Abstract
Enoyl-acyl carrier protein (ACP) reductase catalyzes the last step of the bacterial fatty acid elongation cycle. Enterococcus faecalis is unusual in that it encodes two unrelated enoyl-ACP reductases, FabI and FabK. We recently reported that deletion of the gene encoding FabI results in an unsaturated fatty acid (UFA) auxotroph despite the presence of fabK, a gene encoding a second fully functional enoyl-ACP reductase. By process of elimination, our prior report argued that poor expression was the reason that fabK failed to functionally replace FabI. We now report that FabK is indeed poorly expressed and that the expression defect is at the level of translation rather than transcription. We isolated four spontaneous mutants that allowed growth of the E. faecalis ΔfabI strain on fatty acid-free medium. Each mutational lesion (single base substitution or deletion) extended the fabK ribosome binding site. Inactivation of fabK blocked growth, indicating that the mutations acted only on fabK rather than a downstream gene. The mutations activated fabK translation to levels that supported fatty acid synthesis and hence cell growth. Furthermore, site-directed and random mutagenesis experiments showed that point mutations that resulted in increased complementarity to the 3′ end of the 16S rRNA increased FabK translation to levels sufficient to support growth, whereas mutations that decreased complementarity blocked fabK translation.
INTRODUCTION
The type II or dissociated fatty acid synthesis (FAS II) pathway is responsible for membrane fatty acid synthesis in most bacteria. Fatty acid synthesis is carried out by a series of discrete proteins encoded by separate genes (1, 2) and differs significantly from the type I mammalian and fungal systems, in which multifunctional polypeptides catalyze fatty acid synthesis (3–5). This distinguishing characteristic makes the type II system a prime target for antibacterial agents. Enoyl-acyl carrier protein (ACP) reductases (ENRs) catalyze the last step of the fatty acid elongation cycle, the reduction of trans-2-acyl-ACPs (an enoyl-ACP) to the fully saturated acyl-ACP species. Unlike the other FAS II enzymes, ENRs display an unusual diversity. In bacteria, four ENR isozymes have been identified: the products of the fabI (6, 7), fabL (8), fabV (9, 10), and fabK (11, 12) genes. FabI, FabL, and FabV are diverse members of the short-chain dehydrogenase/reductase (SDR) superfamily, whereas FabK is a TIM barrel flavoprotein (13). Genes encoding homologues of FabI are readily identified in most bacteria. These genes are about 40% identical to Escherichia coli FabI and contain a conserved Tyr-156–(Xaa)6–Lys-163 (E. coli numbering) catalytic dyad (10, 14). For example, E. coli and Francisella tularensis contain a single ENR encoded by fabI that is essential for bacterial viability (7, 15). In contrast, two SDR superfamily ENRs, FabI and FabV, have been characterized in Pseudomonas aeruginosa. Each of the encoding genes can support growth and functionally replace E. coli fabI (10), as also found for the two SDR superfamily ENRs of Bacillus subtilis (8). Thus, it seems that some bacteria have only a single ENR whereas others have two, either of which suffices for growth in the laboratory. However, this is not the case for Enterococcus faecalis, a Gram-positive commensal bacterium that normally inhabits the gastrointestinal tract of humans and other mammals (16, 17). Recently, we characterized two ENRs, FabI and FabK, that play different roles in E. faecalis. FabI is the major ENR of this bacterium, in that deletion of fabI results in unsaturated fatty acid (UFA) auxotrophy, whereas deletion of fabK has no effect on growth, although the traces ENR activity remaining in the ΔfabI strain do alter the cellular fatty acid composition (18). Both FabI and FabK are fully functional in vitro and functionally replace E. coli fabI. Moreover, when expressed from a plasmid construct, FabK restores growth of the ΔfabI strain, indicating that it is poorly expressed in its native chromosomal location.
In this report we demonstrate that the inability of fabK to support growth of E. faecalis is due to inefficient translation. We found that upon plating of E. faecalis fabI null mutants on fatty acid-free medium, colonies arose that no longer required oleate supplementation. The mutations were found to extend the complementarity between the fabK ribosome binding region and the 3′ end of the 16S rRNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. faecalis strains (Table 1) were all derived from FA2-2, a plasmid-free strain (30). Primers used in this study are given in Table 2. Generally, E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 10 g/liter; pH 7.0), whereas E. faecalis strains were maintained at 37°C or 42°C in GM17 medium (31). Antibiotics were added as required at the following concentrations (in mg/liter): sodium ampicillin at 100 for E. coli; kanamycin sulfate at 25 for E. coli; erythromycin at 150 for E. coli and 5 for E. faecalis. Bacterial growth was determined by measuring the optical density at 600 nm (OD600). Oligonucleotide primers were synthesized by Integrated DNA Technologies, and cloned genes were verified by sequencing performed by ACGT, Inc. Gene sequences were amplified using Pfu Turbo DNA polymerase (Stratagene) according to the manufacturer's recommendations. The pCR2.1-TOPO vector (Invitrogen) was applied for PCR cloning, and E. coli strain DH5α was the corresponding recipient host (Table 1). Primers fabKup275 and fabKdo288 were used to amplify fabK fragments from E. faecalis FA2-2 and four spontaneous mutants, and the PCR products were cloned to the pCR2.1-TOPO vector to give plasmids pZL52, pZL53, pZL54, pZL55, and pZL56 for sequencing. These five plasmids were digested with EcoRV and BamHI and inserted into plasmid pTRKL2 to yield plasmids pZL157, pZL158, pZL159, pZL160, and pZL161, respectively, which were linearized and used to transform strain FAZL1. Qiagen provided plasmid isolation and PCR product purification kits.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Δ(argF-lac)169 ϕ80dlacZ58(M15) ΔphoA8 glnV44 deoR481 gyrA96 recA1 endA1 hsdR17 | Lab stock |
| EC1000 | Emr; MC1000 derivative carrying single copy of pWV01repA in glgB | Lab stock |
| E. faecalis strains | ||
| FA2-2 | Rifampin/fusidic acid-resistant mutant derived from plasmid-free strain JH2 | 30 |
| FAZL1 | ΔfabI of E. faecalis FA2-2 | 18 |
| BHK260 | Spontaneous RBS mutant S1 of FAZL1, C-12 to G | This work |
| BHK261 | Spontaneous RBS mutant S2 of FAZL1, C-12 deletion | This work |
| BHK262 | Spontaneous RBS mutant S3 of FAZL1, A-9 to G | This work |
| BHK263 | Spontaneous RBS mutant S4 of FAZL1, T-19 to G | This work |
| FAZL6 | FA2-2 fabK (disruption) | This work |
| FAZL7 | FAZL1 fabK (disruption) | This work |
| FAZL8 | S1 fabK (disruption) | This work |
| FAZL9 | S2 fabK (disruption) | This work |
| FAZL10 | S3 fabK (disruption) | This work |
| FAZL11 | S4 fabK (disruption) | This work |
| Plasmids | ||
| pTRKL2 | Ermr, low-copy-no. E. coli/Lactococcus shuttle vector | 42 |
| pCR2.1 | Ampr Kanr, TOPO TA cloning vector | Invitrogen |
| pZL52 | Ampr Kanr, pCR2.1 carrying E. faecalis FAZL1 fabK plus 275 bp upstream and 288 bp downstream | This work |
| pZL53 | Ampr Kanr, pCR2.1 carrying E. faecalis mutant S1 (strain BHK260) fabK plus 275 bp upstream and 288 bp downstream | This work |
