Abstract
The major phospholipid constituents of Moraxella catarrhalis membranes are phosphatidylglycerol, phosphatidylethanolamine, and cardiolipin (CL). However, very little is known regarding the synthesis and function of these phospholipids in M. catarrhalis. In this study, we discovered that M. catarrhalis expresses a cardiolipin synthase (CLS), termed MclS, that is responsible for the synthesis of CL within the bacterium. The nucleotide sequence of mclS is highly conserved among M. catarrhalis isolates and is predicted to encode a protein with significant amino acid similarity to the recently characterized YmdC/ClsC protein of Escherichia coli. Isogenic mclS mutant strains were generated in M. catarrhalis isolates O35E, O12E, and McGHS1 and contained no observable levels of CL. Site-directed mutagenesis of a highly conserved HKD motif of MclS also resulted in a CL-deficient strain. Moraxella catarrhalis, which depends on adherence to epithelial cells for colonization of the human host, displays significantly reduced levels of adherence to HEp-2 and A549 cell lines in the mclS mutant strains compared to wild-type bacteria. The reduction in adherence appears to be attributed to the absence of CL. These findings mark the first instance in which a CLS has been related to a virulence-associated trait.
INTRODUCTION
Moraxella catarrhalis is a human-specific pathogen of the mucosa and the causative agent of otitis media in children and respiratory infections in adults. After only nontypeable Haemophilus influenzae (NTHi) (∼26%) and Streptococcus pneumoniae (∼23%), M. catarrhalis is a leading bacterial cause of otitis media in children, being responsible for up to 20% of cases (1–4). Nearly 70% of infants are colonized by M. catarrhalis within their first 12 months of life (5). Moraxella catarrhalis is also the second leading cause of bacterial exacerbations of chronic obstructive pulmonary disease (COPD), being responsible for approximately 10% of cases (6). In the United States, COPD currently stands as the fourth leading cause of death (7). Exacerbations of COPD due to M. catarrhalis are responsible for an estimated $2 billion in medical costs and health care each year (8).
Currently, there is no licensed vaccine available to prevent M. catarrhalis infection. A vaccine is desirable, however, due to the high prevalence, antibiotic resistance, and financial burden associated with M. catarrhalis infection. The vast majority of clinical isolates of M. catarrhalis (>95%) are now resistant to the β-lactamase family of antibiotics that was once considered a front-line treatment for the disease (9). This antibiotic resistance was acquired rapidly over a 10- to 15-year period in which the resistance spread from a few isolates to the majority (10–12). In addition, the incidence and prevalence of disease due to M. catarrhalis infection are expected to increase in the United States following the introduction of vaccines against the upper respiratory tract pathogens S. pneumoniae and NTHi (13). Finally, a vaccine to protect against the top three causes of otitis media would save upwards of $1.3 billion annually and substantially improve the overall health status of infants (14).
Moraxella catarrhalis is a Gram-negative, unencapsulated, aerobic diplococcus. Several virulence factors of M. catarrhalis have been identified and characterized, and many of these are transported through the plasma membrane and are either localized to the outer membrane (i.e., outer membrane proteins [OMPs]) or secreted outside the cell. These molecules then mediate processes such as adherence to epithelial cells, complement resistance, biofilm formation, and nutrient acquisition in order to colonize and cause disease in the human host. Many of these traits are multifactorial. For example, M. catarrhalis expresses several adhesins that mediate adherence to human epithelial cells, including UspA1 (15), Hag/MID (16), McaP (17), OMP CD (18), and the FHA-like proteins MhaB1 and MhaB2 (19).
The phospholipase D (PLD) superfamily is composed of a group of proteins with various functions, yet all members contain a signature HXKX4DX6G(G/S) (HKD) motif (20–22). This superfamily includes prokaryotic and eukaryotic PLD, bacterial cardiolipin (CL) synthases (CLSs) and phosphatidylserine synthases (PSSs), Poxviridae envelope proteins, and bacterial endonucleases. PLD, CLSs, and PSSs all contain two HKD motifs that associate to form the active site (23, 24) and catalyze reactions that synthesize or modify phospholipids (PLs). The histidine and lysine residues within the HKD motif are required for the enzymatic activity of the proteins (21, 25). Members of the PLD superfamily have been studied in many bacterial species, and several have been shown to exhibit virulence-associated characteristics. These include PLD superfamily members of the species Neisseria gonorrhoeae (26), Acinetobacter baumannii (27), Chlamydophila pneumoniae (28), Rickettsia prowazekii (29), Yersinia pestis (30), Arcanobacterium haemolyticum (31), and Corynebacterium pseudotuberculosis (32). Furthermore, N. gonorrhoeae and A. haemolyticum express a PLD that has specifically been shown to impact adherence to human epithelial cells (26, 31). To our knowledge, a role for CLSs in microbial pathogenesis has not been reported.
Three types of CLSs have been characterized and identified; all three catalyze the formation of the PL CL, though the substrates differ. Eukaryotic-type CLSs synthesize CL from one molecule of CDP-diacylglycerol and one molecule of phosphatidylglycerol (PG). Though expressed predominantly in eukaryotes, eukaryotic-type CLSs are also expressed by Streptomyces coelicolor and most actinobacteria (33). The eukaryotic-type CLS is not a member of the PLD superfamily, unlike the other two types of CLSs. The first type, classically referred to as the prokaryotic-type CLS, has been characterized in several bacteria, including Escherichia coli (34), Bacillus firmus (35), and Pseudomonas putida (36). This type of CLS catalyzes the condensation of two molecules of PG to form CL (37). Recently, a third type of CLS, termed YmdC/ClsC, has been identified and characterized in E. coli (25). Like the prokaryotic-type CLS, YmdC/ClsC contains two HKD motifs and is a member of the PLD superfamily; however, it generates CL from one molecule of PG and one molecule of phosphatidylethanolamine (PE) (25). Prior to this study, the presence of ymdC/clsC-like CLSs had not been confirmed in any bacterial species outside E. coli, though bioinformatic tools have suggested that YmdC/ClsC-type CLSs are prevalent (25). Some bacteria, including E. coli, express multiple types of CLSs. In these organisms, the regulation of CLS depends on growth conditions (25, 34, 38).
Cardiolipin is an anionic PL present in energy-transducing membranes, primarily the inner mitochondrial membrane of eukaryotes and the plasma membrane of prokaryotes. Along with PG and PE, CL is a major PL constituent of M. catarrhalis membranes (39). Composed of four acyl chains and a head group with two negative charges, CL has high intrinsic curvature and is found predominantly in membrane regions with negative curvature, such as poles or septa. In fact, studies in E. coli and Bacillus subtilis have shown that CL localizes to the polar and septal regions of bacteria, forming microdomains (40, 41). Furthermore, CL is required for the polar localization of the E. coli osmoregulator ProP, which may explain why CLS-deficient bacteria are more susceptible to osmotic stress (42, 43). The presence of CL in bacterial membranes is required for efficient protein translocation across the plasma membrane in both Sec-dependent and Sec-independent mechanisms (44, 45). In E. coli, the CL content of bacterial membranes varies depending on growth conditions. Cardiolipin levels are highest during stationary growth due to the upregulation of both the expression and activity of CLSs (38, 46, 47).
Here we demonstrate that M. catarrhalis expresses a CLS that impacts adherence to human epithelial cells. Nucleotide sequence analysis revealed that M. catarrhalis contains a gene predicted to encode a member of the PLD superfamily displaying significant similarity to the YmdC/ClsC protein of E. coli. This gene, termed mclS, for the Moraxella catarrhalis cardiolipin synthase, is highly conserved throughout M. catarrhalis isolates and is located immediately upstream of a gene predicted to encode a protein with similarity to ProP of E. coli. An M. catarrhalis isogenic mutant strain lacking expression of mclS was found to contain no observable levels of CL, thereby confirming that mclS encodes a CLS. Likewise, a strain of M. catarrhalis with a mutated HKD motif lacks CL, demonstrating that the intact motif is required for CLS activity. Both mutant strains exhibit reduced levels of adherence to human epithelial cell lines, suggesting that the CLS activity and, subsequently, CL are required for wild-type (WT) levels of adherence. This work documents the first instance in which a CLS contributes to a virulence-associated trait.
MATERIALS AND METHODS
Strains, plasmids, tissue culture cell lines, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Moraxella catarrhalis was grown at a temperature of 37°C using Todd-Hewitt (TH) medium (Difco). Where indicated, TH medium was supplemented with the following antibiotics at the indicated concentrations: spectinomycin (Spec; 15 μg/ml), kanamycin (Kan; 20 μg/ml), zeocin (Zeo; 5 μg/ml), chloramphenicol (Cat; 1 μg/ml), and streptomycin (Stm; 75 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Selectable marker | Source |
|---|---|---|---|
| Strains | |||
| M. catarrhalis | |||
| O35E | WT isolate | None | 76 |
| O35E.mclS | mclS isogenic insertion mutant strain of O35E | Spec | This study |
| O35E.mclS repaired | Repaired strain of O35E.mclS | Stm | This study |
| O35E.mclS.K438R | mclS isogenic point mutant strain of O35E | Stm | This study |
| O35E.ZCSM | hag, uspA1, uspA2, and mcaP isogenic mutant strain of O35E | Zeo, Cat, Spec, Kan | 17 |
| O35E.SM100 | rpsL isogenic mutant strain of O35E | Stm | 77 |
| O35E.CD1 | ompcd isogenic mutant strain of O35E | Kan | 18 |
| O35E.MhaB | mhaB1 and mhaB2 isogenic mutant strain of O35E | Spec, Zeo | 19 |
| O12E | WT isolate | None | 15 |
| O12E.mclS | mclS isogenic insertion mutant strain of O12E | Spec | This study |
| O12E.mclS repaired | Repaired strain of O35E.mclS | Stm | This study |
| O12E.12Hg | hag, uspA1, and uspA2 isogenic mutant strain of O12E | Zeo, Cat, Spec | Unpublished data |
| McGHS1 | WT isolate | None | 49 |
| McGHS1.mclS | mclS isogenic insertion mutant strain of McGHS1 | Spec | This study |
| McGHS1.mclS repaired | Repaired strain of McGHS1.mclS | Stm | This study |
| McGHS1.Hag | hag isogenic mutant strain of McGHS1 | Spec | 49 |
| E. coli EPI300 | Cloning strain | None | Epicentre Illumina |
| Plasmids | |||
| pSPECr | Source of Specr cassette | Spec | 78 |
| pCC1 | Cloning vector, replicative in E. coli | Cat | Epicentre Illumina |
| pCC1.mclS | pCC1 containing mclS from O35E | Cat | This study |
| pCC1.mclS.spec | pCC1.mclS with Specr cassette inserted into the mclS ORF | Cat, Spec | This study |
Escherichia coli was grown at a temperature of 37°C using Luria-Bertani (LB) medium (Difco). Where indicated, LB medium was supplemented with Cat and/or Spec at concentrations of 15 μg/ml and 200 μg/ml, respectively. To isolate plasmid DNA from TransforMax EPI300 E. coli harboring a pCC1-based plasmid, strains were grown on LB agar (approximately 20 ml per plate) supplemented with 30 μl of CopyControl induction solution to induce the transition of plasmid maintenance from a single copy to multiple copies.
