Abstract
In this study we tested 39 Lactococcus lactis strains isolated from diverse habitats for their robustness under heat and oxidative stress, demonstrating high diversity in survival (up to 4 log units). Strains with an L. lactis subsp. lactis phenotype generally displayed more-robust phenotypes than strains with an L. lactis subsp. cremoris phenotype, whereas the habitat from which the strains had been isolated did not appear to influence stress survival. Comparison of the stress survival phenotypes with already available comparative genomic data sets revealed that the absence or presence of specific genes, including genes encoding a GntR family transcriptional regulator, a manganese ABC transporter permease, a cellobiose phosphotransferase system (PTS) component, the FtsY protein, and hypothetical proteins, was associated with heat or oxidative stress survival. Finally, 14 selected strains also displayed diversity in survival after spray drying, ranging from 20% survival for the most robust strains, which appears acceptable for industrial application, to 0.1% survival for the least-tolerant strains. The high and low levels of survival upon spray drying correlated clearly with the combined robustness under heat and oxidative stress. These results demonstrate the relevance of screening culture collections for robustness under heat and oxidative stress on top of the typical screening for acidifying and flavor-forming properties.
INTRODUCTION
Based on their spoilage-preventing and flavor-enhancing characteristics, lactic acid bacteria (LAB) have been employed since ancient times in the fermentation of foods, e.g., fruits, vegetables, cereal grains, meat, and milk (1, 2). Nowadays, many of these processes have been industrialized, and fermentation is typically initiated by the addition of starter cultures, which contain high concentrations of one or multiple LAB strains (1, 2). As starter cultures require metabolic activity to contribute to the taste and texture of the fermentation end products, there has been an increasing industrial interest in studying robustness phenotypes during industrial production and processing (1), which involves preservation by either freezing or drying techniques (3–5). The major disadvantages of frozen starter cultures are the inconvenience and costs of transport and storage at low temperature, and, therefore, drying techniques are preferred (3–5). During spray drying, cultures are exposed to severe heat and oxidative stress (6, 7), typically resulting in lower survival rates of starter cultures than freeze-drying (3, 4). Therefore, freeze-drying is currently the most often applied industrial drying method (3–5). However, spray drying appears a more cost-effective and energy-efficient drying alternative for the preservation of starter cultures (3–5), providing strains that display high robustness under the stresses encountered in this process can be identified. This appears feasible, as studies on stress phenotypes typically result in highly strain-specific robustness phenotypes, e.g., for the gastrointestinal survival of Lactobacillus plantarum strains (8) and the robustness of several Lactobacillus strains during acid, alkaline, heat, oxidative, osmotic, detergent, and starvation stresses (9).
Lactococcus lactis is one of the most widely used LAB for industrial food fermentations, including the production of cheese and butter(milk) (2). L. lactis strains used in industry are mainly of dairy origin, and within this group of strains a high diversity has been observed in functional characteristics such as bacteriocin production (10) and proteolytic activity (11). Interest in strains from other habitats has increased over the past decade, as diversity studies including nondairy strains demonstrated even more distinct phenotypes than studies including solely dairy strains, e.g., in flavor formation (12, 13). Furthermore, the potential for the application of nondairy strains in dairy starter cultures was demonstrated by adaptation of the plant-derived L. lactis strain KF147 to a dairy environment by long-term propagation (14). Comparative genomics approaches have pinpointed differences between L. lactis strains with respect to genes predicted to be involved in stress responses (15, 16), suggesting differences in stress survival characteristics between L. lactis strains. Nevertheless, diversity in stress survival phenotypes has been minimally studied for this LAB.
In this study we tested 39 L. lactis strains isolated from diverse habitats for their robustness under heat and oxidative stress and compared these data with robustness during lab scale spray drying. The heat and oxidative stress survival data were also correlated to the habitat and subspecies of the strains and to genomic content data (15) to identify genes associated with robustness, an approach that previously has been successfully employed for L. plantarum (17–19).
MATERIALS AND METHODS
Bacterial isolates and media.
The 39 L. lactis strains used in this study were selected from a large collection of phenotypically and genotypically characterized strains (15, 20) and are listed in Table 1. This set contains L. lactis strains of three different subspecies: L. lactis subsp. lactis, L. lactis subsp. cremoris, and L. lactis subsp. hordniae, which were isolated from dairy as well as nondairy environments. All strains were grown in M17 broth (Oxoid, Basingstoke, United Kingdom) supplemented with 0.5% (wt/vol) glucose (Merck, Darmstadt, Germany) (GM17) at 30°C.
TABLE 1.