| pZL54 | Ampr Kanr, pCR2.1 carrying E. faecalis mutant S2 (strain BHK261) fabK plus 275 bp upstream and 288 bp downstream | This work |
| pZL55 | Ampr Kanr, pCR2.1 carrying E. faecalis mutant S3 (strain BHK262) fabK plus 275 bp upstream and 288 bp downstream | This work |
| pZL56 | Ampr Kanr, pCR2.1 carrying E. faecalis mutant S4 (strain BHK263) fabK plus 275 bp upstream and 288 bp downstream | This work |
| pZL157 | Ermr, EcoRV and BamHI-digested fabK fragment of pZL52 inserted into same sites of pTRKL2 | This work |
| pZL158 | Ermr, EcoRV- and BamHI-digested fabK fragment of pZL53 inserted into same sites of pTRKL2, C-12 to G | This work |
| pZL159 | Ermr, EcoRV- and BamHI-digested fab fragment of pZL54 inserted into same sites of pTRKL2, C-12 deletion | This work |
| pZL160 | Ermr, EcoRV- and BamHI-digested fabK fragment of pZL55 inserted into same sites of pTRKL2, A-9 to G | This work |
| pZL161 | Ermr, EcoRV- and BamHI-digested fabK fragment of pZL56 inserted into same sites of pTRKL2, T-19 to G | This work |
| pBHK368 | Ermr, fabK A-9T of pZL157 | This work |
| pBHK369 | Ermr, fabK A-9C of pZL157 | This work |
| pBHK370 | Ermr, fabK C-12A of pZL157 | This work |
| pBHK371 | Ermr, fabK C-12T of pZL157 | This work |
| pBHK372 | Ermr, fabK T-19A of pZL157 | This work |
| pBHK373 | Ermr, fabK T-19C of pZL157 | This work |
| pMC1871 | Tetr, promoterless lacZ reporter plasmid | 32 |
| pBHK322 | Ermr, lacZ from pMC1871 cloned into BamHI site of pTRKL2, p(lacZ) | This work |
| pBHK323 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, p(WT) | This work |
| pBHK394 | Ermr, pTRKL2 fabK (−275 to +35)::lacZ | This work |
| pBHK324 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, C-12 to G, p(S1) | This work |
| pBHK325 | Ermr, pTRKL2 P32::fabK (−136 to + 35)::lacZ, C-12 deletion, p(S2) | This work |
| pBHK326 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, A-9 to G, p(S3) | This work |
| pBHK327 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, T-19 to G, p(S4) | This work |
| pBHK334 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, A-9 to T | This work |
| pBHK335 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, A-9 to C | This work |
| pBHK336 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, C-12 to A | This work |
| pBHK337 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, C-12 to T | This work |
| pBHK338 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, T-19 to A | This work |
| pBHK339 | Ermr, pTRKL2 P32::fabK (−136 to +35)::lacZ, T-19 to C | This work |
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequencea |
|---|---|
| fabKup275 | ATTAGACGCGGACAGCTTAG |
| fabKdo288 | TACGACATCAGGCGTTAGTC |
| P32-f | ACTCGCCTGCAGAGATTAATAG TTTTAGCTA |
| fabKfusion-r | ATACTTGTCGACTTGATTCCAAGCAACTCACA |
| P32FabK-f | CTAGGTAGGTAAAAAAATATTACATGAAAATAGGGGGCT |
| P32FabK-r | AGCCCCCTATTTTCATGTAATATTTTTTTACCTACCTAG |
| fabKup275-PstI | AATATCTGCAGATTAGACGCGGACAGCTTAG |
| fabK-random-f | TAGTGGATCCATTAGACGCGGACAGCTTAGA |
| fabK-random-r | ATATCTGCAGTTCACTTAGCCCCAACGCTGAT |
| Ef16S-f | CATCATGCCCCTTATGACCTGG |
| Ef16S-r | CCTGCAATCCGAACTGAGAGA |
| lacZ-f | ATGAACGGTCTGGTCTTTGC |
| lacZ-r | ATCGCTATGACGGAACAGGT |
| fabK-RT-f | GGCAATCCAGCCAAATACAT |
| fabK-RT-r | ATGTCCACCAGCTTCCATTC |
| c-12a-f | GAAGTGTACTTATCTTAGAACTAAAGGAAGTATAAAATCAATGAATCAACAGTTATG |
| c-12a-r | CATAACTGTTGATTCATTGATTTTATACTTCCTTTAGTTCTAAGATAAGTACACTTC |
| c-12t-f | GAAGTGTACTTATCTTAGAACTAAAGGATGTATAAAATCAATGAATCAACAGTTATG |
| c-12t-r | CATAACTGTTGATTCATTGATTTTATACATCCTTTAGTTCTAAGATAAGTACACTTC |
| a-9c-f | GTACTTATCTTAGAACTAAAGGACGTCTAAAATCAATGAATCAACAGTTATGT |
| a-9c-r | ACATAACTGTTGATTCATTGATTTTAGACGTCCTTTAGTTCTAAGATAAGTAC |
| a-9t-f | GTACTTATCTTAGAACTAAAGGACGTTTAAAATCAATGAATCAACAGTTATGT |
| a-9t-r | ACATAACTGTTGATTCATTGATTTTAAACGTCCTTTAGTTCTAAGATAAGTAC |
| t-19a-f | CTTGAATGAAGTGTACTTATCTTAGAACAAAAGGACGTATAAAATCAATGAATC |
| t-19a-r | GATTCATTGATTTTATACGTCCTTTTGTTCTAAGATAAGTACACTTCATTCAAG |
| t-19c-f | CTTGAATGAAGTGTACTTATCTTAGAACCAAAGGACGTATAAAATCAATGAATC |
| t-19c-f | GATTCATTGATTTTATACGTCCTTTGGTTCTAAGATAAGTACACTTCATTCAAG |
| fabK inside For | ATGCGAGCTCTGAAGAACAGGTTCCTGTCG |
| fabK inside Rev | ATCGCTGCAGCTGCATCTAAATCACGTGCC |
| pBVGh For (P1) | GTACCTACGTAGGATCGATC |
| pBVGh Rev (P2) | GCTTCCAAGGAGCTAAAGAG |
| fabK check Up (P3) | ATTAGACGCGGACAGCTTAG |
| fabK check Down (P4) | TACGACATCAGGCGTTAGTC |
The underlined italic sequences are the introduced restriction sites.
Construction of translational fusions.
The BamHI-digested DNA fragment containing a promoterless lacZ from the plasmid pMC1871 (32) was ligated to the shuttle plasmid pTRKL2 cut with the same enzyme to give the plasmid pBHK322. The insertion direction was confirmed by DNA sequencing. Overlapping PCR was used to generate the fusion of the promoter P32 from Lactococcus lactis to fabK (−136 to +35 relative to the fabK ATG initiation codon). The promoter P32 from L. lactis and fabK (−136 to +35) from E. faecalis FA2-2 were first amplified from the genomic DNAs by using primer sets P32-f/P32fabKfusion-r and P32fabKfusion-f/fabKfusion-r, respectively. The first-step PCR fragments were purified from agarose gels and mixed to serve as templates to amplify chimera-P32fabK with primer set P32-f/fabKfusion-r. The PCR products after digestion with PstI and SalI were ligated into the plasmid pBHK322 to produce pBHK323. For the construction of the other four translational fusion plasmids, each containing a spontaneous mutation in the ribosome binding site (RBS) region, fabK (−136 to +35) was first amplified from the genomic DNAs of spontaneous mutants BHK260, BHK261, BHK262, and BHK263 and used as the first-step PCR template. The following procedure was the same for the construction of the plasmid pBHK323, and finally the fusion plasmids pBHK324, pBHK325, pBHK326, and pBHK327 were obtained.
Construction of E. faecalis strains having disrupted fabK genes.
E. faecalis strains having disruptions of fabK were constructed using the method described previously (33). A centrally located segment of fabK was obtained by amplification from the genomic DNA of E. faecalis FA2-2 strain with Pfu DNA polymerase and primers (Table 2) containing designed restriction sites (a SacI site in the forward primer and a PstI site in the reverse primer). This fragment was then used to construct plasmid pZL235 by insertion of the PstI/SacI-digested PCR fragment into plasmid pBVGh cut with the same enzymes. The disruption plasmid was transformed into competent cells of E. faecalis wild-type strain FA2-2. After recovery in SGM17 medium at 30°C, the transformed cells were plated on SGM17 agar containing 5 μg/ml of erythromycin plus 100 μg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and incubated for 24 h at 30°C. The resulting blue colonies were cultured overnight at 30°C in GM17 liquid medium containing erythromycin, and then the cultures were plated on GM17 agar medium containing 5 μg/ml of erythromycin and 100 μg/ml of X-Gal and incubated for 24 h at 42°C. To construct fabK disruptions in strain carrying the ΔfabI mutation, 0.1% Tween 40, 0.1% Tween 80, and 0.01% oleate were added to the growth medium. The single-crossover homologous recombinant colonies were screened for disruption of the fabK gene by PCR analysis with pBVGh primers (P1 and P2) and fabK check primers (P3 and P4) and verified by sequencing.