The human cell lines A549 (ATCC CCL-185, type II pneumocytes) and HEp-2 (ATCC CCL-23, laryngeal epithelium) were cultured in Ham's F-12 medium supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco Life Technologies), 0.15% (wt/vol) sodium bicarbonate (Cellgro), and GlutaMAX-I according to the manufacturer's recommendations (Gibco Life Technologies) at a temperature of 37°C with an atmosphere of 92.5% air and 7.5% CO2. In addition, the medium for HEp-2 also contained 1 mM sodium pyruvate (Cellgro) and minimal essential medium nonessential amino acids according to the manufacturer's recommendations (Cellgro).
Recombinant DNA techniques.
Standard molecular biology techniques were performed as previously described (48). Genomic DNA was extracted from M. catarrhalis using an Easy-DNA kit (Invitrogen Life Technologies) according to the manufacturer's recommendations. Amplicons used for cloning, constructing isogenic mutants, and sequencing were generated with Platinum Pfx DNA polymerase (Invitrogen Life Technologies) using genomic DNA extracted from M. catarrhalis. All other DNA fragments were amplified using Taq DNA polymerase (Invitrogen Life Technologies). Plasmids were isolated from bacteria using a QIAprep Spin miniprep kit (Qiagen) according to the manufacturer's recommendations. When necessary, DNA was purified using DNA precipitation solution (Epicentre Illumina) according to the manufacturer's recommendations. For DNA electrophoresis, a 1-kb Plus DNA ladder (Invitrogen Life Technologies) was loaded as the DNA size marker.
Cloning of M. catarrhalis O35E mclS gene in E. coli.
On the basis of the available sequence data from M. catarrhalis strain ATCC 43617, oligonucleotide primers P1 and P2 (Table 2) were designed to amplify the mclS gene from strain O35E, including the upstream region likely containing the regulatory sequence. An amplicon of 1,542 bp was generated by PCR using the aforementioned primers. To generate blunt ends, the mclS amplicon was treated with an End-It DNA end repair kit (Epicentre Illumina). The resulting mclS amplicon was ligated into pCC1 using a CopyControl PCR cloning kit (Epicentre Illumina), per the manufacturer's instructions, yielding plasmid pCC1.mclS. The construct was sequenced to verify that no mutations were introduced during PCR and to determine the orientation of the mclS gene in vector pCC1.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′ → 3′) | Gene | Direction |
|---|---|---|---|
| P1 | ATTTGCCCGATACGCCTCACTTAC | mclS | Forward |
| P2 | AATCGATAGGCATCAGTCCAGCCA | mclS | Reverse |
| P3 | TGGGATCCATTTGATAACAATCATCGCC | mclS | Forward |
| P4 | GGATCTGAGTGACCGTTTAATTT | mclS | Reverse |
| P5 | GCCAACCAATTACCCAGCCAGCA | MCORF 821 | Forward |
| P6 | ACCGCCCAAGGGTCTAGCAA | MCORF 821 | Reverse |
| P7 | ACCAAGCAAGTGGCCAGTGCT | MCORF 819 | Forward |
| P8 | CCAAAAGCGGGCATTCGGCG | MCORF 819 | Reverse |
| P9 | TGGCTGGCTGGACTGATGCC | mclS | Forward |
| P10 | AGGCGTATCGGGCAAATTTTTGCA | mclS | Reverse |
| P11 | TGCCAATGACCAAGCCAATT | mcaP | Forward |
| P12 | TCAGATGCTGGGGTAGTTGA | mcaP | Reverse |
| P13 | CGCGGATCCGCGACTCAAGTGAAAATACGCA | rpsL | Forward |
| P14 | CCGGAATTCCGGACACGACGTCTTGGCATAA | rpsL | Reverse |
| P15 | AGTCTACACGCCAGAGCCTTTGCGGTAGAT | mclS | Forward |
| P16 | ATCTACCGCAAAGGCTCTGGCGTGTAGACT | mclS | Reverse |
| P17 | CACACTCTGAGCGGTCATTT | MCORF 821 | Forward |
| P18 | GAAACTCAATTGCTGGCAGA | MCORF 821 | Reverse |
| P19 | GATGCCAATGATGGAACAAC | mcaP | Forward |
| P20 | GATTTGGGTTTGTGCTGATG | mcaP | Reverse |
Construction of M. catarrhalis mutants.
Plasmid pSPECr was restricted with the endonuclease PstI, and a 1.2-kb DNA fragment corresponding to the spectinomycin resistance (Specr) cassette was purified from agarose gel slices using a High Pure PCR product purification kit (Roche Applied Science). After treatment with the End-It DNA end repair kit (Epicentre Illumina), the Specr cassette was then ligated into a unique NsiI site located near the middle of the mclS open reading frame (ORF) in plasmid pCC1.mclS. The resulting construct, pCC1.mclS.spec, was sequenced to verify insertion of the spectinomycin marker at the intended location.
Plasmid pCC1.mclS.spec was introduced into M. catarrhalis strains O35E, O12E, and McGHS1 by natural transformation to generate mclS isogenic mutants via allelic exchange. This pCC1-based construct does not replicate in M. catarrhalis due to the absence of a suitable origin of replication. We identified isolates that incorporated the inactivated copy of mclS in their genomes via homologous recombination by selecting for transformants resistant to spectinomycin. Proper allelic exchange was confirmed by PCR with oligonucleotide primers P3 and P4 (Table 2), which yielded a DNA fragment of 2.3 kb in mutant strains O35E.mclS, O12E.mclS, and McGHS1.mclS. Of note, the same primers, P3 and P4, yielded a DNA fragment of 1.1 kb in WT isolates O35E, O12E, and McGHS1. This 1.2-kb shift in the size of the PCR products is consistent with disruption of the mclS gene with the Specr cassette in the genome of M. catarrhalis.
The mclS mutant strains were complemented by reintroducing the WT copy of the mclS gene in its original locus, yielding strains O35E.mclS repaired, O12E.mclS repaired, and McGHS1.mclS repaired. This was accomplished per the congression procedure described by Balder et al. (19). Briefly, primers P1 and P2 were used to amplify the 1.5-kb WT copy of mclS from strain O35E. The purified mclS amplicon was then combined with an amplicon (generated using primers P13 and P14) containing the rpsL gene of M. catarrhalis strain O35E.SM100, which specifies resistance to streptomycin. This mixture was introduced into mutant strains O35E.mclS, O12E.mclS, and McGHS1.mclS by natural transformation. Streptomycin-resistant and spectinomycin-sensitive transformants were screened by PCR to verify that the WT copy of the mclS gene had been reintroduced in its original location in the M. catarrhalis genome. This reintroduction was confirmed by sequencing the gene of the complemented strains.
Site-directed mutagenesis of the mclS gene.
A QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) was utilized to introduce a point mutation in the second HKD motif of the mclS gene cloned in plasmid pCC1.mclS. To accomplish this, we designed the 30-mer mutagenesis primers P15 and P16 (Table 2) to introduce guanine in place of adenine at nucleotide position 1313 of the mclS ORF, which changes the codon AAA (which specifies lysine 438) to AGA (which codes for arginine). Mutagenesis, which was performed as recommended by the manufacturer, produced plasmid pCC1.mclS.K438R. The construct was sequenced to verify that only the intended mutation (i.e., the change of the lysine at position 438 to arginine, K438 → R) was introduced in the mclS gene. The point mutation was introduced into the O35E.mclS mutant strain by congression, as described previously (79). Streptomycin-resistant and spectinomycin-sensitive transformants were screened by PCR and confirmed by sequencing the mclS gene.
Sequence analysis.
Plasmids and PCR products were sequenced by the University of Michigan Sequencing Core. Chromatograms were assembled using the Sequencher program, version 5.0 (Gene Codes Corp.). Sequence analysis was performed using the Vector NTI program, version 10.1 (Invitrogen Life Technologies).
BLASTP (NCBI) was used to compare the predicted amino acid sequences of MclS and the gene products of MCORF 819 and MCORF 821 with the sequences of proteins from the online database compiled by GenBank. Protein domains and structural features of the products of the mclS, MCORF 819, and MCORF 821 genes were predicted using a variety of algorithms/programs. The presence of transmembrane helices (TMHs) was predicted by the TM Pred (ExPASy) or TMHMM (version 2.0; Center for Biological Sequence Analysis) program. The presence of signal peptides was predicted by the SignalP (version 4.1; Center for Biological Sequence Analysis) and TatP (version 1.0; Center for Biological Sequence Analysis) programs. The Phobius algorithm (Stockholm Bioinformatics Centre) specifically differentiates between transmembrane topology and signal peptides. The ProSite program (ExPASy) was utilized to identify a variety of protein domains, families, and functional sites.
Protein preparation and Western blot analysis.
Whole-cell lysates were prepared and Western blots were performed as previously described (49). Outer membrane proteins were obtained using the EDTA method outlined by Murphy and Loeb (50). Equivalent proteins loads were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (EMD Millipore) for Western blot analysis. A SeeBlue Plus2 prestained standard (Invitrogen Life Technologies) was loaded as the protein size marker. The membranes were probed with the primary antibodies as described below, followed by goat anti-mouse Ig (H+L)-horseradish peroxidase (HRP) secondary antibody (SouthernBiotech) at 1:10,000. Signals were detected using Luminata Crescendo Western HRP substrate (EMD Millipore) and a Foto/Analyst Luminary/FX imaging system (Fotodyne Inc.).
Flow cytometry.
Flow cytometry was performed as described previously (51), with slight modification. Briefly, a 50-μl aliquot of a 250-Klett-unit suspension of M. catarrhalis was incubated with primary antibody for 30 min at 37°C with shaking. After a series of washes and resuspension in phosphate-buffered saline with 0.15% (wt/vol) gelatin (PBSG), the bacteria were incubated with goat anti-mouse Ig (H+L) conjugated with Alexa Fluor 488 (Invitrogen Life Technologies) at 1:100 in darkness for 30 min at 37°C with shaking. After a series of washes and resuspension in PBSG, an equal volume of PBSG supplemented with 4% (wt/vol) paraformaldehyde was added. Samples were analyzed by a BD LSR II flow cytometer (BD Biosciences) and BD FACSDiva software (BD Biosciences). The Alexa Fluor 488 fluorescence from the labeled bacteria was measured through a 525-nm band-pass filter, in which 50,000 events were measured. The experiments were repeated on two separate occasions.
Antibodies.
The Western blot and flow cytometry procedures detected the M. catarrhalis adhesins UspA1 and OMP CD using the murine monoclonal antibodies (MAbs) 24B5 (52, 53) and 5E8 (54), respectively, as the primary antibodies. The procedures also detected the following proteins using the indicated polyclonal antibodies (pAbs) as the primary antibodies: Hag (mouse serum recognizing the C terminus of Hag [55]), McaP (mouse serum recognizing the region of McaP consisting of amino acids [aa] 51 to 650 [51]), and MhaB1/MhaB2 (mouse serum recognizing a portion of the identical region of MhaB1 and MhaB2, aa 72 to 399 [19]). For Western blotting, membranes were probed with MAbs at 1:100 and pAbs at 1:5,000. For flow cytometry, bacteria were incubated with MAbs at 1:200 and pAbs at 1:5,000.
