Abstract
Methane is an undesirable end product of rumen fermentative activity because of associated environmental impacts and reduced host feed efficiency. Our study characterized the rumen microbial methanogenic community in beef cattle divergently selected for phenotypic residual feed intake (RFI) while offered a high-forage (HF) diet followed by a low-forage (LF) diet. Rumen fluid was collected from 14 high-RFI (HRFI) and 14 low-RFI (LRFI) animals at the end of both dietary periods. 16S rRNA gene clone libraries were used, and methanogen-specific tag-encoded pyrosequencing was carried out on the samples. We found that Methanobrevibacter spp. are the dominant methanogens in the rumen, with Methanobrevibacter smithii being the most abundant species. Differences in the abundance of Methanobrevibacter smithii and Methanosphaera stadtmanae genotypes were detected in the rumen of animals offered the LF compared to the HF diet while the abundance of Methanobrevibacter smithii genotypes was different between HRFI and LRFI animals irrespective of diet. Our results demonstrate that while a core group of methanogen operational taxonomic units (OTUs) exist across diet and phenotype, significant differences were observed in the distribution of genotypes within those OTUs. These changes in genotype abundance may contribute to the observed differences in methane emissions between efficient and inefficient animals.
INTRODUCTION
The rumen is inhabited by a diverse community of microorganisms that act as the primary fermenters of feed which is indigestible by the host. Products of microbe-mediated ruminal fermentation (e.g., volatile fatty acids [VFA]) can be converted to energy precursors and ultimately ATP for the host (1). In the rumen, hydrogen (H2) is one of the major fermentation products (2). High concentrations of H2 in the rumen can slow fermentation (3). Opportunistic rumen methanogens prevent H2 accumulation by utilizing it as an energy source in the reduction of CO2 to methane (CH4) (4, 5), a process known as methanogenesis. However, methanogenesis also has negative connotations for rumen function. Enteric CH4 is one of the main contributors to greenhouse gas emissions globally (6). Additionally, CH4 produced in the rumen represents a significant energy loss in cattle, accounting for up to 15% of dietary gross energy intake (7). Therefore, a reduction in methane emissions from livestock has important environmental and economic implications.
Residual feed intake (RFI) is a measure of feed efficiency which is defined as the difference between the actual feed intake of an animal and its predicted feed intake based on maintenance energy requirement and growth rate (8). Research has shown that feed-efficient cattle (low RFI [LRFI]) produce less daily CH4 (g/day [9]; g/kg body weight [10]) than do their inefficient counterparts (high RFI [HRFI]). However, published data on the association between host feed efficiency and enteric CH4 emissions are ambiguous, with some studies from our own group showing marked differences in CH4 emissions between HRFI and LRFI animals (11) while others show no differences (12). In both studies, the sulfur hexafluoride (SF6) technique, a method for CH4 output analysis, was utilized to measure enteric CH4. Given the high variability in published enteric CH4 measurements using this technique, characterization of rumen methanogens in animals divergently selected for host feed efficiency could prove beneficial in elucidating the relationship between CH4 emissions and methanogens in HRFI and LRFI animals. Indeed, a diet effect and a correlation between RFI and rumen methanogen composition have been reported (13, 14). However, as these authors generated data solely under a feedlot-based high-grain feeding regimen, it is unclear whether these results are consistent across other feeding regimens and diets. Work has shown that the relative ranking of animals selected on the basis of phenotypic RFI may vary when the animals are changed from a low- to a high-energy diet (15). Thus, consideration of the diet offered is warranted when investigating the rumen microbiota of cattle divergent for RFI. Indeed, it is widely accepted that diet type can affect methane emissions (16) and rumen methanogenic diversity (17).
Therefore, the objective of this research was to characterize the ruminal methanogenic community in cattle divergent for RFI across two contrasting diets: a high-forage (HF) diet followed by a low-forage (LF) diet. To achieve this, both traditional, 16S rRNA gene clone libraries and high-throughput, 454-pyrosequencing, molecular biology-based approaches were employed.
MATERIALS AND METHODS
Animal experiment.
All procedures involving animals were approved by the University College Dublin Animal Research Ethics Committee and licensed by the Irish Department of Health and Children in accordance with the European Community Directive 86/609/EC. Details of the animal experiment used in this study were as previously described (18). Dietary ingredients utilized and chemical composition were as previously described (19). Briefly, the HF diet was composed of grass silage only. In contrast, the LF diet was composed of pelleted concentrate and corn silage at a 70:30 concentrate/forage ratio and was offered as a total mixed ration (TMR). Both diets were offered ad libitum. Individual dry matter intake (DMI) and growth were recorded for 86 yearling beef heifers offered a high-energy low-forage (LF) diet over 112 days. All animals were subsequently ranked retrospectively on phenotypic RFI. Fourteen heifers with the highest (HRFI; less efficient) and 14 heifers with the lowest (LRFI; more efficient) RFI coefficients during the study by Kelly et al. (19) were selected for use in the current study. Following selection, animals were reallocated to a high-forage (HF) diet for a 44-day period (period 1). Animals were then turned out to pasture for a 56-day dietary “washout” period. Subsequently, animals were rehoused and reallocated to an LF diet for 35 days (period 2). The experiment was therefore designed to have two factors, (i) RFI phenotype and (ii) diet type. Rumen sampling was performed at the end of both dietary periods using a transesophageal sampling device (Flora rumen scoop; Guelph, ON, Canada) as previously described (18).
DNA extraction and PCR.
A detailed description of the DNA extraction and dilution method utilized has been given elsewhere (18). Approximately 800 bp of the 16S rRNA gene was amplified using the primer set Met 86 forward (5′-GCT CAG TAA CAC GTG G-3′) (20) and Met 915 reverse (5′-GTG CTC CCC CGC CAA TTC CT-3′) (21). All PCR amplifications were optimized and performed as previously described (18).
Clone library construction and sequencing.
