Abstract
The animal gut is perpetually exposed to microorganisms, and this microbiota affects development, nutrient allocation, and immune homeostasis. A major challenge is to understand the contribution of individual microbial species and interactions among species in shaping these microbe-dependent traits. Using the Drosophila melanogaster gut microbiota, we tested whether microbe-dependent performance and nutritional traits of Drosophila are functionally modular, i.e., whether the impact of each microbial taxon on host traits is independent of the presence of other microbial taxa. Gnotobiotic flies were constructed with one or a set of five of the Acetobacter and Lactobacillus species which dominate the gut microbiota of conventional flies (Drosophila with untreated microbiota). Axenic (microbiota-free) flies exhibited prolonged development time and elevated glucose and triglyceride contents. The low glucose content of conventional flies was recapitulated in gnotobiotic Drosophila flies colonized with any of the 5 bacterial taxa tested. In contrast, the development rates and triglyceride levels in monocolonized flies varied depending on the taxon present: Acetobacter species supported the largest reductions, while most Lactobacillus species had no effect. Only flies with both Acetobacter and Lactobacillus had triglyceride contents restored to the level in conventional flies. This could be attributed to two processes: Lactobacillus-mediated promotion of Acetobacter abundance in the fly and a significant negative correlation between fly triglyceride content and Acetobacter abundance. We conclude that the microbial basis of host traits varies in both specificity and modularity; microbe-mediated reduction in glucose is relatively nonspecific and modular, while triglyceride content is influenced by interactions among microbes.
INTRODUCTION
Animals are in constant contact with microorganisms, and this host-associated microbiota impacts nearly every aspect of animal biology (1). In many animals, the largest and most diverse populations of microbes associated with animals reside in the gut, a critical interface for nutrient acquisition and immune recognition (2–4). In humans, the taxonomic composition of the gut microbiota has been linked to obesity (5), diabetes (6), and inflammatory bowel disease (7). Therapies that target the gut microbiota have tremendous potential to improve human health, but their success depends on gaining a better understanding of how the microbiota composition is coupled to its function(s), i.e., impacts on host traits (8–10).
The relationship between the composition and function of host-associated microbiota depends critically on the degree of functional modularity of the microbiota. For a perfectly modular microbiota, the impact of each microbial taxon on host traits is independent of the presence of other microbial taxa. Specifically, the various microorganisms may affect the same or different host traits, but the effect of a community of any composition on host traits can be predicted from the sum of effects of each taxon in isolation. Deviation from modularity would be indicative of interactions among the microorganisms (e.g., metabolites or signaling molecules produced by microbe A may alter the abundance or traits of microbe B, with knock-on consequences for host traits) or interactions between microorganisms and the host (e.g., apparent competition, such as a situation where microbe B is very susceptible to host immune response triggered by microbe A).
A powerful route to study the relationship between the composition and function of host-associated microbial communities, and especially to investigate the degree of functional modularity of the microbiota, is provided by gnotobiotic animals. These are animals experimentally colonized by known sets of microorganisms. Studies of single-species, dual-species, and multispecies microbiota reconstituted in rodents have identified bacterial genes important for microbiota persistence (11), described syntrophy between different species (12), and revealed competitive interactions that shape microbiota metabolism (13, 14). Despite recent progress in scaling up the complexity of gnotobiotic communities in mammals (15, 16), it remains unclear how the many interactions between microbes, and between microbiota and host, sum together to shape an animal's phenotype.
The fruit fly Drosophila melanogaster offers an amenable system to investigate the relationship between composition and function of gut microorganisms. Drosophila has a low-diversity gut microbiota, typically containing <30 taxa and commonly dominated by as few as five species in the genera Acetobacter and Lactobacillus (17, 18), which are readily culturable. These gut bacteria have profound effects on the nutritional traits of Drosophila. Generally, axenic Drosophila (i.e., reared under germfree conditions) display extended larval development time and hyperglycemia, and the traits of conventional Drosophila (i.e., with unmanipulated microbiota) are recapitulated, partially or completely, in axenic hosts reinfected with Acetobacter, Lactobacillus, or undefined microbial inocula derived from conventional flies (19–22). However, the results across the several studies are not fully consistent, perhaps reflecting variations in the diet and genotypes of the host and bacteria.
In principle, the gut microbiota can influence host nutrition in two ways. First, it can modulate the host nutritional signaling networks, thus altering nutritional allocation patterns. For example, the production of acetic acid by Acetobacter pomorum has been determined to promote insulin signaling, thereby promoting larval growth and development and reducing adult lipid and sugar levels (21), and Lactobacillus has been implicated in modulating TOR (target of Rapamycin) signaling, which intersects with insulin signaling in the regulation of growth and metabolism (22). Additionally or alternatively, the bacteria can modify the nutritional inputs to the host by providing supplementary nutrients and consuming dietary constituents. The latter would be deleterious to the host for limiting nutrients and beneficial for nutrients that are in excess. For hosts like Drosophila that shed feces containing live bacteria into the medium on which they feed, bacterial modulation of the food can be displayed by bacteria both in the gut and externally.
The purpose of this study was to assess the degree of functional modularity among the bacteria that contribute to the gut microbiota of Drosophila. We focused on the five species previously identified as dominating the gut community in our laboratory stock of Drosophila (17). First, we measured the development time and nutritional indices of Drosophila in monoassociation with each of these bacteria to determine baseline measures of the function of each bacterium. We then quantified these traits in dual-species associations. Our results reveal that the degree of functional modularity varies across the different microbial functions and that synergistic interactions can be explained by effects of cocolonization on bacterial abundance.
MATERIALS AND METHODS
Cultivation of bacteria and flies.
Wolbachia-free Drosophila melanogaster Canton S flies were reared at 25°C in a 12-h-light/12-h-dark cycle on a Y-G diet (100 g liter−1 Brewer's yeast [inactive; MP Biomedicals], 100 g liter−1 glucose [Sigma], 12 g liter−1 agar [Apex], and preservatives comprising 0.04% phosphoric acid and 0.42% propionic acid [Sigma]). Acetobacter pomorum, Acetobacter tropicalis, Lactobacillus brevis, Lactobacillus fructivorans, and Lactobacillus plantarum were isolated on modified MRS agar from fly guts and identified by sequencing of 16S rRNA genes. Modified MRS (MMRS) contained the following (all from Sigma unless otherwise noted): 1.25% vegetable peptone (Becton Dickinson), 0.75% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% dipotassium hydrogen phosphate, 0.2% triammonium citrate, 0.02% magnesium sulfate heptahydrate, 0.005% manganese sulfate tetrahydrate, 1.2% agar (Apex). All bacteria were maintained on MMRS medium at 30°C, with Lactobacillus plates kept in sealed containers that were gassed with CO2 prior to sealing.