Strains employed for heat and oxidative stress survival screening
| Strain codea | NIZO code | Isolation source | Other information |
|---|---|---|---|
| L. lactis subsp. lactis genotype, L. lactis subsp. lactis phenotype | |||
| ML8 | 20 | Dairy starter | |
| LMG8526 | 26 | Chinese radish seeds | |
| ATCC 19435T | 29 | Milk (dairy starter) | |
| UC317 | 644 | Dairy starter | |
| M20 | 844 | Soil | L. lactis subsp. lactis biovar diacetylactis |
| Li-1 | 1156 | Grass | |
| E34 | 1173 | Silage | |
| DRA4 | 1592 | Dairy starter A | L. lactis subsp. lactis biovar diacetylactis |
| LMG9446 | 2123 | Frozen peas | |
| LMG9447 | 2124 | Frozen peas | |
| K231 | 2199 | White kimchi | |
| K337 | 2202 | White kimchi | |
| P7266 | 2206 | Litter on pastures | |
| P7304 | 2207 | Litter on pastures | |
| NCDO895 | 2211 | Dairy starter | |
| KF7 | 2219 | Alfalfa sprouts | |
| KF24 | 2220 | Alfalfa sprouts | |
| KF67 | 2223 | Grapefruit juice | |
| KF134 | 2226 | Alfalfa and radish sprouts | |
| KF146 | 2229 | Alfalfa and radish sprouts | |
| KF147 | 2230 | Mung bean sprouts | |
| KF196 | 2236 | Japanese kaiware shoots | |
| KF201 | 2238 | Sliced mixed vegetables | |
| NIZO2244B | 3919 | Mustard and cress | |
| KF282 | 3920 | Mustard and cress | |
| N42 | 1230 | Soil and grass | |
| LMG14418 | 2424 | Bovine milk | |
| IL1403 | 2441 | Dairy starter | |
| L. lactis subsp. cremoris genotype, L. lactis subsp. lactis phenotype | |||
| NCDO763 | 643 | Dairy starter | Derivative of NCDO712 |
| V4 | 1157 | Raw sheep milk | |
| N41 | 1175 | Soil and grass | |
| MG1363 | 1492 | Cheese starter | Plasmid-free derivative of NCDO712 |
| KW10 | 2249 | Kaanga wai | |
| L. lactis subsp. cremoris genotype and phenotype | |||
| SK11 | 32 | Dairy starter | |
| AM2 | 33 | Dairy starter | |
| HP | 42 | Dairy starter | Same as LMG6897T but from different collection |
| FG2 | 2252 | Dairy starter | |
| LMG6897T | 2418 | Cheese starter | Same as HP but from different collection |
| L. lactis subsp. hordniae LMG8520T | 24 | Leaf hopper (insect) |
Underlined strains were included in the spray drying analysis.
Heat and oxidative stress survival assays.
From a preculture, a 1% (vol/vol) inoculum was added to 2 ml of fresh GM17 in duplicate in a 96-well Masterblock (Greiner BioOne GmbH, Frickenhausen, Germany) and incubated for 16 h at 30°C. In stationary phase, cells were harvested from 0.5 ml of culture by centrifugation at 1,865 × g for 15 min and resuspended in 1 ml sterile 50 mM sodium phosphate buffer (Merck, Darmstadt, Germany), pH 7.2. To measure heat stress survival, 100 μl of the cell suspensions was incubated at 50°C for 30 min in a 0.1-ml 96-well PCR plate (MicroAmp; Applied BioSystems, Foster City, CA) in a Gene-Amp PCR system 9600 (Applied BioSystems). Controls were left at room temperature for 30 min. For assessment of oxidative stress survival, 1 ml of culture was centrifuged at 1,865 × g for 15 min and resuspended in the same volume of phosphate buffer. Hydrogen peroxide (Merck) in phosphate buffer was added to 0.25 ml of the cell suspensions to a final concentration of 5 mM and an end volume of 0.5 ml, followed by incubation for 3 h at 30°C in a water bath. To the controls, buffer without hydrogen peroxide was added, and these cell suspensions were also incubated for 3 h at 30°C. After incubation, samples were centrifuged at 1,865 × g for 15 min and cells were resuspended in 0.5 ml of phosphate buffer. Survival was measured by spotting serial dilutions in triplicate on GM17 agar plates (21). CFU were assessed after incubation of the plates for 72 h at 30°C.
Genotype-phenotype comparison.
Heat stress and oxidative stress survival data were associated with the gene presence/absence matrix derived from the pangenome-based comparative genome hybridization analysis performed by Siezen et al. (15). The Web tool PhenoLink (22), which applies the Random Forest classification algorithm (23), was used to identify genes associated with stress survival. Default parameters were used, except for the bagging parameter, which was set to 100, and the “ratio of largest phenotype size to smallest phenotype size” parameter (explained below), which was set to 1 for the heat stress data. These parameters were modified from the default setting to deal with imbalance in sample group sizes for some survival parameters measured. Class imbalance can severely skew importance estimation of genes due to overtraining of the classification model to the larger sample group. Briefly, from the larger class (sample group), 100 (bagging parameter) times the number of samples (ratio of 1) from the smallest class were drawn to ensure that, even for very unbalanced sample groups, all samples of the larger group were taken. Three groups of strains were defined (see Fig. 1 and 2), based on the heat and oxidative stress survival phenotypes (robust, intermediate, and sensitive groups). The presence of a gene in 75% of the strains in the robust or sensitive group and the absence of this gene in 75% of the strains in the reciprocal group were used as a minimum cutoff to pinpoint genes possibly associated with robustness under the applied stress.
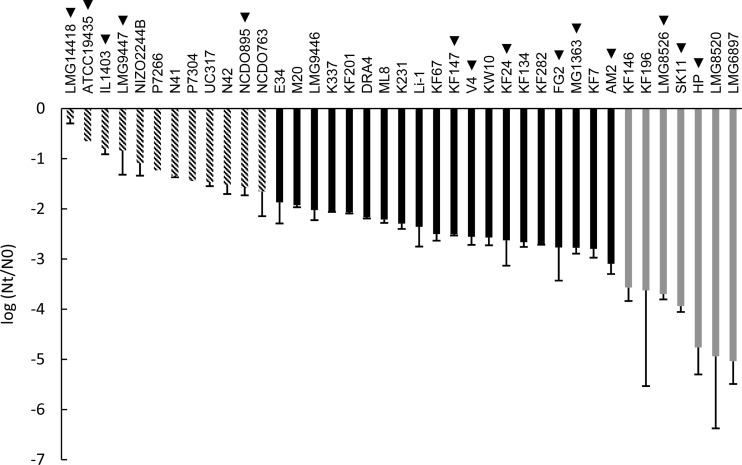
FIG 1.
