Abstract
Anammox bacteria perform anaerobic ammonium oxidation (anammox) and have a unique compartmentalized cell consisting of three membrane-bound compartments (from inside outwards): the anammoxosome, riboplasm, and paryphoplasm. The cell envelope of anammox bacteria has been proposed to deviate from typical bacterial cell envelopes by lacking both peptidoglycan and a typical outer membrane. However, the composition of the anammox cell envelope is presently unknown. Here, we investigated the outermost layer of the anammox cell and identified a proteinaceous surface layer (S-layer) (a crystalline array of protein subunits) as the outermost component of the cell envelope of the anammox bacterium “Candidatus Kuenenia stuttgartiensis.” This is the first description of an S-layer in the phylum of the Planctomycetes and a new addition to the cell plan of anammox bacteria. This S-layer showed hexagonal symmetry with a unit cell consisting of six protein subunits. The enrichment of the S-layer from the cell led to a 160-kDa candidate protein, Kustd1514, which has no homology to any known protein. This protein is present in a glycosylated form. Antibodies were generated against the glycoprotein and used for immunogold localization. The antiserum localized Kustd1514 to the S-layer and thus verified that this protein forms the “Ca. Kuenenia stuttgartiensis” S-layer.
INTRODUCTION
Anammox bacteria are able to perform anaerobic ammonium oxidation, thereby converting ammonium and nitrite to dinitrogen gas (1, 2). Anammox bacteria are applied in wastewater treatment to remove ammonium from wastewater (3) and play an important role in the biological nitrogen cycle (4, 5). They comprise five genera (which all have a “Candidatus” status, since they are described from phylotypes that are not isolated in pure culture) that belong to the phylum Planctomycetes in the order Brocadiales (6). The species “Candidatus Kuenenia stuttgartiensis” is the most extensively studied anammox bacterium, and its genome (7), proteome, and metabolism (8) were described previously. Functional gene analysis remains difficult since no genetic system is available for anammox bacteria.
The phylum Planctomycetes is known for encompassing strikingly complex cell plans involving multiple cellular compartments and extensive membrane invaginations (9). Currently, the cell organization of Planctomycetes is under debate (10–13). Even within this phylum, the cell biology of anammox bacteria is remarkable, since anammox cells are divided into no fewer than three compartments, separated by bilayer membranes (Fig. 1). The inner compartment, the anammoxosome, is a so-called “prokaryotic organelle” (14, 15) in which the anammox reaction is assumed to take place. During the anammox reaction (7, 8, 16), a proton motive force (PMF) is established over the anammoxosome membrane. Membrane-bound ATPases could utilize this PMF for ATP production in the riboplasm. The riboplasm (which is topologically equivalent to the “pirellulosome” compartment in nonanammox planctomycete species) is the compartment that surrounds the anammoxosome, and it contains ribosomes and the nucleoid, thereby resembling the classical bacterial cytoplasm. The function of the outermost, apparently ribosome-free compartment, the paryphoplasm, has not yet been elucidated.
FIG 1.
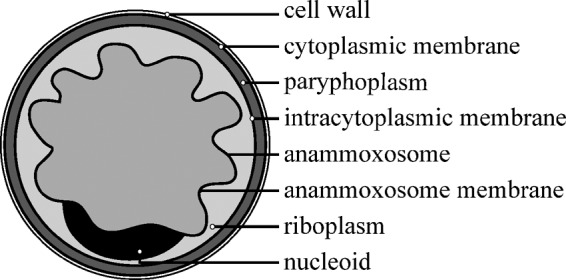
Cell plan of the anammox cell showing the three different compartments and their surrounding membranes. The riboplasm compartment has been defined the pirellulosome in Planctomycetes.
The composition of the cell envelope which encloses the paryphoplasm is unknown but has been proposed to deviate from both the typical Gram-positive and Gram-negative bacterial cell envelope types because it is proposed to lack both peptidoglycan (17) and a typical outer membrane. The proposed lack of peptidoglycan is based on (i) the close relationship of anammox bacteria to other planctomycetes where the cell wall composition has been chemically analyzed (18, 19) and (ii) the fact that not all genes necessary for the biosynthesis of peptidoglycan are present in the genome. Interestingly, most peptidoglycan biosynthesis genes are harbored by the genome, except for those encoding penicillin binding protein (PBP1a) and PBP1b (7, 20), which are required for the insertion of peptidoglycan precursors into polymeric peptidoglycan. Although in the transcriptome, a small number of reads is found for all peptidoglycan genes, none of their respective proteins, besides the protein d-alanine–d-alanine ligase (Ddl), could be detected in the “Ca. Kuenenia stuttgartiensis” proteome (8). Peptidoglycan has been proposed to contribute to the integrity of the cell and, in some cases, to the maintenance of cell shape (21). This raises the question of whether the cell envelope of anammox bacteria contains other structures that help to maintain the integrity of the cell. It is therefore of high interest to investigate the cell envelope of anammox bacteria in more detail.
Another interesting feature of the anammox cell plan is the outermost membrane, which surrounds the paryphoplasm. This membrane has been defined as a cytoplasmic membrane, which is also consistent with the immunogold localization of an ATPase to this membrane (22). However, several outer membrane proteins and key proteins in outer membrane biosynthesis have been detected in both the genomes and proteomes of two anammox species (7, 8, 13, 22), although none of these have yet been localized to any particular cell structure. At the moment, the identity of the cytoplasmic membrane of anammox bacteria remains under debate and needs further investigation.
In Gram-negative and Gram-positive bacteria as well as Archaea, a proteinaceous surface layer (S-layer) can be present as the outermost component of the cell envelope. S-layers constitute a two-dimensional (2D) crystalline array of (usually) identical protein subunits covering the entire cell surface (23, 24). The regular pattern formed by the S-layer can exist in oblique (p1 and p2), square (p4), or hexagonal (p3 and p6) symmetry, which is dictated by the arrangement and number of protein subunits (indicated by the number behind the p) that form the single morphological unit. The (self-)assembly of the proteins into the regular pattern is thought to be driven by entropical forces (25). S-layer proteins have a broad molecular mass range, between 40 and 200 kDa, and isoelectric points of between 3 and 6 (25). These proteins typically consist of 40 to 60% hydrophobic amino acids (26), although S-layer proteins with a predominant amount of hydrophilic amino acids have also been described (27). Many S-layer proteins are glycosylated (i.e., glycoproteins) by either N- or O-glycosylation. In some rare cases, both glycosylation types can be found on the same protein (28). One clear distinction between the various S-layers on different prokaryotic cells is the anchor to the underlying cell envelope component. In Gram-positive bacteria, S-layers are linked to the underlying cell wall components, including peptidoglycan and so-called secondary cell wall polymers (SCWPs) (29–31). In Gram-negative bacteria, S-layers are anchored in the outer membrane, while in Archaea, S-layers are always found to be anchored in the cytoplasmic membrane (32–34).
In the present study, we investigated the cell envelope of the anammox bacterium “Ca. Kuenenia stuttgartiensis.” We found a proteinaceous S-layer as the outermost component of the cell (envelope) using transmission electron microscopy on freeze-etched cells as well as in thin sections of cryofixed, freeze-substituted, and Epon-embedded cells. The S-layer was found to have a hexagonal (p6) symmetry, in which each S-layer motif is formed by six identical proteins. Enrichment of the S-layer led to the identification of a candidate S-layer glycoprotein, which was used to generate specific antibodies. Immunogold localization showed the antibody to bind to the outermost rim of the cell, where the S-layer is located, and thus verified that this protein forms the S-layer.
MATERIALS AND METHODS
S-layer enrichment.