RNA isolation.
Plate-grown M. catarrhalis was used to generate a 230-Klett-unit suspension in TH broth. RNA from M. catarrhalis was extracted by mechanical disruption (Mini Beadbeater-1; Biospec Products) with the RNAprotect bacterial reagent (Qiagen) and an RNeasy minikit (Qiagen). The procedure was then continued per the manufacturer's recommendation. To ensure degradation of genomic DNA, a second DNase I treatment was performed using a RiboPure-Bacteria kit (Ambion) according to the manufacturer's protocols. The quality and quantity of extracted RNA were determined by visualization of the 16S and 23S rRNA bands during gel electrophoresis and spectrophotometric analysis of the absorbance of the samples at 260 nm (A260) and 280 nm (A280) (SmartSpec Plus; Bio-Rad).
RT-PCR and qRT-PCR.
For both reverse transcriptase PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR), RNA was isolated and cDNA was synthesized using the same procedure. cDNA was synthesized from extracted RNA using a ThermoScript RT-PCR system (Invitrogen Life Technologies), as recommended by the manufacturer. The cDNA was synthesized using the provided random hexamer primer to generate a cDNA library from M. catarrhalis. Equal amounts of RNA from each strain were used for cDNA synthesis according to the A260 spectrophotometric readings. In separate parallel reactions, the ThermoScript reverse transcriptase enzyme was withheld to control for DNA contamination, and the isolated RNA was withheld to control for contamination of reagents.
For RT-PCR experiments, the PCR was performed using Taq DNA polymerase (Invitrogen Life Technologies). Genomic DNA obtained from M. catarrhalis served as the template for the positive control for the PCR. Extension times varied depending on the expected fragment size (1 min per kb). Oligonucleotide primers P5 and P10 (Table 2) were designed to determine the transcription linkage between mclS and MCORF 819, while primers P8 and P9 were designed to determine the transcription linkage between mclS and MCORF 821. Oligonucleotide primers P1 and P4 were used to monitor the transcription of mclS. Likewise, primers P5 and P6 were used to monitor the transcription of MCORF 819, and primers P7 and P8 were used to monitor the transcription of MCORF 821. As a control, transcription of mcaP, which is constitutively expressed in M. catarrhalis, was monitored using primers P11 and P12. Each RT-PCR experiment was conducted on two separate occasions.
For qRT-PCR experiments, iQ SYBR green supermix (Bio-Rad) was utilized for the amplification of the cDNA template, as recommended by the manufacturer. Gene expression was measured and analyzed by the Bio-Rad iQ5 optical system and software. Oligonucleotide primers P17 and P18 (Table 2) were designed to monitor the expression of the MCORF 821 gene. The reference control gene mcaP, which is constitutively expressed in M. catarrhalis and shown to be unaffected in the mclS mutants (see Fig. S4 and S5G to I in the supplemental material), was monitored using primers P19 and P20 (Table 2). The qPCRs were performed in triplicate, and the entire qRT-PCR process was conducted on three individual occasions. All primers used for qPCR were validated to ensure minimal dimerization and nonspecific binding.
Extraction of bacterial PL.
Todd-Hewitt broth (30 ml) was seeded with M. catarrhalis, and the bacteria were grown overnight in a shaker (200 rpm) at 37°C. Bacterial cultures were then centrifuged for 10 min at 3,500 × g. The supernatant was discarded, and the pellet was resuspended in 5 ml PBSG. Bacterial suspensions were transferred to polypropylene copolymer (PPCO) centrifuge tubes (Oak Ridge). Phospholipids were then extracted by the method of Bligh and Dyer (56). Briefly, the bacterial suspension was mixed with 18.75 ml of a 1:2 (vol/vol) mixture of chloroform-methanol (Acros Organics; Fisher Scientific) and vortexed. Next, 6.25 ml of chloroform was added to the mixture and the mixture was vortexed. Finally, 6.25 ml of distilled water was added and the mixture was vortexed. After a 5-min centrifugation at 2,500 × g, the bottom phase was transferred to a new tube using a Pasteur pipette. The solvent was evaporated over a stream of nitrogen gas (AirGas). The dried PLs were resuspended in 200 μl of a 1:2 mixture of methanol-chloroform and transferred to glass vials (Supelco Analytical). Phospholipid preparations were stored at −20°C.
TLC.
Phospholipid preparations (10 to 40 μl) were spotted on silica gel high-performance thin-layer chromatography (HP-TLC) plates with inorganic binder (Analtech). Plates were developed in one dimension using a mobile phase consisting of a 65:25:10 mixture of chloroform-methanol-acetic acid (Fisher Scientific). Spots were visualized by spraying plates with the Molybdenum Blue spray reagent (Sigma Life Science). Phospholipid spots were identified by comparison with the following standards: PE, PG, and CL (Sigma Life Science).
Adherence assays.
Quantitative adherence assays were performed as previously described (16). Briefly, a 24-well plate was seeded with epithelial cells (HEp-2 or A549) on the day before the assay to generate a monolayer. On the day of the assay, freshly grown bacteria were resuspended in PBSG to an optical density (OD) of 230 Klett units (∼109 CFU/ml). Portions of the suspension were used to inoculate wells containing epithelial cells at a multiplicity of infection (MOI) of 100:1. Meanwhile, an aliquot of the bacterial suspension was diluted and plated to determine the inoculum. The 24-well plate was centrifuged to facilitate contact between the bacteria and the epithelial cell monolayer. The plate was then incubated for 30 min at 37°C. The wells were washed five times with PBSG to remove nonadherent bacteria. Cells were treated with a saponin solution (5% [wt/vol] saponin [Acros Organics] and 0.85% [wt/vol] EDTA [Fisher Scientific] in PBSG) for 5 min to release the bacteria from the epithelial cells. The bacteria were then diluted and plated on TH agar for enumeration following incubation at 37°C. Percent adherence was calculated by dividing the number of adherent bacteria by the inoculum. Experiments were performed in duplicate or triplicate on at least three occasions.
Survival in PBSG, TC medium, and saponin solution.
Moraxella catarrhalis strains were first cultured onto TH agar plates supplemented with the appropriate antibiotics. Bacterial cultures were diluted, and portions were distributed into tubes containing PBSG, tissue culture (TC) medium (HEp-2 cell culture medium), or saponin solution. Tubes were incubated at 37°C for 15 min (saponin) or 30 min (PBSG, TC medium), with the incubation time corresponding to the period in which the bacteria were exposed to the each reagent in the adherence assay procedure. Aliquots were removed and plated on TH agar to determine survival. Percent survival was calculated by dividing the number of CFU postincubation by the number of CFU preincubation. Survival experiments were performed in duplicate on at least three separate occasions.
Growth rate experiments.
Moraxella catarrhalis strains were first cultured onto TH agar plates supplemented with the appropriate antibiotics. These plate-grown bacteria were used to inoculate sidearm flasks containing 20 ml of TH broth to an OD of 50 Klett units. The cultures were then incubated with shaking (200 rpm) at a temperature of 37°C for up to 7 h. The OD of each culture was determined every 60 min using a Klett colorimeter (Scienceware). To examine the growth of the M. catarrhalis strains under osmotic stress, TH broth was supplemented with either sodium chloride (Fisher Scientific) or d-sorbitol (Sigma Life Science). Growth curves were repeated on at least three separate occasions.
Statistical methods.
All statistical analyses were performed using the Mann-Whitney test and GraphPad Prism (version 4) software (GraphPad Software). P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The nucleotide sequences of mclS from M. catarrhalis isolates O12E and McGHS1 were deposited in the GenBank database in February 2013 under the accession numbers KC692996 and KC692997, respectively.
RESULTS
Identification of an ORF in M. catarrhalis that is predicted to encode a member of the PLD superfamily of proteins.
Analysis of the published genomic sequence of M. catarrhalis isolate ATCC 43617 (GenBank accession numbers AX067426 to AX067466 [57]) using the NCBI BLASTP algorithm identified a gene, MCORF 820, predicted to encode a phospholipase D/transphosphatidylase. Further investigation revealed that the predicted protein contains two HXKXDXG(G/S) (HKD) motifs, which suggests that MCORF 820 belongs to the PLD superfamily of molecules (21). The amino acid residues comprising the HKD motif are required for activity (21, 25, 58). Most members of the PLD superfamily contain two HKD motifs, which associate to form the active site (23, 24). Specifically, the histidine residues within the active site are crucial, as one serves as the nucleophile and forms the phosphoenzyme intermediate, while the other acts as a general acid to cleave the phosphodiester bond (23, 24).
MCORF 820 also displayed significant similarity to the recently characterized YmdC/ClsC protein of E. coli strain K-12 substrain W3110 (25) (E value, 9e−113) and CLSs of various bacterial species. The greatest identity was to a predicted phospholipase of Psychrobacter sp. strain PAMC 21119 (WP 010199650.1), with greater than 50% identity (BLASTP E value, 0.0). Due to the sequence similarity of MCORF 820 to bacterial CLSs, the gene was named the Moraxella catarrhalis cardiolipin synthase (mclS).
The 1,629-bp ORF of mclS is predicted to encode a 542-aa protein with a molecular mass of 62 kDa. According to the ProSite algorithm (ExPASy), the N terminus of MclS is predicted to contain a 23-aa signal peptide followed immediately by a cysteine residue, which serves as a lipid attachment site for either a palmitoyl or a diacylglycerol group (score = 5.000). This is in contrast to the results from the TM Pred service (ExPASy) that indicated a possible N-terminal transmembrane domain of 18 to 20 aa in length (scores = 1,711 and 1,380). Additionally, there were no Sec or Tat signal sequences detected within mclS by the SignalP (version 4.1) and TatP (version 1.0) programs, respectively (59, 60). Figure 1 depicts the structural features identified in MclS.
FIG 1.
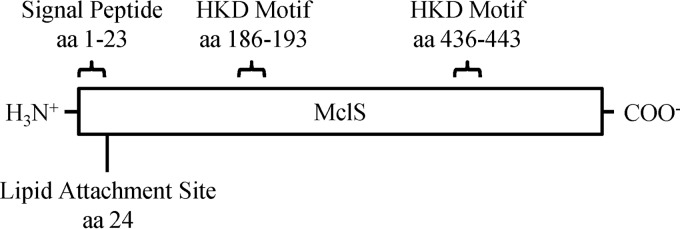
Predicted structure of MclS from M. catarrhalis. Bioinformatic data predict that MCORF 820 (renamed mclS) encodes a CLS. The presence of two HKD motifs (aa 186 to 193 and aa 436 to 443) indicates that MclS is a member of the PLD superfamily. The HKD motifs are thought to associate to form the active site of the enzyme. A signal peptide (aa 1 to 23) and lipid attachment site (aa 24) were predicted at the N terminus, thereby characterizing MclS as a putative lipid-anchored protein.
MclS is conserved among M. catarrhalis isolates.