Preparation of axenic and gnotobiotic flies.
To produce axenic flies, freshly laid eggs (≤18 h old) were collected from grape juice agar plates, surface sterilized by 3 washes with 0.6% hypochlorite (equivalent to 1:10 dilution of Clorox bleach) followed by 3 washes with sterile water, and then aseptically transferred to sterile food. For conventional flies with uncontrolled microbiota, embryos from the same collection were transferred to sterile food directly from the egg collection agar. To introduce defined gut microbiota for gnotobiotic treatments, the inoculum was prepared as follows and added to the food surface immediately following aseptic egg transfer. Optical density (OD) measurements were made on an overnight culture of each bacterial species used. The cells were then pelleted at 7,500 × g for 2 min and resuspended in fresh growth medium at a final cell density of 108 cells per ml. The resuspension volume was determined based on the following empirically determined constants (CFU ml−1 at an OD of 1): A. pomorum, 1.3 × 109; A. tropicalis, 9.5 × 108; L. brevis, 6.5 × 108; L. fructivorans, 1.0 × 109; and L. plantarum, 9.0 × 108. Fifty microliters of resuspended cells was added to each gnotobiotic vial to give 5 × 106 cells per vial. For microbiota of >1 species, each component was added in equal parts to make up the total inoculum (e.g., the 5-species microbiota inoculum contained 1 × 106 cells of each species). This inoculum density was chosen because it yielded 5-species gnotobiotic adults with total CFU counts comparable to those in conventional flies (data not shown).
Insect development.
For the experiment whose results are shown in Fig. 1A and B, observations were made once daily between 8 and 10 h postdawn (h.p.d.) and the times of pupation and eclosion were scored. For the experiment whose results are shown in Fig. 1C and D, observations were made three times daily: at 0, 6, and 11.5 h.p.d. for pupation and at 2, 7, and 11 h.p.d. for eclosion. In each case, data from 5 independent experiments were collected. Data for gnotobiotic treatments were analyzed in R Software for Statistical Computing, version 2.15.3, using the Survival, coxme, and multcomp packages. The first development data were formatted as survival objects for each treatment using the Surv function. Kaplan-Meier plots were generated from survival objects using plot(Survfit(survival object)). A Cox mixed-effects model was applied to survival objects using experiment as a random effect, and pairwise Tukey's tests were made with the general linear hypothesis test (glht) function, correcting P values for multiple comparisons by the single-step method (default procedure in multcomp).
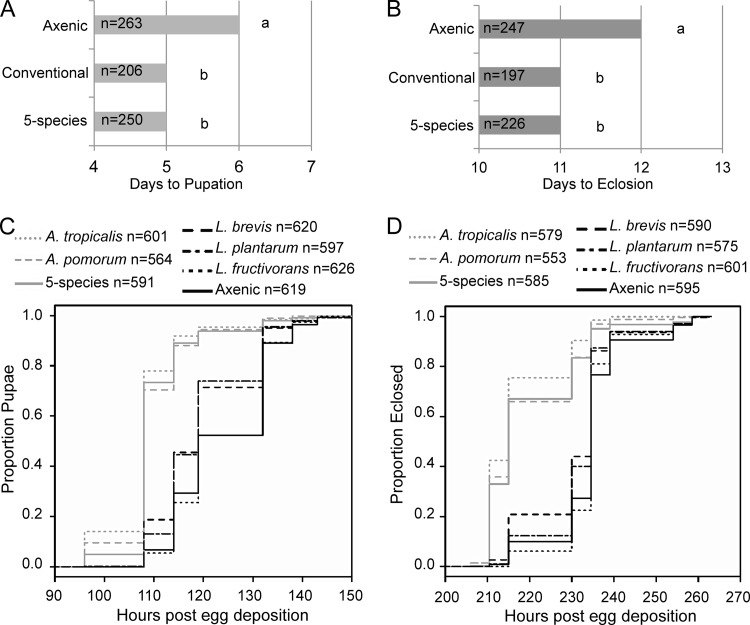
FIG 1.
Gut microbiota impact Drosophila development. (A and B) Times to pupation (A) and to eclosion (B) are shown in days post-egg deposition. The median of medians from 8 independent experiments is displayed, with n indicating the total number of observations. Different letters to the right of the bars indicate statistically significant differences by the Mann-Whitney test at a P value of <0.05. (C and D) Kaplan-Meier plots of times to pupation (C) and eclosion (D), respectively, with a key above indicating the treatments compared and number of observations (n) for each treatment.
Nutritional and performance indices.
Mated females were collected under light CO2 anesthesia 5 to 6 days after eclosion, weighed in groups of 3 to 5 to the nearest microgram using a Mettler Toledo (MX5) microbalance, and then homogenized in 125 μl TET buffer (10 mM Tris, pH 8, 1 mM EDTA, 0.1% Triton X-100) in 1.5-ml tubes with ∼100 μl of lysis matrix D (MP Biomedicals) with shaking for 30 s in a FastPrep-24 instrument with the default settings (MP Biomedicals). Next, tubes were centrifuged for 1 min at 20,000 × g to pellet debris. Twenty-microliters of the resulting supernatant was flash frozen for subsequent protein determination, while 40 μl was heated at 72°C for 20 min to inactivate endogenous enzymes and then flash frozen. The replicates for each treatment in each experiment consisted of 3 groups of flies from 3 different vials. A second group of 3 to 5 flies from each of the 3 vials was collected for CFU determination.
The protein content of each sample was analyzed using the Bio-Rad DC kit according to the manufacturer's instructions. Triglyceride (TAG) was measured using the free glycerol detection kit in combination with triglyceride reagent, following the manufacturer's instructions (Sigma). Glucose content was measured by the glucose oxidase (GO) method (23). The GO reagent contained 1 μg ml−1 GO (product number G7141; Sigma), 1 μg ml−1 horseradish peroxidase (product number P8375; Sigma), and 20 μg ml−1 o-dianisidine (product number D3252; Sigma) in 100 mM Tris, pH 7. In a 96-well plate, 150 μl of GO reagent was added to 5 μl of homogenate and incubated at 37°C for 30 min, and then 150 μl of 6.25 M H2SO4 was added and the absorbance read at 544 nm against glucose standards.