A detailed description of the cloning procedure utilized is provided in Text S1 in the supplemental material. Briefly, individual PCR amplicons were pooled by mixing 4 μl of each sample from HRFI animals (n = 14) while offered the HF diet (library 1, HRFI-HF), LRFI animals (n = 14) while offered the HF diet (library 2, LRFI-HF), and from the same HRFI animals (n = 14) while offered the LF diet (library 3, HRFI-LF) and the same LRFI animals (n = 14) while offered the LF diet (library 4, LRFI-LF) for library construction. Four clone libraries were constructed (library 1, HRFI-HF; library 2, LRFI-HF; library 3, HRFI-LF; and library 4, LRFI-LF) by cloning pooled PCR products into TOP10 vectors (TOPO TA cloning kit; Invitrogen, Carlsbad, CA, USA) by chemical transformation. From each of the four libraries (1, 2, 3, and 4), approximately 300 clones were generated from which 93, 89, 90, and 86 clones were randomly selected, respectively, and subjected to sequence analysis by the dideoxy-chain termination method with an ABI 3730 XL sequencer using the sequencing service provided by Macrogen (Seoul, South Korea) with both M13 forward (CGCCAGGGTTTTCCCAGTCACGAC) and M13 reverse (TTCACACAGGAAACAGCTATGAC) primers according to the manufacturer's instructions.
Clone library analysis.
A detailed description of clone library analysis is provided in Text S2 in the supplemental material. Briefly, in total 571 raw sequence reads were obtained by Sanger sequencing. Following removal of chimeric sequences, sequence reads were trimmed manually. Libraries 1, 2, 3, and 4 yielded 93, 89, 90, and 86 good-quality sequences, respectively, and were searched using the basic local alignment and search tool from NCBI (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) for comparison with sequences available in the GenBank database. Sequences were aligned using MUSCLE (22) and clustered for classification using CD-HIT (23). A phylogenetic tree was constructed from the alignments using PhyML under the GTR model of evolution (24).
Pyrosequencing of methanogen 16S rRNA gene amplicons.
To compare the extremes for both the HRFI and LRFI animals utilized in the subcloning experiment, a subgroup of the top 7 HRFI and the top 7 LRFI animals were selected for tag-encoded amplicon pyrosequencing of their rumen microbial DNA. Barcoded fusion primers were designed to amplify an approximately 550-bp methanogen-specific region of the 16S rRNA gene using forward ARC344 (5′ACGGGGCGCAGCAG3′) (10) and reverse primer ARC915-GC (3′AGGAATTGGCGGGGGAGCAC5′) (21). All PCR amplifications and analysis were performed as previously described (18) using the following program: 95°C for 2 min and 30 cycles of 94°C for 30 s, 80°C for 30 s, 72°C for 1 min, and 72°C for 7 min. PCR products were purified using a magnetic purification kit (Ampure; Agencourt Bioscience Corporation, Massachusetts, USA) according to the manufacturer's directions. Sample concentration was determined using a Qubit fluorimeter (Invitrogen, Carlsbad, CA) and Pico Green assay (Invitrogen). Subsequently, samples were pyrosequenced using the Roche-454 GS FLX titanium platform.
Pyrosequencing analysis and phylogenetic classification.
From all 14 (n = 7 HRFI and 7-LRFI) animals, 2,823 raw sequences were obtained. Raw sequencing reads were screened and quality trimmed as previously described (25), with read lengths no shorter than 500 bp and having a quality score of ≥20 considered for further analysis. Sequences were clustered using CD-HIT-OTU (26) using the following procedure. After filtering for chimeric sequences (of which none were found), all sequences were initially clustered at 100% identity for calculation of a cutoff for low-abundance clusters of identical sequences (clusters with fewer than 3 members). After low-abundance sequences were removed, all remaining sequences were clustered at 98% identity to identify operational taxonomic units (OTUs). The automated phylogenetic tree-based small-subunit taxonomy and alignment pipeline (STAP) (27) was used for taxonomic identification of the OTUs. To identify genotypes, the sequences from each OTU were clustered at 99.5% identity using CD-HIT (28). The 0.5% difference was allowed to reduce the impact of sequencing error and length on the identification of genotypes. 16S type sequences from all available archaeal groups were retrieved from the Ribosomal Database Project (29). These were aligned with representative sequences from each OTU using AQUA (30) and manually refined using Jalview (31). RAxML-HPC (32) was used to construct a bootstrapped phylogeny (100 replicates) under the GTRCAT model of evolution. The distribution of OTUs in the phylogeny was visualized using iTOL (33).
Statistical analysis.
The summed counts of genotypes across phenotype (HRFI versus LRFI) and across diets (HF versus LF) were scaled and tested for significance using a binomial test as implemented in the statistical package R (version 2.15.1) (R Development Core Team, 2009) (using the command “binomial.test”). The null hypothesis used assumed no difference in the distribution of genotypes across either RFI phenotype or diet.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database under accession numbers JQ952744 to JQ952761 with the prefix “AGRIC.” The nucleotide sequences generated from the pyrosequencing have been deposited in the GenBank database under accession numbers KF761676 to KF764482.
RESULTS
Analysis of clone libraries.
In total, 332 sequences out of the 358 were identified as methanogens with one sequence identified as bacteria, specifically Clostridium sp., and the other 25 identified as belonging to a group of archaea, from the genus Thermoplasma. Classification of all sequences revealed 18 possible OTUs on the basis of 97% similarity, 6 of which were unique (see Table S1 in the supplemental material). A summary of the diversity of methanogens according to phenotypic RFI and diet is presented in Table 1.
TABLE 1.
Summary of the number of clones in each library and their nearest identified taxon
| Nearest taxon | No. of clones/library |
|||
|---|---|---|---|---|
| HRFI-HF | LRFI-HF | HRFI-LF | LRFI-LF | |
| Methanosphaera stadtmanae | 6 | 9 | 27 | 11 |
| Methanobrevibacter smithii | 68 | 60 | 50 | 59 |
| Methanobrevibacter ruminantium | 12 | 11 | 8 | 11 |
| Thermoplasma volcanium | 7 | 9 | 4 | 5 |
| Clostridium sp. | 1 | |||
| Total | 93 | 89 | 90 | 86 |
Taxonomic characterization of clone library rumen methanogens.