In order to evaluate conservation of the mclS gene product, we sequenced the ORF from three M. catarrhalis strains commonly used in our laboratory: O35E, O12E, and McGHS1. Additionally, we searched the genomic sequences of 10 M. catarrhalis isolates available through the NCBI genomic BLAST service (61) for the mclS gene. All strains specified a highly conserved mclS gene product. Eight single nucleotide polymorphisms (SNPs) were identified among the 14 sequences analyzed, and three were found to specify amino acid substitutions: C-A at nucleotide (nt) 508 (arginine → serine substitution), G-A at nt 1081 (valine → isoleucine substitution), and C-T at nt 1205 (alanine → valine substitution). No missense SNPs were observed within the HKD motifs, signal sequence, or lipid attachment site. There were no obvious trends between SNPs and the geographical, anatomical, and disease sources of these M. catarrhalis isolates.
Genetic organization of the mclS locus.
Examination of the mclS genetic locus of strain ATCC 43617 revealed that the ORF is flanked upstream by MCORF 819, which is predicted to encode a GTP-cyclohydrolase I (GCYH-I; NCBI BLASTP E value, 4e−99), and downstream by MCORF 821, which is predicted to encode a protein of the major facilitator superfamily (MFS; NCBI BLASTP E value, 1.73e−5), as diagramed in Fig. 2A. The genes corresponding to MCORF 819 and MCORF 821 were also found flanking mclS in all 12 isolates of M. catarrhalis with sequenced genomes.
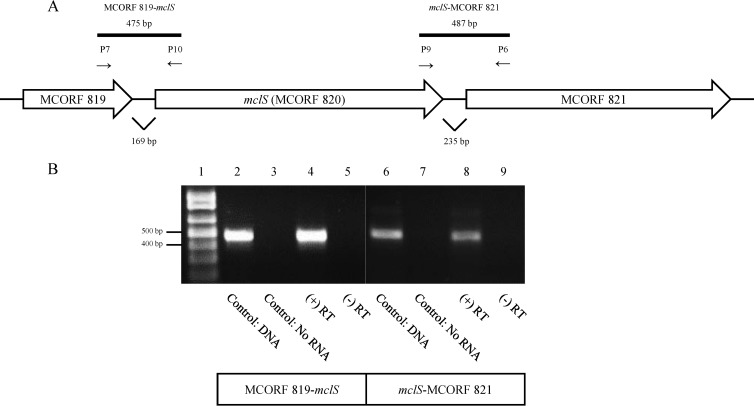
FIG 2.
A transcriptional linkage exists between MCORF 819, mclS, and MCORF 821. (A) Schematic representation of the mclS genetic locus, including MCORF 819 and MCORF 821. The primers used in PCR are indicated by arrows. Solid black bars, expected PCR products. (B) An agarose gel displaying the results of RT-PCR examining the organization of the mclS locus. Primers P7 and P10 were used to amplify the region between MCORF 819 and mclS, while primers P6 and P9 were used to amplify the region between mclS and MCORF 821. Lane 1, DNA size marker; lanes 2 and 6, PCR products from genomic DNA of M. catarrhalis isolate O35E; lanes 3 and 7, RT-PCR products from a reaction with no RNA added; lanes 4 and 5 (MCORF 819-mclS) and lanes 8 and 9 (mclS-MCORF821), RT-PCR products from RNA of M. catarrhalis isolate O35E. During cDNA synthesis, reverse transcriptase enzyme was either added [(+) RT, lanes 4 and 8] or withheld [(−)RT, lanes 5 and 9].
In prokaryotes, GCYH-I catalyzes the hydrolysis of GTP in the first committed step in the biosynthesis of tetrahydrofolate (62). The 201-aa MCORF 819 gene product is predicted to have a molecular mass of 22.5 kDa. No signal peptides or TMH were predicted (SignalP; TatP; Phobius; TMHMM, version 2.0; ProSite). In E. coli, GCYH-I exists as a decamer (a pentamer of dimers) and requires zinc for activity (63, 64).
The MFS is a diverse group of secondary transporters that facilitate transport across cytoplasmic or internal membranes. Substrates can vary but may include ions, sugar phosphates, neurotransmitters, nucleosides, amino acids, and peptides. The 509-aa MCORF 821 gene product is predicted to have a molecular mass of 56.6 kDa. According to the TMHMM algorithm, the MCORF 821 gene product possesses six TMHs at the N terminus, six TMHs at the C terminus, and a cytoplasmic region located between the two transmembrane domains, thereby indicating that it is likely an integral membrane protein. Significant similarity to E. coli ProP (BLASTP E value, 3e−22), an H+ osmoprotectant symporter with the capability of transporting proline and glycine betaine, was observed. Additionally, ProP has been shown to localize at the cell poles, a process that is dependent on CL (43, 65). Due to the relationship between CL and ProP, we examined whether or not mclS is transcriptionally linked to MCORF 821. Though no data that propose a functional relationship between CL and GCYH-I have been published, we also investigated if a transcriptional linkage exists between mclS and upstream gene MCORF 819.
RNA was extracted from M. catarrhalis isolate O35E and analyzed by RT-PCR to determine if the flanking genes were transcriptionally linked to mclS. Primers were specifically designed to span the intergenic regions between ORFs. The presence of amplicons of 475 bp and 487 bp in size is consistent with both MCORF 819 and MCORF 821, respectively, being transcriptionally linked to mclS (Fig. 2B). The results demonstrate that mclS is cotranscribed with both MCORF 819 and MCORF 821.
Construction of M. catarrhalis strains that lack expression of a WT mclS gene product.
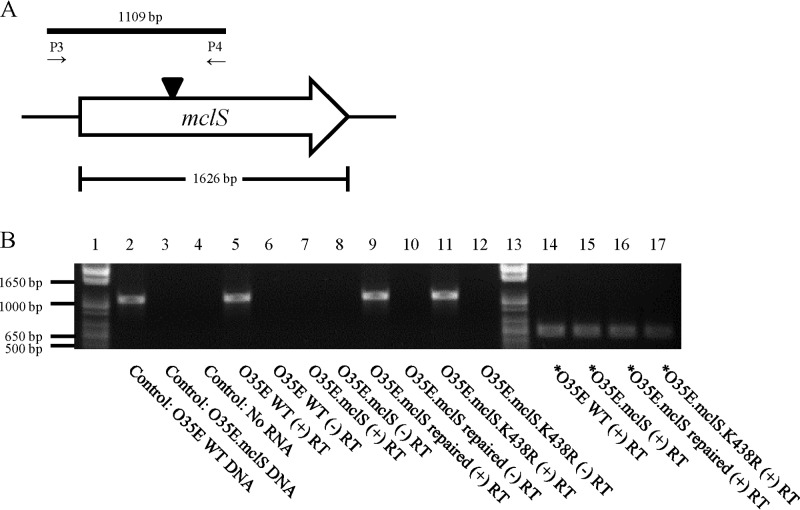
To investigate the biological role of MclS, a mutation was engineered in the mclS gene of M. catarrhalis WT isolates O35E, O12E, and McGHS1. This was accomplished by inserting a spectinomycin resistance cassette near the middle of the ORF and introducing the disrupted mclS gene into the M. catarrhalis genome via homologous recombination. The approach yielded isogenic mutant strains O35E.mclS, O12E.mclS, and McGHS1.mclS (Table 1). To verify that the mutant strains lacked expression of the mclS gene, RT-PCR was utilized, and the results of these experiments are shown in Fig. 3. Using oligonucleotide primers internal to the mclS transcript (P1 and P4; Fig. 3A), a DNA fragment of the expected size was amplified in WT isolate O35E (Fig. 3B, lane 5) but was absent in isogenic mutant strain O35E.mclS (Fig. 3B, lane 7). Using control primers specific for the mcaP gene, RT-PCR analysis demonstrated that equivalent amounts of RNA from the WT and mutant strains were analyzed (Fig. 3B, lanes 14 to 17). Similar results were obtained when analyzing expression of the mclS gene in M. catarrhalis strains O12E, O12E.mclS, McGHS1, and McGHS1.mclS (data not shown). Taken together, the data indicate that our mutagenesis approach successfully abrogated expression of the mclS gene product in mutant strains O35E.mclS, O12E.mclS, and McGHS1.mclS. Each of these mutants was also complemented by reintroducing the WT copy of the mclS gene into its original genomic locus, yielding strains O35E.mcls repaired, O12E.mclS repaired, and McGHS1.mclS repaired (Table 1). Proper allelic expression was verified by PCR (data not shown) and sequencing of the mclS gene in the repaired strains. In addition, RT-PCR experiments using internal primers specific for the mclS transcript confirmed that the repaired strains expressed the mclS gene (Fig. 3B, lane 9, for O35E.mclS repaired; data not shown for O12E.mclS repaired and McGHS1.mclS repaired).
FIG 3.
RT-PCR validates the M. catarrhalis O35E isogenic mclS mutant strains. (A) Schematic representation of the mclS genetic locus. Expression of mclS was examined by RT-PCR using the indicated oligonucleotide primers. Solid black bar, the expected PCR product in the WT strain of M. catarrhalis isolate O35E; black triangle, location of the spectinomycin resistance cassette which interrupts the mclS ORF in the mclS insertion mutants. (B) An agarose gel displaying the results of RT-PCR examining the transcription of mclS in the WT, mclS insertion mutant, mclS repaired, and mclS point mutant strains of M. catarrhalis isolate O35E. Oligonucleotide primers P3 and P4 were used to amplify a portion of mclS; in the mclS insertion mutant strains, this region was interrupted by a spectinomycin resistance cassette. Lanes 1 and 13, DNA size markers; lanes 2 and 3, PCR products from the genomic DNA of the M. catarrhalis O35E isolate WT strain and the mclS insertion mutant strain, respectively; lane 4, RT-PCR product from a reaction with no RNA added; lanes 5 to 12, RT-PCR products derived from WT M. catarrhalis isolate O35E (lanes 5 and 6), the mclS insertion mutant strain (O35E.mclS; lanes 7 and 8), the mclS repaired strain (O35E.mclS repaired; lanes 9 and 10), and the mclS point mutant strain (O35E.mclS.K438R; lanes 11 and 12). During cDNA synthesis, reverse transcriptase enzyme was either added (lanes 5, 7, 9, and 11) or withheld (lanes 6, 8, 10, and 12). Lanes 14 to 17, RT-PCR products derived using the mcaP-specific primers P11 and P12 to ensure that equal amounts of RNA were analyzed in lanes 5 to 12. Asterisks signify that mcaP-specific primers were used for PCR amplification of cDNA.
MclS is responsible for the synthesis of CL in M. catarrhalis.