Cells were harvested from a “Ca. Kuenenia stuttgartiensis” single-cell membrane bioreactor at an optical density at 600 nm (OD600) of 1.1 and were centrifuged for 10 min at 4,000 × g to concentrate them 40-fold in their original growth medium (35). Cells were then stored at −80°C and thawed just before the S-layer enrichment procedure. The procedure of freezing and thawing already partially disrupts the cells. The concentrated cells were resuspended in 20 mM HEPES buffer (pH 7.5) (including 15 mM NaHCO3, 2 mM CaCl2, and 0.8 mM MgSO4), after which the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) and DNase II were added to final concentrations of 24 mg liter−1 and 6.0 × 10−5 mg ml−1, respectively. The cells were then further disrupted by using a Potter homogenizer (50 strokes), and the disrupted cells were left at room temperature (RT) for 20 min (DNase incubation time). After this incubation, the detergent Triton X-100 was added to a final concentration of 0.5% (vol/vol), and the disrupted cells were incubated for 30 min at RT. The enriched S-layers were then pelleted by centrifugation at 31,000 × g for 20 min. The pellet was resuspended in the HEPES buffer described above and washed three times by centrifugation at 20,800 × g for 15 min and resuspension in HEPES buffer each time. The final pellets were resuspended in a small amount of buffer. This sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as well as transmission electron microscopy (TEM) after freeze-etching using a Philips CM 12 instrument (FEI, Eindhoven, the Netherlands) operated at 120 kV. Dominant bands in the SDS-PAGE gel were cut out to be analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Freeze-etching.
Freeze-etching was performed, as described previously (36, 37), on concentrated “Ca. Kuenenia stuttgartiensis” cells, taken from the single-cell membrane bioreactor and centrifuged for 4 min at 12,900 × g. The freeze-etched replicas were cleaned with 70% H2SO4 for 2 to 16 h and then twice with MilliQ water for 10 min each, picked up on copper grids, and investigated via TEM (as described above).
Freeze-drying.
Samples obtained after S-layer enrichment were visualized by freeze-drying, in order to investigate the presence of S-layers. After the application of 5 μl of sample onto a piece of freshly cleaved mica (Baltic Preparation, Niesgrau, Germany), the mica was blotted briefly onto filter paper (Whatman, Dassel, Germany) and plunge frozen in liquid nitrogen. The sample was then inserted in a Cressington CFE-50 freeze-etch machine at <−170°C with a pressure of <0.1 Pa. The sample was heated up to −80°C and held at that temperature for 60 to 120 min in order to sublimate the water from the samples, after which the samples were shadowed with approximately 2 nm Pt-C (angle, 45°) and approximately 15 nm C (angle, 90°). The replicas were floated off the mica with 70% H2SO4, incubated with the acid for 1 h, and then washed twice with MilliQ water for 10 min each, picked up on copper grids, and investigated by using TEM (as described above).
Cryofixation, freeze-substitution, Epon embedding, and sectioning.
Cryofixation, freeze-substitution, Epon embedding, sectioning, and imaging via TEM were performed as described previously (36).
Image processing.
Fast Fourier transform (FFT) power spectra of freeze-etched “Ca. Kuenenia stuttgartiensis” cells displaying S-layers were acquired by using the real-time FFT option in EM-MENU (version 4; Tietz Video & Image Processing Systems GmbH, Gauting, Germany). Correlation averaging images, showing the noise-reduced S-layer lattice, and relief reconstruction images, representing the height distribution of a small piece of S-layer lattice, were made by using the SEMPER software package (38) according to previously described methods (39) or by using Animetra Crystals software (40).
SDS-PAGE.
Samples obtained by S-layer enrichment were denatured by incubation of the proteins for 7 min at 100°C with 158 mM Tris-HCl buffer (pH 7) containing 5% β-mercaptoethanol, 2.6% SDS, and 16% glycerol. SDS-PAGE was performed on 8% slab gels in running buffer as described previously (41). After separation of the proteins, gels were stained with either Coomassie brilliant blue (G250) to visualize the proteins or periodic acid-Schiff's (PAS) reagent to visualize glycoproteins (42). Gels for PAS reagent were first fixed in 40% ethanol and 5% acetic acid for 30 min, followed by an oxidation step in 0.7% periodic acid in 5% acetic acid for 120 min and a reduction step in 0.2% sodium metabisulfite in 5% acetic acid for 30 min. Staining was performed for 18 h with Schiff's reagent (Carl Roth, Karlsruhe, Germany) and rendered the possible glycoproteins in magenta.
Library preparation, sequencing, and data analysis.
Sequencing of the “Ca. Kuenenia stuttgartiensis” continuous culture was performed on occasion, in order to check for changes after the initial genome sequencing (7). All kits mentioned in this paragraph were obtained from Life Technologies (Carlsbad, CA, USA). Genomic DNA was sheared for 5 min by using the Ion Xpress Plus fragment library kit according to the manufacturer's instructions. Further library preparation was performed by using the Ion Plus fragment library kit according to the manufacturer's instructions. Size selection of the library was performed by using an E-gel 2% agarose gel, resulting in a median fragment size of 331 bp. Emulsion PCR was performed by using the Onetouch 200-bp kit, and sequencing was performed on an IonTorrent PGM using the Ion PGM 200-bp sequencing kit and an Ion 316 chip. The resulting 2.33 million reads with an average length of 187 bp were quality trimmed and assembled using default settings of the CLC genomics workbench (v6.04; CLCbio, Aarhus, Denmark). Contigs were assigned to “Ca. Kuenenia stuttgartiensis” based on coverage. Comparison with the reference draft genome (NCBI Bioproject number PRJNA16685) was performed by using read mapper and BLASTn as implemented in the CLC genomics workbench (v6.04; CLCbio, Aarhus, Denmark).
MALDI-TOF MS and LC-MS/MS.
Bands cut out of the SDS-PAGE gels were prepared for MS analysis by alternately washing the gel pieces in acetonitrile and then in 50 mM ammonium bicarbonate, followed by reduction in a 10 mM dithiothreitol (DTT) solution and alkylation in 50 mM 2-chloroacetamide (43) in 50 mM ammonium bicarbonate. Trypsin digestion was performed overnight by using 12.5 ng μl−1 trypsin in 50 mM ammonium bicarbonate. The peptides were extracted from the gel pieces by using a mix of 0.05% trifluoroacetic acid and 50% acetonitrile. For MALDI-TOF MS, the samples were applied onto a MALDI plate using α-cyano-hydroxy cinnamic acid as matrix and analyzed by using a Bruker Biflex III MALDI-TOF MS instrument (44). For LC-MS/MS, the gel pieces were analyzed as described previously (45). Proteins were identified by using the MASCOT search tool (Matrix Science, London, United Kingdom) and a database of the predicted “Ca. Kuenenia stuttgartiensis” proteome available at the Genoscope website (https://www.genoscope.cns.fr/agc/microscope/export/export.php?format=Prot&S_id=260).
Antibody generation.
The antiserum containing the antibodies against the putative S-layer protein Kustd1514 was generated against protein bands cut out of an SDS-PAGE gel. For this purpose, a sample obtained after S-layer enrichment was loaded onto an 8% SDS-PAGE gel that was stained and destained with solutions that included ethanol instead of methanol. The band corresponding to a protein of approximately 250 kDa (Kustd1514) was cut out and sent to Davids Biotechnology (Regensburg, Germany) for immunization of rabbits. To verify the contents of this protein band, one band from the same gel was analyzed by LC-MS/MS.
Immunoblotting.