All OTUs were subjected to BLAST analysis to determine the closest taxonomic relative. All OTUs classified as methanogenic belonged to the family Methanobacteriaceae (61.1% of total OTUs), while all other OTUs classified as archaeal belonged to the family Thermoplasmataceae (33.3% of total OTUs). The majority of clones from each library belonged to the genus Methanobrevibacter, with 237 sequences showing similarities of 94 to 98% to the species Methanobrevibacter smithii and 42 sequences showing similarities of 94 to 99% to the species Methanobrevibacter ruminantium. In total, 53 sequences were similar (94 to 97%) to Methanosphaera stadtmanae while the remaining minor proportion (25 sequences) were closest to Thermoplasma volcanium but at a similarity of only 79 to 80%. Members of the Methanobrevibacter and Methanosphaera genera were predominant in all libraries but were found at different distributions (Table 1).
Phylogenetic analysis of the sequenced libraries was performed based on the representative OTU sequences generated from CD-HIT using interactive tree of life (iTOL) (http://itol.embl.de/) (33) (see Fig. S1 in the supplemental material). Phylogenetic analysis divided the 18 OTUs into two clusters, one including all methanogenic phylotypes (order Methanobacteriales) and the second with all the archaeal phylotypes (uncultured archaea) with Clostridiaceae (AGRIC 0) as an outgroup (see Fig. S1). To test the reliability of the tree, bootstrap resampling was employed. Bootstrap data firmly supported both clades at 99 to 100%. All methanogenic OTUs clustered with their closest classification while OTUs identified at low similarity (ca. 80%) as Thermoplasma spp. clustered with uncultured archaeal clones from the BLAST database. The majority of sequences (93%) were placed within the Methanobacteriales clade. Within this clade, the three methanogenic species formed monophyletic assemblages, with a similar finding observed for sequences within the uncultured archaea clade. All of the sequences identified as similar to Methanosphaera stadtmanae clustered together. The AGRIC 9 phylotype clustered at 100% bootstrap confidence with Methanosphaera and 71% bootstrap confidence with Methanobrevibacter smithii PS. Phylotypes AGRIC 2, 5, 6, and 7 all clustered closely with Methanobrevibacter spp. with high bootstrap values. Sequences within the Methanobacteriales clade presented sequence similarity values ranging from 94 to 99% with Methanosphaera stadtmanae, Methanobrevibacter smithii, and Methanobrevibacter ruminantium, respectively, and may represent distinct species or strains of Methanosphaera and Methanobrevibacter.
Clone library richness and diversity.
The expected number of unique OTUs was plotted against the number of clones (see Fig. S2 in the supplemental material). The calculated rarefaction curves of HRFI-HF and LRFI-HF OTUs showed almost complete saturation (asymptote); however, curves for HRFI-LF and LRFI-LF OTUs did not reach saturation (see Fig. S2).
Methanogen community evenness, coverage, and diversity of the clone libraries were assessed using the estimators of evenness (J′), Good's coverage (C) (34), Shannon-Weaver diversity index (H), Chao1, and Simpson's diversity index (1/D) based on the phylotypes formed at 97% similarity (Table 2). Species evenness was 0.52, 0.60, 0.59, and 0.52 in the HRFI-HF, LRFI-HF, HRFI-LF, and LRFI-LF libraries, respectively. Shannon-Weaver and Simpson diversity indices were highest for the HRFI-LF and LRFI-HF libraries, indicating that the methanogen communities in these groups were more diverse than HRFI-HF and LRFI-LF libraries. The Chao1 minimum richness estimates indicate that the HRFI-LF and LRFI-LF libraries did not plateau with the current sampling depth, as the estimates for this index were higher than the actual number of OTUs observed (Table 2). However, the Chao1 estimates for the LRFI-HF (9.0) and the HRFI-HF (8.5) libraries were close to the number of OTUs (8) estimated at the 97% similarity level. Indeed, rarefaction analysis of these libraries revealed that these samples were approaching a plateau, indicating that the OTU diversity in these groups was almost completely covered.
TABLE 2.
Summary of the number of clones analyzed, observed richness (OTUs), estimated OTU richness (ACE and Chao1), evenness, diversity indices (Shannon and Simpson), and estimated sample coverage of clone libraries
| Clone library | No. of clones | No. of OTUs | Shannon | Evennessa | Chao1b | Simpsonc | Coveraged (%) |
|---|---|---|---|---|---|---|---|
| HRFI-HF | 93 | 8 | 1.07 | 0.52 | 8.5 (8.03, 16.26) | 1.88 | 97.8 |
| LRFI-HF | 89 | 8 | 1.25 | 0.60 | 9 (8.07, 21.87) | 2.31 | 97.7 |
| HRFI-LF | 90 | 14 | 1.55 | 0.59 | 18.2 (14.78, 36.33) | 3 | 92.2 |
| LRFI-LF | 86 | 9 | 1.14 | 0.52 | 12 (9.39, 31.99) | 2.04 | 95.3 |
Calculation based on the formula by Pielou (61) (J′ = H′/H′max), where H′ is the Shannon diversity index and H′max is the logarithm of the number of species.
Values in parentheses are 95% confidence intervals.
Calculated as Simpson's reciprocal index (1/D).
Calculation based on the formula by Good (34) [C = 1 − (n/N)], where n is the number of unique clones and N is the total number of clones.
Methanogen tag-encoded pyrosequencing analysis.