Robustness of L. lactis strains upon exposure to heat stress expressed as the difference between log CFU/ml after heat stress (Nt) and control (N0). For genotype-phenotype matching, the strains were divided into groups of robust (striped bars), intermediate (black bars), and sensitive (gray bars) strains. Strain names indicated with a triangle represent strains that were selected for the spray drying analysis (see Fig. 4). The data represent averages of two biological replicates. Error bars indicate standard deviations.
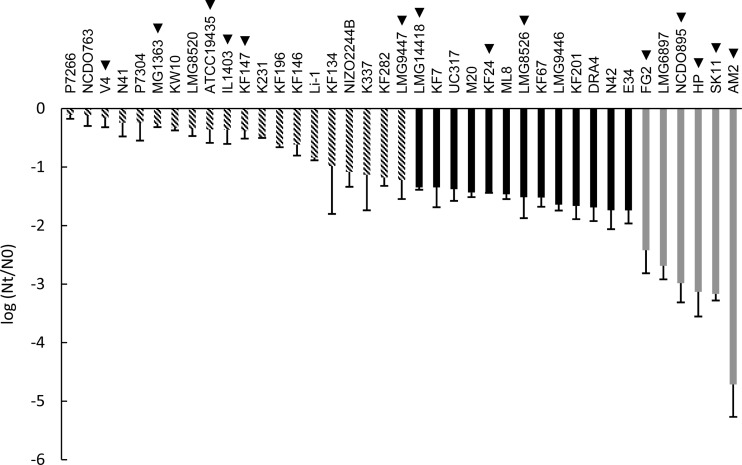
FIG 2.
Robustness of L. lactis strains upon oxidative stress exposure expressed as the difference between log CFU/ml after oxidative stress (Nt) and control (N0). For genotype-phenotype matching, groups were made up of robust (striped bars), intermediate (black bars), and sensitive (gray bars) strains. Strain names indicated with a triangle represent strains that were selected for the spray drying analysis (see Fig. 4). The data represent averages of two biological replicates. Error bars indicate standard deviations.
Spray drying.
From the set of 39 L. lactis strains, 14 strains (LMG8526, ATCC 19435, LMG9447, NCDO895, KF24, KF147, LMG14418, IL1403, V4, MG1363, SK11, AM2, HP, and FG2) were selected to determine survival during spray drying. Strains were incubated in duplicate at 30°C in 200 ml GM17, and stationary-phase cells were harvested by centrifugation at 3,315 × g for 7 min and dissolved in 200 ml 20% (wt/vol) skim milk powder. Cell suspensions were dried in a mini-lab-scale spray dryer (model B-290; Büchi Labortechnik AG, Flawil, Switzerland) by using an inlet temperature of 200°C and an outlet temperature of 100°C. Ice water was continuously used to cool the nozzle. The moisture content of the resulting powder was determined in duplicate by measuring weight loss during incubation at 102°C for 3 h. To determine survival rates, the generated powders were rehydrated in water (1% [wt/vol]) for 1 h at room temperature. The rehydrated cell suspension and the feed cell suspension were serially diluted in duplicate and spotted in triplicate on GM17 agar plates. After 3 days of incubation at 30°C, CFU were assessed. Dry weight of the cell suspensions was determined in triplicate by measuring the weight of 5 ml of sample after incubation at 55°C for 5 days.
Statistical analysis.
Significance of the differences in robustness of groups with different origins, genotypes, or phenotypes was assessed with a t test. Significance of the correlations of survival during heat stress, oxidative stress, and spray drying was assessed by a linear model. All statistic calculations were done in R (version 2.15; R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org]).
RESULTS
Heat and oxidative stress survival phenotypes are highly diverse.
As LAB robustness can be highly strain specific (8, 9), we assessed the heat and oxidative stress survival of a collection of 39 L. lactis strains of diverse origins (Table 1). The collection of L. lactis strains displayed highly variable heat survival characteristics at 50°C, with the decrease in viability ranging from 0.2 log unit for the most robust strain (LMG14418) to 5.0 log units for the least robust strain (LMG6897) (Fig. 1). Moreover, the phenotype did not appear binary, as a continuum of intermediate survival levels was observed.
The same collection of strains was assessed for their oxidative stress tolerance (Fig. 2). Similar to results for heat stress, high diversity was observed (more than 4 log units). Strain P7266 displayed the highest robustness under oxidative stress, with a viability loss of 0.1 log unit. By contrast, strain AM2 was by far the most sensitive to oxidative stress, with a viability loss of 4.7 log units, approximately 1 log unit more than the second-most-sensitive strain (SK11). The stress analyses revealed that strains HP and LMG6897, which represent the same strain derived from different culture collections, displayed similar sensitivities to both heat stress (5.0- and 4.8-log-unit decreases, respectively) and oxidative stress (3.1- and 2.7-log-unit decreases, respectively), confirming the reproducibility of both assays and demonstrating that the strain-specific survival differences observed are far greater than the technical variability in our assays. Furthermore, statistical analysis revealed that the robustness of each of the three groups (as represented in Fig. 1 and 2) was significantly different from the robustness of the other two groups of strains (P < 0.001).
Robustness under heat stress and robustness under oxidative stress are correlated.
Combining the data generated in both stress analyses, two strains, ATCC 19435 and IL1403, appeared relatively robust under both heat and oxidative stress (less than 1 log unit loss of viability). LMG6897, HP, and SK11 were among the most sensitive strains upon exposure to both heat and oxidative stress. However, some strains showed larger differences in their robustness under the two stresses. For example, LMG8520 was robust under oxidative stress (0.3 log unit), whereas this strain displayed a larger decrease in viability (4.9 log units) when exposed to heat stress. Other strains, like NCDO895, were more robust under heat stress (1.6 log units) than under oxidative stress (3.0 log units). Nevertheless, a correlation between responses to heat stress and oxidative stress was observed (P < 0.05).