CFU determination.
For CFU determination, each sample of 3 to 5 female flies was homogenized as described above except that MMRS medium was employed instead of TET buffer. The resulting homogenate was diluted to 1 ml and assayed for bacterial abundance by spiral plating (on a WASP-2 instrument, Microbiology International) on MMRS. Differential detection of each species from multispecies gnotobiotes was achieved by plating the sample onto two different MMRS plates; one contained 10 μg per ml ampicillin to exclude the growth of lactobacilli but permit the growth of acetobacters, and the other was placed under a CO2 atmosphere to exclude the growth of acetobacters but permit the growth of lactobacilli. The species were further differentiated by colony morphology, as L. plantarum makes large, yellow colonies, while L. brevis makes smaller white colonies, and A. pomorum makes rough, opaque colonies with a distinctive copper color, while A. tropicalis makes smooth, translucent colonies that are highly reflective. L. fructivorans grows slowly in vitro and could not be scored in mixed-bacterial samples. CFU counts were made with the Protocol 3 colony counter (Microbiology International).
Statistical analyses.
All statistics were performed in R, version 2.15.3. When analysis of variance (ANOVA) indicated significant differences, a linear mixed-effects model was implemented using the multcomp and lme4 packages with experiment as a random effect. This approach allowed us to account for any “block” variation among experiments, which were performed at different times in the calendar year. Pairwise comparisons were made via Tukey's test (glht function in multcomp, correcting P values for multiple comparisons by the single-step method). Mann-Whitney (MW) pairwise tests were made with the wilcox.test function, and P values were adjusted for multiple comparisons by the Bonferroni correction. Correlations were tested by Spearman's method using the cor.test function.
RESULTS
Effects of defined microbiota on development time.
Axenic Drosophila have a prolonged developmental period relative to that of conventional Drosophila (with unmanipulated microbiota) (20–22). To assess the performance of gnotobiotic flies colonized with our gut microbiota isolates, we first examined the effect of a defined, 5-species microbiota (A. pomorum, A. tropicalis, L. brevis, L. fructivorans, and L. plantarum) on development time to pupation and eclosion compared to the times for axenic and conventional controls. Axenic flies pupated 6 days post-egg deposition (p.e.d.), a significant increase over the time for conventional and gnotobiotic flies, which formed pupae 5 days p.e.d. (MW comparing the medians of gnotobiotic and axenic treatments from 8 experiments, U = 2.5, n = 8, P < 0.05) (Fig. 1A). Axenic flies eclosed 12 days p.e.d., while gnotobiotic and conventional flies eclosed significantly sooner, at 11 days p.e.d. (MW, gnotobiotic to axenic, U = 7, n = 8, P < 0.05) (Fig. 1B). These results indicate that our defined, 5-species microbiota can support development times matching those of conventional Drosophila (MW comparing gnotobiotic to conventional flies for pupation, U = 12.5, n = 8, P > 0.05, and for eclosion, U = 24.5, n = 8, P > 0.05).
Next, we generated monoassociated flies with each of the 5 species and scored the development times, increasing the frequency of observations from the previous experiment. Flies colonized with A. pomorum had the same larval development time as 5-species gnotobiotes, while A. tropicalis significantly accelerated development relative to that observed for the 5-species microbiota (P < 0.01) (development statistics can be found in Table 1). All three of these treatments promoted larval development compared to that of the Lactobacillus gnotobiotes and axenic flies (P < 0.001). L. plantarum and L. brevis gnotobiotes had similar larval development times, which were significantly shorter than the larval development time of axenic flies (P < 0.001), while L. fructivorans gnotobiotes were no different than axenics (P > 0.05). In a separate experiment, we found that larvae reared with all three Lactobacillus species did not develop significantly faster than axenic larvae (data not shown).
TABLE 1.
Multiple comparisons of Cox mixed-effects survival modelsa
| Microbiota compared | Cox statistic for time to: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pupation |
Eclosion |
|||||||
| Estimateb | SE | z value | P value | Estimateb | SE | z value | P value | |
| A. pomorum–axenic | 1.327 | 0.059 | 22.324 | <0.001 | 1.178 | 0.060 | 19.634 | <0.001 |
| A. tropicalis–axenic | 1.476 | 0.059 | 25.114 | <0.001 | 1.538 | 0.060 | 25.595 | <0.001 |
| 5-Species–axenic | 1.250 | 0.059 | 21.324 | <0.001 | 0.974 | 0.059 | 16.507 | <0.001 |
| L. brevis–axenic | 0.389 | 0.057 | 6.797 | <0.001 | 0.334 | 0.059 | 5.694 | <0.001 |
| L. fructivorans–axenic | −0.007 | 0.057 | −0.119 | 1 | 0.013 | 0.058 | 0.225 | 1 |
| L. plantarum–axenic | 0.352 | 0.058 | 6.105 | <0.001 | 0.266 | 0.059 | 4.521 | <0.001 |
| A. tropicalis–A. pomorum | 0.150 | 0.059 | 2.537 | 0.14 | 0.360 | 0.060 | 6.034 | <0.001 |
| 5-Species–A. pomorum | −0.077 | 0.059 | −1.307 | 0.84 | −0.204 | 0.060 | −3.412 | 0.01 |
| L. brevis–A. pomorum | −0.937 | 0.059 | −15.826 | <0.001 | −0.844 | 0.060 | −14.087 | <0.001 |
| L. fructivorans–A. pomorum | −1.334 | 0.059 | −22.493 | <0.001 | −1.165 | 0.060 | −19.534 | <0.001 |
| L. plantarum–A. pomorum | −0.975 | 0.060 | −16.380 | <0.001 | −0.912 | 0.060 | −15.175 | <0.001 |
| 5-Species–A. tropicalis | −0.227 | 0.058 | −3.889 | 0.001 | −0.564 | 0.060 | −9.466 | <0.001 |
| L. brevis–A. tropicalis | −1.087 | 0.058 | −18.608 | <0.001 | −1.204 | 0.060 | −20.191 | <0.001 |
| L. fructivorans–A. tropicalis | −1.483 | 0.059 | −25.303 | <0.001 | −1.525 | 0.060 | −25.538 | <0.001 |
| L. plantarum–A. tropicalis | −1.125 | 0.059 | −19.080 | <0.001 | −1.272 | 0.060 | −21.216 | <0.001 |
| L. brevis–5-species | −0.860 | 0.059 | −14.670 | <0.001 | −0.640 | 0.059 | −10.784 | <0.001 |
| L. fructivorans–5-species | −1.257 | 0.058 | −21.484 | <0.001 | −0.961 | 0.059 | −16.358 | <0.001 |
| L. plantarum–5-species | −0.898 | 0.059 | −15.254 | <0.001 | −0.708 | 0.060 | −11.907 | <0.001 |
| L. fructivorans–L. brevis | −0.396 | 0.057 | −6.928 | <0.001 | −0.321 | 0.058 | −5.497 | <0.001 |
| L. plantarum–L. brevis | −0.038 | 0.057 | −0.656 | 0.99 | −0.068 | 0.059 | −1.159 | 0.90 |
| L. plantarum–L. fructivorans | 0.358 | 0.058 | 6.232 | <0.001 | 0.253 | 0.059 | 4.316 | <0.001 |
The Cox model makes a global comparison of development observations, using experimental replicate as a random effect. Pairwise comparisons of the Cox models were made by Tukey's HSD test implemented in the multcomp package for R, adjusting P values for multiple comparisons using the default (single-step) method.