In total, 2,807 good-quality sequences were recovered, with an average read length of 536 bp and a range between 508 bp and 611 bp. It is well documented that in high-throughput sequencing experiments most of the Pyrotags that form the rare biosphere are likely to be small sequencing errors (35, 36). Therefore, it was necessary to filter the data for low-abundance clusters (<3 sequences per cluster) and for chimeric sequences, resulting in the removal of 2,036 and 0 sequences, respectively. The remaining 772 sequences were clustered at 98% identity, resulting in 6 OTUs being identified. Each individual rumen sample was represented by an average of 28 sequences from, on average, 3 OTUs with a range of 15 to 50 sequences per sample. The HRFI and LRFI animals shared a high degree of similarity in terms of their rumen methanogenic communities, with all OTUs detected in both phenotypic groups (Table 3). All OTUs except 1 (similar to Methanobacterium smithii) were also found in both diets (Table 3). At the OTU level, however, two genotypes of Methanobrevibacter smithii were found to be significantly overrepresented (P = 0.05 and P = 0.01) in HRFI compared to LRFI animals. Two genotypes were found to be significantly overrepresented in the HF diet, one from Methanobacterium sp. (P = 0.03) and one from Methanobrevibacter smithii 1 (P = 0.02). Two genotypes were found to be significantly overrepresented in the LF diet, one from Methanobrevibacter smithii 1 (P < 0.01) and one from Methanosphaera stadtmanae 1 (P < 0.01), with the latter the only genotype identified in Methanosphaera stadtmanae 1 and representing the entire OTU (Table 3). A further 12 genotypes from across all other OTUs showed no overabundance in either phenotype or diet (Table 3).
TABLE 3.
Distribution of OTUs and genotypes across phenotype and dieta
| Species (OTU) | Genotype | Diet (no. of samples) |
P value | Phenotype (no. of samples) |
P value | ||
|---|---|---|---|---|---|---|---|
| HF | LF | HRFI | LRFI | ||||
| Methanobrevibacter smithii | 1 | 226 | 162 | 0.08 | 187 | 201 | 0.72 |
| 2 | 118 | 64 | <0.01 | 86 | 96 | 0.71 | |
| 3 | 5 | 6 | 1.00 | 4 | 7 | 0.54 | |
| 4 | 10 | 2 | 0.06 | 6 | 6 | 1.00 | |
| 5 | 8 | 6 | 1.00 | 11 | 3 | 0.05 | |
| 6 | 2 | 23 | <0.01 | 10 | 15 | 0.42 | |
| 7 | 7 | 7 | 1.00 | 12 | 2 | 0.01 | |
| 8 | 0 | 5 | 0.06 | 3 | 2 | 1.00 | |
| 9 | 3 | 2 | 1.00 | 2 | 3 | 1.00 | |
| 10 | 0 | 5 | 0.06 | 2 | 3 | 1.00 | |
| 11 | 3 | 1 | 0.62 | 1 | 3 | 0.62 | |
| Methanosphaera stadtmanae 1 | 12 | 16 | 50 | <0.01 | 33 | 33 | 0.90 |
| Methanobacterium sp. | 13 | 6 | 0 | 0.03 | 4 | 2 | 0.68 |
| Methanosphaera stadtmanae 2 | 14 | 1 | 4 | 0.37 | 2 | 3 | 1.00 |
| 15 | 1 | 3 | 0.62 | 2 | 2 | 1.00 | |
| Thermoplasma sp. | 16 | 1 | 3 | 0.62 | 1 | 3 | 0.62 |
| Methanobrevibacter ruminantium | 17 | 5 | 13 | 0.06 | 10 | 8 | 0.81 |
| 18 | 2 | 2 | 1.00 | 3 | 1 | 0.62 | |
Values in bold represent OTUs with significantly (P < 0.05) different numbers of genotypes in the rumen contents of cattle across phenotype (HRFI versus LRFI) and across diets (HF versus LF).
Pyrosequencing richness and diversity.
Rarefaction analysis was used to compare species richness between the OTUs obtained from HRFI and LRFI animals (see Fig. S3a in the supplemental material) and from the HF and LF diets (see Fig. S3b). Rarefaction analysis revealed that there was no difference in methanogen richness between HRFI and LRFI animals; however, methanogen richness was higher when animals were offered the HF diet than when offered the LF diet. In addition, methanogen richness and diversity were estimated with ACE, Chao1, Shannon, and Simpson procedures using EstimateS (Table 4). For both phenotypes and dietary groups analyzed, the number of OTUs detected was in good agreement with the total number of OTUs estimated by ACE and Chao1 richness indices. Good's coverage of the sequences calculated for both phenotypes and both diets was used as an indicator of the completeness of sampling. The coverage ranged from ca. 99.8 to 100% for all four groups. Using the Shannon diversity index (H′), methanogen diversity from the OTU data of each group of sequences was estimated (Table 4). Rarefaction analysis of these estimates showed that the diversity of each group had reached a stable value (see Fig. S4).
TABLE 4.
Summary of the number of sequences analyzed, observed richness (OTUs), estimated OTU richness (ACE and Chao1), evenness, diversity indices (Shannon and Simpson), and estimated sample coverage of pyrosequences
| Group | No. of reads | No. of OTUs | ACE | Chao1b | Shannon | Evennessa | Simpsonc | Coveraged |
|---|---|---|---|---|---|---|---|---|
| LF | 358 | 5 | 5 | 5 (5, 5) | 0.70 | 0.44 | 1.55 | 100.0 |
| HF | 414 | 6 | 6 | 6 (6, 6) | 0.37 | 0.21 | 1.17 | 100.0 |
| HRFI | 379 | 6 | 6 | 6 (6, 6) | 0.58 | 0.32 | 1.35 | 100.0 |
| LRFI | 393 | 6 | 7 | 6 (6, 6) | 0.53 | 0.30 | 1.32 | 99.8 |
Calculation based on the formula by Pielou (61) (J′ = H′/H′max), where H′ is the Shannon diversity index and H′max is the logarithm of the number of species.
Values in parentheses are 95% confidence intervals.
Calculated as Simpson's reciprocal index (1/D).
Calculation based on the formula by Good (34) [C = 1 − (n/N)], where n is the number of unique clones and N is the total number of clones.
Taxonomic classification of methanogen pyrosequences.