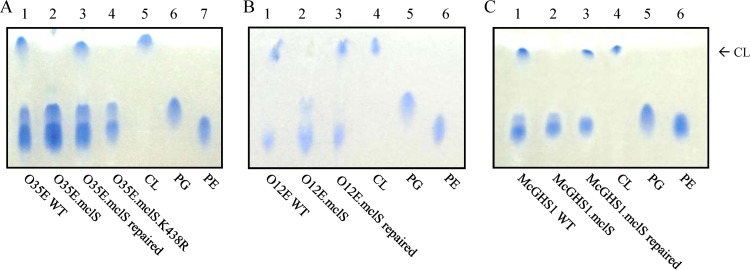
Beebe and Wlodkowski previously reported that CL is a major component of the M. catarrhalis cell envelope, constituting 5 to 30% of the total membrane PL (39). Since mclS is the only gene in the genome predicted to encode a CLS, we hypothesized that mclS is solely responsible for the production of CL in M. catarrhalis. To address this, PLs were extracted from our panel of M. catarrhalis WT and mutant strains, separated by high-performance thin-layer chromatography (HP-TLC), and visualized by staining with the Molybdenum Blue spray reagent. Purified CL, PG, and PE were used as standards in these experiments in order to identify the various PLs present in the cell envelope of the M. catarrhalis strains. Figure 4A clearly demonstrates that CL was produced by the WT (lane 1) and repaired (lane 3) strains, while it was missing from the mutant O35E.mclS (lane 2). Similar results were obtained when analyzing PLs produced by the O12E (Fig. 4B) and McGHS1 (Fig. 4C) strain sets.
FIG 4.
Moraxella catarrhalis requires mclS for synthesis of CL. Analysis of the PL profile of the WT, mclS insertion mutant, mclS repaired, and mclS point mutant strains of M. catarrhalis isolates O35E (A), O12E (B), and McGHS1 (C) by HP-TLC. Phospholipids were extracted by the method of Bligh and Dyer (56) and developed in one dimension using a mobile phase of chloroform-methanol-acetic acid (65:25:10). Phospholipid spots were visualized by staining with the Molybdenum Blue spray reagent. Purified CL, PG, and PE were used as standards to identify the PLs of M. catarrhalis. Arrow, location to which CL migrates upon HP-TLC analysis.
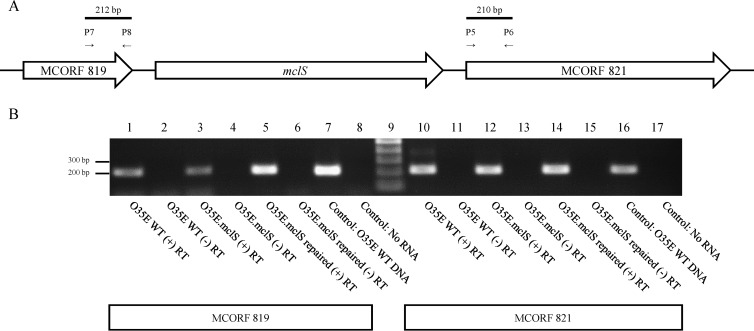
It is possible that the mclS mutation in strains O35E.mclS, O12E.mclS, and McGHS1.mclS results in polar effects on expression of the cotranscribed genes MCORF 819 and MCORF 821 (Fig. 2), which in turn abrogate the accumulation of CL in the cell envelope of M. catarrhalis. To rule out this possibility, we examined expression of MCORF 819 and MCORF 821 in mutant strain O35E.mclS by use of RT-PCR. As shown in Fig. 5B, amplicons of 212 bp (lane 3) and 210 bp (lane 12) indicating transcription of MCORF 819 and MCORF 821, respectively, were generated in the mclS mutant strain. Furthermore, qRT-PCR analysis confirmed that the mclS insertion mutant strain expressed MCORF 821 at levels equal to or even slightly higher than those at which the WT and repaired strains of O35E expressed the gene (see Fig. S1 in the supplemental material). These results indicate that transcription of the flanking genes was not markedly affected by the mutation in mclS.
FIG 5.
Transcription of MCORF 819 and MCORF 821 is not markedly affected by an mclS insertion mutation in M. catarrhalis. (A) Schematic representation of the mclS genetic locus, including MCORF 819 and MCORF 821. The primers used in PCR are indicated by arrows. (B) An agarose gel displaying the results of RT-PCR examining the transcription of MCORF 819 and MCORF 821 in the WT, mclS mutant, and repaired strains of M. catarrhalis isolate O35E. Primers P7 and P8 were used to amplify a portion of MCORF 819, while primers P5 and P6 were used to amplify a portion of MCORF 821. Lane 9, DNA size marker; lanes 1 to 8 and lanes 10 to 17, RT-PCR products of MCORF 819 and MCORF 821, respectively. For RT-PCR, RNA was extracted from M. catarrhalis isolate O35E (WT; lanes 1, 2, 10, 11) and the mclS insertion mutant (O35E.mclS; lanes 3, 4, 12, 13) and mclS repaired (O35E.mclS repaired; lanes 5, 6, 14, 15) strains. During cDNA synthesis, reverse transcriptase enzyme was either added (lanes 1, 3, 5, 10, 12, 14) or withheld (lanes 2, 4, 6, 11, 13, 15). Lanes 7 and 16, PCR products from the genomic DNA of M. catarrhalis isolate O35E; lanes 8 and 17, RT-PCR products from a reaction with no RNA added.
Sung et al. previously demonstrated that proteins of the PLD superfamily require the lysine residue in the C-terminal HKD motif for enzymatic activity (21). It was reported that even the most conservative amino acid change, lysine to arginine, abolished its activity in human PLD. The expression, localization, and protein conformation of PLD were not affected by the mutation. Therefore, a point mutation (lysine to arginine at amino acid position 438, K438 → R) was engineered in the C-terminal HKD motif of the mclS gene product (Fig. 1) of isolate O35E, and the mutated ORF was reintroduced in its original genomic locus, yielding strain O35E.mclS.K438R (Table 1). Confirmation that the mutated mclS gene was transcribed at levels equivalent to those for the WT and repaired strains was attained by RT-PCR (compare lane 11 to lanes 5 and 9 in Fig. 3B) and qRT-PCR (see Fig. S1 in the supplemental material). The PL profile of strain O35E.mclS.K438R was examined by HP-TLC, and these experiments revealed that expression of the mutated mclS gene product abolished accumulation of CL (Fig. 4A, lane 4). Taken together, our data conclusively demonstrate that MclS is a CLS responsible for the production of CL in the cell envelope of M. catarrhalis and that this function is conserved among isolates of various clinical and geographical origins. Moreover, site-directed mutagenesis of the mclS gene product demonstrated that the lysine at position 438 is necessary for CLS activity. This is the first study where the enzymatic activity of a bacterial CLS is abolished due to the generation of a point mutation. This result further validates the claim by Sung et al. that mutagenesis of the lysine residue of the second HKD motif abolishes the enzymatic activity of members of the PLD superfamily (21).
Cardiolipin-deficient M. catarrhalis exhibits WT growth in complete medium and under osmotic stress.
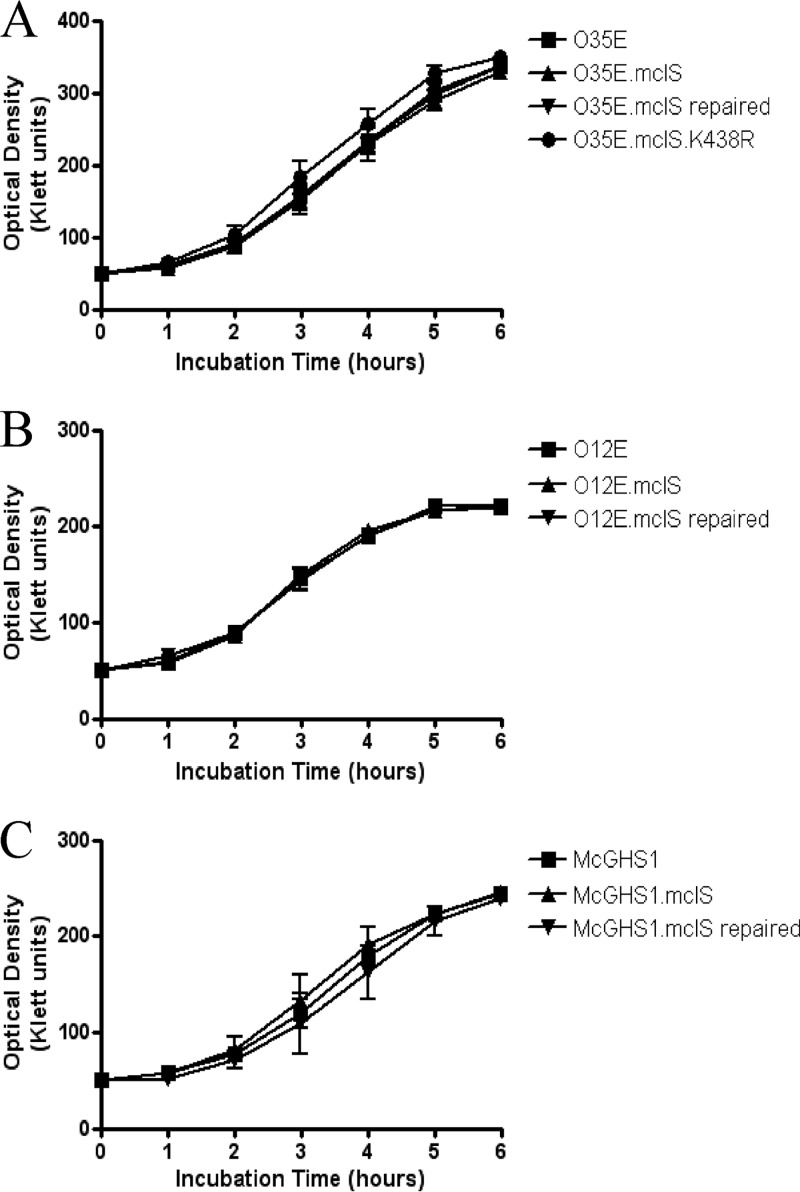
It has previously been shown that CLS is required for viability/optimal growth in vitro for some bacterial species, including E. coli (34, 38). Therefore, we tested whether or not the mclS mutant strains exhibit growth defects by culturing the bacteria in TH broth and monitoring the OD of the cultures over a period of 6 h. As shown in Fig. 6, mutant strains O35E.mclS (Fig. 6A), O12E.mclS (Fig. 6B), and McGHS1.mclS (Fig. 6C) did not display growth defects compared to the growth of the WT and repaired strains. Additionally, the M. catarrhalis strain expressing the lipolytically inactive form of mclS (O35E.mclS.K438R) grew at WT levels (Fig. 6A). These results demonstrate that the mclS gene product is not required for optimal growth of M. catarrhalis in vitro.
FIG 6.
The mclS mutant strains do not exhibit growth defects in vitro. Growth curves of the WT, mclS insertion mutant, mclS repaired, and mclS point mutant strains of M. catarrhalis isolates O35E (A), O12E (B), and McGHS1 (C) are shown.
Previous studies have demonstrated that CL-deficient E. coli displays reduced viability when grown under conditions of osmotic stress (42, 43). To see if CL-deficient M. catarrhalis is also sensitive to osmotic pressure, we examined the growth rates of the WT and mclS mutant strains of O35E in broth containing high concentrations of either sodium chloride or sorbitol. There was no significant difference between WT and CL-deficient M. catarrhalis strains when grown in medium with a high concentration of sodium chloride (see Fig. S2A in the supplemental material). Likewise, there was no significant difference between growth rates when they were grown in media with high concentrations of sorbitol (see Fig. S2B in the supplemental material). Therefore, we conclude that the viability of M. catarrhalis, unlike that of E. coli, does not depend on the presence of CL under conditions of high osmotic stress.
