erysipelotrichaceae), and Negativicutes (family Veillonellaceae) classes; (B) isolates in the Clostridia class and Lachnospiraceae family; (C) isolates in the Clostridia class and Peptostreptococcaceae, Clostridiaceae 1, Clostridiales incertae sedis XI, and Eubacteriaceae families. The genus and species names and GenBank (NCBI) accession number are included for the reference strains. Bacteria with ≥99% similarity are grouped in order to simplify the dendrogram. The line at the top represents percent similarity across isolates; Schlesneria paludicola (Plantomycetes) is used as the outgroup.
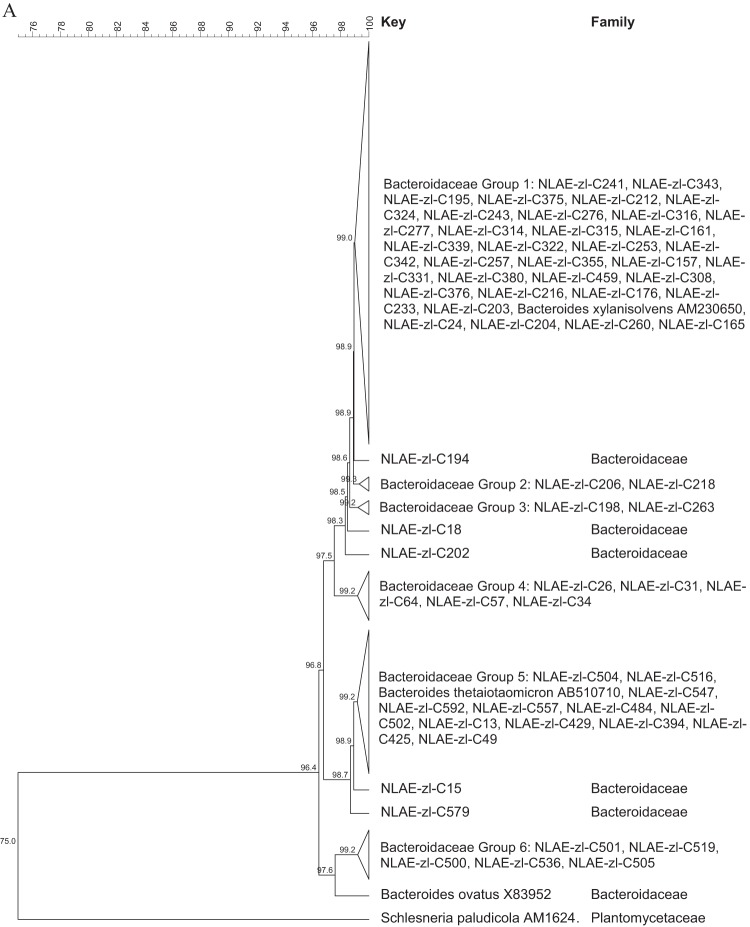
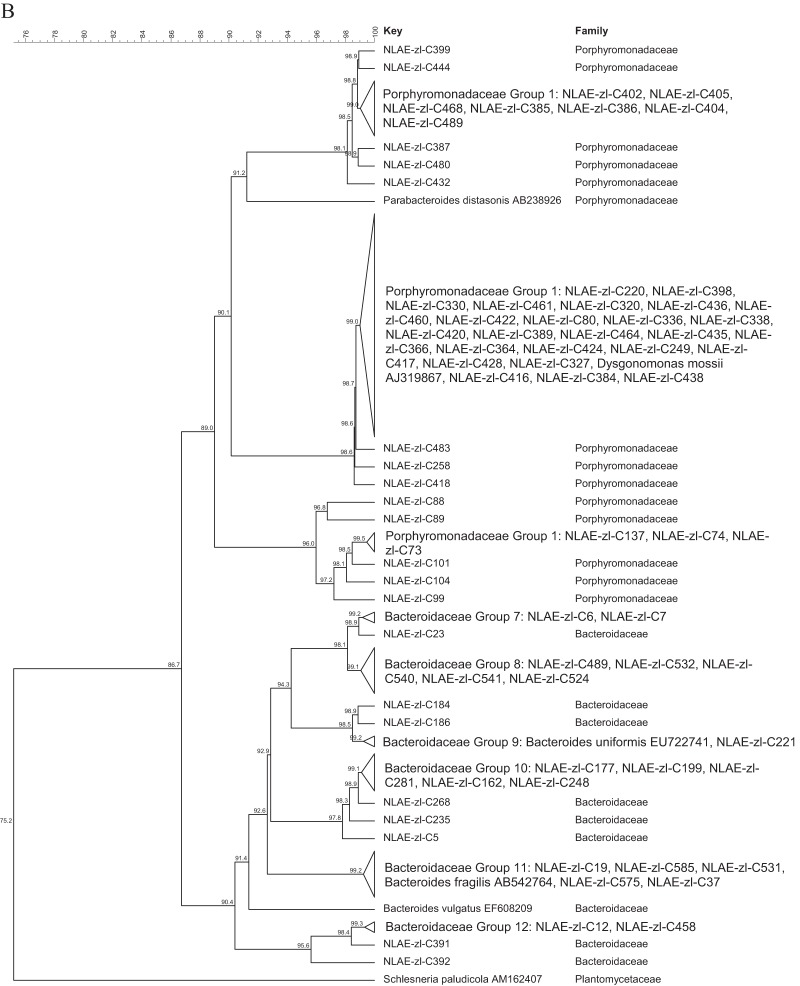
The bacterial isolates identified as Bacteroidetes are included in Fig. 2 as the Bacteroides thetaiotaomicron-B. ovatus-B. xylanisolvens grouping (45.8% of the Bacteroidetes) (Fig. 2A) and the Porphyromonadaceae (33.8% of the Bacteroidetes) and Bacteroidaceae (other than those in Fig. 2A, 20.4% of the Bacteroidetes) family groupings (Fig. 2B). All of the 65 bacterial isolates in the B. thetaiotaomicron-B. ovatus-B. xylanisolvens grouping (Fig. 2A) had greater than 96.4% identity. The two largest groupings, at ≥99% identity, were for bacterial isolates identified as B. thetaiotaomicron (Bacteroidaceae group 5) and B. xylanisolvens (Bacteroidaceae group 1). The Porphyromonadaceae isolates (Fig. 2B) grouped within the genera Dysgonomonas (28 isolates) and Parabacteroides (12 isolates); the other Bacteroidaceae isolates were most closely related to B. uniformis, B. fragilis, and B. vulgatus.
FIG 2.
Cluster analysis with an unrooted dendrogram of bacterial isolates from cellulose and xylan-pectin enrichments of cow feces in the phylum Bacteroidetes. (A) Isolates in the B. thetaiotaomicron- B. xylanisolvens-B. ovatus assemblage of the family Bacteroidaceae; (B) isolates in the Bacteroidaceae (other than those in the group shown in panel A) and Porphyromonadaceae families. The genus