Restricted maximum likelihood estimate of the coefficient.
The durations of the pupal period were similar across all treatments (Fig. 1C and D), suggesting that differences in time to eclosion result from changes in larval development time. The absence of Acetobacter retarded the time to eclosion relative to the times for treatments containing Acetobacter (Fig. 1D; Table 1). Collectively, the development data suggest that individual Acetobacter species are necessary and sufficient to produce development times comparable to those observed for the 5-species gut microbiota community. Additionally, some Lactobacillus species tested can accelerate development relative to that of axenic Drosophila, while others cannot. Subsequent experiments found no difference between the development times of dual-species gnotobiotes (one species of Acetobacter and one species of Lactobacillus, described below) and 5-species gnotobiotes (data not shown).
Effects of defined microbiota on nutritional indices of Drosophila.
To examine the effects of our single- and 5-species microbiota on host nutrient allocation, we compared flies with these treatments to conventional and axenic flies in the following measures: adult weight and protein, glucose, glycogen, and TAG contents. We found no significant effect of microbiota treatment on fresh weight (ANOVA, F7,232 = 0.741, P > 0.05), protein level (ANOVA, F7,232 = 1.475, P > 0.05), or glycogen level (ANOVA, F7,112 = 0.751, P > 0.05) (data not shown). Subsequently, the values of glucose and TAG contents were normalized to the protein content.
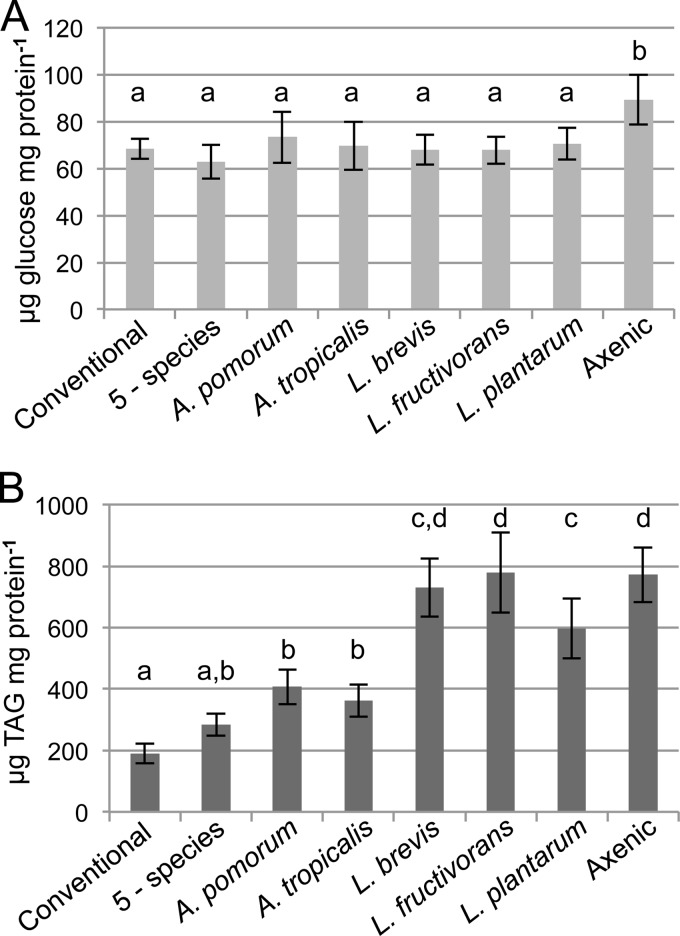
All microbiota treatments reduced the host glucose concentration to that of conventional flies (Fig. 2A). TAG levels, by contrast, showed a more varied response to microbiota treatment. The flies in the axenic and Lactobacillus treatment groups contained over 3× the level of TAG found in conventional flies, and of the species tested, only the L. plantarum treatment showed a significant reduction in host TAG relative to that in axenic flies (Fig. 2B). Acetobacter species reduced TAG relative to the levels in flies with Lactobacillus and axenic treatments but still resulted in levels significantly greater than that in conventional flies. Only the 5-species microbiota produced flies with TAG levels comparable to the level in conventional flies (Fig. 2B), suggesting that microbiota with >1 species may have a distinct effect on host nutrient allocation.
FIG 2.
Effects of single-species gut microbiota on host nutritional indices. Glucose (A) and triglyceride (TAG) (B) contents were determined for the indicated treatment groups and are reported here as the means of 10 independent experiments ± standard errors, normalized to the protein content of the same samples. Treatments that do not share a letter above the bars had significantly different results by Tukey's honestly significant difference (HSD) test at a P value of <0.05, corrected for multiple comparisons.
Impacts of dual-species microbiota.
Since hosts colonized with a multispecies microbiota had lower TAG levels than monocolonized hosts, we hypothesized that interaction(s) between microbiota species could impact host phenotypes, with the implication that the bacteria may not be functionally modular with respect to host TAG content. To test this hypothesis, we examined all possible pairwise Acetobacter-Lactobacillus combinations and compared them to the 5-species treatment and axenic controls.
