Abstract
The genome of Thermococcus kodakarensis, along with those of most Thermococcus and Pyrococcus species, harbors five paralogous genes encoding putative α subunits of nucleoside diphosphate (NDP)-forming acyl coenzyme A (acyl-CoA) synthetases. The substrate specificities of the protein products for three of these paralogs have been clarified through studies on the individual enzymes from Pyrococcus furiosus and T. kodakarensis. Here we have examined the biochemical properties of the remaining two acyl-CoA synthetase proteins from T. kodakarensis. The TK0944 and TK2127 genes encoding the two α subunits were each coexpressed with the β subunit-encoding TK0943 gene. In both cases, soluble proteins with an α2β2 structure were obtained and their activities toward various acids in the ADP-forming reaction were examined. The purified TK0944/TK0943 protein (ACS IIITk) accommodated a broad range of acids that corresponded to those generated in the oxidative metabolism of Ala, Val, Leu, Ile, Met, Phe, and Cys. In contrast, the TK2127/TK0943 protein exhibited relevant levels of activity only toward 2-(imidazol-4-yl)acetate, a metabolite of His degradation, and was thus designated 2-(imidazol-4-yl)acetyl-CoA synthetase (ICSTk), a novel enzyme. Kinetic analyses were performed on both proteins with their respective substrates. In T. kodakarensis, we found that the addition of histidine to the medium led to increases in intracellular ADP-forming 2-(imidazol-4-yl)acetyl-CoA synthetase activity, and 2-(imidazol-4-yl)acetate was detected in the culture medium, suggesting that ICSTk participates in histidine catabolism. The results presented here, together with those of previous studies, have clarified the substrate specificities of all five known NDP-forming acyl-CoA synthetase proteins in the Thermococcales.
INTRODUCTION
Hyperthermophilic archaea that belong to the genera Thermococcus and Pyrococcus are capable of utilizing peptides/amino acids for cell growth (1). The breakdown of amino acids in these organisms is considered to be composed of three steps: (i) a deamination reaction forming 2-oxo acids, (ii) coenzyme A (CoA)-dependent oxidative decarboxylation leading to CoA-thioester compounds, and (iii) hydrolysis of the thioester bond releasing a carboxylic acid and CoA accompanied by substrate-level phosphorylation. Step i is catalyzed by either glutamate dehydrogenase (GDH) in the case of Glu or multiple aminotransferases coupled with GDH for other amino acids (2). Step ii is catalyzed by various ferredoxin-dependent oxidoreductases. A number of enzymes with distinct substrate specificities, including pyruvate:ferredoxin oxidoreductase (POR) (3, 4), 2-oxoisovalerate:ferredoxin oxidoreductase (VOR) (5), indolepyruvate:ferredoxin oxidoreductase (IOR) (6), and 2-oxoglutarate:ferredoxin oxidoreductase (KGOR) (7), have been identified in Pyrococcus furiosus, and closely related genes are also present in other members of the Thermococcales, including T. kodakarensis (8). The final step (step iii) is made possible by various nucleoside diphosphate (NDP)-forming acyl-CoA synthetases. These enzymes catalyze the following reversible reaction: acyl-CoA + NDP + Pi ⇄ acid + CoA + NTP.
The archaeal NDP-forming acyl-CoA synthetases were first identified in P. furiosus and were designated ADP-forming acetyl-CoA synthetase I (ACS IPf) and ACS IIPf (9–15). The enzymes exhibit activity not only toward acetyl-CoA but also toward branched-chain acyl-CoAs (ACS IPf/ACS IIPf) and aryl-CoAs (ACS IIPf) (12). ADP-forming ACSs have also been identified in Archaeoglobus fulgidus (16, 17), Methanocaldococcus jannaschii (17), Pyrobaculum aerophilum (18), Thermococcus kodakarensis (ACS IITk) (19), and the halophilic Haloarcula marismortui (18). The enzymes display a common structure and are comprised of two α and two β subunits (α2β2) or are homodimers of α-β fusion proteins. They have been proposed to be members of the nucleoside diphosphate (NDP)-forming acyl-CoA synthetase superfamily (20).