and species names and GenBank (NCBI) accession number are included for the reference strains. Bacteria with ≥99% similarity are grouped in order to simplify the dendrogram. The line at the top represents percent similarity across isolates; Schlesneria paludicola (Plantomycetes) is used as the outgroup.
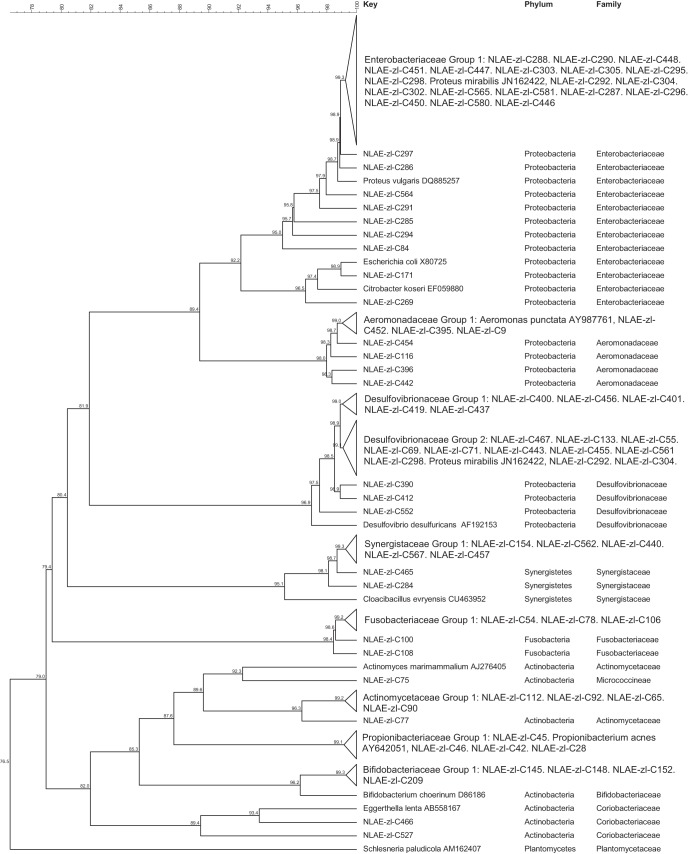
Bacteria in the Proteobacteria phylum were Gammaproteobacteria (35 isolates) from the Proteus (25 isolates), Aeromonas (7 isolates), Escherichia/Shigella (2 isolates), and Morganella (1 isolates) genera and Deltaproteobacteria of the Desulfovibrionaceae family (16 isolates) (Fig. 3). Figure 3 also contains bacterial isolates from the remaining 3 phyla. The Fusobacteria were closely related, and all 5 isolates had greater than 98.4% identity. The Synergistetes isolates were also closely related (>98.1% identity). Within the Actinobacteria phylum, the bacterial isolates were identified to be members of 5 families: Actinomycetaceae (33.3% of Actinobacteria), Micrococcineae (6.3%), Propionibacteriaceae (2.5%), Bifidobacteriaceae (2.5%), and Coriobacteriaceae (1.3%).
FIG 3.
Cluster analysis with an unrooted dendrogram of bacterial isolates from cellulose and xylan-pectin enrichments of cow feces in the phyla Proteobacteria; Gammaproteobacteria class, Enterobacteriaceae family; Deltaproteobacteria class, Desulfovibrionaceae family; Synergistetes; Fusobacteria; and Actinobacteria. The genus and species names and GenBank (NCBI) accession number are included for the reference strains. Bacteria with ≥99% similarity are grouped in order to simplify the dendrogram. The line at the top represents percent similarity across isolates; Schlesneria paludicola (Plantomycetes) is used as the outgroup.
DISCUSSION