All sequences could be classified at the class level, 99.5% of which were classified as Methanobacteria. At the genus level, 98% of sequences were classifiable, and out of these, 89% were classified as Methanobrevibacter and 9% were classified as Methanosphaera. Within these genera, 87% were identified as Methanobrevibacter smithii, 10% were identified as Methanosphaera stadtmanae, and 3% were identified as Methanobrevibacter ruminantium. At the genus level, 3% of sequences were classified as Thermoplasma; however, when queried against the NCBI database using the blastn algorithm (37), they were only between 74 and 83% similar to the best Thermoplasma sequence. Therefore, it is probable that the sequences represent uncultured methanogenic or other archaeal species which have yet to be identified.
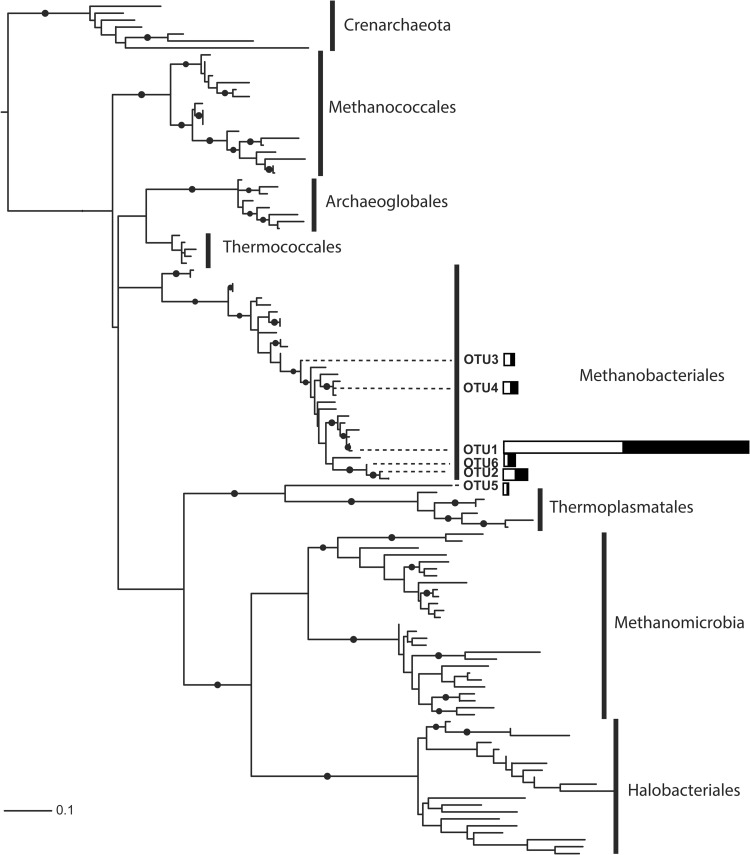
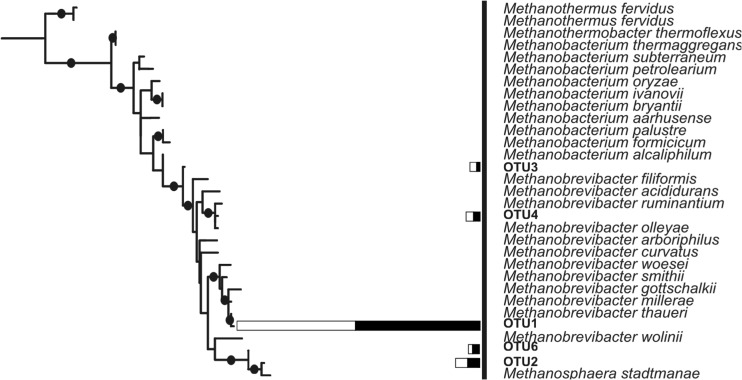
Phylogenetic analysis of pyrosequences was performed based on the representative OTU sequences generated from CD-HIT-OTU using interactive tree of life (iTOL) (http://itol.embl.de/) (33) (Fig. 1). Phylogenetic analysis clustered the 5 methanogenic OTUs together, placing them within the Methanobacteriales clade (Fig. 1). To test the reliability of the tree, bootstrap resampling was employed. Bootstrap data firmly supported both clades at 99 to 100%. A second tree was constructed to assess the phylogeny of methanogenic OTUs within the Methanobacteriales order (Fig. 2). Five OTUs clustered with Methanobrevibacter spp., while one OTU (OTU 2) clustered with Methanosphaera stadtmanae.
FIG 1.
Phylogeny of the V3 region of archaeal 16S rRNA type sequences from the Ribosomal Database Project (29). The phylogenetic positions of the OTUs discovered from pyrosequencing are indicated. The bars represent the abundances of each of the OTUs in HRFI samples (white) and LRFI samples (black). Dots at internal branches indicate bootstrap support greater than 80%. A detailed version of the full phylogeny with all species names is shown in Fig. S5 in the supplemental material. The relationships from only within the Methanobacteriales are shown in Fig. 2.
FIG 2.
Phylogeny of the V3 region of Methanobacteriales 16S rRNA type sequences from the Ribosomal Database Project (29). The phylogenetic positions of the OTUs discovered from pyrosequencing are indicated. The bars represent the abundance of each of the OTUs in HRFI samples (white) and LRFI samples (black). Dots at internal branches indicate bootstrap support greater than 80%.
DISCUSSION
It has been suggested that specific rumen methanogens may be associated with host feed efficiency (13, 14); however, dietary chemical composition can greatly influence methanogenesis in the rumen (38) due to the change in dietary substrates available for fermentation. The aim of our study was to characterize the rumen methanogenic community in cattle divergent in phenotypic RFI across two contrasting diets using both traditional molecular subcloning and next-generation sequencing technologies.