Blots were made from 8% SDS-PAGE gels containing cell extracts of “Ca. Kuenenia stuttgartiensis” cells. These were prepared by harvesting “Ca. Kuenenia stuttgartiensis” cells by centrifugation, after which the cell pellet was resuspended in 1 volume of 20 mM potassium phosphate buffer (pH 7.0). The cells were passed through a French press at 138 MPa in three passages and centrifuged at 4°C for 15 min at 2,200 × g. The resulting supernatant was the cell-free extract containing “Ca. Kuenenia stuttgartiensis” proteins. This cell-free extract was boiled for 7 min in SDS sample buffer (as described above), and 20 μg protein per lane was loaded onto 8% SDS-PAGE gels. After separation, the proteins were transferred from the gel onto a Protran nitrocellulose transfer membrane with a pore size of 0.45 μm (Whatman, Dassel, Germany) with a semidry transfer cell blotting system (Bio-Rad, Veenendaal, the Netherlands). Two different blotting buffers were used, both consisting of 48 mM Tris and 39 mM glycine. The buffer in which the gel was incubated had an additional 0.05% SDS, and the one that was used for the membrane contained 20% methanol. The blotting was performed at 50 mA for 60 min at room temperature, and afterwards, dried blots were stored at 4°C.
Immunoblotting was performed on blots that were incubated in deionized water (dH2O) for 30 min and afterwards for 30 min in protein-free (Tris-buffered saline [TBS]) blocking buffer (Thermo Scientific, Rockford, IL, USA). The blots were then incubated for 60 min in antiserum diluted 1,000-fold in blocking buffer. Two negative controls were performed; instead of antiserum, one was incubated in blocking buffer, and the other was incubated in preimmune serum diluted 1,000-fold in blocking buffer. The blots were then washed three times for 10 min in TBS containing 0.05% Tween and incubated for 60 min in monoclonal mouse anti-rabbit IgG alkaline phosphatase conjugate (Sigma, Zwijndrecht, The Netherlands) diluted 150,000-fold in blocking buffer. The blots were then washed two times for 10 min in TBS containing 0.05% Tween and two times for 10 min in 10 mM TBS containing 8% NaCl and 0.2% KCl and finally incubated with a 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium (NBT) liquid substrate system (Sigma, Zwijndrecht, The Netherlands) for 9 min and rinsed for 10 min in dH2O. All lanes were imaged with the same settings.
Immunogold localization.
Samples for immunogold localization were prepared by using a rehydration method (46), as described previously (22). In short, high-pressure frozen cells were freeze-substituted in acetone containing 0.5% glutaraldehyde, 0.1% uranyl acetate, and 1% H2O; rehydrated in a graded acetone series on ice; embedded in gelatin; cut into small cubes; infiltrated with sucrose; and frozen in liquid nitrogen. Ultrathin cryosections (65 nm) were cut by using a UC7/FC7 cryo-ultramicrotome (Leica Microsystems, Vienna, Austria), picked up with a drop of 1% methylcellulose and 1.15 M sucrose in PHEM buffer [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2 (pH 6.9)], and transferred onto carbon-Formvar-coated grids.
Grids containing ultrathin cryosections of “Ca. Kuenenia stuttgartiensis” cells were washed for 30 min at 37°C on PHEM buffer and for 10 min at room temperature in drops of PHEM buffer containing 20 mM glycine. Blocking was achieved by incubation on drops of PHEM buffer containing 1% bovine serum albumin (BSA) for 15 min, after which the grids were incubated for 60 min with antiserum diluted 100-fold in PHEM buffer containing 1% BSA. Negative controls were incubated in PHEM containing 1% BSA without antiserum for 60 min. As an additional control, grids were incubated with preimmune serum instead of antiserum. After this incubation, the grids were washed for 11 min on drops of PHEM buffer with 1% BSA and incubated for 20 min with the secondary antibody, protein A coupled to 10-nm gold (PAG-10; CMC UMC Utrecht), diluted 70-fold in PHEM buffer with 1% BSA. The grids were then washed for 5 min on drops of PHEM buffer with 1% BSA and for 10 min on drops of PHEM buffer. The cryosections on the grids were fixed by incubation for 5 min on drops of 1% glutaraldehyde in PHEM buffer and were consequently washed for 10 min on drops of MilliQ water. Poststaining was performed by incubation for 5 min in 2% uranyl acetate in 0.15 M oxalic acid set to pH 7.0 with 30% ammonium hydroxide, after which the grids were quickly washed on 2 drops of water. The grids were then immediately washed with two drops of 1.8% methylcellulose containing 0.4% aqueous uranyl acetate on ice. The sections were embedded by incubation for 5 min on ice on a drop of methylcellulose containing uranyl acetate. After the sections were air dried, the grids containing labeled cryosections were investigated at 100 kV in a JEOL (Tokyo, Japan) JEM-1010 TEM instrument. Images were recorded by using a SIS Mega View III camera (Olympus, Münster, Germany).
RESULTS
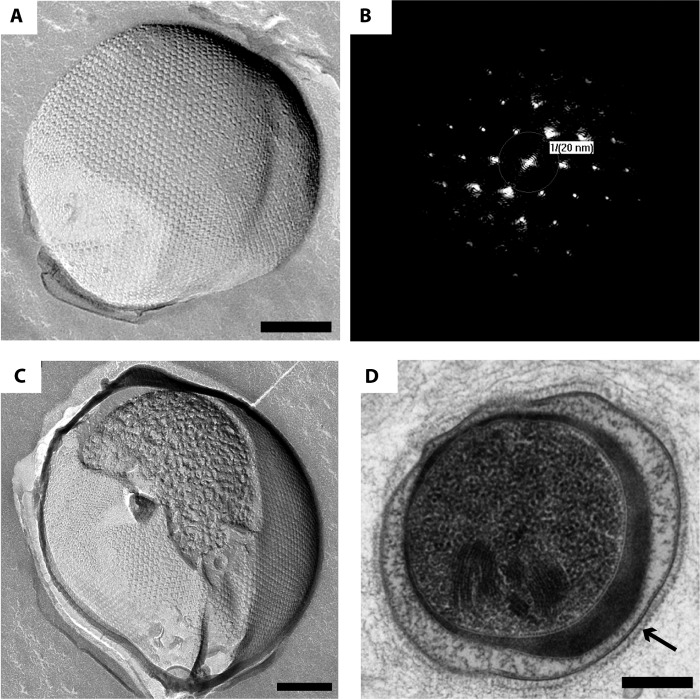
“Ca. Kuenenia stuttgartiensis” single cells were freeze-etched and visualized via transmission electron microscopy (TEM) to investigate the outermost layer of the cell. The freeze-etched “Ca. Kuenenia stuttgartiensis” cells clearly showed S-layers with a hexagonal symmetry (Fig. 2A and C) on top of the cytoplasmic membrane (Fig. 2D). The fast Fourier transform (FFT) power spectra confirmed the regular structure of the S-layer (Fig. 2B) and the hexagonal symmetry. Analysis of the FFT power spectra showed a center-to-center spacing between the S-layer unit cells of about 20 nm, which fits in the range of 2.5 to 35 nm that is typical for S-layers (25). With the use of correlation averaging, the S-layer fine structure and lattice can be visualized with a higher signal-to-noise ratio (39). Through these analyses (Fig. 3A), it became apparent that each S-layer unit cell consisted of six protein densities surrounding a central pore. This was further visualized by a relief reconstruction (47), resulting in a three-dimensional model of the surface of the S-layer (Fig. 3B). The relief reconstruction showed that the protein densities appear cylindrical. Both the correlation averaging and the relief reconstruction are consistent with hexagonal p6 symmetry, although p3 symmetry cannot be excluded at the present level of resolution.
FIG 2.