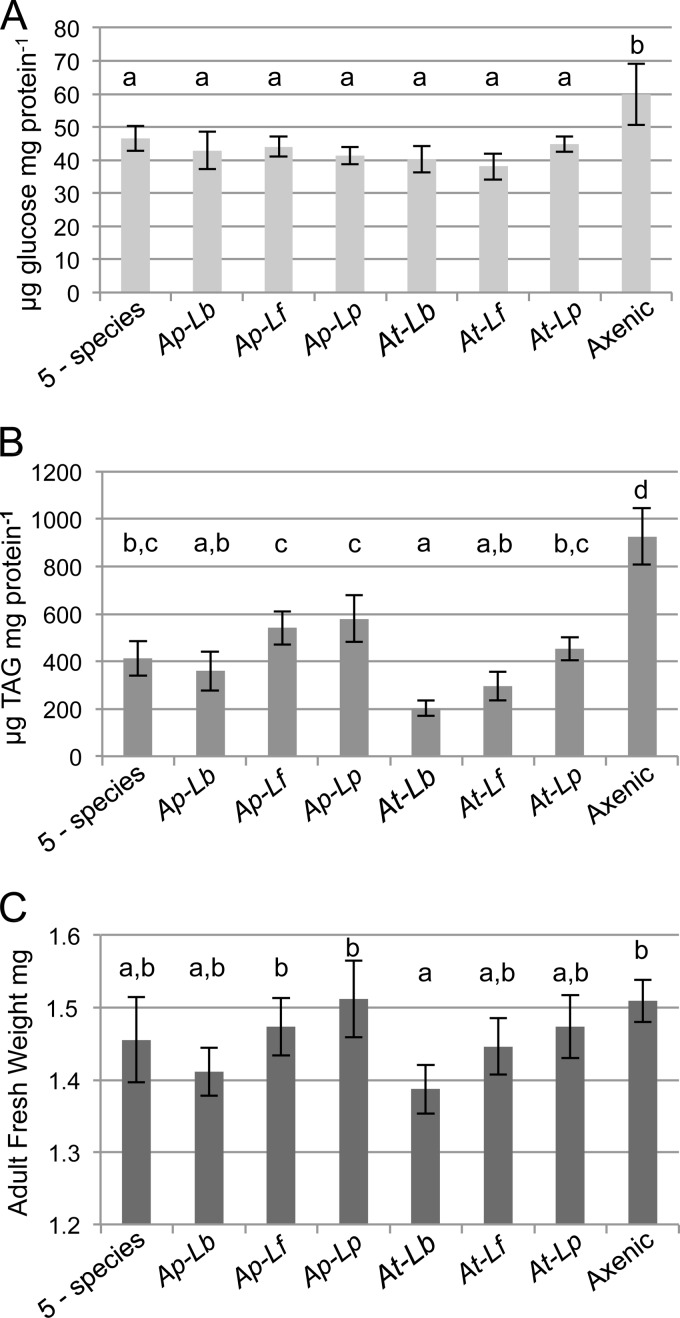
Each dual-species treatment reduced the glucose levels relative to the level in axenic flies (Fig. 3A), consistent with the results for single-species microbiota. Assessment of TAG contents showed that all dual-species microbiota produced flies with reduced levels compared to the level in axenic flies and all but one dual-species microbiota yielded levels comparable to the level observed for the 5-species treatment (Fig. 3B). Uniquely, the A. tropicalis–L. brevis treatment reduced TAG to levels significantly lower than those observed for the 5-species microbiota and some of the other dual-species gnotobiotes (Fig. 3B).
FIG 3.
Effects of dual-species gut microbiota on host nutritional indices. (A and B) Glucose (A) and triglyceride (TAG) (B) contents were determined for the indicated treatment groups and are reported here as the means of 5 independent experiments ± standard errors, normalized to the protein concentration determined for the same samples. (C) Fresh weights of female flies, 5 days after eclosion, are reported as means ± standard errors from 5 independent experiments. Treatments that do not share a letter above the bars had significantly different results by Tukey's HSD test at a P value of <0.05, corrected for multiple comparisons. Ap, A. pomorum; At, A. tropicalis; Lb, L. brevis; Lf, L. fructivorans; Lp, L. plantarum.
These data highlight that dual-species pairs containing A. tropicalis and L. brevis had particularly potent effects on TAG levels in the host. Each dual-species combination that contained one of these bacteria produced a lower TAG level than the corresponding pairs that did not contain it. For example, the TAG value for A. pomorum–L. brevis microbiota flies was lower than those for other treatments containing A. pomorum, and the same was true for A. tropicalis–L. brevis compared to other combinations with A. tropicalis (Fig. 3B). The TAG content of flies with each A. tropicalis-containing dual-species pair was lower than that in flies with the corresponding A. pomorum-containing pair (Fig. 3B). Although not all of these differences were statistically significant after correcting for multiple comparisons, the trend was consistent.
The dual-species microbiota treatments had no significant effect on host protein content (ANOVA, F7,110 = 1.13, P > 0.05), but unlike the gnotobiotic treatments tested previously, adult weight varied significantly with the microbiota (ANOVA, F7,112 = 3.19, P < 0.01). Here again, the A. tropicalis–L. brevis treatment was distinctive, significantly lowering weight relative to that of axenic and A. pomorum–L. plantarum gnotobiotic flies (Fig. 3C). While these were the only differences that were statistically significant, dual-species microbiota containing A. tropicalis or L. brevis yielded consistently lower weights than those without, a trend matching that observed in the TAG data.
Impact of interspecies interactions on bacterial abundance.
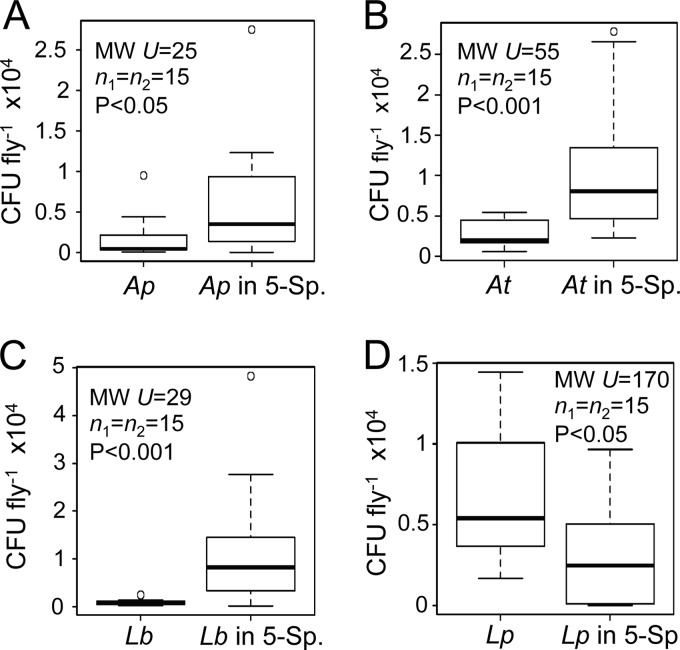
Several studies utilizing gnotobiotic animals have indicated that interspecies interactions can impact microbial metabolism and/or abundance (11–14). To establish whether the abundance of the gut bacteria in Drosophila is influenced by interactions among microbiota members, we quantified the numbers of bacteria in the various gnotobiotic flies. Our investigation was initiated with an analysis of the 5-species gnotobiotes. If interactions had no impact on abundance, each species would be expected to be equally abundant in the single-species gnotobiotes and 5-species community. Inconsistent with this prediction, A. pomorum, A. tropicalis, and L. brevis exhibited significantly more CFU per fly in the 5-species gnotobiotes than in monocolonized hosts (Fig. 4A, B, and C). The L. plantarum levels were significantly reduced in the 5-species gnotobiotes compared to the L. plantarum level in monoassociated flies (Fig. 4D). L. fructivorans was not tested because its low growth rate in vitro precluded quantification of its abundance in the presence of other Lactobacillus species. These observations suggest that positive and negative interactions affect the abundance of microbiota members.