In the rumen, archaea are believed to account for less than 4% of all microbes (39), making any analysis of their diversity difficult. Our data suggest that we have surveyed the full extent of the methanogenic microbial communities within these animals at this depth of sequencing as evidenced by the asymptotes generated in the rarefaction curves. This showed that 6 archaeal phylotypes were present in all animals at the 98% similarity level irrespective of phenotype or diet type offered. Furthermore, our data suggest that no further sampling of the archaeal population is required as there was no difference between the observed number of OTUs and the ACE or Chao1 estimates. In total, we sequenced 2,807 methanogen-specific reads and detected 6 phylotypes with the 98% cutoff value. We demonstrated that at this level of similarity all 6 different methanogenic phylotypes were constituent in both phenotypic groups. Both approaches revealed a conserved methanogen population represented by Methanobrevibacter smithii, Methanobrevibacter ruminantium, and Methanosphaera stadtmanae. This is consistent with previous analyses (13, 40, 41, 42 43, 44), and although methanogens were once thought to be the only archaeal representatives in the rumen, Thermoplasma-like sequences have been identified (45), which is consistent with our findings. Research has shown that Thermoplasma bacteria in the bovine rumen represent a novel group of methylotrophic methanogens which can be reduced via dietary CH4 mitigation strategies (46). However, in our study the sequences identified as Thermoplasma were only about 80% similar to the nearest Thermoplasma sequence in GenBank, suggesting that they may represent previously uncharacterized methanogens or other archaea. Furthermore, consistent with previous investigations (40, 47), we failed to detect any members of the Methanomicrobium or Methanosarcina genus using either the clone library or the pyrosequencing approach, suggesting that their absence is not due to library preparation or sequencing artifacts (48). However, it must be acknowledged that the lower depth of sequencing achieved coupled with the necessity to adhere to stringent data filtering could also explain the absence of these methanogens in our animals. In addition, specificity of the primer sequences to these targets could also contribute to these methanogens being undetected.
Both experimental approaches confirmed the diversity of methanogens in our rumen samples. Clone libraries are thought to be nonrandom and prone to PCR and cloning biases (48); however, our results suggest a role for clone library analysis for investigating microbial diversity, but without accurate quantification. However, it can be appreciated that as only a subgroup of animals from each group used in the subcloning experiment were subjected to pyrosequencing analysis, data could not be directly compared. Sequences obtained from the pyrosequencing approach provided an interesting insight into the relative distribution of methanogens in the rumen. In total, 18 distinct genotypes (classified at the 99.5% identity level) from 6 different OTUs (classified at the 98% identity level) were identified. All OTUs were found in both phenotypes; however, two distinct genotypes of Methanobrevibacter smithii were significantly overrepresented in the HRFI group. This finding is consistent with previous reports of differences in methanogen communities between efficient and inefficient cows (13).
Low-abundance microbes have been shown to play important roles in different environments (49, 50, 51), so even though these genotypes represent only 10% of all genotypes found and consist of only 3% of all sequences recovered, they may play an important role in metabolic efficiency, ultimately leading to variation in host feed efficiency and host methane emissions. Similarly, 4 genotypes were found to be significantly differentially abundant in either the HF (1 genotype) or the LF (3 genotypes) group; however, they consisted of a much larger proportion of all the sequences analyzed (36%) than those found to be different between the phenotypes (3%), indicating that diet has a much larger effect on the microbial community than does phenotype.
Similar to our results in the rumen, Methanobrevibacter smithii has been described as the dominant archaeon in the human gut (52). As Methanobrevibacter smithii can produce CH4 from CO2, H2, and formate, its energy metabolism is less restricted than that of Methanosphaera stadtmanae, whose ability to produce methane is only via reduction of methanol with H2. Several studies have reported a reduction in methane emissions (53, 54, 55) when the diet shifted from HF to LF. This is because change in diet alters the fermentation pattern in the rumen, due to a change in the available fermentable substrate, causing a shift in volatile fatty acid (VFA) production from acetate and butyrate (on an HF diet) to propionate (on an LF diet). Propionate synthesis is considered to be a rival pathway to methanogenesis as it directly competes for hydrogen in the rumen during formation (4). Despite this reduction in CH4 emissions, previous studies have shown that methanogen abundance appeared to be unaffected by the change from a low- to a high-energy diet (13, 14, 17). In agreement, our pyrosequencing data revealed that within the archaeal population, total methanogens (represented by Methanobacteria) were found to be unaffected by the type of diet offered; however, at the genotype level there were significant changes.
Methanosphaera stadtmanae is the only member of the order Methanobacteriales with the ability to produce methane via the reduction of methanol with H2. In the rumen, methanol is a product of pectin hydrolysis by protozoa and esterase activity of bacteria (1). Grass is a relatively poor source of dietary pectin, while some constituents of concentrate feeds such as citrus and beet pulp are abundant (56). Previous studies have reported increases in protozoa when the concentrate portion of the diet is increased (18, 57). Thus, given the greater potential for increased availability of methanol, it was unsurprising that the most abundant genotype of Methanosphaera stadtmanae was significantly more abundant in the LF diet. Similarly, the effect of diet on the relative abundance of Methanobrevibacter smithii was most likely a consequence of the aforementioned change in fermentation pattern and the inevitable decrease in H2 availability for methanogenesis on the LF diet.
While pyrosequencing confirmed Methanobrevibacter as the dominant methanogen genus in the rumen, Methanobrevibacter ruminantium was in lowest abundance. Strictly speaking, this strain of methanogen has an obligate requirement for the key methanogenesis factor coenzyme M for growth (58), as it is unable to synthesize this coenzyme due to a lack of required genes for coenzyme M biosynthesis (59). Therefore, Methanobrevibacter ruminantium must obtain this key factor exogenously (60), which may be facilitated by other methanoarchaea in the system. These results present a potential explanation for the differences in methane emissions between efficient and inefficient animals previously reported (10, 53). Further research is warranted to identify dietary manipulation strategies which could exploit these differences between phenotypic groups.
Conclusion.