“Ca. Kuenenia stuttgartiensis” cells display S-layers as observed by TEM after multiple types of sample preparation. (A) “Ca. Kuenenia stuttgartiensis” cells are covered by a hexagonal S-layer, as observed after freeze-etching. (B) The FFT power spectrum of a part of the S-layer seen in panel A reflects the regular pattern of the S-layer. The p6 symmetry and center-to-center spacing of 20 nm for the S-layer of “Ca. Kuenenia stuttgartiensis” are reflected by the FFT power spectrum. (C) A freeze fracture through the S-layer gives an inside view into the “Ca. Kuenenia stuttgartiensis” cell underneath. (D) The S-layer (indicated by the arrow) forms a zigzag layer on top of the cytoplasmic membrane in “Ca. Kuenenia stuttgartiensis” cells that were cryofixed; freeze-substituted in acetone containing 2% osmium tetroxide, 0.2% uranyl acetate, and 1% water; Epon embedded; and thin sectioned. Scale bars, 200 nm.
FIG 3.
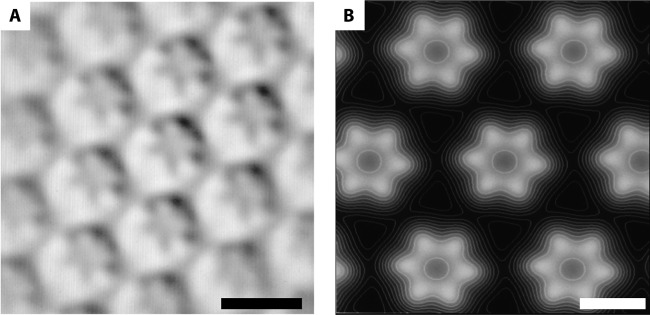
The S-layer of “Ca. Kuenenia stuttgartiensis” visualized by image processing. (A) Correlation averaging shows that the S-layer of “Ca. Kuenenia stuttgartiensis” has hexagonal symmetry with a unit cell consisting of six protein densities surrounding a central pore. Scale bar, 20 nm. (B) Relief reconstruction gives an impression of the surface of the S-layer in three dimensions. White represents high and black represents low areas. Scale bar, 10 nm.
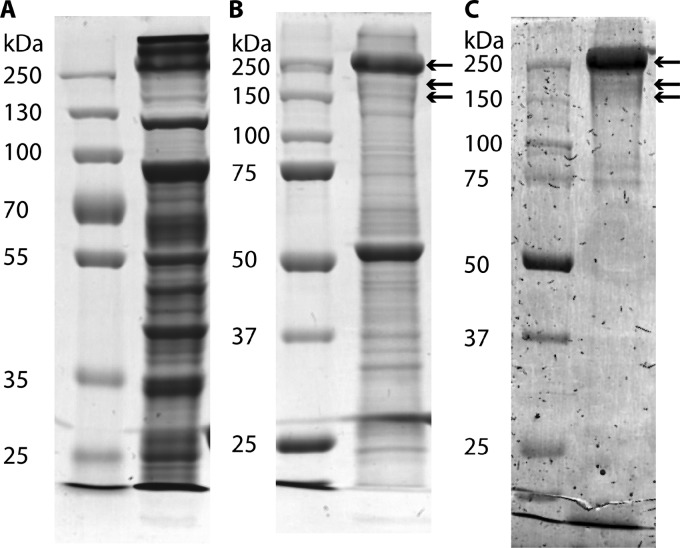
To investigate which protein forms the observed S-layer of “Ca. Kuenenia stuttgartiensis,” the S-layer was enriched from whole cells. After treatment of the cells with a Potter homogenizer and the detergent Triton X-100, (patches of) S-layers were present, as visualized by TEM of freeze-dried samples (Fig. 4). In addition to the S-layers, membrane patches were also present, as seen after negative staining (data not shown). The protein composition of the sample obtained by S-layer enrichment was analyzed by SDS-PAGE and subsequent MALDI-TOF MS and LC-MS/MS analyses. When comparing the crude extract (Fig. 5A) to the S-layer enrichment (Fig. 5B), only two major proteins and two less abundant proteins were detected (next to some minor bands) in the S-layer enrichment (Fig. 5B). MALDI-TOF MS and LC-MS/MS analyses and subsequent searches (Mascot; Matrix Science) resulted in significant matches. The protein at about 250 kDa was identified as Kustd1514 (for contig d from “Ca. Kuenenia stuttgartiensis”), and the protein at about 55 kDa was identified as the putative outer membrane protein (OMP) Kustd1878, which has a predicted β-barrel structure, which is characteristic of OMPs (13). The protein bands at approximately 160 and 210 kDa were both found to contain significant amounts of Kustd1514 as well. Sequencing of the “Ca. Kuenenia stuttgartiensis” enrichment culture (which was continuously run in bioreactor systems over the period described) used for all experiments described here was performed at two time points, namely, in 2002 (NCBI Bioproject 16685) (7) and in 2012 (NCBI Bioproject number PRJEB4259, which is the sequence obtained as described in Materials and Methods and is thus first reported in this publication). There are indications that during cultivation, the Kustd1514 protein sequence has undergone a sequence change at the amino acid level in part of the “Ca. Kuenenia stuttgartiensis” population since the initial metagenome analysis. Compared to the original sequence (7), the 43 amino acids at the N terminus and 759 amino acids at the C terminus of the new sequence are identical. For the remaining 672 amino acids between N and C termini and for the 116 amino acids that are at the end of the C terminus, identities of 43% and 77%, respectively, have been determined. Currently, proteins with both Kustd1514 sequences (74% protein sequence identity for the total sequence, as determined by the PIR pairwise alignment tool [http://pir.georgetown.edu/pirwww/search/pairwise.shtml]) are present in the “Ca. Kuenenia stuttgartiensis” membrane bioreactor and were detected in all three Kustd1514 protein bands from the S-layer enrichment, as determined by LC-MS/MS analysis. Both proteins are similar concerning their characteristics (as listed below), and therefore, the protein encoded by the original sequence (7) was used for further (bioinformatics) analysis.
FIG 4.
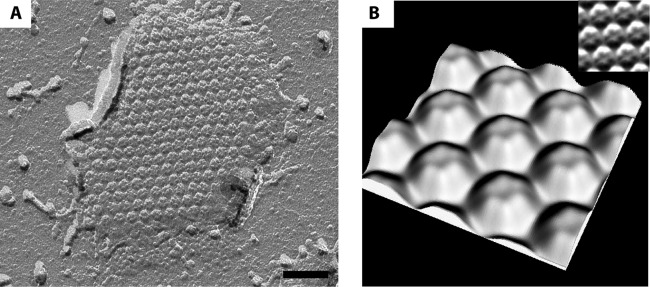
S-layer patches are present after S-layer enrichment of “Ca. Kuenenia stuttgartiensis.” (A) S-layer patch with typical hexagonal symmetry observed by freeze-drying. (B) A three-dimensional model of the isolated S-layer shows the same relief as observed for the S-layers present on “Ca. Kuenenia stuttgartiensis” cells. (Inset) Correlation averaging used to determine the three-dimensional representation. Scale bar, 70 nm.
FIG 5.
Analysis of crude extract and enriched S-layers of “Ca. Kuenenia stuttgartiensis” by SDS-PAGE. (A) SDS-PAGE gel stained with Coomassie G shows all proteins present in the “Ca. Kuenenia stuttgartiensis” crude extract. (B) SDS-PAGE gel stained with Coomassie G shows the proteins present after S-layer enrichment. (C) SDS-PAGE gel stained with periodic acid-Schiff's reagent shows glycoproteins present after S-layer enrichment. Arrows indicate protein bands in which Kustd1514 was detected via LC-MS/MS analysis.