Robustness is related to L. lactis subsp. lactis or L. lactis subsp. cremoris phenotype but not to the habitat from which strains originated.
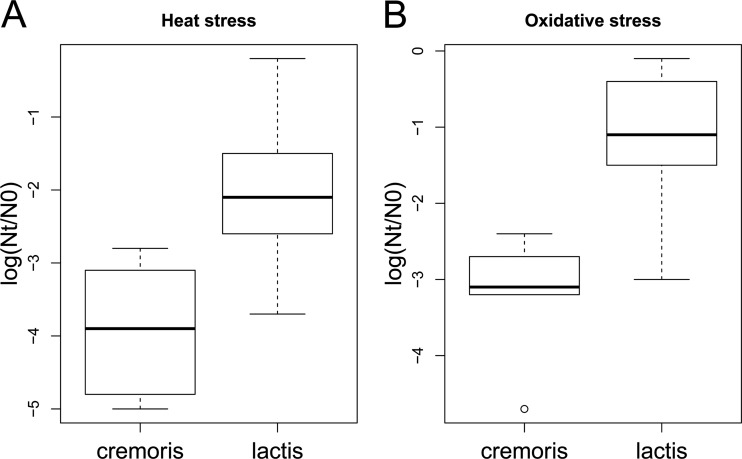
Differences in robustness under both heat and oxidative stress between the different phenotypic groups were assessed. Strains with an L. lactis subsp. lactis phenotype displayed more robustness under heat as well as oxidative stress than strains with an L. lactis subsp. cremoris phenotype (P < 0.01) (Fig. 3A and B). Besides the phenotype-based division, currently the species is divided genotypically into L. lactis subsp. cremoris and L. lactis subsp. lactis (24, 25). These genotypic groups are not clearly reflected in the phenotype-based grouping (Table 1). The groups based on genotype differ less in stress survival phenotype; strains with an L. lactis subsp. cremoris genotype are more sensitive to heat stress than strains with an L. lactis subsp. lactis genotype (P < 0.05) but respond similarly to oxidative stress (P = 0.3).
FIG 3.
Box plots of heat stress (A) and oxidative stress (B) survival phenotypes of strains with an L. lactis subsp. cremoris or L. lactis subsp. lactis phenotype. Strains with an L. lactis subsp. lactis phenotype display more robustness than strains with an L. lactis subsp. cremoris phenotype (P < 0.01).
Similarly, we assessed differences in stress tolerance phenotypes of isolates from different origins. Dairy and nondairy isolates did not significantly differ in robustness under heat stress (P = 0.9) or oxidative stress (P = 0.054).
Identification of genes associated with robustness under heat stress.
A genotype-phenotype comparison was made by matching the heat stress survival data with the gene presence/absence matrix available from a pangenome-based comparative genome hybridization analysis (15). Based on their stress survival phenotypes, the set of strains was manually divided into three groups (robust, intermediate, and sensitive) (Fig. 1). If a gene resulting from the PhenoLink analysis (22) was present in 75% of the strains in the robust or sensitive group and absent in 75% of the strains in the opposite group, this gene was considered to be correlated or anticorrelated, respectively, with robustness under heat stress.
Matching the genotype with the heat stress survival phenotype revealed that the presence of a gene (LLKF_1440 [ortholog, LACR_1472]) encoding a GntR family transcriptional regulator negatively correlated with robustness under heat stress (Table 2). This gene is genetically linked (i.e., part of a group of genes that are adjacent and colinear and that have intergenic spacing smaller than 100 bp) to genes encoding a sugar transporter (LLKF_1444 [LACR_1477], LLKF_1445 [LACR_1478]), a beta-glucosidase (LLKF_1441 [LACR_1473]), and a hypothetical protein (LLKF_1442 [LACR_1475]) that are also negatively correlated with robustness under heat stress. The presence of a gene encoding part of a manganese ABC transporter (mtsC) positively correlated with robustness under heat stress. Although this gene is present in the heat-sensitive strain SK11, it appears to be absent in all the other heat-sensitive strains (15). Moreover, the presence of a gene encoding an ABC transporter ATP-binding protein (yabE) and two genes encoding hypothetical proteins (yliD and ymgH) positively correlated with robustness under heat stress. Furthermore, some genes that were associated with robustness under heat stress encode phage-related functions, which are highly variable among strains (26) and which were not regarded as possible robustness markers and therefore were excluded from Table 2.
TABLE 2.