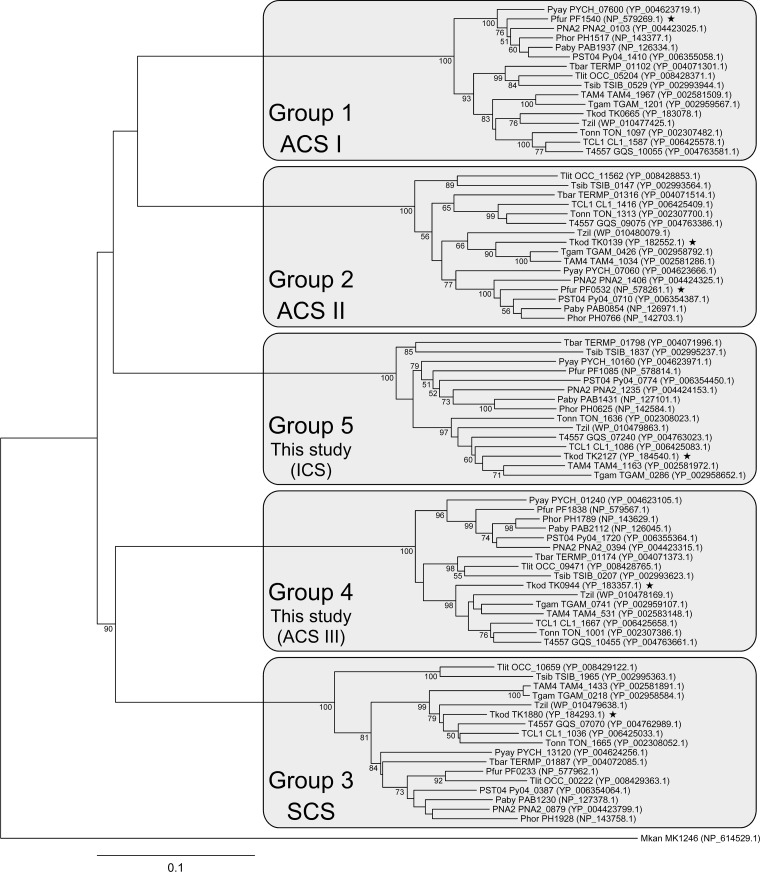
Almost all genomes of Thermococcales members harbor five α subunit and two β subunit paralogs. It has been suggested that the two β subunits can assemble in T. kodakarensis with multiple α subunits and that the α subunits are responsible for the substrate specificity of the enzymes in terms of acid/acyl-CoA compounds (19). As described below, the α subunits in Thermococcales can be phylogenetically divided into five groups (Fig. 1). One group is represented by ACS IPf (α subunit; PF1540), and another group includes ACS IIPf (α subunit; PF0532) and ACS IITk (α subunit; TK0139), while a third group includes the α subunit of an ADP-forming acyl-CoA synthetase specific to succinate/succinyl-CoA (succinyl-CoA synthetase [SCSTk]) (α subunit; TK1880) from T. kodakarensis (19). The enzyme has recently been shown to be involved in Glu catabolism and succinate generation (2).
Fig 1.
Phylogenetic analysis of NDP-forming acyl-CoA synthetase α subunits of the Thermococcales. The five NDP-forming acyl-CoA synthetase α subunit homologs present on the genomes of various members of the Thermococcales were aligned, and a phylogenetic tree was constructed with the neighbor-joining method. Species are Thermococcus kodakarensis (Tkod), Thermococcus barophilus (Tbar), Thermococcus gammatolerans (Tgam), Thermococcus litoralis (Tlit), “Thermococcus onnurineus” (Tonn), Thermococcus sibiricus (Tsib), Thermococcus zilligii (Tzil), Thermococcus sp. AM4 (TAM4), Thermococcus sp. CL1 (TCL1), Thermococcus sp. 4557 (T4557), Pyrococcus furiosus (Pfur), “ Pyrococcus abyssi” (Paby), Pyrococcus horikoshii (Phor), Pyrococcus yayanosii (Pyay), Pyrococcus sp. NA2 (PNA2), and Pyrococcus sp. ST04 (PST04). Both locus tags and accession numbers are shown except for the proteins from T. zilligii, which are identified only with accession numbers. An NDP-forming acyl-CoA synthetase α subunit homolog from Methanopyrus kandlerii (Mkan, MK1246) was used as the outgroup. Bootstrap values above 50 are shown. Stars indicate proteins that have been examined experimentally.
In this study, we examined the biochemical properties of the NDP-forming acyl-CoA synthetases from T. kodakarensis whose α subunits are composed of TK0944 and TK2127, with TK0943 as the β subunit. These proteins are included in the two remaining groups of ACS α subunits, whose members have not yet been biochemically characterized. While TK0944/TK0943 recognized a relatively broad range of substrates, we found that TK2127/TK0943 displayed strict substrate specificity, leading us to conclude that the enzyme is an ADP-forming 2-(imidazol-4-yl)acetyl-CoA synthetase.
MATERIALS AND METHODS
Phylogenetic tree analysis.
Sequences of the ACS α subunits from all members of the Thermococcales available from NCBI were collected and aligned using the ClustalW program provided by the DNA Databank of Japan (DDBJ). Multiple sequence alignment was performed with the following default parameters: Protein Weight Matrix, Gonnet; GAP OPEN, 10; GAP EXTENSION, 0.20; GAP DISTANCES, 5; NO END GAPS, no; ITERATION, none; NUMITER, 1; and CLUSTERING, NJ. The phylogenetic tree was constructed by the neighbor-joining method (21). Bootstrap resampling was performed 1,000 times.