A wide range of bacteria was successfully isolated from the enrichments used in this study, and while the results were not strictly quantitative, a rough estimation of the abundance of the different phyla could be obtained. As expected, the Firmicutes and Bacteroidetes isolates predominated at 82.8% of isolates, followed by the Proteobacteria, Actinobacteria, Synergistetes, and Fusobacteria (11.1%, 1.1%, 3.5%, and 1.5%, respectively). Results reported by Ley et al. (1) support the abundance of the isolates from these enrichments in different phyla, with similar results across feces from 60 different mammalian species being obtained for Firmicutes and Bacteroidetes (82% of nearly 20,000 classified sequences) and the other phyla (14.2% Proteobacteria, Fusobacteria, and Actinobacteria). The similarity in the abundance of the phyla in this study to that of the study of Ley et al. (1) was surprising because the isolations used both selective enrichment and selective isolation agar. It appears that even with enrichment the microbial community maintains broad diversity and that there is a lack of selectivity with the agars. These results demonstrate that the large intestinal bacteria in cattle are similar to those in other mammals, on the basis of analysis of feces. While the rumen microbiota are also dominated by the Firmicutes and Bacteroidetes (7, 22, 23), the majority of organisms present in the rumen differ from those present in the feces (12, 13, 24).
Over 98% of the isolates (n = 450) were less than 98.5% similar to cultured bacteria (in RDP) and represent new species and/or genera, using 98.5% identity as a species cutoff and 95% as a genus cutoff (25, 26). Historically, fewer bacteria were cultured from the feces of ruminants because the majority of their nutritional needs are supplied by rumen microbial fermentation. While the use of selective media introduces bias, in general, easy-to-grow bacteria are isolated, while many others remain to be detected; the fact that most of these isolates represent previously uncultured bacteria indicates that many uncultured intestinal bacteria could be recovered by using a broader range of nutrient medium formulations.