M. catarrhalis lacking CL exhibits reduced levels of adherence to human epithelial cells.
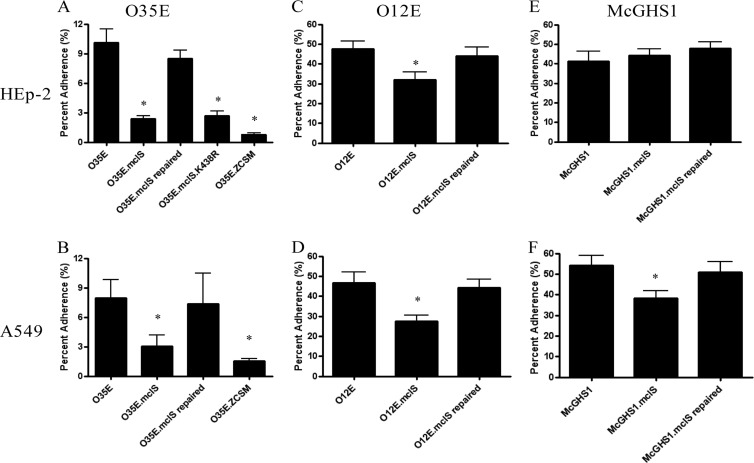
Some members of the PLD superfamily have been shown to affect bacterial adherence to epithelial cells (26, 66). Alteration of the levels of PLs, specifically, PE, has been shown to result in a reduction in virulence-associated traits (67, 68). To date, no CLS has been associated with adherence or virulence in general. For these reasons, we tested our panel of M. catarrhalis strains for adherence to cells of the HEp-2 (laryngeal epithelium) and A549 (type II pneumocytes) human epithelial cell lines. For isolate O35E, adherence to HEp-2 cells was reduced by 76% for the mclS insertion mutant strain compared to that for the WT strain (Fig. 7A). The level of adherence for the mclS insertion mutant was nearly reduced to that for the O35E.ZCSM strain, which lacks expression of three known M. catarrhalis adhesins (UspA1, Hag, and McaP). Adherence to A549 cells by the mclS insertion mutant strain was also significantly reduced (62%) (Fig. 7B). Furthermore, the mclS mutant strains of isolates O12E and McGHS1 displayed comparable reductions in adherence (Fig. 7C to F), except for McGHS1.mclS, which adhered to HEp-2 cells at a WT level. For each isolate, adherence of the repaired strains was restored to WT levels (Fig. 7A to F). To ensure that the adherence defect of the mclS mutant strains was authentic and not simply an artifact of the adherence assay procedure, we monitored the survival of the O35E strains in the reagents used in the procedure: PBSG, tissue culture (TC) medium, and saponin solution. There was no difference in survival between the WT and mclS mutant strains of O35E, therefore demonstrating that the adherence defect is genuine (see Fig. S3 in the supplemental material).
FIG 7.
Moraxella catarrhalis requires CL to exhibit WT levels of adherence to human epithelial cells. Shown are the results of the quantitative adherence assays of M. catarrhalis isolates O35E (A, B), O12E (C, D), and McGHS1 (E, F). The results are expressed as the mean percentage (± standard error) of inoculated bacteria binding to HEp-2 (A, C, E) or A549 (B, D, F) epithelial cell monolayers after 30 min incubation. Asterisks, the reduction in adherence, compared to that of the WT, was found to be statistically significant (P < 0.05, Mann-Whitney test).
Taken together, our data indicate that expression of the mclS gene product impacts the adherence of M. catarrhalis to human epithelial cells. The results, however, do not specify if MclS is an adhesin that directly mediates binding to the surface of epithelial cells. It is possible that the effect of mclS expression on M. catarrhalis adherence is indirect. The absence of CL in the M. catarrhalis membrane may perturb the proper surface display of adhesins, which in turn reduces binding to epithelial cells. To address this, we examined the adherence of strain O35E.mclS.K438R, which expresses a mutated mclS gene product that is lipolytically inactive and does not support the accumulation of CL in the M. catarrhalis membrane (Fig. 4A, lane 4). These experiments revealed that the binding of O35E.mclS.K438R to cells of the HEp-2 and A549 cell lines was equivalent to that of mutant strain O35E.mclS, which lacks expression of the CLS (Fig. 7A). Hence, the data support the hypothesis that the contribution of mclS to adherence is indirect, possibly by modulating the proper surface display of M. catarrhalis adhesins on the bacterial surface through its CLS activity.
To determine if the expression and/or localization of a known adhesin is affected by the absence of CL, we examined whole-cell lysates and outer membrane preparations of the CL-deficient strains of O35E by Western blotting. The following adhesins were expressed at WT levels in the CL-deficient strains of O35E and detected in the whole-cell lysates: Hag, UspA1, McaP, OMP CD, and MhaB1/MhaB2 (see Fig. S4A in the supplemental material). Furthermore, the adhesins were localized to the outer membrane (see Fig. S4B in the supplemental material), as shown by detection of the proteins in the outer membrane preparations. Analysis of the strains by flow cytometry further confirmed not only that the adhesins UspA1, Hag, and McaP are expressed at WT levels in the mclS mutant strains but also that they are localized and displayed on the outer membrane (see Fig. S5 in the supplemental material). By using monoclonal antibody (MAb) 24B5, we were able to detect a single epitope of UspA1 in both O35E and O35E.mclS, suggesting that UspA1 is folded and displayed correctly (see Fig. S5A to C in the supplemental material). Flow cytometry using polyclonal antibodies (pAbs) also detected Hag and McaP in both O35E and O35E.mclS (see Fig. S5D to I in the supplemental material). The negative control, O35E.ZCSM, which lacks Hag, UspA1, and McaP, displayed considerably reduced fluorescence for each of the antibodies (see Fig. S5C, F, and I in the supplemental material). Combined, the results demonstrate that the CL-deficient strains express and display five major M. catarrhalis adhesins (Hag, UspA1, McaP, OMP CD, and MhaB1/MhaB2) on the outer membrane at WT levels.
DISCUSSION
In this study, we have shown that M. catarrhalis expresses a CLS, termed MclS, that contributes to the ability of the bacterium to adhere to human epithelial cells. The mclS gene is predicted to encode a member of the PLD superfamily and displays significant similarity to ymdC/clsC of E. coli. Sequence analysis of numerous M. catarrhalis isolates revealed that mclS is highly conserved throughout the species at both the nucleotide and amino acid levels. Mutant strains of M. catarrhalis isolates O35E, O12E, and McGHS1 that lacked expression of the mclS gene product were constructed. In addition, a mutant strain of isolate O35E that contained a point mutation in one of the two signature HKD motifs of mclS was generated. The mclS insertion and point mutant strains were unable to synthesis CL, thus verifying that MclS is a CLS. The mclS mutant strains also displayed significantly reduced levels of adherence to human epithelial cells compared to their WT counterparts. As expected, the adherence of the mclS-repaired strains was restored to WT levels. The reduction in adherence was attributed to the absence of endogenous CL, as both the mclS insertion mutant strain and the mclS point mutant strain adhered at similar levels. We hypothesize that the absence of CL from the membranes of M. catarrhalis results in altered expression and/or display of adhesins on the outer membrane, thereby contributing to the reduction in adherence to epithelial cells.
The sequence and role of mclS appear to be conserved among M. catarrhalis isolates. The mclS gene was present in all 14 M. catarrhalis isolates examined. Within the 1.6-kb ORF of mclS, only eight SNPs were identified. Three of these SNPs resulted in amino acid changes, none of which occurred in the signal peptide, lipid attachment site, or HKD motifs (Fig. 1). In addition, mutation of mclS in three isolates of M. catarrhalis (O35E, O12E, and McGHS1) yielded strains that were unable to produce CL and displayed reduced levels of adherence to human epithelial cell lines compared to WT strains (Fig. 4 and 7). These data lead us to conclude that the sequence and role of mclS are conserved among M. catarrhalis species.
MclS appears to be unique among CLSs, in that it is predicted to be a lipoprotein, and this property may aid in determining the subcellular localization of the protein within M. catarrhalis. The ProSite algorithm predicts the mclS gene product to be a lipid-anchored protein consisting of an N-terminal signal peptide followed by a cysteine residue at amino acid position 24 serving as the lipid attachment site (Fig. 1). The YmdC/ClsC protein, which has significant similarity with MclS, is not predicted to be a lipoprotein. Instead, YmdC/ClsC contains an N-terminal signal peptide and requires expression of a second protein, YmdB, in order to exhibit phospholipolytic activity (25). Yamaguchi et al. have shown that the amino acid residue immediately following the lipid attachment site is a strong indicator of the localization of lipoproteins in E. coli (69). An aspartic acid at this position targets the lipoprotein for the plasma membrane, while the presence of any other amino acid results in localization at the outer membrane (69). The BRO β-lactamases of M. catarrhalis (BRO-1 and BRO-2) are expressed as lipoproteins and are localized to the periplasmic leaflet of the outer membrane (70). Like BRO-1 and BRO-2, MclS possesses a lysine residue following the lipidated cysteine residue, suggesting outer membrane localization (70). However, a prior study demonstrated that ClsA of E. coli is localized to the periplasmic leaflet of the plasma membrane (71). Cardiolipin, the PL product of CLS, is predominantly found in the plasma membrane of bacteria, though it has been detected in smaller amounts in the outer membrane of E. coli (72). Moraxella catarrhalis does contain higher levels of CL than most bacteria, so it is possible that CL is present at comparatively higher levels in the outer membrane (39, 72). Further experiments must be conducted to determine the subcellular localization of both CL and MclS within M. catarrhalis.
Examination of the genetic organization of the mclS locus by RT-PCR revealed that mclS is transcriptionally linked to both MCORF 819 upstream and MCORF 821 downstream (Fig. 2). Interestingly, the downstream gene MCORF 821 is predicted to encode a protein of the MFS with similarity to ProP, an osmosensory transporter that requires CL for polar localization in E. coli (43, 65). In the E. coli genome, however, proP is not located near any of the three CLS genes. Under conditions of osmotic stress, CL-deficient B. subtilis suffers reduced viability, suggesting a role for CL (direct or indirect) in the regulation of osmotic pressure (42). We found that CL-deficient M. catarrhalis does not display reduced viability when grown in broth with high concentrations of sodium chloride or sorbitol (see Fig. S2 in the supplemental material). This may be due to the possibility that MCORF 821 does not encode a protein that is a functional homolog of ProP. It is also likely that M. catarrhalis expresses multiple osmoregulators whose functions may be unaffected by CL, thereby allowing CL-deficient strains to display WT viability under osmotic pressure. While no functional relationship between the products of the mclS and MCORF 819 genes is apparent, we are currently investigating how the transcriptional linkage between the two genes may impact their roles in M. catarrhalis.
The impact of CL on viability is one example of how the characteristics of CL-deficient bacteria differ between M. catarrhalis and E. coli. As demonstrated by the growth curves of isolates O35E, O12E, and McGHS1 (Fig. 6), there were no significant differences in viability between the WT and CL-deficient strains of M. catarrhalis. The growth curves were extended for several days to include stationary phase, and still no significant differences were observed between the WT and mclS mutant strains (data not shown). In contrast, clsA is required by E. coli to achieve WT growth rates during long-term incubation in stationary phase (38). However, the role of clsB and ymdC/clsC under such conditions has not been investigated, to our knowledge.