Genes associated with robustness against heat and oxidative stress resulting from the genotype-phenotype comparison
| Locus(i)a | Gene | Product | Presence of gene in robust phenotype |
|---|---|---|---|
| Heat stress | |||
| LACR_1479 (LLKF_1446) | Hypothetical protein | Absent | |
| LLKF_1440 (LACR_1472) | GntR family transcriptional regulator | Absent | |
| L183932 | yliD | Hypothetical protein | Present |
| LLKF_1406 (llmg_1137, LACR_1439, L149891) | mtsC | Manganese ABC transporter permease | Present |
| LLKF_1445 (LACR_1478) | Sugar ABC transporter | Absent | |
| LACR_1475 (LLKF_1442) | Hypothetical protein | Absent | |
| LLKF_1441 (LACR_1473) | Beta-glucosidase | Absent | |
| L66209 | ymgH | Hypothetical protein | Present |
| LLKF_0011 (L15262) | yabE | ABC transporter ATP-binding protein | Present |
| LACR_1477 (LLKF_1444) | Sugar ABC transporter permease | Absent | |
| Oxidative stress | |||
| L31294 (LLKF_0836) | yidB | Cellobiose-specific PTS IIC component | Present |
| LACR_0155 | Hypothetical protein | Absent | |
| LACR_1321 | Hypothetical protein | Absent | |
| llmg_1634 (LACR_0981) | ABC transporter permease | Absent | |
| LACR_0651 | Surface antigen | Absent | |
| LACR_1297 | Saccharopine dehydrogenase-related protein | Absent | |
| LACR_1259 | Hypothetical protein | Absent | |
| LACR_0861 | Hypothetical protein | Absent | |
| LACR_1215 | Type I restriction-modification system methyltransferase subunit | Absent | |
| LACR_1347 | Transcriptional regulator | Absent | |
| LACR_1992 | Hypothetical protein | Absent | |
| LACR_2213 | Hypothetical protein | Absent | |
| LACR_2214 | Hypothetical protein | Absent | |
| LACR_0503 | Hypothetical protein | Absent | |
| LACR_0994 | Lipopolysaccharide biosynthesis glycosyltransferase | Absent | |
| LACR_0995 | Lipopolysaccharide biosynthesis glycosyltransferase | Absent | |
| LACR_1262 | Hypothetical protein | Absent | |
| LACR_1322 | Hypothetical protein | Absent | |
| LACR_1393 | Hypothetical protein | Absent | |
| LACR_0354 | Molybdopterin/thiamine biosynthesis dinucleotide-utilizing protein | Absent | |
| LACR_1164 | Hypothetical protein | Absent | |
| LACR_1257 | Glycosyltransferase | Absent | |
| LACR_1261 | Hypothetical protein | Absent | |
| LACR_1963 | Hypothetical protein | Absent | |
| LACR_2216 | Hypothetical protein | Absent | |
| LACR_2218 | Hypothetical protein | Absent | |
| LACR_2480 | Hypothetical protein | Absent | |
| LACR_1318 | Hypothetical protein | Absent | |
| LACR_1319 | ADP-ribose pyrophosphatase | Absent | |
| LACR_1348 | Arabinose efflux permease | Absent | |
| LACR_1930 | Hypothetical protein | Absent | |
| LACR_0803 | Hypothetical protein | Absent | |
| LACR_1260 | Hypothetical protein | Absent | |
| LACR_2551 | XRE family transcriptional regulator | Absent | |
| LACR_2447 | LacI family transcription regulator | Absent | |
| LACR_0292 | Hypothetical protein | Absent | |
| LACR_1300 | Hypothetical protein | Absent | |
| LACR_2549 | Serine/threonine protein kinase | Absent | |
| LLKF_0831 (L0206, llmg_1744) | ftsY | Signal recognition particle docking protein FtsY | Present |
| LACR_0846 | Hypothetical protein | Absent | |
| LACR_1296 | Putative intracellular protease/amidase | Absent | |
| LACR_1931 | Hypothetical protein | Absent | |
| LACR_2086 | Glycosyltransferase | Absent | |
| LACR_0154 | Cell surface protein | Absent | |
| LACR_1038 | Hypothetical protein | Absent | |
| LACR_1264 | Hypothetical protein | Absent | |
| LACR_1388 | Hypothetical protein | Absent | |
| LACR_2121 (llmg_0814) | ps323 | Hypothetical protein | Absent |
| LACR_0157 | XRE family transcriptional regulator | Absent | |
| LACR_1834 (LLKF_1826) | yrbH | Hypothetical protein | Absent |
| LLKF_2283 (llmg_1258, L66407) | ymgI | Hypothetical protein | Present |
| llmg_1259 (LLKF_0279, LLKF_2282) | ymgH | Hypothetical protein | Present |
Loci with prefixes of LACR, LLKF, llmg, and L represent strains SK11, KF147, MG1363, and IL1403, respectively.
Identification of genes associated with robustness under oxidative stress.
Gene presence and absence patterns were also compared with oxidative stress survival phenotypes using the same criteria as described above for heat stress. Genes that positively correlated with robustness under oxidative stress included a gene (L31294 [LLKF_0836]) encoding part of a cellobiose-specific phosphotransferase system (PTS) (Table 2). Oxidative stress survival was also associated with the presence of the gene ftsY, encoding a signal recognition particle docking protein. Furthermore, the neighboring genes ymgH and ymgI, encoding hypothetical proteins, were associated with oxidative stress survival. Gene ymgH of strain IL1403 (L66209), which was associated with heat stress, is orthologous (defined by the orthology prediction program InParanoid [27]) to the MG1363 gene llmg_1259 and the KF147 genes LLKF_0279 and LLKF_2282, which appeared in the genotype-phenotype matching results for the oxidative stress phenotypes. These genes were not assigned to the same orthologous group (15) because strict criteria were required for adequate orthology grouping due to the high similarity of the strains. All genes in a group were required to be orthologs of one another, and any gene in that group should not have other orthologs. In contrast to the other genes, L66209 was not an ortholog of LACR_2147 and, therefore, was excluded from the group.
Most genes associated with oxidative stress anticorrelated with robustness, and these genes were often found to encode hypothetical proteins (Table 2). However, genes encoding four transcriptional regulators and four glycosyltransferases were also found to negatively correlate with robustness under oxidative stress.
Heat and oxidative stress survival predicts spray drying robustness.