This study is the first to extensively investigate the rumen methanogen microbiota of cattle divergent for phenotypic RFI across two contrasting diets using molecular culture-independent methods. Additionally, this study represents the first bovine ruminal barcoded pyrosequencing effort to investigate specifically methanogenic microorganisms. While the number of sequences analyzed was low, we were still able to confirm that a core set of OTUs from different methanogenic phyla exist irrespective of host feed efficiency or diet. Furthermore, the novel approach of identifying genotypes within the OTUs revealed that the abundance of different genotypes can vary widely and be significantly associated with host feed efficiency and diet. It is possible that different genotypes of the same methanogen species may associate with different strains of hydrogen-producing organisms. However, deeper sequencing of these rare genotypes in conjunction with the key microbial groups, such as protozoa, with which they are known to interact will be necessary to obtain a greater understanding of the role that they play in host feed efficiency and methanogenesis. Such an approach will better facilitate the development of strategies focused on methane abatement without compromising ruminal function and animal performance, which is key to producer adoption as well as the future sustainability of the agri-food sector.
Supplementary Material
ACKNOWLEDGMENTS
Funding for the development and main work of this research was provided under the National Development Plan, through the Research Stimulus Fund, administered by the Department of Agriculture, Fisheries & Food, Ireland, RSF 05 224. The pyrosequencing work on this project was jointly funded by Teagasc projects RMIS 5781 and 6004. Christopher J. Creevey is funded from the Science Foundation Ireland (SFI) Stokes Lectureship Programme (reference no. 07/SK/B1236A).
Footnotes
Published ahead of print 8 November2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03131-13.
REFERENCES
- 1.Hobson PN, Stewart CS. (ed). 1997. The rumen microbial ecosystem. Blackie Academic & Professional, London, United Kingdom [Google Scholar]
- 2.Demeyer DI. 1991. Quantitative aspects of microbial metabolism in the rumen and hindgut, p 217–237 In Jouany JP. (ed), Rumen microbial metabolism and ruminant digestion. INRA, Paris, France [Google Scholar]
- 3.Wolin MJ, Miller TL, Stewart CS. 1997. Microbe-microbe interactions, p 467–491 In Hobson PN, Stewart CS. (ed), The rumen microbial ecosystem. Blackie Academic & Professional, London, United Kingdom [Google Scholar]
- 4.Moss AR, Jouany J-P, Newbold J. 2000. Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49:231–253. 10.1051/animres:2000119 [DOI] [Google Scholar]
- 5.Zinder SH. 1993. Physiological ecology of methanogens, p 128–206 In Ferry JG. (ed), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman & Hall, New York, NY [Google Scholar]
- 6.Steinfeld H, Gerber P, Wassenaar TD, Castel V, Haan C. 2006. Livestock's long shadow: environmental issues and options. Food and Agriculture Organization of the United Nations, Rome, Italy [Google Scholar]
- 7.Van Nevel CJ, Demeyer DI. 1996. Control of rumen methanogenesis. Environ. Monit. Assess. 42:73–97. 10.1007/BF00394043 [DOI] [PubMed] [Google Scholar]
- 8.Crews DH. 2005. Genetics of feed utilization and national cattle evaluation: a review. Genet. Mol. Res. 4:152–165 [PubMed] [Google Scholar]
- 9.Nkrumah JD, Okine EK, Mathison GW, Schmid K, Li C, Basarab JA, Price MA, Wang Z, Moore SS. 2006. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 84:145–153 [DOI] [PubMed] [Google Scholar]
- 10.He Z, Xiao S, Xie X, Zhong H, Hu Y, Li Q, Gao F, Li G, Liu J, Qiu G. 2007. Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine. Extremophiles 11:305–314. 10.1007/s00792-006-0044-z [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimons C, Kenny DA, Deighton MH, Fahey AG, McGee M. 2013. Methane emissions, body composition, and rumen fermentation traits of beef heifers differing in residual feed intake. J. Anim. Sci. 91:5789–5800. 10.2527/jas.2013-6956 [DOI] [PubMed] [Google Scholar]
- 12.McDonnell R, Hart KJ, Boland TM, Kelly AK, McGee M, Kenny DA. 2009. Effect of ranking on phenotypic residual feed intake and diet on ruminal methane emissions from beef heifers, p 42 In Proceeding of the Agricultural Research Forum, Tullamore, Ireland, 12 to 13 March 2009 [Google Scholar]
- 13.Zhou M, Hernandez-Sanabria E, Guan LL. 2009. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 75:6524–6533. 10.1128/AEM.02815-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Hernandez-Sanabria E, Guan LL. 2010. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 76:3776–3786. 10.1128/AEM.00010-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durunna ON, Mujibi FDN, Goonewardene L, Okine EK, Basarab JA, Wang Z, Moore SS. 2011. Feed efficiency differences and reranking in beef steers fed grower and finisher diets. J. Anim. Sci. 89:158–167. 10.2527/jas.2009-2514 [DOI] [PubMed] [Google Scholar]
- 16.McAllister TA, Cheng KJ, Okine EK, Mathison GW. 1996. Dietary, environmental and microbiological aspects of methane production in ruminants. Can. J. Anim. Sci. 76:231–243. 10.4141/cjas96-035 [DOI] [Google Scholar]
- 17.Popova M, Martin C, Eugene M, Mialon MM, Doreau M, Morgavi DP. 2011. Effect of fibre- and starch-rich finishing diets on methanogenic Archaea diversity and activity in the rumen of feedlot bulls. Anim. Feed Sci. 166:113–121. 10.1016/j.anifeedsci.2011.04.060 [DOI] [Google Scholar]
- 18.Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78:4949–4958. 10.1128/AEM.07759-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly AK, McGee M, Crews DH, Jr, Fahey AG, Wylie AR, Kenny DA. 2010. Effect of divergence in residual feed intake on feeding behavior, blood metabolic variables, and body composition traits in growing beef heifers. J. Anim. Sci. 88:109–123. 10.2527/jas.2009-2196 [DOI] [PubMed] [Google Scholar]
- 20.Wright AD, Pimm C. 2003. Improved strategy for presumptive identification of methanogens using 16S riboprinting. J. Microbiol. Methods 55:337–349. 10.1016/S0167-7012(03)00169-6 [DOI] [PubMed] [Google Scholar]