Differences in the types of bacteria that were isolated from the different carbohydrate enrichments were evident in the Firmicutes and Proteobacteria phyla. More specific differences were detected at the genus level across all phyla. Feedlot cattle fed diets based on unprocessed grain, processed grain, or forage had distinct differences in the abundance of families within a phylum for each diet. This reflected the different types and concentrations of polysaccharides in the diets (15). de Menezes et al. (27) found few differences in the ruminal microbiota of dairy cattle fed pasture or total mixed rations at the phylum level; however, at the family level, diet did alter the abundance of organisms, with the greatest differences being noted for the Fibrobacteriaceae, Coriobacteriaceae, Lachnospiraceae, and Veillonellaceae families. In the current study, the families Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, Parabacteroidaceae, Enterococcaceae, and Desulfovibrionaceae had the greatest differences in the number of isolates obtained from the 2 carbohydrate enrichments. The differences in the distribution of isolates among the various genera warrant further exploration, especially with regard to isolates from the genera Clostridium, Anaerosporobacter, Proteus, and Bacteroides.
Members of the Lachnospiraceae family were the most frequently isolated bacteria, accounting for 45.0% of Firmicutes and 23.3% of total isolates. This family includes clostridial groups XIVa and XIVb (28), groups that predominate in the intestinal tracts of most mammals (29). Many of the Firmicutes found in the intestinal tract are strains with demonstrated cellulose and hemicellulose degradation enzymes, including members of the Lachnospiraceae and Clostridiaceae I families, with a few isolates from the Eubacteriaceae and Bacillaceae 1 families also being detected (30, 31). Two examples among the isolates were 10 bacteria closely related to Clostridium celerecrescens, a cellulolytic Lachnospiraceae bacterium (32), and 17 isolates clustered near the cellulolytic rumen bacterium Ruminococcus albus (5, 30), but at only 88.5% identity. An additional 15 isolates of the Ruminococcaceae were isolated, and all were ≥97% similar to Oscillibacter valericigenes. This bacterium, as well as Desulfovibrio, Faecalibacterium, Anaerostipes, and Prevotella species, can utilize the exopolysaccharides produced by Bifidobacterium species (33), indicating that they may play a supporting role for polysaccharide fermenters. For a number of the species related to the isolates, especially those in the Eubacteriaceae and Bacillaceae, information on plant cell wall utilization was not readily available, and this will need to be explored in greater detail. An additional niche that some Firmicutes may fill in these enriched communities is fermentation of peptides and amino acids. Cotta et al. (34) identified a number of Clostridium, Enterococcus, and Staphylococcus species isolated from pig feces and stored manure with this capacity.
Members of the Bacteroidaceae family have been demonstrated to be able to utilize a wide range of carbohydrates, including plant cell wall carbohydrates (cellulose, xylan, pectins, and β-glucans and galactans) and host mucopolysaccharides and glycoproteins (35). Genomic analysis of Bacteroides species confirms the enrichment for genes for enzymes targeting carbohydrates and that the range of these enzymes is quite broad (36). Active enzymes targeting plant cell wall carbohydrates have been demonstrated in Bacteroides ovatus, B. thetaiotaomicron, B. xylanisolvens, B. uniformis, and B. fragilis (37–39). Among the isolates, the majority of the Bacteroidetes were closely related to B. ovatus, B. thetaiotaomicron, and B. xylanisolvens; both B. ovatus and B. thetaiotaomicron have multiple polysaccharide utilization loci that encode a broad range of enzymes targeting various plant cell wall components (39). While the Bacteroides are some of the most studied intestinal bacteria, 95.1% of the Bacteroidetes isolates had less than 97% identity to previously cultured strains. While some high-throughput sequencing has shown Prevotella species to be present in bovine feces (15, 40), no Prevotella species were isolated from any enrichment culture, similar to the findings of analysis of 16S rRNA gene clone libraries by Ozutsumi et al. (24). Interestingly, the Porphyromonadaceae were 36.3% of the Bacteroidetes, and 12 isolates were similar (91%) to Parabacteroides distasonis, a human gut species that has been shown to convert aromatic amino acids to phenolic acids (41). Another 28 Porphyromonadaceae isolates were similar to Dysgonomonas mossii, which has been isolated from diseased human intestine (42) and microbial fuel cells (43); however, little is known about its function in the intestine.
Most of the Proteobacteria isolated (68.6% of total Proteobacteria) were Gammaproteobacteria in the Enterobacteriaceae family. Enterobacteriaceae bacteria are members of the mammalian intestinal microbiota and make up 7 to 8% of the microbiota (1), with similar results found in cattle (15). The majority (89.3%) of the Enterobacteriaceae isolates were classified as Proteus. In addition to their ability to utilize a wide range of less complex substrates (sugars, yeast extract, peptones, etc.), members of the Enterobacteriaceae with the ability to utilize plant cell wall substrates have been isolated previously (44). Sixteen strains of the Desulfovibrionaceae (Deltaproteobacteria, 31.4% of Proteobacteria) were isolated, even though the media used were not designed to grow sulfate-reducing bacteria.