FIG 4.
Bacterial species abundance in single- and multispecies microbiota. CFU counts from whole flies are shown for each bacterial species indicated. In each plot, the left bar presents data from single-species gnotobiotes and the right bar shows data from 5-species gnotobiotes. Each box delineates the first and third quartiles, the dark line is the median, and the whiskers show the range minus outliers, which are shown as open circles. Statistics reported are from Mann-Whitney tests. Ap, A. pomorum; At, A. tropicalis; Lb, L. brevis; Lp, L. plantarum; 5 Sp., 5-species treatment.
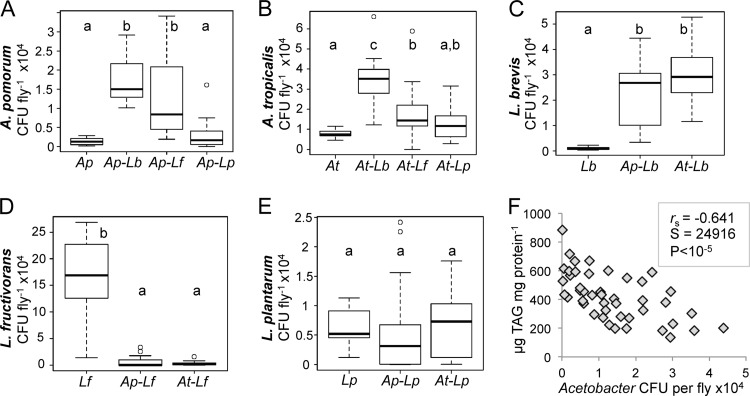
Next, the various dual-species treatments were compared to single-species controls to clarify which bacterial species pairs exhibit positive or negative interactions. Pairings of each Acetobacter species with L. brevis showed reciprocal, positive interactions, with both bacteria reaching a higher abundance in the host when colonizing together than when colonizing alone (Fig. 5A, B, and C). Acetobacter abundance also increased significantly during cocolonization with L. fructivorans, but the benefit was not mutual. L. fructivorans levels dropped by >30-fold under cocolonization conditions, indicating a negative interaction with Acetobacter (Fig. 5D). L. plantarum did not show significant reciprocal effects on abundance in dual-species microbiota (Fig. 5A, B, and E).
FIG 5.
Bacterial species abundance in dual-species gnotobiotic flies. (A to E) CFU counts from whole flies for each bacterial species, indicated on the y axis, from the gnotobiotic treatment indicated on the x axis. Each box delineates the first and third quartiles, the dark line is the median, and the whiskers show the range minus outliers, which are shown as open circles. Treatments that do not share letters above the bars had significantly different results by Mann-Whitney test at a P value of <0.05 after Bonferroni correction. Ap, A. pomorum; At, A. tropicalis; Lb, L. brevis; Lf, L. fructivorans; Lp, L. plantarum. (F) Correlation between Acetobacter abundance and TAG content across all gnotobiotic treatments (single, dual, and 5 species); values are matched on the basis of vial (technical replicate), and correlation statistics are from Spearman's rank-order test.
Correlation between Acetobacter abundance and host phenotypes.
The experiments described above revealed that the impacts of the microbiota on host TAG content differed between single- and dual-species treatments, indicating that the contributions of the bacteria to reductions in host TAG were not additive. For example, the TAG levels in flies monocolonized with L. brevis and L. plantarum were not significantly different (Fig. 2B), yet TAG was significantly lower in Acetobacter–L. brevis dual-species gnotobiotes than in flies with the corresponding Acetobacter–L. plantarum pairs (Fig. 3B). One explanation for this nonadditive effect is that the density of Acetobacter was greater in Acetobacter–L. brevis hosts than in Acetobacter–L. plantarum hosts (Fig. 5A and B), and increased levels of Acetobacter led to a corresponding decrease in host TAG. Consistent with this hypothesis, the median CFU of Acetobacter species was significantly negatively correlated with host TAG levels across all gnotobiotic treatments (Fig. 5F). These observations suggest a dose-dependent relationship between Acetobacter abundance and reductions in host TAG content.