The detection of Kustd1514 at different apparent molecular masses in the SDS-PAGE gel suggested that this protein might be present in multiple posttranslationally modified states. This is supported by the predicted molecular mass for Kustd1514 (using the ExPASy compute pI/Mw tool [http://web.expasy.org/compute_pi/]). Kustd1514 is predicted to consist of 1,591 amino acids (aa) (Uniprot) for the total protein, and the predicted molecular mass is 160 kDa for the protein after processing of the predicted 35-aa-long signal peptide (predicted by SignalP 4.1 [48]). This predicted molecular mass of 160 kDa matches the lowest of the three Kustd1514-containing bands observed in the SDS-PAGE gel. Glycosylation is the most common posttranslational modification for S-layer proteins (49). Therefore, glycan-detecting periodic acid-Schiff's (PAS) staining (42) was performed on an SDS-PAGE gel containing enriched S-layers, which confirmed glycosylation of Kustd1514 (Fig. 5C).
The Kustd1514 protein shows no primary sequence similarity to other known (S-layer) proteins, as indicated by the lack of significant hits using BLAST (50) and PSI-BLAST (51) searches: all hits with an E value of <10−10 are from “Ca. Kuenenia stuttgartiensis” and have a maximum query coverage of 25%. The amino acid composition of Kustd1514 is in many aspects different from the typical S-layer protein: 49.5% of the amino acids are hydrophilic (due to the abundance in serine [12.4%] and threonine [14.5%]), and only 29.7% are hydrophobic (the typical value for S-layer proteins would be 40 to 60%). The predicted pI of Kustd1514 of 4.39 (using the ExPASy compute pI/Mw tool) fits with the typical values for S-layer proteins (pIs of between 3 and 6) (25). When comparing the predicted secondary structure of Kustd1514 to other proteins via HHpred (52, 53), the only hits that were found target the last 300 amino acids of the protein. When looking at the full-length secondary structure prediction of Kustd1514 using PSIPRED (54), it is noteworthy that only 2.1% of the structure is predicted to consist of α-helices, which is clearly lower than the average 20% that is reported for S-layer proteins (26). The percentage of predicted β-sheets is 44.8%, which is close to the average S-layer value of 40%. No transmembrane regions were predicted by using TMHMM (55, 56). It thus becomes apparent that the S-layer protein of “Ca. Kuenenia stuttgartiensis” shows some of the global characteristics of a typical S-layer protein but is not similar to any known protein in primary or secondary structure.
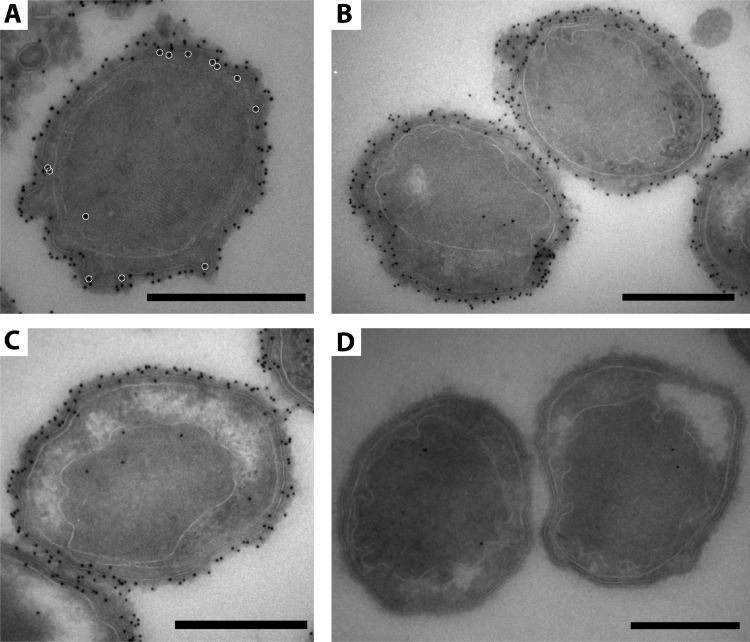
The glycoprotein band at around 250 kDa obtained from the S-layer enrichment was used to immunize a rabbit in order to generate antibodies against the putative S-layer glycoprotein Kustd1514. The affinity and specificity of the antibody for the glycoprotein Kustd1514 were confirmed by immunoblotting on a blot containing a “Ca. Kuenenia stuttgartiensis” cell extract. As expected, a specific band at approximately 250 kDa was observed after immunoblotting with the Kustd1514 antiserum (Fig. 6). This band was absent when incubations were performed with preimmune serum or with secondary antibody only. Immunogold localization was performed by using the Kustd1514 antiserum on “Ca. Kuenenia stuttgartiensis” cryosections of cells prepared via the rehydration method (46). This immunogold labeling localized the Kustd1514 protein to the electron-dense S-layer that forms the outermost rim of cells of “Ca. Kuenenia stuttgartiensis” (Fig. 7).
FIG 6.

Immunoblot analysis of the antiserum directed against the “Ca. Kuenenia stuttgartiensis” S-layer glycoprotein Kustd1514 tested against a “Ca. Kuenenia stuttgartiensis” cell extract. Lane 1, marker; lane 2, incubation with preimmune serum; lane 3, incubation with anti-Kustd1514. The arrow indicates the expected target size (about 250 kDa).
FIG 7.
(A to C) Immunogold localization of the antiserum directed at Kustd1514 localizes the protein to the S-layer surrounding the cells in rehydrated “Ca. Kuenenia stuttgartiensis” cryosections. In panel A, for clarification, white circles indicate gold labels that are localized inside the cytoplasmic membrane (so not directly on the S-layer). Considering the length of the antibody-protein A-gold complex (25 nm), these gold labels most likely also correspond to S-layer labeling (except for the one in the anammoxosome). (D) Negative control incubated with preimmune serum instead of antiserum. Scale bars, 500 nm.
DISCUSSION
Here, we have identified a glycoprotein S-layer as the outermost layer of the anammox bacterium “Ca. Kuenenia stuttgartiensis,” which forms a new addition to the cell plan of anammox bacteria. The S-layer has six protein subunits per unit cell, which means that the symmetry is hexagonal (p6). The S-layer was enriched from “Ca. Kuenenia stuttgartiensis” cells, leading to the identification of a putative S-layer protein. Antibodies against this Kustd1514 protein were raised and used to localize the Kustd1514 glycoprotein to the S-layer via immunogold localization. This verified that Kustd1514 indeed forms the S-layer in “Ca. Kuenenia stuttgartiensis.”
After S-layer enrichment, membrane patches were observed together with the S-layer. In the LC-MS/MS analysis, one particular protein, the putative OMP Kustd1878 (13), was shown to be highly abundant in the S-layer enrichment. This finding suggests that the “Ca. Kuenenia stuttgartiensis” S-layer is relatively strongly anchored in the cytoplasmic membrane of the anammox cell. In this respect, it would be an interesting future experiment to see if the S-layer can self-assemble from isolated Kustd1514 monomers (as in reference 57) without a membrane(-like) lattice underneath. The presence of Kustd1878 in the S-layer enrichment also suggests that Kustd1878 is located in or on the cytoplasmic membrane of “Ca. Kuenenia stuttgartiensis,” but the location and function of this putative OMP need further investigation. Another interesting question to investigate is whether the S-layer proteins interact directly with Kustd1878 or instead with other proteins or molecules associated with the cytoplasmic membrane.
The intense signal in the PAS staining indicated that the Kustd1514 protein is highly glycosylated, which is a common feature of several S-layer proteins (49). In bacteria, O-glycosylation seems most abundant, although some N-glycosylated (S-layer) proteins have also been described (28, 58). The Kustd1514 protein contains both potential O- and N-glycosylation sites, as found by a manual search of the protein sequence for strict “consensus” sequences and by using the GlycoPP prediction server (59). Consensus sequences used were D/E-Y-NY-S/T (where Y can be all amino acids except P) for N-glycosylation (60) and D-S/T-A/I/L/V/M/T for O-glycosylation (61). Further studies need to elucidate which of these sites are in use and if the S-layer protein is O- or N-glycosylated or if both types of glycosylation occur on the same protein. In addition to the posttranslational modification, the protein sequence shift of Kustd1514 itself during cultivation is under investigation in our laboratory.