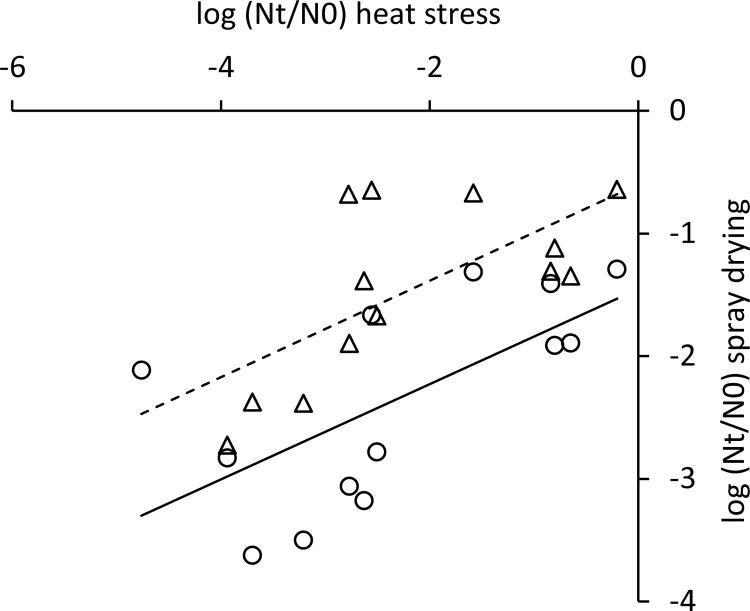
During spray drying, cultures are exposed to a combination of heat stress and oxidative stress, which both lead to loss of viability (6, 7). To assess whether robustness under heat and oxidative stress can therefore predict robustness under spray drying, stationary-phase cells of 14 L. lactis strains with diverse responses to heat and oxidative stress (Fig. 1 and 2) were subjected to lab scale spray drying. Inlet and outlet temperatures were optimized to obtain powders with a moisture content that did not exceed 4%, which is required for powder stability and spoilage prevention (28, 29). Survival was determined by comparing the viability of the rehydrated powder with the viability of the feed and was expressed in CFU per gram of dry weight. Relative survival compared to strain IL1403 was calculated to compensate for technical variability. The most robust strains (MG1363, LMG14418, and NCDO895) displayed a more-than-200-fold-better survival during spray drying than the most sensitive strains (SK11, AM2, and LMG8526) (Fig. 4). Most strains with a sensitive or intermediate response to both heat and oxidative stress displayed a larger decrease of viability during spray drying than strains with a robust phenotype under at least one of the stresses, which is in line with the fact that during spray drying cells are exposed to heat as well as oxidative stress (6, 7). Of the individual stresses, robustness under heat stress appeared to have the highest correlation with survival during spray drying (Fig. 5). To further analyze whether the robustness under spray drying could be predicted by measuring robustness under heat and oxidative stress, we employed linear modeling using spray drying as the response variable and measurement series, robustness under heat stress, and robustness under oxidative stress as explanatory variables. Survival upon spray drying appeared to be related to robustness under heat stress (R2 is 0.59, P < 0.001) and to robustness under oxidative stress (R2 is 0.41, P < 0.01). Combining heat and oxidative stress robustness in a single model improved the predictive power of the model (R2 is 0.61, P < 0.001), demonstrating the feasibility of our relatively simple, high-throughput heat and oxidative stress assays to predict spray drying robustness, which can be assessed only in more-tedious experiments.
FIG 4.
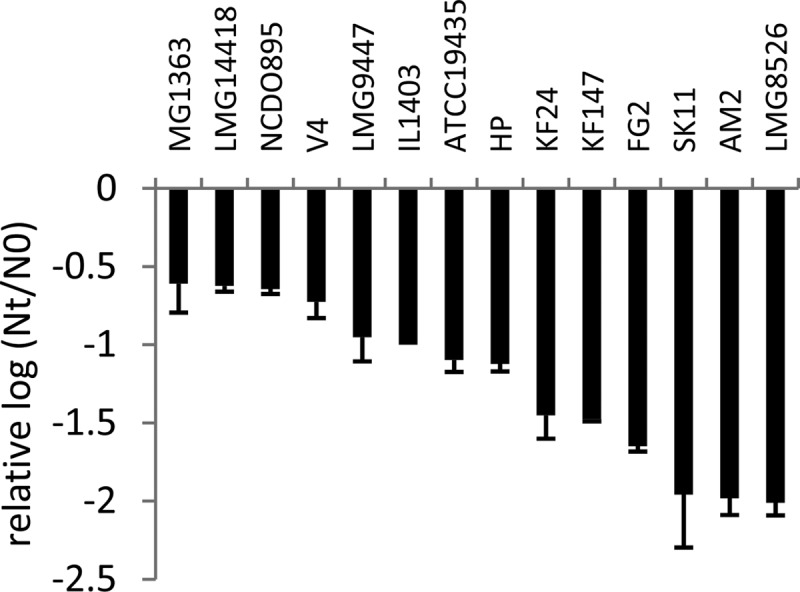
Viability of L. lactis strains after spray drying expressed as the relative difference between log CFU/g dry weight after spray drying (Nt) and before spray drying (N0), compared to that of strain IL1403. The data represent averages of two biological replicates. Error bars indicate standard deviations.
FIG 5.
Correlation of spray drying survival and heat stress survival. Different markers indicate the two series of spray drying analyses.
DISCUSSION
High, strain-specific diversity in stress responses and functional characteristics makes it worthwhile to identify novel (more-robust) strains for application in starter cultures. The diversity of the 39 L. lactis strains employed in this study, which were selected from a larger collection and are anticipated to be a good representation of the L. lactis species as a whole (20), appeared high (up to 4 log units) in both heat and oxidative stress survival assays. The heat stress survival level of strain IL1403 corresponds with results in a study of Hartke et al., in which heat resistance of IL1403 at 52°C was assessed (30). Although specific strains displayed different responses to the two stresses, overall correlation between the response to heat stress and oxidative stress was observed, suggesting that heat and oxidative stress responses are partly driven by general stress mechanisms, as has been shown in strain MG1363 (31, 32). Moreover, these observations also corroborate earlier studies demonstrating that preexposure of L. lactis strains to a specific stress provides cross-protection to another stress condition, e.g., increased robustness under both heat and oxidative stress during carbohydrate starvation (30) and mildly acid conditions (33).