Isolates identified in the remaining 3 phyla (Fusobacteria, Actinobacteria, and Synergistetes) have been detected in mammalian intestinal tracts (1, 45). While it not clear what function isolates from these phyla carry out in the microbial communities of cellulose and xylan-pectin enrichments, they have a wide range of fermentative capacities. They may support the plant cell wall-utilizing bacteria by actively fermenting nitrogen compounds; bacteria in both the Actinobacteria and Fusobacteria phyla have those properties (45). All of the Fusobacteria isolated had less than 95% identity to previously cultured strains. Although only 9 isolates were identified as Actinobacteria, they had broad phylogenetic diversity. Actinomycetaceae accounted for 31.2% of the Actinobacteria, but their role in the large intestine is not clear. A large amount of data on Bifidobacterium species (25% of Actinobacteria isolates), in particular, data on their selective growth in the large intestine when oligosaccharides (inulin, fructo-oligosaccharides, and galacto-oligosaccharides) are included in the diet of humans, are available (46). Propionibacterium acnes and other species contribute to the proteolysis and peptidase activity in the large intestine (47, 48). Only 7 isolates of the Synergistetes were isolated, and while their presence in the intestinal tract of mammals has been documented, their function within the intestinal microbiota is unknown (45, 49).
This is the first report of extensive isolations of bacteria from plant cell wall-degrading microbial communities in ruminant feces, evidenced by 98% of the isolates having less than 98.5% phylogenetic identity to cultured bacteria in the RDP. These bacteria provide new opportunities to study the genomic and metabolic capacities of these members of the complex intestinal microbiota. The microbial communities that arose in these enrichment cultures of cow feces had broad bacterial diversity that resembled that in the feces of other mammals. Many of the bacteria isolated are closely related to species demonstrated to produce enzymes that ferment the cellulose and hemicellulose fractions of the plant cell wall, but a significant number of the isolates likely fill metabolic roles, such as amino acid synthesis, that support the plant cell wall fermenters. The diversity and the fermentation capabilities of these bacterial isolates confirm the complexity of the interactions among microorganisms from a community, even in a highly specialized community such as that which grew in these carbohydrate enrichment cultures. Although not all representatives of the microbes in these enrichment communities have been isolated, metagenomic analysis of the total microbiome in these communities could be used to gain further insight into and a comprehension of total plant cell wall-fermenting microbial communities.
ACKNOWLEDGMENTS
This research was supported by grants from the Defense Advanced Research Projects Agency as part of the Intestinal Fortitude and Crystalline Cellulose Conversion to Glucose programs.
I thank Lance Baumgard for access to cows and Todd Atherly, Kerrie Franzen, and John Tenhundfeld for technical analyses.
Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
Footnotes
Published ahead of print 8 November 2013
REFERENCES
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6:776–788. 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lineback DR. 1999. The chemistry of complex carbohydrates, p 115–129 In Cho SS, Prosky L, Dreher M. (ed), Complex carbohydrates in foods. Marcel Dekker, New York, NY [Google Scholar]
- 4.Bergman EN. 1990. Entergy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590 [DOI] [PubMed] [Google Scholar]
- 5.Van Soest PJ. 1994. Nutritional ecology of the ruminant. 2nd ed. Comstock Publishing Associates, Ithaca, NY [Google Scholar]
- 6.Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Martins Dos Santos VAP, Strompl C, Elborough K, Jarvis G, Neef A, Yakimov MM, Timmis KN, Golyshin PN. 2005. Novel hydrolase diversity retrieved from a metagemone library of bovine rumen microflora. Environ. Microbiol. 7:1996–2010. 10.1111/j.1462-2920.2005.00920.x [DOI] [PubMed] [Google Scholar]