Transformation of E. faecalis.
Electrocompetent cells were prepared as previously described (31) with minor changes. A single colony of E. faecalis was grown overnight in GM17 medium and then diluted 1:100 into 100 ml of SGM17 medium (GM17 with 0.5 M sucrose and 8% glycine) in a 500-ml flask with vigorous shaking at 37°C. When the OD600 reached 0.4 to 0.6, the cultures were chilled on ice for 30 min and then collected by centrifugation at 3,600 × g for 12 min at 4°C. The harvested pellets in each tube were suspended in 100 ml of ice-cold electroporation buffer (0.5 M sucrose and 10% glycerol) and then centrifuged at 3,800 × g for 16 min at 4°C. Following three rounds of washing the cells with electroporation buffer, the bacterial pellets were suspended in 1 ml of ice-cold electroporation buffer and divided into small aliquots, which were frozen on dry ice and kept at −80°C until use. For the preparation of strain FAZL1 (the ΔfabI mutant strain of E. faecalis FA2-2) electrocompetent cells, 0.1% Tween 40, 0.1% Tween 80, and 0.01% oleate were added to the growth medium.
Prior to electroporation, the frozen cell suspensions were thawed on ice, mixed with plasmid DNA (50 to 100 ng for delivery of plasmids), and then placed on ice for no more than 30 min prior to transfer into a chilled 2-cm gap cuvette (Bio-Rad). Electroporation parameters for the Gene Pulser apparatus (Bio-Rad) were 25-μF capacitance, 200-Ω resistance, and a 2.5-kV electrical pulse to give a field strength of 12.5 kV cm−1, with an exponential decay constant of approximately 4 to 5 ms. After immediate placement on ice for 5 min, the pulsed cells were quickly diluted with 1 ml of SGM17MC medium (31), grown at 37°C for 2 h, and plated on SR agar plates containing the appropriate antibiotics. The plates were incubated at 37°C for 18 to 36 h. Transformants containing plasmids were verified by plasmid isolation.
Acyl-ACP preparations.
The ACP thioester of trans-2-decenoic acid was synthesized as described previously (34). Briefly, a typical reaction mixture consisted of 25 μM holo-ACP, 200 μM fatty acid, and 170 nM Vibrio harveyi AasS in a buffer containing 100 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 1 mM tris(2-carboxyethyl) phosphine HCl, and 10 mM ATP in a reaction volume of 1 ml. The reaction mixtures were incubated at 37°C for 4 h, and reactions were stopped by addition of 2 volumes of acetone; the proteins were allowed to precipitate at −20°C overnight. The precipitates were pelleted at 20,000 × g for 30 min at 4°C and then washed twice with 3 volumes of acetone. The pellets were air dried and resuspended in 20 mM Tris-HCl (pH 7.4) containing 1 mM Tris(2-carboxyethyl) phosphine HCl. The final samples were concentrated with an Amicon ultracentrifugation filter device from Millipore (5,000 molecular weight cutoff). Acyl-ACP synthesis was verified by electrophoresis on 18% gels containing 2.5 M urea (2, 35) and by electrospray mass spectrometry. Note that the ACP preparations retained the N-terminal methionine.
NADH oxidation assay.
ENR activity was monitored spectrophotometrically based on the decrease in the absorbance at 340 nm and using an NADH extinction coefficient of 6,220 M−1 (9). Cultures grown to mid-log phase in 100 ml of GM17 medium were collected by centrifugation, washed twice with 0.9% KCl, repelleted, and treated for 30 min with lysozyme (15 mg/g, wet weight) in an osmotically protected medium consisting of 0.4 M NaCl, 0.05 M phosphate buffer (pH 7.2), and 0.5 M sucrose. The cyclopropane fatty acid cis-9,10-methylenehexadecanic acid (0.005%) was added to GM17 medium to support the growth of the ΔfabI strain FAZL1. After lysozyme treatment, the cells were resuspended in 1 ml of 100 mM Tris-HCl (pH 8.0) containing 400 mM NaCl and lysed by passaging three times through a French pressure cell press at 11,000 lb/in2. The crude extract was collected on ice and cleared by centrifugation, and the protein concentration was determined by the Bio-Rad Bradford assay, using a standard curve of bovine serum albumin (36). Crude extract protein preparations (1 to 10 μg) were added directly to UV-transparent microcuvettes containing the assay reaction mixture (volume, 100 μl) consisting of 0.1 M sodium phosphate buffer (pH 7.0), 100 μM trans-2-decenoyl-ACP, 150 μM NADH, and 0.1 M LiCl. Kinetic constants were determined using GraphPad Prism software, version 4.
Radioactive labeling, phospholipid extraction, and fatty acid analysis.
Fatty acid biosynthesis was analyzed based on [1-14C]acetate incorporation. E. faecalis strains were cultured in GM17 medium (containing 0.005% cis-9,10-methylenehexadecanic acid for growth of the ΔfabI mutant strain FAZL1) at 37°C overnight. Cells were washed twice with water and resuspended in the same volume of GM17 medium, and 0.1 ml of this suspension was added to 5 ml of GM17 containing 5 μCi of sodium [1-14C]acetate. These cultures were grown at 37°C to mid-log phase, and the cells were collected by centrifugation. The phospholipids were extracted by using the method of Bligh and Dyer (37), and labeled fatty acid methyl esters were prepared and analyzed by using thin-layer chromatography and autoradiography as described previously (38).
β-Galactosidase assays.
The cultures were grown overnight, then diluted 1:100 into fresh medium having the same composition, and grown to the mid-log phase before β-galactosidase assays were performed. Mid-log-phase cultures were collected by centrifugation, washed twice with Z buffer, and assayed for β-galactosidase activity after lysis with sodium dodecyl sulfate-chloroform (39). The data were obtained in triplicate in more than three independent experiments.
RNA isolation and reverse transcription-PCR.
Total RNA preparations were isolated from the mid-log-phase cells of E. faecalis FA2-2 and its derivatives grown in GM17 medium by using the RNeasy bacterial RNA isolation kit (Qiagen). The contaminating DNA was digested using RNase-free DNase set (Qiagen) according to the manufacturer's protocol. The isolated RNA samples were analyzed by agarose gel electrophoresis to assess the quality of the rRNA preparations. To further rule out the possibility of trace DNA contamination of the RNA, a general PCR-based detection was also conducted by using total RNA as the template with primers Ef16S-f plus Ef16S-r (Table 2). The concentration and purity of RNA were determined by using a NanoDrop 2000C spectrophotometer (Thermo Scientific). RNA preparations were stored at −80°C until use. Synthesis of cDNA was performed in 20 μl of a mixture and using the Omniscript reverse transcription (RT) kit (Qiagen) according to the manufacturer's protocol. The resulting cDNA (0.5 μl) served as the template for PCR amplification of the fatty acid synthesis genes (fab genes), using specific primers (Table 2) and an Eppendorf thermal cycler.
Real-time quantitative RT-PCR.
To test if the mRNA levels of fabK were altered in the spontaneous mutants, real-time quantitative PCR (qPCR) assays were conducted by using the SYBR green dye method as previously reported (40). The qPCR mixture (20 μl) contained the following components: 10 μl of iQ SYBR green supermix, 1 μl each of the primers, 1 μl of 50-fold-diluted cDNA sample, and 8 μl of sterile water. Data analyses were done in triplicate on a Mastercycler ep realplex instrument (Eppendorf), using a program of a denaturing cycle at 95°C for 2 min, 45 cycles comprising 94°C for 20 s, 53°C for 20 s, and 72°C for 20 s, and a final step in which the temperature was elevated on a gradient from 60°C to 90°C for dissociating double-stranded DNA products. The mRNA levels relative to those of the wild type were calculated using the method of Livak and Schmittgen (41) with the 16S rRNA as an internal control gene, and water functioned as a blank control to monitor cross-contamination of the cDNA samples.