To gain a better understanding of the S-layer in relation to the underlying cell wall components of “Ca. Kuenenia stuttgartiensis,” it would be of interest to identify to which structure the S-layer attaches and via which mechanism. The genome organization around Kustd1514 gives no clues about possible glycosylation, secretion, and attachment mechanisms. In the case of Gram-negative bacteria, much research is still required to find out common processes involved in attachment of the S-layer to the cell surface. In Caulobacter crescentus and Campylobacter fetus, the S-layer specifically attaches to lipopolysaccharides (LPSs) (the O-antigens are crucial in the case of C. crescentus) via an N-terminal stretch (62, 63). A bit more is known about attachment mechanisms of S-layers in Gram-positive bacteria, where the S-layer attaches to secondary cell wall polymers or the peptidoglycan itself (64). Many Gram-positive S-layer proteins contain a so-called S-layer homology (SLH) motif at their N terminus that is involved in anchoring the S-layer to the secondary cell wall polymers (29, 65). In Gram-positive S-layer proteins without SLH domains, an N-terminal motif (having, for instance, a net positive charge) or, in some cases, a C-terminal motif seems to play a role in attachment (63, 64, 66). When a BLAST search with Kustd1514 was performed against the S-layer SLH consensus motif proposed previously by Engelhardt and Peters (32), no significant hits were found. Since peptidoglycan is probably absent from “Ca. Kuenenia stuttgartiensis” (20), the attachment mechanism of the S-layer might more resemble the attachment mechanisms for Gram-negative organisms. However, the identity (in terms of structure and function) of the outermost membrane of anammox bacteria as either an outer membrane typical of Gram-negative bacteria or a cytoplasmic membrane remains under investigation. If the outermost membrane of the cell is indeed a typical cytoplasmic membrane, and the S-layer would anchor into this membrane, this would be a unique case for Bacteria. Anchoring of the S-layer into the cytoplasmic membrane has thus far been described only for Archaea. It is thus clear that much more research is needed, focusing on possible N- or C-terminal modifications of Kustd1514 or, for instance, lipid modifications which might be involved in membrane anchoring. In addition, it would be important to further investigate the composition of the entire cell envelope of anammox bacteria.
To our knowledge, the S-layer of “Ca. Kuenenia stuttgartiensis” is the first S-layer described for a cultured planctomycete. However, this finding fits data from previous reports (18, 19) showing that several Planctomycetes have cell walls that consist predominantly of protein. If the proteinaceous cell walls in the described Planctomycetes (18, 19) might be S-layers, they are probably quite different from the “Ca. Kuenenia stuttgartiensis” S-layer. While cell walls of the described Planctomycetes were enriched by incubation in 10% SDS at 100°C, no S-layers have been observed by electron microscopy investigations (19, 67). In addition, boiling of “Ca. Kuenenia stuttgartiensis” cells in 10% SDS yielded a sample without cell walls or S-layers (M. C. F. van Teeseling, unpublished results). Furthermore, the previously reported proteinaceous cell walls were enriched in proline and cysteine (19), and the “Ca. Kuenenia stuttgartiensis” S-layer did not contain large amounts of these amino acids but was enriched in serine and threonine instead. It would therefore be very interesting to investigate by rigorous electron microscopy studies if S-layers are present in other Planctomycetes as well.
As often seen for S-layer proteins, which of course have to cover the complete cell surface, Kustd1514 is very abundant. In the proteome of membrane preparations from “Ca. Kuenenia stuttgartiensis” cells, Kustd1514 is one of the most abundant proteins (8). A significant effort of the “Ca. Kuenenia stuttgartiensis” cells is thus invested in synthesizing S-layer proteins, and it is therefore obvious to raise the question of which function the S-layer has for this anammox bacterium. Multiple functions for S-layers in prokaryotes have been proposed (65), including protection against predation (68, 69), adhesion of cell-associated exoenzymes (70), osmoprotection (71), and maintaining cell shape and integrity (58, 64). The latter function seems especially interesting in the case of anammox bacteria, since they are proposed to lack peptidoglycan and might therefore be in need of a structure that maintains cell shape and integrity. In the case of Archaea, which also lack peptidoglycan (23, 72), S-layers are indeed often assumed to have a function in maintaining cellular integrity, which is also substantiated by the fact that S-layer-deficient mutants in Archaea have not been found (64). Loss of S-layers in laboratory strains is a common feature of Bacteria, which probably occurs when the cells do not need their S-layers under culturing conditions, in which case S-layer-deficient mutants might outgrow S-layer-containing cells (58, 73). If the S-layer indeed plays a role in maintaining the integrity of the cell, this could also explain why the S-layers have not been lost in our “Ca. Kuenenia stuttgartiensis” culture even though it has been continuously cultivated in the laboratory for over 10 years (taking an average generation time of 2 weeks, this would mean over 260 generations).
Future research will have to show if, as hypothesized, cellular integrity truly is the (only) function of the S-layer for “Ca. Kuenenia stuttgartiensis.” Since no genetic system is available for “Ca. Kuenenia stuttgartiensis,” it is unfortunately not possible to make a knockout mutant of the S-layer to assess the function of the S-layer. A first test of this hypothesis, however, would be to assess if S-layers are present in other anammox bacteria as well. It has been proposed that all anammox bacteria lack peptidoglycan, and therefore, following the above-mentioned hypothesis, all anammox bacteria would need an S-layer or other type of (proteinaceous) cell wall component. Further experiments thus have to show whether the S-layer that is described here is indeed the component of the cell envelope that gives these highly interesting compartmentalized cells their structural integrity.
ACKNOWLEDGMENTS
We thank Rob Mesman and Elly van Donselaar for their assistance with and advice on the electron microscopy techniques. We acknowledge Hans Wessels for his LC-MS/MS analysis of the protein bands. We acknowledge Jennifer Flechsler and Thomas Heimerl for their help in preparing the protein sample for the immunization procedure and Karl Hermann Fuchs for Fig. 4B.
Muriel C. F. van Teeseling and Mike S. M. Jetten are supported by the European Research Council (ERC232937); Naomi M. de Almeida and Laura van Niftrik are supported by the Netherlands Organization for Scientific Research (ALW grant 818.02.105 and VENI grant 863.09.009, respectively); Daan R. Speth is supported by BE-Basic fp0702; Andreas Klingl is supported by the LOEWE program of the state of Hessen, Germany (LOEWE Research Centre for Synthetic Microbiology); and Reinhard Rachel is supported by the German Research Foundation (DFG) (grant HU 703/2-2).