Our data indicate that MclS is the only CLS expressed by M. catarrhalis. As shown in Fig. 4, the mclS mutant strains of M. catarrhalis did not contain detectable levels of CL. Moreover, a BLASTP search yielded no additional CLS homologs in the M. catarrhalis genome. Interestingly, previous reports have shown that other bacteria, including E. coli and B. subtilis, express up to three CLSs, which differentially contribute to CL synthesis, depending on the growth phase of the bacteria (25, 38, 41). It has been documented that the CL content of several bacteria, including E. coli, is the highest during stationary phase (38, 73). We did not observe any significant changes in CL content, despite the use of various conditions and evaluations in different growth phases (data not shown). Our data agree with the initial findings by Beebe and Wlodkowski that CL levels in M. catarrhalis remain constant throughout logarithmic and stationary growth (39). However, it is possible that our detection system (HP-TLC with the Molybdenum Blue spray reagent for visualization) was not sensitive enough to perceive relatively small changes in CL content. In contrast to the levels in E. coli, CL levels in M. catarrhalis remained constant throughout exponential and stationary growth and were dependent on only a single CLS enzyme.
Recently, Tan et al. found that not all prokaryotic-type CLSs utilize PG as the sole substrate for CL synthesis, prompting us to examine the substrates of MclS (25). On the basis of the results of the HP-TLC experiments (Fig. 4), it is not apparent whether or not PG and PE levels are altered in the mclS mutant strains compared to those in WT isolates in each instance. Again, it is possible that the detection methods are not sensitive enough to detect relatively small changes in PL levels. It did appear that the intensity of the spot corresponding to PG was increased in the mclS insertion mutant strain compared to that in the WT and repaired strains of O12E (Fig. 4B, lanes 1 to 3). However, this observation alone cannot allow us to conclude which substrates that MclS utilizes for CL synthesis. Based on the similarity of MclS to YmdC/ClsC, we sought to determine if PE is a substrate for MclS by generating strains of M. catarrhalis which lack enzymes specific to the PE synthetic pathway. A BLASTP search of the genome of M. catarrhalis isolate ATCC 43617 identified genes (MCORF 174 and MCORF 700) predicted to encode a phosphatidylserine synthase (PssA) and a phosphatidylserine decarboxylase (Psd), respectively. Generation of pssA and psd mutant strains of M. catarrhalis is under way. We plan to utilize HP-TLC to determine if CL is present in these mutant strains.
This study established that the lysine residue within the HKD motif is required for CLS activity. Site-directed mutagenesis of the lysine residue in the C-terminal HKD motif rendered MclS enzymatically inactive and unable to catalyze the synthesis of CL (Fig. 4). Previously, Tan et al. generated a point mutation in the histidine residues of both HKD motifs of YmdC/ClsC that abolished the phospholipolytic activity of the CLS (25). Together, it appears that the amino acid residues in HKD motifs of CLSs are crucial for activity. This appears to be true for all members of the PLD superfamily, considering that human PLD, yeast PLD, and a vaccinia viral protein were rendered catalytically inactive through mutagenesis of the HKD motif. The lysine-to-arginine mutation within the HKD motif was originally described by Sung et al. using human PLD. The mutation, however, did not alter the expression or localization of the protein (21).
We also conclude that M. catarrhalis requires CL to exhibit WT levels of adherence to human epithelial cells (Fig. 7) and that MclS itself is not an adhesin. Adherence to cells of both the A549 and HEp-2 human epithelial cell lines was significantly reduced in the mclS mutant strains of isolates O35E, O12E, and McGHS1 compared to the WT and repaired strains (Fig. 7). In O35E, adherence to human epithelial cells was reduced when MclS was knocked out (mclS insertion mutant) and rendered enzymatically inactive (mclS point mutant). These results suggest that MclS is not an adhesin itself. The one instance in which adherence was not reduced in the CL-deficient strain (the adherence of McGHS1 to HEp-2 cells; Fig. 7E) may be explained by variation in the usage, expression, or composition of adhesins in different isolates of M. catarrhalis.
The adherence phenotype of the mclS mutant strains may be explained by specific interactions between CL and proteins within M. catarrhalis membranes. In E. coli, CL is known to impact the localization and activity of several proteins, including the SecYEG and SecA components of the Sec translocon. Researchers have shown that CL associates with and stimulates the SecYEG components of the Sec translocon and is required for the proper subcellular distribution of the SecYEG complex (45). It is conceivable that, as in E. coli, M. catarrhalis requires CL for the proper localization and activity of many proteins, including ProP and the Sec translocon. In the absence of CL, the Sec translocon may no longer efficiently transport proteins, including adhesins, through the inner membrane. It is possible that the expression, localization, or display of a known adhesin is altered in the CL-deficient strains, thereby causing the adherence phenotype. According to data obtained by Western blotting and flow cytometry, the adhesins Hag, UspA1, McaP, OMP CD, and MhaB1/MhaB2 are all expressed and localized to the outer membrane. Adherence is a multifactorial process that involves many adhesins, so it is possible that the expression or localization of other adhesins not examined in this study (e.g., lipooligosaccharide) is altered in the CL-deficient strains (15–19, 74, 75). A better understanding of the relationship between CL and these proteins will require characterization of the Sec translocon in M. catarrhalis.
In summary, we discovered that M. catarrhalis expresses a single CLS, MclS, which contributes to the ability of the bacterium to adhere to human epithelial cells. The mclS gene is present in all M. catarrhalis isolates and conserved at greater than 99% identity. The mclS gene product is predicted to be lipid modified, likely promoting an association with the membrane. While MclS displays significant similarity to YmdC/ClsC of E. coli, we have yet to determine if it utilizes PE, in addition to PG, as a substrate for CL synthesis. Like other members of the PLD superfamily, MclS requires intact HKD motifs for catalytic activity. The reduction in adherence observed in the mclS mutant strains can be attributed to the absence of CL. The MclS protein itself is not an adhesin but instead may be necessary for the expression, localization, and display of other M. catarrhalis adhesins. This study marks the first instance in which CL is shown to contribute to virulence-associated traits.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AIO51477 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, awarded to E.R.L.
We thank the Silvia Moreno laboratory in the Department of Cellular Biology at the University of Georgia for their assistance with TLC, Jennifer Willingham Lane for her help with qRT-PCR, and Jamie Barber for his assistance with flow cytometry. Also, we thank Laura Wiese for mutant construction and Teresa Shaffer and Shawn Zimmerman for their helpful comments on the manuscript.
Footnotes
Published ahead of print 18 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00298-13.
REFERENCES
- 1.Verduin CM, Hol C, Fleer A, van Dijk H, van Belkum A. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125–144. 10.1128/CMR.15.1.125-144.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi S, Evans N, Grant BJB, Murphy TF. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465–471. 10.1056/NEJMoa012561 [DOI] [PubMed] [Google Scholar]
- 3.Kilpi T, Herva E, Kaijalainen T, Syrjanen R, Takala AK. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654–662. 10.1097/00006454-200107000-00004 [DOI] [PubMed] [Google Scholar]
- 4.Murphy TF, Parameswaran GI. 2009. Moraxella catarrhalis, a human respiratory tract pathogen. Clin. Infect. Dis. 49:124–131. 10.1086/599375 [DOI] [PubMed] [Google Scholar]
- 5.Faden H. 2001. The microbiologic and immunologic basis for recurrent otitis media in children. Eur. J. Pediatr. 160:407–413. 10.1007/s004310100754 [DOI] [PubMed] [Google Scholar]
- 6.Murphy TF, Brauer AL, Grant BJB, Sethi S. 2005. Moraxella catarrhalis in Chronic obstructive pulmonary disease burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195–199. 10.1164/rccm.200412-1747OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez A, Shibuya K, Rao C, Mathers C, Hansell A, Held L, Schmid V, Buist S. 2006. Chronic obstructive pulmonary disease: current burden and future projections. Eur. Respir. J. 27:397–412. 10.1183/09031936.06.00025805 [DOI] [PubMed] [Google Scholar]
- 8.Dewan NA, Rafique S, Kanwar B, Satpathy H, Ryschon K, Tillotson GS, Niederman MS. 2000. Acute exacerbation of COPD: factors associated with poor treatment outcome. Chest J. 117:662–671. 10.1378/chest.117.3.662 [DOI] [PubMed] [Google Scholar]
- 9.Levy F, Walker E. 2004. BRO β-lactamase alleles, antibiotic resistance and a test of the BRO-1 selective replacement hypothesis in Moraxella catarrhalis. J. Antimicrob. Chemother. 53:371–374. 10.1093/jac/dkh063 [DOI] [PubMed] [Google Scholar]
- 10.Davies B, Maesen F. 1986. Epidemiological and bacteriological findings on Branhamella catarrhalis respiratory infections in The Netherlands. Drugs 31:28–33. 10.2165/00003495-198600313-00008 [DOI] [PubMed] [Google Scholar]
- 11.Boyle FM, Georghiou PR, Tilse MH, McCormack JG. 1991. Branhamella (Moraxella) catarrhalis: pathogenic significance in respiratory infections. Med. J. Aust. 154:592–596 [DOI] [PubMed] [Google Scholar]
- 12.Malmvall BOE, Brorsson JE, Johnsson J. 1977. In vitro sensitivity to penicillin V and β-lactamase production of Branhamella catarrhalis. J. Antimicrob. Chemother. 3:374–375. 10.1093/jac/3.4.374 [DOI] [PubMed] [Google Scholar]
- 13.Omeñaca F, Merino JM, Tejedor JC, Constantopoulos A, Papaevangelou V, Kafetzis D, Tsirka A, Athanassiadou F, Anagnostakou M, François N. 2011. Immunization of preterm infants with 10-valent pneumococcal conjugate vaccine. Pediatrics 128:e290–e298. 10.1542/peds.2010-1184 [DOI] [PubMed] [Google Scholar]
- 14.O'Brien MA, Prosser LA, Paradise JL, Ray GT, Kulldorff M, Kurs-Lasky M, Hinrichsen VL, Mehta J, Colborn DK, Lieu TA. 2009. New vaccines against otitis media: projected benefits and cost-effectiveness. Pediatrics 123:1452–1463. 10.1542/peds.2008-1482 [DOI] [PubMed] [Google Scholar]
- 15.Lafontaine ER, Cope LD, Aebi C, Latimer JL, McCracken GH, Hansen EJ. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364–1373. 10.1128/JB.182.5.1364-1373.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm MM, Vanlerberg SL, Sledjeski DD, Lafontaine ER. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977–4984. 10.1128/IAI.71.9.4977-4984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timpe JM, Holm MM, Vanlerberg SL, Basrur V, Lafontaine ER. 2003. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71:4341–4350. 10.1128/IAI.71.8.4341-4350.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm MM, Vanlerberg SL, Foley IM, Sledjeski DD, Lafontaine ER. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906–1913. 10.1128/IAI.72.4.1906-1913.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balder R, Hassel J, Lipski S, Lafontaine ER. 2007. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect. Immun. 75:2765–2775. 10.1128/IAI.00079-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin EV. 1996. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21:242–243 [PubMed] [Google Scholar]