Site-directed mutagenesis of fabK.
Plasmids pBHK368, pBHK369, pBHK370, pBHK371, pBHK372, and pBHK373, each carrying a single mutation within the RBS sequence, were obtained by using the QuikChange site-directed mutagenesis kit (Stratagene) with pZL157 as the PCR template, following the manufacturer's instructions. The primers used in PCRs and in mutagenesis are listed in Table 2. The constructed plasmids were transformed into E. coli DH5α by CaCl2 treatment. Plasmids pBHK334, pBHK335, pBHK336, pBHK337, pBHK338, and pBHK339, each carrying a single mutation within the RBS sequence, were obtained by using the QuikChange site-directed mutagenesis kit (Stratagene) with pBHK323 as the PCR template, following the manufacturer's instructions. All constructs were verified by DNA sequencing.
Random mutagenesis of fabK and isolation of mutant plasmids for complementation.
Random mutagenesis of the fabK gene plus 275 bp upstream was performed with the GeneMorph II random mutagenesis kit using primers fabK-random-f and fabK-random-r (Table 2) and plasmid pZL157 as the template at a concentration of 500 ng in a total reaction volume of 50 μl. The mutagenesis condition designed to give a low mutation frequency was used, because we expected that single mutations would restore growth of E. faecalis fabI mutants in the absence of oleate. The PCR products of the mutagenesis reactions were then digested with BamHI and PstI and inserted into vector pTRKL2 digested with the same enzymes. The ligation products were then transformed into E. coli DH5α with selection on brain heart infusion plates for resistance to erythromycin. The transformants of about 15,000 clones were collected, and the mixture of mutant plasmids was purified using the Qiagen Spin miniprep kit. The plasmids were electroporated into the E. faecalis fabI mutant strain FAZL1 as described above. The 10,000 clones obtained were screened for growth on GM17 medium without fatty acid supplementation. The plasmids of the growth-complemented strains were then isolated and again transformed to the strain FAZL1 to confirm the growth complementation. Finally, the confirmed complementing plasmids were sequenced to identify the mutations.
RESULTS
Spontaneously arising mutants that restore fatty acid synthesis to the ΔfabI strain contain point mutations within the fabK ribosome binding site region.
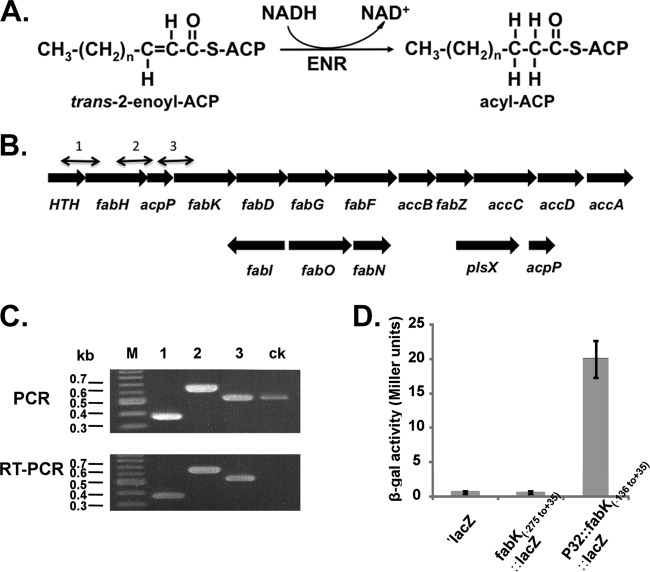
The two E. faecalis proteins catalyzing the ENR reaction (Fig. 1A) are located in different genome neighborhoods. The fabK gene is found within a putative operon responsible for saturated fatty acid synthesis, whereas fabI is located in a smaller cluster with two genes involved in unsaturated fatty acid synthesis (19) (Fig. 1B). The ΔfabI strain FAZL1 is severely defective in fatty acid synthesis and is unable to grow on GM17 medium unless the medium is supplemented with an unsaturated fatty acid, such as oleic acid (18). A culture of strain FAZL1 was plated on GM17 medium lacking oleate supplementation at 37°C. Colonies appeared at a frequency of approximately 10−9, and purified isolates were shown to retain the ΔfabI mutation. Four isolates, strains BHK260, BHK261, BHK262, and BHK263, were chosen for further study.
FIG 1.
The ENR reaction, the fabK and fabI genome neighborhoods, and transcriptional analyses of the operon containing fabK. (A) The enoyl-acyl-ACP reductase reaction. (B) Genetic organization of the E. faecalis fatty acid biosynthetic genes (fab). The numbered short lines (1, 2, and 3) represent the specific PCR amplicons observed in the PCR and RT-PCR assays shown in panel C. The second acpP gene (EF_3111 in strain V583) located downstream of plsX is conserved in all extant E. faecalis genome sequences. (C) PCR and RT-PCR analyses of the fab operon. The results for the four genes at the 5′ end of the operon are shown. The helix-turn-helix-encoding gene probably encodes a homologue of the FabT transcription factor found in other Firmicutes bacteria (43, 44). The primer numbering system is the same as that for panel B. Both PCR products obtained from genomic DNA and those obtained by RT-PCR were separated by electrophoresis on a 1.5% agarose gel. The ck designation denotes two neighboring genes transcribed from the opposite strand that were included as controls. (D) β-Galactosidase activities of fabK::lacZ translational fusions. The wild-type strain FA2-2 carrying each of the fusion plasmids was grown in GM17 medium to mid-log phase, and β-galactosidase activities were measured in more than three independent experiments. The error bars indicate standard deviations. The plasmids carried a promoterless lacZ (′lacZ), the −275 to +35 fragment, which includes the first 35 bp of the fabK coding sequence fused to lacZ (fabK-lacZ), or the −136 to +35 fragment driven by the P32 promoter. The fusion plasmids were pBHK322, pBHK394, and pBHK323, respectively.
Since overproduction of FabK alleviates the growth defect of a fabI null mutant strain (18), the most likely explanation for the spontaneous mutations was increased FabK expression. We first investigated how fabK is transcribed. The cluster of fatty acid synthesis genes in which fabK is located appears to be an operon, although there is an unusually large intergenic region (136 bp) between the acpP and fabK genes. To test if acpP and fabK are cotranscribed together with the upstream genes, we performed RT-PCR analyses using primers bracketing each intergenic region (Fig. 1C). The same patterns were seen in both RT-PCR and PCR analyses, indicating that at least these four genes are cotranscribed. ACP is generally produced at much higher levels than the Fab proteins, because it acts as a coenzyme rather than an enzyme. Hence, acpP would be expected to be transcribed at a higher level than the other genes, which we did not observe. However, E. faecalis has a second acpP gene (Fig. 1) that could be expressed at high levels. We are currently studying the relative contributions of the two acpP genes.
We also investigated if the acpP-fabK intergenic region contained a promoter by using the intergenic sequence to drive expression of a fabK-lacZ translational fusion carried on a plasmid (Fig. 1D). The intergenic sequence gave no increase in β-galactosidase activity over that seen with the reporter construct lacking a promoter, whereas a construct driven by the Lactococcus lactis P32 promoter gave high levels of β-galactosidase activity.