Footnotes
Published ahead of print 18 October 2013
REFERENCES
- 1.van de Graaf AA, Mulder A, de Bruijn P, Jetten MSM, Robertson LA, Kuenen JG. 1995. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61:1246–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM. 1999. Missing lithotroph identified as a new planctomycete. Nature 400:446–449. 10.1038/22749 [DOI] [PubMed] [Google Scholar]
- 3.Kartal B, Kuenen JG, van Loosdrecht MCM. 2010. Sewage treatment with anammox. Science 328:702–703. 10.1126/science.1185941 [DOI] [PubMed] [Google Scholar]
- 4.Jetten MSM, van Niftrik L, Strous M, Kartal B, Keltjens JT, Op den Camp HJM. 2009. Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44:65–84. 10.1080/10409230902722783 [DOI] [PubMed] [Google Scholar]
- 5.Lam P, Kuypers MMM. 2011. Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar. Sci. 3:317–345. 10.1146/annurev-marine-120709-142814 [DOI] [PubMed] [Google Scholar]
- 6.Jetten MSM, Op den Camp HJM, Kuenen JG, Strous M. 2010. Family I. “Candidatus Brocadiaceae” fam. nov, p 596–602 In Krieg NR, Staley JT, Brown DR, Hedlund B, Paster BJ, Ward N, Ludwig W, Whitman WB. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 4 Springer, New York, NY [Google Scholar]
- 7.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Médigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi H, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes H-W, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794. 10.1038/nature04647 [DOI] [PubMed] [Google Scholar]
- 8.Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, Op den Camp HJM, Harhangi H, Janssen-Megens EM, Francoijs K-J, Stunnenberg HG, Keltjens JT, Jetten MSM, Strous M. 2011. Molecular mechanism of anaerobic ammonium oxidation. Nature 479:127–130. 10.1038/nature10453 [DOI] [PubMed] [Google Scholar]
- 9.Lindsay MR, Webb RI, Strous M, Jetten MSM, Butler MK, Forde RJ, Fuerst JA. 2001. Cell compartmentalization in planctomycetes: novel types of structural organization for the bacterial cell. Arch. Microbiol. 175:413–429. 10.1007/s002030100280 [DOI] [PubMed] [Google Scholar]
- 10.Lieber A, Leis A, Kushmaro A, Minsky A, Medalia O. 2009. Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus. J. Bacteriol. 191:1439–1445. 10.1128/JB.01513-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yee B, Sagulenko E, Morgan GP, Webb RI, Fuerst JA. 2012. Electron tomography of the nucleoid of Gemmata obscuriglobus reveals complex liquid crystalline cholesteric structure. Front. Microbiol. 3:326. 10.3389/fmicb.2012.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP. 2013. Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol. 11:e1001565. 10.1371/journal.pbio.1001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speth DR, van Teeseling MCF, Jetten MSM. 2012. Genomic analysis indicates the presence of an asymmetric bilayer outer membrane in Planctomycetes and Verrucomicrobia. Front. Microbiol. 3:304. 10.3389/fmicb.2012.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Niftrik LA, Fuerst JA, Sinninghe Damsté JS, Kuenen JG, Jetten MSM, Strous M. 2004. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol. Lett. 233:7–13. 10.1016/j.femsle.2004.01.044 [DOI] [PubMed] [Google Scholar]
- 15.van Teeseling MCF, Neumann S, van Niftrik L. 2013. The anammoxosome organelle is crucial for the energy metabolism of anaerobic ammonium oxidizing bacteria. J. Mol. Microbiol. Biotechnol. 23:104–117. 10.1159/000346547 [DOI] [PubMed] [Google Scholar]
- 16.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJM, Jetten MSM, Keltjens JT. 2013. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37:428–461. 10.1111/1574-6976.12014 [DOI] [PubMed] [Google Scholar]
- 17.Jetten MSM, Sliekers O, Kuypers M, Dalsgaard T, van Niftrik L, Cirpus I, van de Pas-Schoonen K, Lavik G, Thamdrup B, Le Paslier D, Op den Camp HJM, Hulth S, Nielsen LP, Abma W, Third K, Engström P, Kuenen JG, Jørgensen BB, Canfield DE, Sinninghe Damsté JS, Revsbech NP, Fuerst J, Weissenbach J, Wagner M, Schmidt I, Schmid M, Strous M. 2003. Anaerobic ammonium oxidation by marine and freshwater planctomycete-like bacteria. Appl. Microbiol. Biotechnol. 63:107–114. 10.1007/s00253-003-1422-4 [DOI] [PubMed] [Google Scholar]
- 18.König E, Schlesner H, Hirsch P. 1984. Cell wall studies on budding bacteria of the Planctomycetes/Pasteuria group and on a Prosthecomicrobium sp. Arch. Microbiol. 138:200–205. 10.1007/BF00402120 [DOI] [Google Scholar]
- 19.Liesack W, König H, Schlesner H, Hirsch P. 1986. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomycetes group. Arch. Microbiol. 145:361–366. 10.1007/BF00470872 [DOI] [Google Scholar]
- 20.Neumann S, van Teeseling MCF, van Niftrik L. 2013. Cell biology of anaerobic ammonium-oxidizing bacteria: unique prokaryotes with an energy-conserving intracellular compartment, p 89–123 In Fuerst JA. (ed), Planctomycetes: cell structure, origins and biology. Springer, Humana Press, New York, NY [Google Scholar]
- 21.van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R. 10.1093/glycob/11.3.25R [DOI] [PubMed] [Google Scholar]
- 22.van Niftrik L, van Helden M, Kirchen S, van Donselaar EG, Harhangi HR, Webb RI, Fuerst JA, Op den Camp HJM, Jetten MSM, Strous M. 2010. Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium “Candidatus Kuenenia stuttgartiensis.” Mol. Microbiol. 77:701–715. 10.1111/j.1365-2958.2010.07242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingl A, Flechsler J, Heimerl T, Rachel R. 2013. Archaeal cells. eLS. 10.1002/9780470015902.a0000383.pub2 [DOI] [Google Scholar]
- 24.König H, Rachel R, Claus H. 2007. Proteinaceous surface layers of Archaea: ultrastructure and biochemistry, p 315–340 In Cavicchioli R. (ed), Archaea: molecular and cell biology. ASM Press, Washington, DC [Google Scholar]
- 25.Sleytr UB, Sára M. 1997. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 15:20–26. 10.1016/S0167-7799(96)10063-9 [DOI] [PubMed] [Google Scholar]
- 26.Sleytr UB. 1997. Basic and applied S-layer research: an overview. FEMS Microbiol. Rev. 20:5–12. 10.1016/S0168-6445(97)00039-9 [DOI] [Google Scholar]
- 27.Sumper M, Berg E, Mengele R, Strobel I. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schäffer C, Graninger M, Messner P. 2001. Prokaryotic glycosylation. Proteomics 1:248–261. [DOI] [PubMed] [Google Scholar]
- 29.Schäffer C, Messner P. 2005. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology 151:643–651. 10.1099/mic.0.27749-0 [DOI] [PubMed] [Google Scholar]
- 30.Mesnage S, Fontaine T, Mignot T, Delpierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484. 10.1093/emboj/19.17.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sára M. 2001. Conserved anchoring mechanisms between crystalline cell surface S-layer proteins and secondary cell wall polymers in Gram-positive bacteria. Trends Microbiol. 9:47–49. 10.1016/S0966-842X(00)01905-3 [DOI] [PubMed] [Google Scholar]
- 32.Engelhardt H, Peters J. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276–302. 10.1006/jsbi.1998.4070 [DOI] [PubMed] [Google Scholar]
- 33.Haft DH, Payne SH, Selengut JD. 2012. Archaeosortase and exosortases are widely distributed systems linking membrane transit with posttranslation modifications. J. Bacteriol. 194:36–48. 10.1128/JB.06026-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdul Halim MF, Pfeiffer F, Zou J, Frisch A, Haft D, Wu S, Tolić N, Brewer H, Payne SH, Paša-Tolić L, Pohlschroder M. 2013. Haloferax volcanii archaeosortase required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol. Microbiol. 88:1164–1175. 10.1111/mmi.12248 [DOI] [PubMed] [Google Scholar]