- 7.Brulc JM, Antonopoulos DA, Miller ME, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagemomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U. S. A. 106:1948–1953. 10.1073/pnas.0806191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Ligget CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14:169–181. 10.1093/dnares/dsm018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamendella R, Santo Domingo SW, Ghosh S, Martinson J, Oerther DB. 2011. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11:103. 10.1186/1471-2180-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, Konkel ME, Angly F, Dinsdale EE, Edwards RA, Nelson KE, White BA. 2008. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One 3:e2945. 10.1371/journal.pone.0002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey JC, Pell AN, Berthiume R, Lapierre H, Lee S, Ha JK, Mendell JE, Angert ER. 2010. Comparative studies of microbial populations in the rumen, duodenum, ileum and faeces of lactating dairy cows. J. Appl. Microbiol. 108:1982–1993. 10.1111/j.1365-2672.2009.04602.x [DOI] [PubMed] [Google Scholar]
- 13.Michelland RJ, Monteils V, Zened A, Combes S, Cauquil L, Gidenne T, Hamelin J, Fortun-Lamothe L. 2009. Spatial and temporal variations of the bacterial community in the bovine digestive tract. J. Appl. Microbiol. 107:1642–1650. 10.1111/j.1365-2672.2009.04346.x [DOI] [PubMed] [Google Scholar]
- 14.Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, Tringe SG, Brodie EL, Dominguez-Bello MG. 2012. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 6:531–541. 10.1038/ismej.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 77:2992–3001. 10.1128/AEM.02988-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao S, Zhang R, Wang D, Zhu W. 2012. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet. Res. 8:237. 10.1186/1746-6148-8-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durso LM, Harhay GP, Bono JL, Smith TPL. 2011. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Methods 84:278–282. 10.1016/j.mimet.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 18.Leedle JAZ, Hespell RB. 1980. Differential carbohydrate media and anaerobic replica plating techniques in delineating carbohydrate-utilizing subgroups in rumen bacterial populations. Appl. Environ. Microbiol. 39:709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lederberg J, Lederberg EM. 1952. Replicate plating and indirect selection of bacterial mutants. J. Bacteriol. 63:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–163 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 21.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrel DM, Garrity GM, Tiedje JM. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294–D296. 10.1093/nar/gki038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jami E, Mizrahi I. 2012. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:e33306. 10.1371/journal.pone.0033306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross EM, Moate PJ, Bath CR, Davidson SE, Sawbridge TI, Guthridge KM, Cocks BF, Hayes BJ. 2012. High throughput whole rumen metagenome profiling using untargeted massively parallel sequencing. BMC Genet. 13:53. 10.1186/1471-2156-13-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozutsumi Y, Hayashi H, Sakamoto M, Itabashi H, Benno Y. 2005. Culture-independent analysis of fecal microbiota in cattle. Biosci. Biotechnol. Biochem. 69:1793–1797. 10.1271/bbb.69.1793 [DOI] [PubMed] [Google Scholar]
- 25.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer K-H, Ludwig W, Glöcner FO, Rosselló-Móra R. 2008. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250. 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 26.Konstantinidis KT, Stackebrandt E. 2013. Defining taxonomic ranks. In Rosenberg E, DeLong EF, Stackebrandt E, Lory S, Thompson F. (ed), The prokaryotes—prokaryotic biology and symbiotic associations. Springer-Verlag, Berlin, Germany [Google Scholar]
- 27.de Menezes AB, Lewis E, O'Donovan M, O'Neill BF, Clipson N, Doyle EM. 2011. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 78:256–265. 10.1111/j.1574-6941.2011.01151.x [DOI] [PubMed] [Google Scholar]
- 28.Collins MD, Lawson PA, Willems A, Cordoba JJ, Frenandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826. 10.1099/00207713-44-4-812 [DOI] [PubMed] [Google Scholar]
- 29.Furet J-P, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, Doré J, Corthier G. 2009. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68:351–352. 10.1111/j.1574-6941.2009.00671.x [DOI] [PubMed] [Google Scholar]
- 30.Schwarz WH. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634–649. 10.1007/s002530100710 [DOI] [PubMed] [Google Scholar]