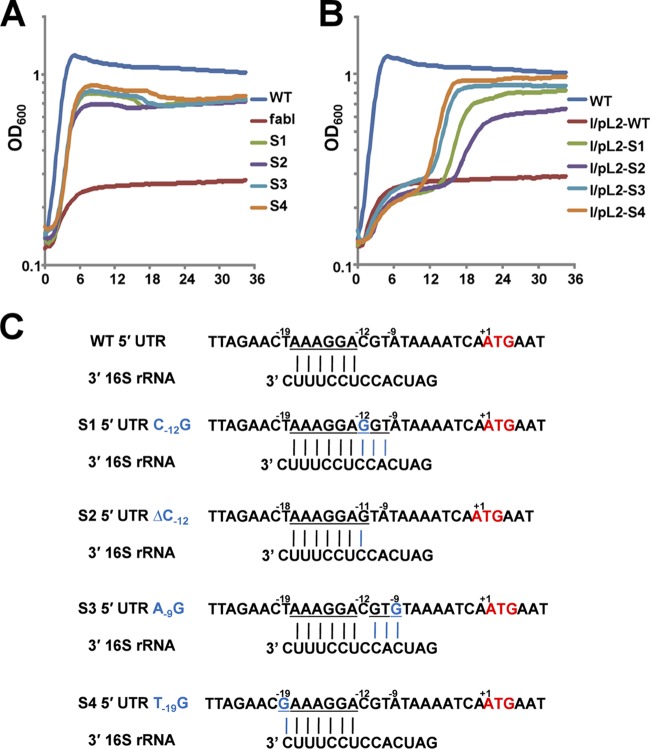
Given the results of these background experiments, we asked if growth of these four spontaneous mutants (Fig. 2A) was mediated by FabK. This was first tested by insertion of the fabK gene (plus about 300 bp of upstream and downstream sequence) from each of the spontaneous mutants into the low-copy-number shuttle vector pTRKL2. All four plasmids allowed the ΔfabI mutant strain to grow on GM17 medium, whereas the plasmid carrying the wild-type genomic segment failed to support growth (Fig. 2B). We then sequenced the fabK regions of each of the spontaneous mutants. All four mutant strains were found to carry a unique point mutation within the fabK ribosome binding region, each of which would extend base pairing with the 3′ end of the 16S rRNA (Fig. 2C). The mutations were C-12G (strain BHK260), a ΔC12 single base deletion (strain BHK261), A-9G (strain BHK262), and T-19G (strain BHK263) (Fig. 2C). These data raised the question of whether our wild-type strain FA2-2 is representative of E. faecalis. It seemed possible that our strain had acquired mutations in this region that resulted in decreased fabK translation and that these were overcome or reversed by the spontaneous mutations. However, the sequence of the strain FA2-2 genomic segment located between the end of the acpP coding sequence and the beginning of the fabK sequence is either completely or almost completely conserved in the 295 complete and draft E. faecalis genome sequences currently available. Those few sequences that do not completely match the strain FA2-2 sequence have only a single base change located 63 bp upstream of the fabK initiation codon and hence are well removed from the ribosome binding region. Moreover, the strain FA2-2 fabK coding sequence is at least 99% conserved at both the nucleotide and protein levels in the 295 genome sequences.
FIG 2.
Growth and sequences of the spontaneously arising mutant strains and complementation of the ΔfabI strain by plasmid-borne fragments that included the fabK mutant or wild-type alleles. (A) Growth of the four spontaneously arising mutant ΔfabI strains (S1 to S4) on GM17 medium in the absence of oleate supplementation. (B) Complementation of the ΔfabI strain FAZL1 by plasmids (Table 1) carrying a mutant fabK allele plus about 300 bp of upstream and downstream sequence. Each growth assay was carried out in triplicate in GM17 medium at 37°C, and the averages are shown. Strain FAZL1 in panel A and the strain carrying wild-type fabK in panel B were used as negative controls. Panels A and B have the same color coding for the mutant fabK alleles. (C) Alignments of 5′ untranslated regions of the wild-type gene fabK or of the four spontaneously arising mutant strains with the 3′ end of the E. faecalis 16S rRNA. The fabK genes of wild-type strain FAZL1 and the four spontaneous mutants plus about 300 bp upstream and downstream were sequenced. Each point mutation expanded the fabK ribosome binding site. The underlined letters indicate bases capable of Watson-Crick base pairing with the 16S rRNA. +1 indicates the A of the initiation ATG codon.
Inactivation of fabK eliminates growth of the mutant strains.
Although it seemed likely that growth of the spontaneously arising mutants of the ΔfabI strain was due to increased fabK expression per se, it was possible that the effects of the mutations were indirect. If the entire cluster of genes in which fabK is located is indeed an operon, then binding of ribosomes to the fabK sequence of the polycistronic mRNA might protect the mRNA, resulting in increased expression of the downstream genes. We tested this possibility by nonpolar disruption of the fabK coding sequence. If increased FabK ENR activity were responsible for growth, then fabK disruption would block growth in the absence of oleate, whereas if increased downstream expression were the important factor, growth should proceed. A suicide plasmid carrying an internal fabK fragment was inserted into fabK by single-crossover homologous recombination events, resulting in two disrupted copies of fabK bracketing the inserted plasmid (Fig. 3A and B). Upon insertion of the disruption plasmid, all four spontaneous mutants lost the ability to grow without oleate (Fig. 3C), whereas the wild-type strain grew well (due to the presence of FabI), thereby demonstrating the lack of a polar effect on expression of the downstream fatty acid synthetic genes.
FIG 3.
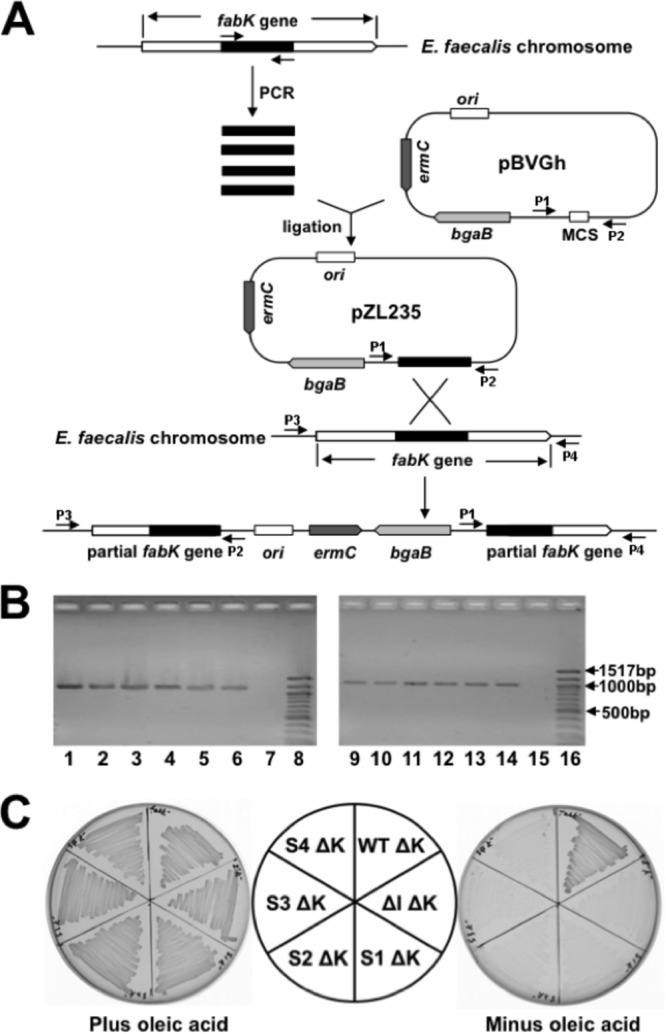
Construction, characterization, and growth of fabK disruption mutant strains. (A) Strategy for isolation of erythromycin-marked chromosomal E. faecalis fabK disruption mutant strains via plasmid insertion. P1, P2, P3, and P4 were the PCR primers used for characterization of the fabK disruption mutant strains. (B) Characterization of the fabK disruption strains by PCR analyses. Lanes 1 to 6, PCR products amplified from lane 1, the fabK disruption strain (FAZL6); lane 2, the fabK disruption ΔfabI double mutant strain (FAZL7); lanes 3 to 6, the S1 to S4 fabK strains in which fabK was disrupted (strains FAZL8 to -11, respectively). Genomic DNA preparations were used as templates with primer sets pBVGh For (P1) and fabK check Down (P4). Lanes 9 to 14 are the PCR products amplified using the same template DNAs as for lanes 1 to 6 but with primers pBVGh Rev (P2) and fabK check Up (P3). Lanes 7 and 15 are the PCR products formed from the genomic DNA of the wild-type strain FA2-2. Lanes 8 and 16 are molecular size standards. (C) Growth of E. faecalis mutant strains on GM17 medium plus 5 μg/ml of erythromycin at 42°C with (left) or without (right) oleic acid. Cell growth proceeded at 42°C for 36 h. ΔK, the disrupted fabK; WT, E. faecalis wild-type strain; ΔI, the E. faecalis ΔfabI gene deletion strain; S1, S2, S3, and S4, the spontaneously arising suppressors the of E. faecalis fabI deletion. The normal growth in the absence of fatty acid supplementation of the disrupted fabK derivative of the wild-type strain indicated that the disruption cassette is not polar on the downstream fatty acid synthesis genes.