- 21.Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht JA, Frohman MA. 1997. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16:4519–4530. 10.1093/emboj/16.15.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponting C, Kerr I. 2008. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5:914–922. 10.1002/pro.5560050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Z, Ho WT, Exton JH. 2000. Association of the N- and C-terminal domains of phospholipase D. J. Biol. Chem. 275:24962–24969. 10.1074/jbc.M909745199 [DOI] [PubMed] [Google Scholar]
- 24.Stuckey JA, Dixon JE. 1999. Crystal structure of a phospholipase D family member. Nat. Struct. Mol. Biol. 6:278–284. 10.1038/6716 [DOI] [PubMed] [Google Scholar]
- 25.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CRH, Guan Z. 2012. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U. S. A. 109:16504–16509. 10.1073/pnas.1212797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards JL, Entz DD, Apicella MA. 2003. Gonococcal phospholipase D modulates the expression and function of complement receptor 3 in primary cervical epithelial cells. Infect. Immun. 71:6381–6391. 10.1128/IAI.71.11.6381-6391.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78:1952–1962. 10.1128/IAI.00889-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciervo A, Mancini F, Cassone A. 2007. Transcription, expression, localization and immunoreactivity of Chlamydophila pneumoniae phospholipase D protein. Microb. Pathog. 43:96–105. 10.1016/j.micpath.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 29.Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 73:6668–6673. 10.1128/IAI.73.10.6668-6673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolph AE, Stuckey JA, Zhao Y, Matthews HR, Patton WA, Moss J, Dixon JE. 1999. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 274:11824–11831. 10.1074/jbc.274.17.11824 [DOI] [PubMed] [Google Scholar]
- 31.Lucas E, Billington S, Carlson P, McGee D, Jost B. 2010. Phospholipase D promotes Arcanobacterium haemolyticum adhesion via lipid raft remodeling and host cell death following bacterial invasion. BMC Microbiol. 10:270. 10.1186/1471-2180-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKean SC, Davies JK, Moore RJ. 2007. Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis, is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology 153:2203–2211. 10.1099/mic.0.2007/005926-0 [DOI] [PubMed] [Google Scholar]
- 33.Sandoval-Calderón M, Geiger O, Guan Z, Barona-Gómez F, Sohlenkamp C. 2009. A eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. J. Biol. Chem. 284:17383–17390. 10.1074/jbc.M109.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishijima S, Asami Y, Uetake N, Yamagoe S, Ohta A, Shibuya I. 1988. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J. Bacteriol. 170:775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo D, Tropp BE. 1998. Cloning of the Bacillus firmus OF4 cls gene and characterization of its gene product. Biochim. Biophys. Acta 1389:34–42. 10.1016/S0005-2760(97)00086-6 [DOI] [PubMed] [Google Scholar]
- 36.Von Wallbrunn A, Heipieper H, Meinhardt F. 2002. Cis/trans isomerisation of unsaturated fatty acids in a cardiolipin synthase knock-out mutant of Pseudomonas putida P8. Appl. Microbiol. Biotechnol. 60:179–185. 10.1007/s00253-002-1080-y [DOI] [PubMed] [Google Scholar]
- 37.Hirschberg CB, Kennedy EP. 1972. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 69:648–651. 10.1073/pnas.69.3.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiraoka S, Matsuzaki H, Shibuya I. 1993. Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 336:221–224. 10.1016/0014-5793(93)80807-7 [DOI] [PubMed] [Google Scholar]
- 39.Beebe J, Wlodkowski T. 1976. Lipids of Branhamella catarrhalis and Neisseria gonorrhoeae. J. Bacteriol. 127:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renner LD, Weibel DB. 2011. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. U. S. A. 108:6264–6269. 10.1073/pnas.1015757108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. 2004. Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 186:1475–1483. 10.1128/JB.186.5.1475-1483.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López CS, Alice AF, Heras H, Rivas EA, Sánchez-Rivas C. 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152:605–616. 10.1099/mic.0.28345-0 [DOI] [PubMed] [Google Scholar]
- 43.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64:1455–1465. 10.1111/j.1365-2958.2007.05727.x [DOI] [PubMed] [Google Scholar]
- 44.Ridder ANJA, Kuhn A, Killian JA, de Kruijff B. 2001. Anionic lipids stimulate Sec-independent insertion of a membrane protein lacking charged amino acid side chains. EMBO Rep. 2:403–408. 10.1093/embo-reports/kve087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gold VAM, Robson A, Bao H, Romantsov T, Duong F, Collinson I. 2010. The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. U. S. A. 107:10044–10049. 10.1073/pnas.0914680107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heber S, Tropp BE. 1991. Genetic regulation of cardiolipin synthase in Escherichia coli. Biochim. Biophys. Acta 1129:1–12. 10.1016/0167-4781(91)90206-2 [DOI] [PubMed] [Google Scholar]
- 47.Shibuya I. 1992. Metabolic regulations and biological functions of phospholipids in Escherichia coli. Prog. Lipid Res. 31:245–299. 10.1016/0163-7827(92)90010-G [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49.Bullard B, Lipski SL, Lafontaine ER. 2005. Hag directly mediates the adherence of Moraxella catarrhalis to human middle ear cells. Infect. Immun. 73:5127–5136. 10.1128/IAI.73.8.5127-5136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy TF, Loeb MR. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159–174. 10.1016/0882-4010(89)90066-1 [DOI] [PubMed] [Google Scholar]
- 51.Lipski SL, Akimana C, Timpe JM, Wooten RM, Lafontaine ER. 2007. The Moraxella catarrhalis autotransporter McaP is a conserved surface protein that mediates adherence to human epithelial cells through its N-terminal passenger domain. Infect. Immun. 75:314–324. 10.1128/IAI.01330-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cope LD, Lafontaine ER, Slaughter CA, Hasemann CA, Jr, Aebi C, Henderson FW, McCracken GH, Jr, Hansen EJ. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aebi C, Lafontaine ER, Cope LD, Latimer JL, Lumbley SL, McCracken GH, Jr, Hansen EJ. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarwar J, Campagnari A, Kirkham C, Murphy T. 1992. Characterization of an antigenically conserved heat-modifiable major outer membrane protein of Branhamella catarrhalis. Infect. Immun. 60:804–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bullard B, Lipski S, Lafontaine E. 2007. Regions important for the adhesin activity of Moraxella catarrhalis Hag. BMC Microbiol. 7:65. 10.1186/1471-2180-7-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 57.Wang W, Reitzer L, Rasko DA, Pearson MM, Blick RJ, Laurence C, Hansen EJ. 2007. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect. Immun. 75:4959–4971. 10.1128/IAI.00073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE. 1998. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc. Natl. Acad. Sci. U. S. A. 95:9202–9207. 10.1073/pnas.95.16.9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 60.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. 10.1186/1471-2105-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davie JJ, Earl J, de Vries SPW, Ahmed A, Hu FZ, Bootsma HJ, Stol K, Hermans PWM, Wadowsky RM, Ehrlich GD. 2011. Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genomics 12:70. 10.1186/1471-2164-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yim JJ, Brown GM. 1976. Characteristics of guanosine triphosphate cyclohydrolase I purified from Escherichia coli. J. Biol. Chem. 251:5087–5094 [PubMed] [Google Scholar]
- 63.Nar H, Huber R, Auerbach G, Fischer M, Hösl C, Ritz H, Bracher A, Meining W, Eberhardt S, Bacher A. 1995. Active site topology and reaction mechanism of GTP cyclohydrolase I. Proc. Natl. Acad. Sci. U. S. A. 92:12120–12125. 10.1073/pnas.92.26.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Auerbach G, Herrmann A, Bracher A, Bader G, Gütlich M, Fischer M, Neukamm M, Garrido-Franco M, Richardson J, Nar H. 2000. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc. Natl. Acad. Sci. U. S. A. 97:13567–13572. 10.1073/pnas.240463497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM. 2010. Protein localization in Escherichia coli cells: comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J. Bacteriol. 192:912–924. 10.1128/JB.00967-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucas EA, Billington SJ, Carlson P, McGee DJ, Jost BH. 2010. Phospholipase D promotes Arcanobacterium haemolyticum adhesion via lipid raft remodeling and host cell death following bacterial invasion. BMC Microbiol. 10:270. 10.1186/1471-2180-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bukata L, Altabe S, De Mendoza D, Ugalde RA, Comerci DJ. 2008. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J. Bacteriol. 190:8197–8203. 10.1128/JB.01069-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi W, Bogdanov M, Dowhan W, Zusman D. 1993. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J. Bacteriol. 175:7711–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamaguchi K, Yu F, Inouye M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423–432. 10.1016/0092-8674(88)90162-6 [DOI] [PubMed] [Google Scholar]
- 70.Bootsma HJ, Aerts PC, Posthuma G, Harmsen T, Verhoef J, Van Dijk H, Mooi FR. 1999. Moraxella (Branhamella) catarrhalis BRO β-lactamase: a lipoprotein of gram-positive origin? J. Bacteriol. 181:5090–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibuya I, Yamagoe S, Miyazaki C, Matsuzaki H, Ohta A. 1985. Biosynthesis of novel acidic phospholipid analogs in Escherichia coli. J. Bacteriol. 161:473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raetz CR, Kantor GD, Nishijima M, Newman KF. 1979. Cardiolipin accumulation in the inner and outer membranes of Escherichia coli mutants defective in phosphatidylserine synthetase. J. Bacteriol. 139:544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Short SA, White DC. 1971. Metabolism of phosphatidylglycerol, lysylphosphatidylglycerol, and cardiolipin of Staphylococcus aureus. J. Bacteriol. 108:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luke NR, Jurcisek JA, Bakaletz LO, Campagnari AA. 2007. Contribution of Moraxella catarrhalis type IV pili to nasopharyngeal colonization and biofilm formation. Infect. Immun. 75:5559–5564. 10.1128/IAI.00946-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spaniol V, Heiniger N, Troller R, Aebi C. 2008. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis. Microbes Infect. 10:3–11. 10.1016/j.micinf.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 76.Unhanand M, Maciver I, Ramilo O, Arencibia-Mireles O, Argyle JC, McCracken GH, Hansen EJ. 1992. Pulmonary clearance of Moraxella catarrhalis in an animal model. J. Infect. Dis. 165:644–650. 10.1093/infdis/165.4.644 [DOI] [PubMed] [Google Scholar]
- 77.Pearson MM, Laurence CA, Guinn SE, Hansen EJ. 2006. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect. Immun. 74:1588–1596. 10.1128/IAI.74.3.1588-1596.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whitby PW, Morton DJ, Stull TL. 2006. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol. Lett. 158:57–60. 10.1111/j.1574-6968.1998.tb12800.x [DOI] [PubMed] [Google Scholar]
- 79.Attia AS, Lafontaine ER, Latimer JL, Aebi C, Syrogiannopoulos GA, Hansen EJ. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400–2410. 10.1128/IAI.73.4.2400-2410.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.