- 21.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann RI, Zeyer J. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185–192. 10.1007/s002030050486 [DOI] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. 10.1093/bioinformatics/btq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559. 10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Sullivan Ó, Coakley M, Lakshminarayanan B, Claesson MJ, Stanton C, O'Toole PW, Ross RP. 2011. Correlation of rRNA gene amplicon pyrosequencing and bacterial culture for microbial compositional analysis of faecal samples from elderly Irish subjects. J. Appl. Microbiol. 111:467–473. 10.1111/j.1365-2672.2011.05067.x [DOI] [PubMed] [Google Scholar]
- 26.Li W, Fu L, Niu B, Wu S, Wooley J. 2012. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief. Bioinform. 13:656–668. 10.1093/bib/bbs035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, Hartman A, Ward N, Eisen JA. 2008. An automated phylogenetic tree-based small subunit rRNA taxonomy and alignment pipeline (STAP). PLoS One 3:e2566. 10.1371/journal.pone.0002566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 29.Maidak BL, Olsen GJ, Larsen N, Overbeek R, McCaughey MJ, Woese CR. 1996. The Ribosomal Database Project (RDP). Nucleic Acids Res. 24:82–85. 10.1093/nar/24.1.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller J, Creevey CJ, Thompson JD, Arendt D, Bork P. 2010. AQUA: automated quality improvement for multiple sequence alignments. Bioinformatics 26:263–265. 10.1093/bioinformatics/btp651 [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 33.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 34.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. 10.2307/2333344 [DOI] [Google Scholar]
- 35.Reeder J, Knight R. 2009. The ‘rare biosphere': a reality check. Nat. Methods 6:636–637. 10.1038/nmeth0909-636 [DOI] [PubMed] [Google Scholar]
- 36.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898. 10.1111/j.1462-2920.2010.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Krasnov S, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Karsch-Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. 2012. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40:D13–D25. 10.1093/nar/gkr1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin C, Morgavi DP, Doreau M. 2010. Methane mitigation in ruminants: from microbe to the farm scale. Animal 4:351–365. 10.1017/S1751731109990620 [DOI] [PubMed] [Google Scholar]
- 39.Lin C, Raskin L, Stahl DA. 1997. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol. Ecol. 22:28l–294 [Google Scholar]
- 40.Whitford MF, Teather RM, Forster RJ. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. 10.1186/1471-2180-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen PH, Kirs M. 2008. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74:3619–3625. 10.1128/AEM.02812-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright A-D, Ma X, Obispo N. 2008. Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb. Ecol. 56:390–394. 10.1007/s00248-007-9351-x [DOI] [PubMed] [Google Scholar]
- 43.Hook SE, Wright A, Denis G, McBride BW. 2010. Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010:945785. 10.1155/2010/945785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei C, Sheng-Yong M, Cheng Y, Zhu W. 2010. Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal 4:20–29. 10.1017/S1751731109990681 [DOI] [PubMed] [Google Scholar]
- 45.Gu M-J, Alam MJ, Kim S-H, Jeon CO, Chang MB, Oh YK, Lee SC, Lee SS. 2011. Analysis of methanogenic archaeal communities of rumen fluid and rumen particles from Korean black goats. Anim. Sci. J. 82:663–672. 10.1111/j.1740-0929.2011.00890.x [DOI] [PubMed] [Google Scholar]
- 46.Poulsen M, Schwab C, Borg Jensen B, Engberg RM, Spang A, Canibe N, Hojberg O, Milinovich G, Fragner L, Schleper C, Weckwerth W, Lund P, Schramm A, Urich T. 2013. Methylotrophic methanogenic thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 4:1428. 10.1038/ncomms2432 [DOI] [PubMed] [Google Scholar]
- 47.Skillman LC, Evans PN, Strompl C, Joblin KN. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222–228. 10.1111/j.1472-765X.2005.01833.x [DOI] [PubMed] [Google Scholar]
- 48.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71:8966–8969. 10.1128/AEM.71.12.8966-8969.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120. 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, Holland T, Cotton D, Hauser L, Keller M. 2007. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 73:3205–3214. 10.1128/AEM.02985-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. U. S. A. 108:4599–4606. 10.1073/pnas.1000071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson KA, Johnson DE. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483–2492 [DOI] [PubMed] [Google Scholar]
- 54.Moss AR, Givens DI, Garnsworthy PC. 1995. The effect of supplementing grass silage with barley on digestibility, in sacco degradability, rumen fermentation and methane production in sheep at two levels of intake. Anim. Feed Sci. Technol. 55:9–33. 10.1016/0377-8401(95)00799-S [DOI] [Google Scholar]
- 55.Doreau M, van der Werf HMG, Micol D, Dubroeucq H, Agabriel J, Rochette Y, Martin C. 2011. Enteric methane production and greenhouse gases balance of diets differing in concentrate in the fattening phase of a beef production system. J. Anim. Sci. 89:2518–2528. 10.2527/jas.2010-3140 [DOI] [PubMed] [Google Scholar]
- 56.McDonald P, Edwards RA, Greenhalgh JFD, Morgan CA. 2002. Animal nutrition, 6th ed. Longman, Harlow, United Kingdom [Google Scholar]
- 57.Dennis SM, Arambel MJ, Bartley EE, Dayton AD. 1983. Effect of energy concentration and source of nitrogen on numbers and types of rumen protozoa. J. Dairy Sci. 66:1248–1254. 10.3168/jds.S0022-0302(83)81931-6 [DOI] [PubMed] [Google Scholar]
- 58.Taylor CD, McBride BC, Wolfe RS, Bryant MP. 1974. Coenzyme M is essential for growth of rumen strain of Methanobacterium ruminantium. J. Bacteriol. 120:974–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, Li D, Kong Z, McTavish S, Sang C, Lambie SC, Janssen PH, Dey D, Attwood GT. 2010. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5:e8926. 10.1371/journal.pone.0008926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rouvière PE, Wolfe RS. 1988. Novel biochemistry of methanogenesis. J. Biol. Chem. 263:7913–7916 [PubMed] [Google Scholar]
- 61.Pielou EC. 1966. Shannon's formula as a measure of species diversity: its use and misuse. Am. Nat. 100:463–465. 10.1086/282439 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.