Strains carrying a chromosomal or plasmid-borne mutant allele contain increased ENR activity.
Our previous results showed that E. faecalis FabI catalyzes an essential step in fatty acid biosynthesis, and deletion of fabI results in almost complete loss of ENR activity (18). To directly test if growth of the spontaneous mutant strains was due to restoration of ENR activity, the ENR activities in cells from cultures grown on GM17 medium were assayed. The four spontaneous mutant strains had ENR activities that were 26 to 38% that of the FabI-containing wild-type strain FA2-2, whereas the ENR activities of the derivatives of the ΔfabI strain carrying the plasmid-borne mutant segments were restored to 9% to 16% that of the wild-type strain (Fig. 4A). We also used [1-14C]acetate labeling and argentation thin-layer chromatographic analysis of the de novo-synthesized fatty acids to demonstrate ENR activity in vivo (Fig. 4B), and we found that the spontaneously arising mutants synthesized levels of saturated and unsaturated fatty acids comparable to those of the wild-type strain. However, consistent with their lower ENR activities and poorer growth, the strains carrying the plasmid-borne mutated segments incorporated less [14C]acetate into fatty acids than the wild-type strain (Fig. 4B), and no increase in ENR activity or [14C]acetate incorporation was seen when the plasmid carried the wild-type genome segment. Hence, the increased ENR activity due to mutations within the fabK RBS sequence was sufficient to restore growth and fatty acid synthesis to the ΔfabI strain.
FIG 4.
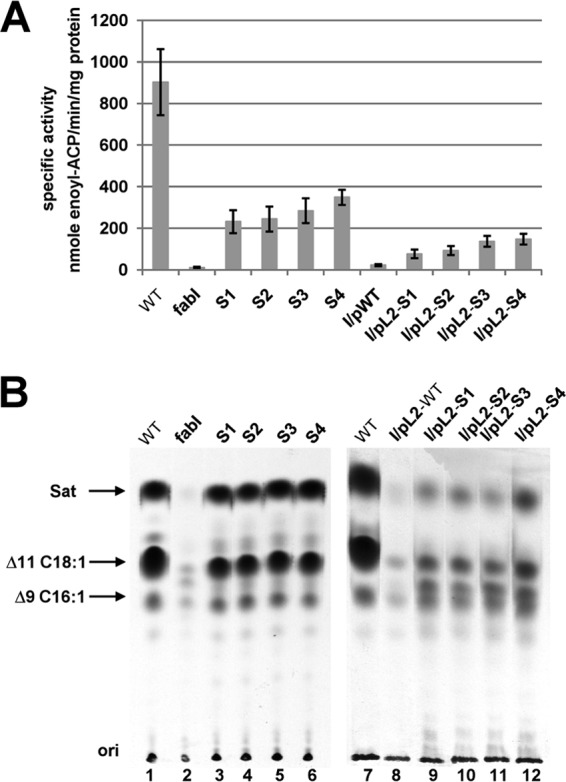
The spontaneous mutations increased expression of the FabK ENR. (A) ENR activities of the wild-type FA2-2, spontaneous mutants, and the complementing plasmid-bearing strains. The 100-μl ENR reaction mixtures contained 0.1 M sodium phosphate buffer (pH 7.0), 0.15 mM NADH, and 2 μg of cell extract protein, and NADH-dependent ENR activity was assayed. The reactions were initiated by addition of 100 μM trans-2-decenoyl-ACP. The data are from three independent experiments and are expressed as means ± standard deviations. (B) Incorporation of [1-14C]acetate into the membrane phospholipids of the wild-type strain FA2-2, the spontaneous mutant strains, and the ΔfabI strain FAZL1 transformed with the wild-type fabK gene or the mutant alleles. The strains used here are the same with those in Fig. 2. The fatty acid methyl esters were prepared and analyzed by argentation thin-layer chromatography as described in Materials and Methods. Designations: Sat, saturated fatty acids; Δ9C16:1, palmitoleic (cis-9-hexadecenoic) acid; Δ11C18:1, cis-vaccenic (cis-11-octadecenoic) acid.
The spontaneous mutations increase fabK translation.
Although the location of the nucleotide changes in the mutant strains argued in favor of increased translation rather than increased transcription, we measured the mRNA levels of fabK in the mutant and wild-type strains. No significant change in mRNA levels was observed (Fig. 5A), and thus the primary effect of the mutations was indeed at the translational level. We constructed fabK-lacZ translational fusions that contained the acpP-fabK intergenic 136 bp plus the first 35 bp of the fabK coding sequence under the control of Lactococcus lactis promoter P32 and then measured fabK translation based on the β-galactosidase activity. The β-galactosidase levels in cultures expressing fusions with the spontaneous fabK mutant sequences were 4- to 7-fold higher than that of the fusion plasmid carrying the wild-type genomic segment (Fig. 5B), whereas the lacZ mRNA levels were essentially the same for the wild-type and mutant constructs (Fig. 5A). Hence, the spontaneous mutations overcome the normally weak translational initiation of fabK. Note that the 136-bp intergenic DNA fragment chosen for construction of the translational fusions was based on two considerations. The observed three intergenic amplicons from RT-PCR analysis (Fig. 1C) showed that at least the first four genes of the fab cluster comprise a transcription unit and hence constitute an operon. We also performed RNA ligase-mediated RACE, an improved version of 5′-RACE for amplification of the 5′ end of the cDNA, and we failed to amplify a PCR product (data not shown). Therefore, as in Fig. 1D, we were unable to detect a promoter between acpP and fabK.
FIG 5.
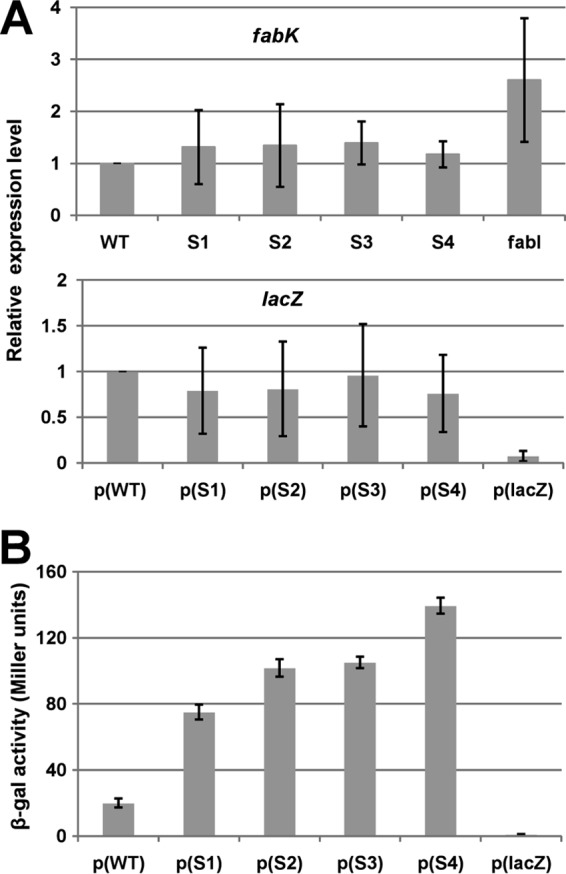
Effects of the spontaneous mutations on fabK transcription and translation. (A) qRT-PCR analyses of the effects on fabK and lacZ transcription. Transcription of fabK was analyzed in the wild-type strain and the spontaneous mutant strains, whereas lacZ transcription was analyzed for the wild-type strains carrying the translational fusion plasmid. Cells were grown in GM17 medium to mid-log phase, and RNA was isolated as described in Materials and Methods. The qRT-PCR data were from no less than four independent tests and are expressed as means ± standard deviations. The p(lacZ) designation denotes the promoterless ′lacZ vector. (B) Effects of the spontaneous mutations on β-galactosidase expression from a plasmid carrying a P32::fabK::lacZ translational fusion. The wild-type strain FA2-2 carrying the fusion plasmids was grown in GM17 medium to mid-log phase, and β-galactosidase activities were measured from more than three independent experiments. The error bars indicate standard deviations. p(WT), p(S1), p(S2), p(S3), p(S4), and p(lacZ) indicate fusion plasmids pBHK323, pBHK324, pBHK325, pBHK326, pBHK327, and pBHK322 carrying the −136 to +35 fragments of wild-type fabK, one of the four spontaneous fabK mutants, or the empty ′lacZ vector, respectively.