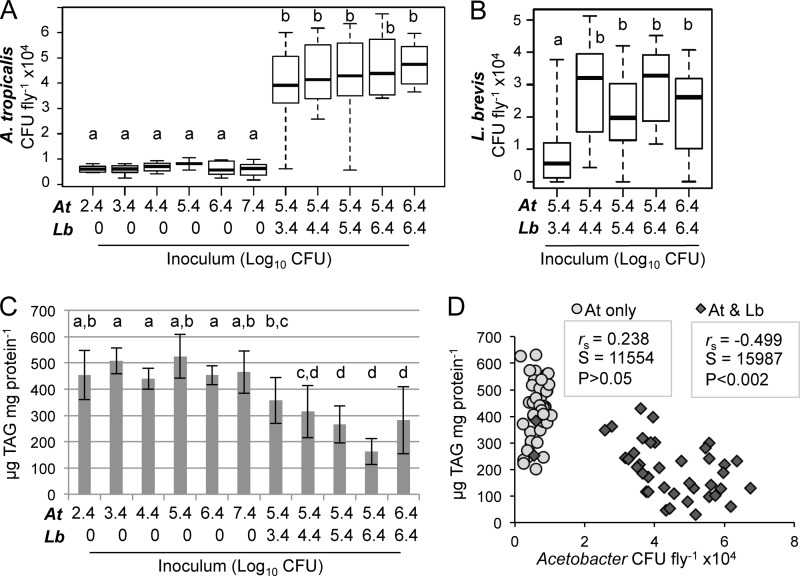
We next sought to determine whether the host TAG level was responsive to the inoculum of Acetobacter alone or whether the observed relationship between Acetobacter abundance and TAG levels was only apparent in multispecies microbiota. We varied the inoculum over 5 orders of magnitude (2.5 × 102 to 2.5 × 107 cells per vial) and observed no significant change in host TAG level or Acetobacter abundance (Fig. 6A), revealing that A. tropicalis density in the fly was not determined by inoculum density. However, when a constant Acetobacter inoculum of 2.5 × 105 cells per vial was introduced in combination with various densities of L. brevis, we observed a stepwise increase in the median Acetobacter abundance (Fig. 6A) and a statistically significant increase of L. brevis abundance as its inoculum density was increased from 2.5 × 103 to 2.5 × 106 cells per vial (Fig. 6B). Unlike what we observed for A. tropicalis monoassociations, the TAG content of dual-species gnotobiotes decreased significantly with increasing bacterial density in the inoculum (Fig. 6C). Acetobacter abundance showed a significant negative correlation with host TAG levels in the case of A. tropicalis–L. brevis treatments but not in the case of treatments with only A. tropicalis (Fig. 6D). L. brevis abundance did not significantly correlate with TAG levels.
FIG 6.
Effect of bacterial inoculum on Acetobacter abundance and host triglyceride content. (A and B) A. tropicalis (A) and L. brevis (B) abundance are shown on a linear scale for the indicated gnotobiotic treatments. Treatments consisted of A. tropicalis (At) alone or in combination with L. brevis (Lb); the approximate number of cells added per vial is indicated at the bottom using a Log10 scale. Each box delineates the first and third quartiles, the dark line is the median, and the whiskers show the range from 3 independent experiments with 5 to 9 replicates each. Treatments that do not share a letter had significantly different results by Mann-Whitney test at a P value of <0.05 after Bonferroni correction. (C) Whole-fly triglyceride (TAG) content for the samples whose CFU counts are shown in panels A and B. Treatments that do not share a letter above the bars had significantly different results by Tukey's HSD test at a P value of <0.05, implemented with a linear mixed-effects model treating experiment as a random effect and corrected for multiple comparisons. (D) Correlation between A. tropicalis abundance and TAG content in the experiment whose results are shown in panel A; values are matched on the basis of vial, and correlation statistics are from Spearman's rank order test.
DISCUSSION
In this study, we investigated whether gut microbiota-dependent traits of Drosophila are mediated by bacterial species acting independently, such that the microbiota is functionally modular, or whether some functions are the product of interactions among bacterial species. Our data suggest that the gut bacteria are modular with respect to their impact on host glucose levels (mediated by all Acetobacter and Lactobacillus species tested) and development time (mediated by Acetobacter only) but interactive with respect to their impact on TAG (Acetobacter-mediated reduction of host TAG is promoted by cocolonization with Lactobacillus species that have no effect on host TAG in monoassociation). These results are not predicted from current understanding of the association. The three microbiota-dependent traits are regulated by insulin- and TOR-dependent signaling pathways, which are known to be influenced by microbiota (21, 22), and the simplest expectation would be that these traits would be coupled, i.e., to vary in concert. Our data raise the possibility that the outputs from insulin and TOR signaling that regulate the three traits may have different response thresholds to the microbiota. Furthermore, the three traits are affected by multiple but not identical host processes (24–27), and differences in the responsiveness of these various processes to the microbiota may also contribute to the variation in their specificity and modularity. Further research is required to establish the molecular basis of this predicted variation in the response of different host traits to the microbiota.
Another indication of differences in the mechanistic basis of the microbiota-mediated reduction in fly glucose and TAG levels comes from the difference in their response to bacterial abundance. Microbiota-dependent reduction of fly glucose content is evident at low bacterial densities, for example, 400 CFU in flies monoassociated with A. pomorum (this study) and 120 CFU in antibiotic-treated flies (19). The microbiota-dependent reduction in TAG content is, in contrast, strongly dependent on the abundance of Acetobacter. Indeed, the dependence of this effect on a certain minimal density of Acetobacter in the microbiota may contribute to the greater variation among studies in the effect of eliminating the microbiota on the TAG level than on glucose content of Drosophila (20, 21; also this study). Other contributory factors could include variation among studies in diet and Drosophila genotype.
More generally, these findings of nondeleterious density-dependent effects of gut microbiota differ from previous research, which has focused primarily on the negative consequences of large bacterial populations. In humans and biomedical rodent models, so-called “blooms” of particular bacteria have been associated with intestinal disease (28–30), immunodeficiency (31), and negative consequences of antibiotic treatments (32, 33). Similarly, experimental conditions that induce high densities of specific gut bacteria in Drosophila can impair gut function and depress fly life span, for example, Gluconobacter morbifer in flies with an overactive immune system (34) and a monoassociation of Lactobacillus brevis bacteria that induces dual oxidase-mediated production of reactive oxygen species (ROS) (35). Interestingly, the L. brevis strain used in this study displayed no detectable deleterious effects on the flies, possibly indicative of variation among strains of L. brevis.
The promotion of microbiota-mediated reduction in host TAG by cocolonization by Acetobacter and Lactobacillus can be explained by, first, the variation in TAG content with Acetobacter abundance and, second, the promotion of Acetobacter abundance by cocolonization with Lactobacillus. In other words, reduced host TAG is a nonmodular function of the microbiota because Lactobacillus stimulates Acetobacter abundance and not because Lactobacillus and Acetobacter interact synergistically with the host regulation of nutrient allocation to TAG reserves. The basis of the positive effect of Lactobacillus on Acetobacter remains to be determined and may involve the provisioning of Lactobacillus-derived nutrients to Acetobacter and Lactobacillus-mediated modification of conditions in the host gut to favor Acetobacter. Metabolite cross-feeding among bacteria with complementary metabolic capabilities is a well-established phenomenon in the mammalian gut (36), and microbe-specific impacts on the gut environment through modulation of immune function (e.g., induction of antimicrobial peptides and ROS) and gut epithelial turnover are also known for both mammals and Drosophila (34, 35, 37–41).