Furthermore, robustness under both heat and oxidative stress appeared to be related to phenotype and only partly to genotype. Therefore, the phenotype-based nomenclature of L. lactis strains appears more helpful for selection of robust strains than the nowadays often applied genotype-based nomenclature. Robustness under both heat and oxidative stress appeared unrelated to the habitat from which strains were originally isolated, despite the fact that genomic analyses have demonstrated the presence of additional genes involved in stress response in nondairy strains (15, 16).
Correlation of robustness under heat and oxidative stress and the gene absence/presence pattern of the collection of L. lactis strains revealed several genes associated with robustness phenotypes. Robustness under heat stress was associated with the presence of a gene encoding a manganese transporter (mtsC). Interestingly, manganese is often associated with stress survival, specifically oxidative stress (34, 35). The presence of genes encoding a cellobiose transporter (yidB), a signal recognition particle docking protein (ftsY), and two hypothetical proteins (ymgH and ymgI) was associated with oxidative stress survival. Involvement in the stress response of these genes was also suggested by the facts that in L. plantarum a transporter of cellobiose was upregulated in an oxidative-stress-sensitive trxB1 deletion mutant compared to the wild-type strain (36), a Streptococcus mutans ftsY deletion mutant was sensitive to both acid and salt stress (37), and the neighboring genes ymgH and ymgI, encoding hypothetical proteins, are located near the general stress-inducible gene gls24 (38). Mutants of strain MG1363 with deletions of the gene encoding part of a manganese ABC transporter (mtsC) and the genes ymgH and ymgI, encoding hypothetical proteins, however, did not display an altered robustness phenotype (data not shown), indicating that not all genes associated with the robustness phenotypes are indeed required for improved robustness. Neither in strain MG1363 nor in IL1403 could the gene ftsY be deleted (data not shown), suggesting that this gene is essential in both strains, as it is in Escherichia coli (39). Taking these results together, we have not been able to confirm the involvement of these genes in robustness phenotypes by a gene deletion approach. Possibly, complementary genes that take over the function originally performed by the gene targeted by deletion are present in strain MG1363. Furthermore, besides the presence or absence of genes, inactivation or differential regulation of conserved genes could be the basis for the high diversity in robustness. These differences in gene activity intrinsically cannot be revealed by genotype-phenotype matching but rather require a transcriptome-phenotype matching approach, as was recently demonstrated in L. plantarum (40). Nevertheless, although the selected genes associated with robustness could not be established as genetic robustness biomarkers, the gene presence/absence profile of the entire group of robustness-associated genes, resulting from the genotype-phenotype matching, might be indicative of robustness under heat and oxidative stress.
As expected from the observed high diversity of L. lactis strains in robustness under heat and oxidative stress, a high diversity in robustness under spray drying was displayed. The strains that were most robust under spray drying displayed survival levels (10 to 20% survival) which are similar to those of commercial L. lactis starter bacteria after freeze-drying (41) and thus are acceptable for practical applications. This suggests that by selection of L. lactis strains for robustness under heat and oxidative stress, spray drying could become a feasible method for preservation of selected strains for dairy starter cultures. Our results not only demonstrate the importance of selection of starter culture strains for robustness characteristics along with acidifying and flavor-forming properties but also provide relatively simple methods for assessment of strains for spray drying-related robustness, which ultimately should aid industry in the identification of novel strains with optimal combinations of industrially relevant traits.
ACKNOWLEDGMENT
This project was carried out within the research program of the Kluyver Center for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print 8 November 2013
REFERENCES
- 1.Bron PA, Kleerebezem M. 2011. Engineering lactic acid bacteria for increased industrial functionality. Bioeng. Bugs 2:80–87. 10.4161/bbug.2.2.13910 [DOI] [PubMed] [Google Scholar]
- 2.Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67–78. 10.1016/j.tifs.2003.09.004 [DOI] [Google Scholar]
- 3.Peighambardoust SH, Golshan Tafti A, Hesari J. 2011. Application of spray drying for preservation of lactic acid starter cultures: a review. Trends Food Sci. Technol. 22:215–224. 10.1016/j.tifs.2011.01.009 [DOI] [Google Scholar]
- 4.Santivarangkna C, Kulozik U, Foerst P. 2007. Alternative drying processes for the industrial preservation of lactic acid starter cultures. Biotechnol. Prog. 23:302–315. 10.1021/bp060268f [DOI] [PubMed] [Google Scholar]
- 5.Silva J, Freixo R, Gibbs P, Teixeira P. 2011. Spray-drying for the production of dried cultures. Int. J. Dairy Technol. 64:321–335. 10.1111/j.1471-0307.2011.00677.x [DOI] [Google Scholar]
- 6.Ghandi A, Powell IB, Howes T, Chen XD, Adhikari B. 2012. Effect of shear rate and oxygen stresses on the survival of Lactococcus lactis during the atomization and drying stages of spray drying: a laboratory and pilot scale study. J. Food Eng. 113:194–200. 10.1016/j.jfoodeng.2012.06.005 [DOI] [Google Scholar]
- 7.Santivarangkna C, Kulozik U, Foerst P. 2008. Inactivation mechanisms of lactic acid starter cultures preserved by drying processes. J. Appl. Microbiol. 105:1–13. 10.1111/j.1365-2672.2008.03744.x [DOI] [PubMed] [Google Scholar]
- 8.van Bokhorst-van de Veen H, van Swam I, Wels M, Bron PA, Kleerebezem M. 2012. Congruent strain specific intestinal persistence of Lactobacillus plantarum in an intestine-mimicking in vitro system and in human volunteers. PLoS One 7:e44588. 10.1371/journal.pone.0044588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parente E, Ciocia F, Ricciardi A, Zotta T, Felis GE, Torriani S. 2010. Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: a multivariate screening study. Int. J. Food Microbiol. 144:270–279. 10.1016/j.ijfoodmicro.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 10.de Vuyst L. 1994. Nisin production variability between natural Lactococcus lactis subsp. lactis strains. Biotechnol. Lett. 16:287–292. 10.1007/BF00134627 [DOI] [Google Scholar]