Effects of site-directed and random mutagenesis of the RBS sequence on fabK expression and cell growth.
The increased complementarity of the fabK RBS to the 3′ end of the 16S rRNA (Fig. 2C) seemed very likely to be responsible for the increased ENR activity of the spontaneous mutants. If so, then substitution of the mutant base with another base should result in loss of growth of the fabI null strain in the absence of oleic acid. Therefore, the mutated base was replaced with other nucleotides and, as expected, this resulted in loss of the abilities of plasmids carrying these constructs to complement growth of the ΔfabI strain (Fig. 6A) and to increase fabK translation (Fig. 6B).
FIG 6.
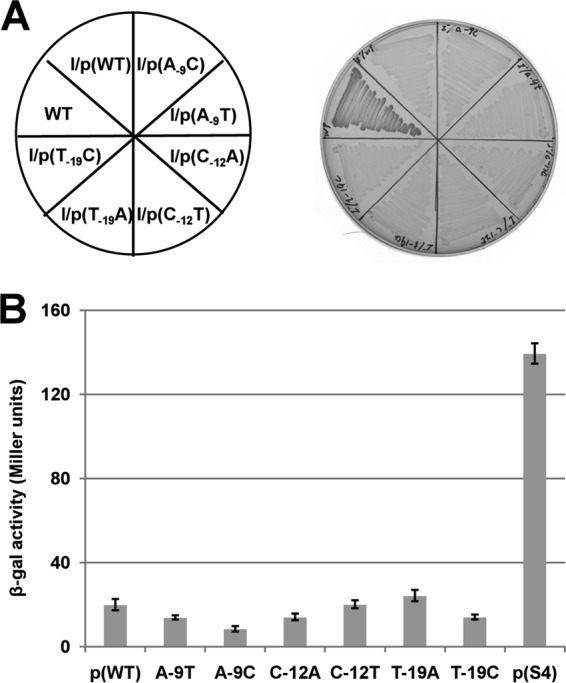
16S rRNA-mRNA complementarity is required for growth of the spontaneous mutants. (A) Growth was tested on GM17 plates incubated for 36 h at 37°C. In three of the spontaneous mutants, the unique nucleotide of the mutation was changed to two other nucleotides. Each site-directed fabK gene was cloned into the vector pTRKL2 and then transformed to the ΔfabI strain, FAZL1. (B) Effects of the nucleotide changes on β-galactosidase expression from a plasmid carrying a P32::fabK::lacZ translational fusion. The wild-type strain FA2-2 carrying the fusion plasmids was grown in GM17 medium to mid-log phase, and β-galactosidase activities were measured from more than three independent experiments. The error bars indicate standard deviations. The designations p(WT), A-9T, A-9C, C-12A, C-12T, T-19A, T-19C, and p(S4) indicate the fusion plasmids pBHK323, pBHK334, pBHK335, pBHK336, pBHK337, pBHK338, pBHK339, and pBHK327, respectively. The p(WT) and p(S4) plasmids were used as negative and positive controls, respectively.
We also randomly mutagenized the wild-type fabK coding sequence plus the upstream 275 bp by using error-prone PCR and isolated plasmids that suppressed the fabI phenotype. A total of 13 colonies were isolated, and their plasmids were sequenced. Two isolates had the A-9G mutation found in the spontaneous mutant BHK262. Another 11 isolates had the A-9G or C-12G mutations, found in the spontaneously isolated strains, and also contained phenotypically silent coding sequence mutations that were generally conservative residue substitutions (Table 3). The reisolation of the mutations found in the spontaneously arising strains confirmed their abilities to increase the efficiency of mRNA-ribosome pairing.
TABLE 3.
Results of error-prone PCR mutagenesis of fabK and selection for complementation of the ΔfabI strain FAZL1
| Isolate | FabK mutation(s) |
|---|---|
| Ran2a | A-9G |
| Ran9 | A-9G, L9 (CUU to CUC) |
| Ran16 | A-9G, D78G, L154I, I163V |
| Ran26 | A-9G, I285V |
| Ran12 | C-12G, A185V, I220V, I261V |
| Ran13a | C-12G, V84I, K124R, L148 (UUA to CUA) |
| Ran15 | C-12G, M281L |
| Ran17 | C-12G, K250 Stop (AAA to UAA) |
Note that in Ran2 and Ran13, two synonymous mutations occurred for residues L9 and L148, respectively. In Ran17, a nonsense mutation truncated the protein at residue K250, suggesting that the last 69 FabK residues are not essential for ENR activity.
DISCUSSION
Selection for growth of the E. faecalis ΔfabI strain in the absence of fatty acid supplementation gave four spontaneous mutant strains. The mutations in these strains were four different single-base changes (three single-base substitutions and one single-base deletion) in the ribosome binding region of fabK, which encodes a fully functional but physiologically cryptic ENR. The discrete nature of our mutations is strikingly different from previously reported spontaneous translation-activating mutants, in which large DNA fragments carrying RBS sequences have been inserted or deleted (20–22).Translation begins with the association between a 30S ribosomal subunit and the mRNA. Translational efficiency is in part determined by the RBS interaction, i.e., the base pairing of the 3′ end of 16S rRNA to a stretch of complementary nucleotides in the messenger, located upstream of the initiation codon (23, 24). Indeed, as a result of the C-12 single-base change of mutant strain BHK260, the substitution with G extended the pairing potential with the 3′ end of the 16S rRNA by 3 bp (Fig. 2C) and increased fabK expression (Fig. 5B). Moreover, site-directed mutagenesis, in which A or T was substituted for C-12, resulted in decreased fabK translation (Fig. 6B). The other three single mutations appeared to have similar effects on base pairing and translation. Moreover, mutagenesis by error-prone PCR produced two of the same mutations that were isolated spontaneously (Table 3). Previous studies suggested that Gram-positive ribosomes require a stronger interaction between 16S rRNA and mRNA than Gram-negative ribosomes in order to initiate translation (21, 25). Our findings are also in agreement with a proposed scheme for the movement of mRNA at different stages of translation, in which a lengthening of the RBS duplex takes place in order to contact ribosomal protein S2 following translation initiation (26). The increased extent of complementarity between the RBS sequence and the 16S rRNA 3′ end (Fig. 2C) resulting from the mutations aids the mRNA-16S rRNA interaction and perhaps also aids the lengthening process.
The broad question is why all E. faecalis genomes encode a fully functional FabK when the enzyme activity is both redundant and is not expressed at physiologically useful levels (at least under laboratory conditions). There seems no selection for retention of this silent gene, because FabI can fully support E. faecalis fatty acid synthesis (18). Given this situation, the fabK sequence would be expected to first degenerate to encode inactive proteins and finally be lost (as may have been the case in the related bacterium Enterococcus faecium). Instead, the fabK sequence has been retained intact and still encodes an active ENR. The only straightforward explanation for this puzzling situation is that FabK expression must be activated by physiological conditions that are not duplicated in the laboratory. However, our data argue that the RBS-16S rRNA interaction is hardwired and, if that is the case, how can FabK expression be activated? Increased transcription from a cryptic promoter is a possible means, although since fabK is in an operon, increased expression of downstream genes would be expected, and in other bacteria overproduction of some of the encoded proteins is toxic. A common means of translational control is sequestration of the RBS by base pairing to an mRNA sequence located upstream of the RBS. The sequestration is released upon synthesis of a small RNA that base pairs with the inhibitory sequence and thereby releases the RBS for rRNA interaction and translational initiation. Another possibility is activation of a translational enhancer sequence, such as those studied in E. coli (26–29). E. coli enhancers are pyrimidine-rich sequences located upstream of the RBS that interact with ribosomal protein S1, and their effects can be augmented by tracts of adenosine bases between the RBS and enhancer (28). To our knowledge, no translational enhancer sequences have been reported in Firmicutes bacteria, but this is not surprising, considering the dearth of data on translational initiation in these bacteria. However, even in the E. coli system the details of enhancer function remain unclear, because the S1 protein is not present in the extant mRNA/ribosome crystal structures. If an E. faecalis translational enhancer were present upstream of fabK, then it could be sequestered by base pairing with a complementary sequence in the mRNA (RNA folding programs predict a high degree of base pairing within the acpP-fabK intergenic region transcript). The putative sequestration by base pairing could be disrupted by a small RNA (or perhaps a protein) that binds the complement of the enhancer sequence and thereby frees the enhancer to bind ribosomal protein S1.