- 35.Kartal B, Geerts W, Jetten MSM. 2011. Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria, p 89–108 In Klotz MG. (ed), Methods in enzymology, vol 486 Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 36.Wu ML, van Teeseling MCF, Willems MJR, van Donselaar EG, Klingl A, Rachel R, Geerts WJC, Jetten MSM, Strous M, van Niftrik L. 2012. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera,” a novel polygon-shaped bacterium. J. Bacteriol. 194:284–291. 10.1128/JB.05816-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rachel R, Meyer C, Klingl A, Gürster S, Heimerl T, Wasserburger N, Burghardt T, Küper U, Bellack A, Schopf S, Wirth R, Huber H, Wanner G. 2010. Analysis of the ultrastructure of Archaea by electron microscopy, p 47–69 In Müller-Reichert T. (ed), Methods in cell biology, vol 96 Elsevier, Philadelphia, PA: [DOI] [PubMed] [Google Scholar]
- 38.Saxton WO. 1996. Semper: distortion compensation, selective averaging, 3-D reconstruction, and transfer function correction in a highly programmable system. J. Struct. Biol. 116:230–236. 10.1006/jsbi.1996.0035 [DOI] [PubMed] [Google Scholar]
- 39.Engelhardt H. 1988. Correlation averaging and 3-D reconstruction of 2d crystalline membranes and macromolecules, p 357–413 In Mayer F. (ed), Methods in microbiology, vol 20 Academic Press, London, United Kingdom [Google Scholar]
- 40.Fuchs KH, Tittmann P, Krusche K, Gross H. 1995. Reconstruction and representation of surface data from two-dimensional crystalline, biological macromolecules. Bioimaging 3:12–24. [DOI] [Google Scholar]
- 41.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 15:680–685 [DOI] [PubMed] [Google Scholar]
- 42.Segrest JP, Jackson RL. 1972. Molecular weight determination of glycoproteins by polyacrylamide gel electrophoresis in sodium dodecyl sulfate, p 54–63 In Ginsburg V. (ed), Methods in enzymology, vol 28 Academic Press, San Diego, CA [Google Scholar]
- 43.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Man M. 2008. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5:459–460. 10.1038/nmeth0608-459 [DOI] [PubMed] [Google Scholar]
- 44.Farhoud MH, Wessels HJCT, Steenbakkers PJM, Mattijssen S, Wevers RA, van Engelen BG, Jetten MSM, Smeitink JA, van den Heuvel LP, Keltjens JT. 2005. Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol. Cell. Proteomics 4:1653–1663. 10.1074/mcp.M500171-MCP200 [DOI] [PubMed] [Google Scholar]
- 45.Wessels HJCT, Vogel RO, van den Heuvel L, Smeitink JA, Rodenburg RJ, Nijtmans LG, Farhoud MF. 2009. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics 9:4221–4228. 10.1002/pmic.200900157 [DOI] [PubMed] [Google Scholar]
- 46.van Donselaar E, Posthuma G, Zeuschner D, Humbel BM, Slot JW. 2007. Immunogold labeling of cryosections from high-pressure frozen cells. Traffic 8:471–485. 10.1111/j.1600-0854.2007.00552.x [DOI] [PubMed] [Google Scholar]
- 47.Guckenberger R. 1985. Surface reliefs derived from heavy-metal-shadowed specimens—Fourier space techniques applied to periodic objects. Ultramicroscopy 16:357–370. 10.1016/0304-3991(85)90104-4 [DOI] [Google Scholar]
- 48.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 49.Ristl R, Steiner K, Zarschler K, Zayni S, Messner P, Schäffer C. 2011. The S-layer glycome—adding to the sugar coat of bacteria. Int. J. Microbiol. 2011:pii=127870. 10.1155/2011/127870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20–W25. 10.1093/nar/gkh435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- 53.Hildebrand A, Remmert M, Biegert A, Södling J. 2009. Fast and accurate automatic structure prediction with HHpred. Proteins 77(Suppl 9):128–132. 10.1002/prot.22499 [DOI] [PubMed] [Google Scholar]
- 54.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. 2010. Protein annotation and modeling servers at University College London. Nucleic Acids Res. 38:W563–W568. 10.1093/nar/gkq427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 56.Sonnhammer ELL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p 175–182 In Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C. (ed), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 57.Pum D, Sára M, Sleytr UB. 1989. Structure, surface charge, and self-assembly of the S-layer lattice from Bacillus coagulans E38-66. J. Bacteriol. 171:5296–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klingl A, Moissl-Eichinger C, Wanner G, Zweck J, Huber H, Thomm M, Rachel R. 2011. Analysis of the surface proteins of Acidithiobacillus ferrooxidans strain SP5/1 and the new, pyrite-oxidizing Acidithiobacillus isolate HV2/2, and their possible involvement in pyrite oxidation. Arch. Microbiol. 193:867–882. 10.1007/s00203-011-0720-y [DOI] [PubMed] [Google Scholar]
- 59.Chauhan JS, Bhat AH, Raghava GPS, Rao A. 2012. GlycoPP: a webserver for prediction of N- and O-glycosites in prokaryotic protein sequences. PLoS One 7:e40155. 10.1371/journal.pone.0040155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, Wacker M, Aebi M. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957–1966. 10.1038/sj.emboj.7601087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Messner P. 2009. Prokaryotic protein glycosylation is rapidly expanding from “curiosity” to “ubiquity.” Chembiochem 10:2151–2154. 10.1002/cbic.200900388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Awram P, Smit J. 2001. Identification of lipopolysaccharide O antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology 147:1451–1460 http://mic.sgmjournals.org/content/147/6/1451.long [DOI] [PubMed] [Google Scholar]
- 63.Dworkin J, Tummuru MK, Blaser MJ. 1995. A lipopolysaccharide-binding domain of the Campylobacter fetus S-layer protein resides within the conserved N terminus of a family of silent and divergent homologs. J. Bacteriol. 177:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engelhardt H. 2007. Are S-layers exoskeletons? The basic function of protein surface layer revisited. J. Struct. Biol. 160:115–124. 10.1016/j.jsb.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 65.Sára M, Sleytr UB. 2000. S-layer proteins. J. Bacteriol. 182:859–868. 10.1128/JB.182.4.859-868.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Z, Kong J, Hu S, Kong W, Lu W, Liu W. 2013. Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl. Microbiol. Biotechnol. 97:1941–1952. 10.1007/s00253-012-4044-x [DOI] [PubMed] [Google Scholar]
- 67.Stackebrandt E, Wehmeyer U, Liesack W. 1986. 16S ribosomal RNA- and cell wall analysis of Gemmata obscuriglobus, a new member of the order Planctomycetales. FEMS Microbiol. Lett. 37:289–292. 10.1111/j.1574-6968.1986.tb01810.x [DOI] [Google Scholar]
- 68.Tarao M, Jezbera J, Hahn MW. 2009. Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria. Appl. Environ. Microbiol. 75:4720–4726. 10.1128/AEM.00251-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chanyi RM, Ward C, Pechey A, Koval SF. 2013. To invade or not to invade: two approaches to a prokaryotic predatory life cycle. Can. J. Microbiol. 59:273–279. 10.1139/cjm-2013-0041 [DOI] [PubMed] [Google Scholar]
- 70.Egelseer E, Schocher I, Sára M, Sleytr UB. 1995. The S-layer from Bacillus stearothermophilus DSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J. Bacteriol. 177:1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engelhardt H. 2007. Mechanism of osmoprotection by archaeal S-layers: a theoretical study. J. Struct. Biol. 160:190–199 [DOI] [PubMed] [Google Scholar]
- 72.Albers S-V, Meyer BH. 2011. The archaeal cell envelope. Nat. Rev. Microbiol. 9:414–426. 10.1038/nrmicro2576 [DOI] [PubMed] [Google Scholar]
- 73.Baldermann C, Lupas A, Lubieniecki J, Engelhardt H. 1998. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J. Bacteriol. 180:3741–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]