- 11.Boutrou R, Sepulchre A, Gripon JC, Monnet V. 1998. Simple tests for predicting the lytic behavior and proteolytic activity of lactococcal strains in cheese. J. Dairy Sci. 81:2321–2328. 10.3168/jds.S0022-0302(98)70121-3 [DOI] [Google Scholar]
- 12.Ayad EHE, Verheul A, de Jong C, Wouters JTM, Smit G. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9:725–735. 10.1016/S0958-6946(99)00140-5 [DOI] [Google Scholar]
- 13.Bachmann H, Starrenburg MJ, Dijkstra A, Molenaar D, Kleerebezem M, Rademaker JL, van Hylckama Vlieg JE. 2009. Regulatory phenotyping reveals important diversity within the species Lactococcus lactis. Appl. Environ. Microbiol. 75:5687–5694. 10.1128/AEM.00919-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann H, Starrenburg MJ, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22:115–124. 10.1101/gr.121285.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siezen RJ, Bayjanov JR, Felis GE, van der Sijde MR, Starrenburg M, Molenaar D, Wels M, van Hijum SA, van Hylckama Vlieg JE. 2011. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb. Biotechnol. 4:383–402. 10.1111/j.1751-7915.2011.00247.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siezen RJ, Starrenburg MJ, Boekhorst J, Renckens B, Molenaar D, van Hylckama Vlieg JE. 2008. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74:424–436. 10.1128/AEM.01850-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. 2010. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One 5:e10632. 10.1371/journal.pone.0010632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187:6128–6136. 10.1128/JB.187.17.6128-6136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE. 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 12:758–773. 10.1111/j.1462-2920.2009.02119.x [DOI] [PubMed] [Google Scholar]
- 20.Rademaker JL, Herbet H, Starrenburg MJ, Naser SM, Gevers D, Kelly WJ, Hugenholtz J, Swings J, van Hylckama Vlieg JE. 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128–7137. 10.1128/AEM.01017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieuwerts S, de Bok FA, Mols E, de Vos WM, van Hylckama Vlieg JE. 2008. A simple and fast method for determining colony forming units. Lett. Appl. Microbiol. 47:275–278. 10.1111/j.1472-765X.2008.02417.x [DOI] [PubMed] [Google Scholar]
- 22.Bayjanov JR, Molenaar D, Tzeneva V, Siezen RJ, van Hijum SA. 2012. PhenoLink—a web-tool for linking phenotype to ∼omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics 13:170. 10.1186/1471-2164-13-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. 2001. Random forests. Mach. Learn. 45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 24.Godon JJ, Delorme C, Ehrlich SD, Renault P. 1992. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 58:4045–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly W, Ward L. 2002. Genotypic vs. phenotypic biodiversity in Lactococcus lactis. Microbiology 148:3332–3333 [DOI] [PubMed] [Google Scholar]
- 26.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270. 10.1128/JB.01768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remm M, Storm CE, Sonnhammer EL. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314:1041–1052. 10.1006/jmbi.2000.5197 [DOI] [PubMed] [Google Scholar]
- 28.Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. 2005. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J. Appl. Microbiol. 99:493–501. 10.1111/j.1365-2672.2005.02648.x [DOI] [PubMed] [Google Scholar]
- 29.Gardiner GE, O'Sullivan E, Kelly J, Auty MA, Fitzgerald GF, Collins JK, Ross RP, Stanton C. 2000. Comparative survival rates of human-derived probiotic Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl. Environ. Microbiol. 66:2605–2612. 10.1128/AEM.66.6.2605-2612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. 1994. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL1403. Appl. Environ. Microbiol. 60:3474–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilstrup M, Jacobsen S, Hammer K, Vogensen FK. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rallu F, Gruss A, Ehrlich SD, Maguin E. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517–528. 10.1046/j.1365-2958.2000.01711.x [DOI] [PubMed] [Google Scholar]
- 33.O'Sullivan E, Condon S. 1997. Intracellular pH is a major factor in the induction of tolerance to acid and other stresses in Lactococcus lactis. Appl. Environ. Microbiol. 63:4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, Maguin E. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216. 10.1023/A:1020631532202 [DOI] [PubMed] [Google Scholar]
- 35.Archibald FS, Fridovich I. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145:442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serrano LM. 2008. Oxidative stress response in Lactobacillus plantarum WCFS1: a functional genomics approach. Ph.D. thesis Wageningen University and Research Center, Wageningen, The Netherlands [Google Scholar]
- 37.Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. 2005. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. U. S. A. 102:17466–17471. 10.1073/pnas.0508778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giard JC, Verneuil N, Auffray Y, Hartke A. 2002. Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol. Lett. 206:235–239. 10.1111/j.1574-6968.2002.tb11015.x [DOI] [PubMed] [Google Scholar]
- 39.Zelazny A, Seluanov A, Cooper A, Bibi E. 1997. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc. Natl. Acad. Sci. U. S. A. 94:6025–6029. 10.1073/pnas.94.12.6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Bokhorst-van de Veen H, Lee IC, Marco ML, Wels M, Bron PA, Kleerebezem M. 2012. Modulation of Lactobacillus plantarum gastrointestinal robustness by fermentation conditions enables identification of bacterial robustness markers. PLoS One 7:e39053. 10.1371/journal.pone.0039053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broadbent JR, Lin C. 1999. Effect of heat shock or cold shock treatment on the resistance of Lactococcus lactis to freezing and lyophilization. Cryobiology 39:88–102. 10.1006/cryo.1999.2190 [DOI] [PubMed] [Google Scholar]