- 31.Maki M, Leung KT, Qin W. 2009. The prospects of cellulose-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 5:500–516. 10.7150/ijbs.5.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palop MLL, Valles S, Piñaga F, Flors A. 1989. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium celerecrescens sp. nov. Int. J. Syst. Bacteriol. 39:68–71. 10.1099/00207713-39-1-68 [DOI] [Google Scholar]
- 33.Salazar N, Gueimonde M, Hernández-Barranco AM, Ruas-Madiedo P, de los Reyes-Gavilán CG. 2008. Exopolysaccharides produced by intestinal Bifidobacterium strains as fermentable substrates for human intestinal bacteria. Appl. Environ. Microbiol. 74:4737–4745. 10.1128/AEM.00325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotta MA, Whitehead TR, Zeltwanger RL. 2003. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 5:737–745. 10.1046/j.1467-2920.2003.00467.x [DOI] [PubMed] [Google Scholar]
- 35.Salyers AA. 1979. Energy sources of major intestinal fermentative anaerobes. Am. J. Clin. Nutr. 32:158–163 [DOI] [PubMed] [Google Scholar]
- 36.Karlsson FH, Ussery DW, Nielsen J, Nookaew I. 2011. A closer look at Bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microb. Ecol. 61:473–485. 10.1007/s00248-010-9796-1 [DOI] [PubMed] [Google Scholar]
- 37.Salyers AA, Vercellotti JR, West SHE, Wilkens TD. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirande C, Mosoni P, Béra-Maillet C, Bernalier-Donadille A, Forano E. 2010. Characterization of Xyn10A, a highly active xylanase from the human gut bacterium Bacteroides xylanisolvens XB1A. Appl. Microbiol. Biotechnol. 87:2097–2105. 10.1007/s00253-010-2694-0 [DOI] [PubMed] [Google Scholar]
- 39.Martens EC, Low EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. 10.1371/journal.pbio.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durso LM, Harhay GP, Smith TPL, Bono JL, DeSantis TZ, Harhay DM, Andersen GL, Keen JE, Laegreid WW, Clawson ML. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76:4858–4862. 10.1128/AEM.00207-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ. 2013. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 57:523–535. 10.1002/mnfr.201200594 [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto T, Kawakami Y, Oana K, Honda T, Yamauchi K, Okimura Y, Siohara M, Kauga E. 2006. First isolation of Dysgonomonas mossii from intestinal juice of a patient with pancreatic cancer. Arch. Med. Res. 37:914–916. 10.1016/j.arcmed.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 43.Kodama Y, Shimoyama T, Watanabe K. 2012. Dysgonomonas oryzarvi sp. nov., isolated from a microbial fuel cell. Int. J. Syst. Evol. Microbiol. 62:3055–3059. 10.1099/ijs.0.039040-0 [DOI] [PubMed] [Google Scholar]
- 44.Van Laere KMJ, Hartemink R, Bosveld M, Schols HA, Voragen AGJ. 2000. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food Chem. 48:1644–1652. 10.1021/jf990519i [DOI] [PubMed] [Google Scholar]
- 45.Shah HN, Olsen I, Bernard K, Finegold SM, Gharbia S, Gupta RS. 2009. Approaches to the study of the systematics of anaerobic, Gram-negative, non-sporeforming rods: current status and perspectives. Anaerobe 15:179–194. 10.1016/j.anaerobe.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 46.Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, Wolvers D, Watzi B, Szajewska H, Stahl B, Guamer F, Respondek F, Whelan K, Coxam V, Davicco M-J, Léotoing L, Wittrant Y, Delzenne NM, Cani PD, Neyrinck AM, Meheust A. 2010. Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104(Suppl 2):S1–S63. 10.1017/S0007114510003363 [DOI] [PubMed] [Google Scholar]
- 47.Macfarlane GT, Allison C, Gibson SAW, Cummings JH. 1988. Contribution of the microflora to proteolysis in the human large intestine. J. Appl. Bacteriol. 64:37–46. 10.1111/j.1365-2672.1988.tb02427.x [DOI] [PubMed] [Google Scholar]
- 48.Zoetendal EG, Collier CT, Koike S, Mackie RI, Gaskins HR. 2004. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 134:465–472 [DOI] [PubMed] [Google Scholar]
- 49.Marchandin H, Damay A, Roudière L, Teyssier C, Zorgniotti I, Dechaud H, Jean-Pierre H, Jumas-Bilak E. 2010. Phylogeny, diversity and host specialization in the phylum Synergistetes with emphasis on strains and clones of human origin. Res. Microbiol. 161:91–100. 10.1016/j.resmic.2009.12.008 [DOI] [PubMed] [Google Scholar]
- 50.Allison MJ, Robinson IM, Bucklin JA, Booth GD. 1979. Comparison of bacterial populations of the pig cecum and colon based upon enumeration with specific energy sources. Appl. Environ. Microbiol. 37:1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]