The variation in dual-species associations involving the different Lactobacillus species (Fig. 5) indicates that multiple processes contribute to the Acetobacter-Lactobacillus interaction. Not only do the Lactobacillus species vary in their promotion of Acetobacter abundance, but this variation is accompanied by dramatic differences in the effect of Acetobacter on Lactobacillus. While the association between Acetobacter and L. brevis is mutualistic, yielding increased abundance of both species in the fly, the relationship with L. fructivorans is antagonistic, involving a 30-fold reduction in this Lactobacillus species. A possible explanation for the latter observation could be that the presence of either Lactobacillus species promotes Acetobacter colonization of the gut but only L. brevis cooperates metabolically with Acetobacter, while L. fructivorans competes. Alternatively or additionally, there could be a temporal dimension to these interactions, e.g., L. fructivorans could increase Acetobacter abundance by promoting its growth in larvae, followed by a competitive interaction in adult flies from which Acetobacter emerges as the victor. Lastly, the survival or proliferation of each bacterial species in the diet could be impacted by the presence of others, which would influence their abundance in the fly via ingestion. Further research will investigate the mechanisms responsible for interspecies interactions in the Drosophila gut microbiota, taking into account their spatiotemporal dimensions.
Like the human gut microbiota, the gut microbiota of conventionally reared Drosophila flies exhibits variation among individuals in species content, as well as compositional changes over time (18, 42, 43). Unlike in humans, however, the Drosophila gut community is of low taxonomic diversity such that its content can be comprehensively determined, enabling us to draw conclusions about how compositional changes impact microbiota function. By utilizing Drosophila with gnotobiotic gut microbiota, we found that not all microbial species are functionally equivalent and, furthermore, that their impacts on certain host traits are not modular but shaped by interactions among species. One important implication of our findings is that natural variation in microbiota content across conventionally reared Drosophila may result in substantial phenotypic inconsistency. This would be especially true for fly stocks lacking Acetobacter, which we observed in a previous survey (42). Even with an Acetobacter species in common between two hosts, differences in the Lactobacillus species present could cause significant variation in host TAG content.
In conclusion, we found that the microbial basis of host traits varies in both specificity and modularity. The microbe-mediated reduction in glucose is relatively nonspecific and modular, while the reduction of TAG content is influenced by interspecies interactions. One output of these interactions is an increased abundance of Acetobacter in the presence of some Lactobacilli. Further research will uncover the basis of these interactions and provide a more mechanistic understanding of how the microbiota functions in this experimental system.
ACKNOWLEDGMENTS
This work was supported by a Ruth L. Kirschstein National Research Service Award to P.D.N. (grant F32GM099374) from the National Institute of General Medical Sciences (NIGMS) and National Institutes of Health (NIH) grant 1R01GM095372 to A.E.D., as well as the Sarkaria Institute for Insect Physiology and Toxicology.
We thank J. Chaston and A. Dobson for constructive discussions and thoughtful reading of the manuscript and J. Chaston, G. Kim, D. Sannino, C. N. A. Wong, and Leanne Donahue for technical assistance.
The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the NGIMS or the NIH.
Footnotes
Published ahead of print 15 November 2013
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110:3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 3.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. 10.1126/science.1224203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71–92. 10.1146/annurev.ento.49.061802.123416 [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 7.Loh G, Blaut M. 2012. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 3:544–555. 10.4161/gmic.22156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cani PD, Delzenne NM. 2011. The gut microbiome as therapeutic target. Pharmacol. Ther. 130:202–212. 10.1016/j.pharmthera.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 9.Holmes E, Li JV, Marchesi JR, Nicholson JK. 2012. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell metabolism. 16:559–564. 10.1016/j.cmet.2012.10.007 [DOI] [PubMed] [Google Scholar]
- 10.Flint HJ. 2011. Obesity and the gut microbiota. J. Clin. Gastroenterol. 45(Suppl):S128–S132. 10.1097/MCG.0b013e31821f44c4 [DOI] [PubMed] [Google Scholar]
- 11.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. U. S. A. 103:10011–10016. 10.1073/pnas.0602187103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285:22082–22090. 10.1074/jbc.M110.117713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A. 108:6252–6257. 10.1073/pnas.1102938108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. 2013. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. U. S. A. 110:13582–13587. 10.1073/pnas.1312524110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong CN, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13:1889–1900. 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7:e1002272. 10.1371/journal.pgen.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridley EV, Wong AC, Douglas AE. 2013. Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster. Appl. Environ. Microbiol. 79:3209–3214. 10.1128/AEM.00206-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridley EV, Wong AC, Westmiller S, Douglas AE. 2012. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One 7:e36765. 10.1371/journal.pone.0036765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- 22.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14:403–414. 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 23.Bergmeyer H-U. 1965. Methods of enzymatic analysis, vol 1 Verlag Chemie, Weinheim, Germany [Google Scholar]
- 24.Arrese EL, Soulages JL. 2010. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55:207–225. 10.1146/annurev-ento-112408-085356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz MW, Gade G. 2009. Hormonal regulation of energy metabolism in insects as a driving force for performance. Integr. Comp. Biol. 49:380–392. 10.1093/icb/icp019 [DOI] [PubMed] [Google Scholar]
- 26.Mirth CK, Riddiford LM. 2007. Size assessment and growth control: how adult size is determined in insects. Bioessays 29:344–355. 10.1002/bies.20552 [DOI] [PubMed] [Google Scholar]
- 27.Oldham S. 2011. Obesity and nutrient sensing TOR pathway in flies and vertebrates: functional conservation of genetic mechanisms. Trends Endocrinol. Metab. 22:45–52. 10.1016/j.tem.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter SE, Lopez CA, Baumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14:319–327. 10.1038/embor.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haag LM, Fischer A, Otto B, Plickert R, Kuhl AA, Gobel UB, Bereswill S, Heimesaat MM. 2012. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One 7:e35988. 10.1371/journal.pone.0035988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, Clemente JC, Cullender TC, Barnich N, Darfeuille-Michaud A, Vijay-Kumar M, Knight R, Ley RE, Gewirtz AT. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12:139–152. 10.1016/j.chom.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80:62–73. 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayres JS, Trinidad NJ, Vance RE. 2012. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18:799–806. 10.1038/nm.2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777–782. 10.1126/science.1149357 [DOI] [PubMed] [Google Scholar]
- 35.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ. 2013. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153:797–811. 10.1016/j.cell.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 36.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, Lambeth JD, Denning PW, Neish AS. 18 October 2013. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 10.1038/emboj.2013.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23:2333–2344. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. 2010. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 8:152. 10.1186/1741-7007-8-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7:1922–1932. 10.1038/ismej.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]