However, it should be noted that our mutational data argue against the above scenarios, because no mutations, either spontaneous or those obtained by error-prone PCR, mapped in the region upstream of the RBS. The PCR mutagenesis produced 15,000 clones, and those that survived the selection for both increased fabK translation and preservation of FabK enzymatic activity all contained two of the RBS-extending mutations we had isolated spontaneously. Moreover, the fact that several of the error-prone PCR mutants had multiple base changes within the coding sequence despite the fact that FabK function was required for their isolation indicates that the mutagenesis was robust.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant AI15650 from the National Institute of Allergy and Infectious Diseases (to J.E.C.) and grants from the National Natural Science Foundation of China (30870036 and 31200028) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (20104404110005) (to H.W.).
We thank Carin Vanderpool for valuable comments on the manuscript.
Footnotes
Published ahead of print 25 October 2013
REFERENCES
- 1.Lu YJ, Zhang YM, Rock CO. 2004. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol. 82:145–155. 10.1139/o03-076 [DOI] [PubMed] [Google Scholar]
- 2.Cronan JE, Thomas J. 2009. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459:395–433. 10.1016/S0076-6879(09)04617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronan JE., Jr 2004. The structure of mammalian fatty acid synthase turned back to front. Chem. Biol. 11:1601–1602. 10.1016/j.chembiol.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 4.Cronan JE. 2006. Remarkable structural variation within fatty acid megasynthases. Nat. Chem. Biol. 2:232–234. 10.1038/nchembio0506-232 [DOI] [PubMed] [Google Scholar]
- 5.Schweizer E, Hofmann J. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68:501–517. 10.1128/MMBR.68.3.501-517.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. 1994. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J. Biol. Chem. 269:5493–5496 [PubMed] [Google Scholar]
- 7.Turnowsky F, Fuchs K, Jeschek C, Hogenauer G. 1989. envM genes of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:6555–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath RJ, Su N, Murphy CK, Rock CO. 2000. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275:40128–40133. 10.1074/jbc.M005611200 [DOI] [PubMed] [Google Scholar]
- 9.Massengo-Tiasse RP, Cronan JE. 2008. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J. Biol. Chem. 283:1308–1316. 10.1074/jbc.M708171200 [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Lin J, Ma J, Cronan JE, Wang H. 2010. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 54:689–698. 10.1128/AAC.01152-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath RJ, Rock CO. 2000. A triclosan-resistant bacterial enzyme. Nature 406:145–146. 10.1038/35018162 [DOI] [PubMed] [Google Scholar]
- 12.Marrakchi H, Dewolf WE, Jr, Quinn C, West J, Polizzi BJ, So CY, Holmes DJ, Reed SL, Heath RJ, Payne DJ, Rock CO, Wallis NG. 2003. Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK). Biochem. J. 370:1055–1062. 10.1042/BJ20021699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massengo-Tiasse RP, Cronan JE. 2009. Diversity in enoyl-acyl carrier protein reductases. Cell. Mol. Life Sci. 66:1507–1517. 10.1007/s00018-009-8704-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath RJ, White SW, Rock CO. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467–497. 10.1016/S0163-7827(01)00012-1 [DOI] [PubMed] [Google Scholar]
- 15.Kingry LC, Cummings JE, Brookman KW, Bommineni GR, Tonge PJ, Slayden RA. 2012. The Francisella tularensis FabI enoyl-acyl carrier protein reductase gene is essential to bacterial viability and is expressed during infection. J. Bacteriol. 195:351–358. 10.1028/JB.01957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jett BD, Huycke MM, Gilmore MS. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, Bi H, Ma J, Hu Z, Zhang W, Cronan J, Wang H. 2013. The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 4(5):e00613–13. 10.1128/mBio.00613-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Cronan JE. 2004. Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J. Biol. Chem. 279:34489–34495. 10.1074/jbc.M403874200 [DOI] [PubMed] [Google Scholar]
- 20.Anderson C, Potter AA, Gerlach GF. 1991. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect. Immun. 59:4110–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CK, Goldfarb DS, Doi RH, Rodriguez RL. 1985. Mutations that affect the translation efficiency of Tn9-derived cat gene in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 82:173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moniruzzaman M, Lai X, York SW, Ingram LO. 1997. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl. Environ. Microbiol. 63:4633–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui A, de Boer HA. 1987. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 84:4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shine J, Dalgarno L. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 71:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stallcup MR, Sharrock WJ, Rabinowitz JC. 1976. Specificity of bacterial ribosomes and messenger ribonucleic acids in protein synthesis reactions in vitro. J. Biol. Chem. 251:2499–2510 [PubMed] [Google Scholar]
- 26.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. 2006. Structural basis for messenger RNA movement on the ribosome. Nature 444:391–394. 10.1038/nature05281 [DOI] [PubMed] [Google Scholar]
- 27.Komarova AV, Tchufistova LS, Dreyfus M, Boni IV. 2005. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 187:1344–1349. 10.1128/JB.187.4.1344-1349.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi S, Furusawa H, Ueda T, Okahata Y. 2013. Translation enhancer improves the ribosome liberation from translation initiation. J. Am. Chem. Soc. 135:13096–13106. 10.1021/ja405967h [DOI] [PubMed] [Google Scholar]
- 29.Vimberg V, Tats A, Remm M, Tenson T. 2007. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 8:100. 10.1186/1471-2199-8-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clewell DB, Tomich PK, Gawron-Burke MC, Franke AE, Yagi Y, An FY. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shepard BD, Gilmore MS. 1995. Electroporation and efficient transformation of Enterococcus faecalis grown in high concentrations of glycine. Methods Mol. Biol. 47:217–226 [DOI] [PubMed] [Google Scholar]
- 32.Casadaban MJ, Martinez-Arias A, Shapira SK, Chou J. 1983. Beta-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 100:293–308. 10.1016/0076-6879(83)00063-4 [DOI] [PubMed] [Google Scholar]
- 33.Blancato VS, Magni C. 2010. A chimeric vector for efficient chromosomal modification in Enterococcus faecalis and other lactic acid bacteria. Lett. Appl. Microbiol. 50:542–546. 10.1111/j.1472-765x.2010.02815 [DOI] [PubMed] [Google Scholar]
- 34.Bi H, Christensen QH, Feng Y, Wang H, Cronan JE. 2012. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol. Microbiol. 83:840–855. 10.1111/j.1365-2958.07968.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Chan CH, Cronan JE. 2006. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45:10008–10019. 10.1021/bi060842w [DOI] [PubMed] [Google Scholar]
- 36.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 37.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 38.Feng Y, Cronan JE. 2009. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284:29526–29535. 10.1074/jbc.M109.023440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 40.Feng Y, Cronan JE. 2009. A new member of the Escherichia coli fad regulon: transcriptional regulation of fadM (ybaW). J. Bacteriol. 191:6320–6328. 10.1128/JB.00835-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 42.O'Sullivan DJ, Klaenhammer TR. 1993. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227–231. 10.1016/0378-1119(93)90011-Q [DOI] [PubMed] [Google Scholar]
- 43.Jerga A, Rock CO. 2009. Acyl-acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J. Biol. Chem. 284:15364–15368. 10.1074/jbc.C109.002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu YJ, Rock CO. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59:551–566. 10.1111/j.1365-2958.2005.04951.x [DOI] [PubMed] [Google Scholar]