Microorganisms, plasmids, and media.
Thermococcus kodakarensis KOD1 (22, 23) cells were grown under anaerobic conditions at 85°C in a nutrient-rich growth medium (ASW-YT-S0 medium). ASW-YT-S0 medium contains 5.0 g liter−1 yeast extract, 5.0 g liter−1 tryptone, 16 g liter−1 NaCl, 2.4 g liter−1 MgCl2·6H2O, 4.8 g liter−1 MgSO4·7H2O, 0.8 g liter−1 (NH4)2SO4, 0.16 g liter−1 NaHCO3, 0.24 g liter−1 CaCl2·2H2O, 0.4 g liter−1 KCl, 0.336 g liter−1 KH2PO4, 40 mg liter−1 NaBr, 16 mg liter−1 SrCl2·6H2O, 8 mg liter−1 ferric ammonium citrate, with 2.0 g liter−1 elemental sulfur (S0). The medium was supplemented with 0.1 g liter−1 Na2S and 0.25 mg liter−1 resazurin. Escherichia coli DH5α and plasmid pUC118 were used for general DNA manipulation and sequencing. E. coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) and pET21a(+) (Novagen, Madison, WI) were used for gene expression. E. coli strains were cultivated in Luria-Bertani (LB) medium (10 g liter−1 tryptone, 5 g liter−1 yeast extract, 10 g liter−1 NaCl, pH 7.0) at 37°C. Ampicillin was added to the medium at a concentration of 100 μg ml−1. Unless mentioned otherwise, all chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
DNA manipulation and sequencing.
KOD Plus polymerase (Toyobo, Osaka, Japan) was used for PCR. Restriction and modification enzymes were purchased from Toyobo. Plasmid DNA was isolated with a Qiagen Plasmid Mini Kit (Qiagen, Hilden, Germany) or a Quantum Prep Mini Kit (Bio-Rad, Hercules, CA). DNA fragments were recovered from agarose gels with a Wizard SV gel and PCR clean-up system (Promega, Madison, WI). DNA sequencing was performed with a BigDye Terminator cycle sequencing kit (v.3.1) and a model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Cloning and expression of genes TK0944 and TK2127 with gene TK0943 in E. coli.
The TK0943, TK0944, and TK2127 genes were amplified from T. kodakarensis genomic DNA with the following primers: for TK0943, F0943 (5′-CCCTCTAGAATTCAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGCGCCAAAGAAGAGGCCC-3′) and R0943 (5′-CTTGTCGACTCACTCTTTCTTTTCTGGAGCTTTC-3′) (underlined sequences indicate EcoRI and SalI sites); for TK0944, F0944 (5′-CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGTCAGAGAAAATCGTCGAAGAACTC-3′) and R0944 (5′-TTAGAATTCGGAACCTCATTCCCCCTTAACCTTGCGG-3′) (underlined sequences indicate XbaI and EcoRI sites); and for TK2127, F2127 (5′-CCTTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGCCTAGACTTCTTTTTCTACCCG-3′) and R2127 (5′-TTCGAATTCGGAACCCTATCTCATCGAGAGCCACTTC-3′) (underlined sequences indicate XbaI and EcoRI sites). The amplified fragments were individually inserted into pUC118 and sequenced. After confirming the absence of unintended mutations, the EcoRI-SalI-digested fragment of TK0943 was inserted into pET21a(+), resulting in plasmid pET-TK0943. The XbaI-EcoRI-digested fragments of TK0944 and TK2127 were inserted into pET-TK0943 upstream of TK0943, resulting in plasmids pET-TK0944/0943 and pET-TK2127/0943, respectively. In these plasmids, besides the ribosomal binding site upstream of TK0944 and TK2127, another ribosomal binding site is inserted upstream of TK0943. After sequence confirmation, the plasmids were introduced into E. coli BL21-CodonPlus(DE3)-RIL and grown in LB medium at 37°C to an optical density (at 660 nm) of 0.5. Gene expression was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside at the mid-exponential-growth phase followed by further incubation for 4 h. Cells were harvested by centrifugation (6,000 × g, 4°C, 15 min), washed with 50 mM Tris-HCl buffer (pH 7.5), and centrifuged again (4,500 × g, 4°C, 20 min).
Purification of recombinant TK0944/TK0943 and TK2127/TK0943 proteins.
The collected cells were resuspended in 50 mM Tris-HCl buffer (pH 7.5) and sonicated for 10 min (output:pause = 0.5:0.5 s; output power = ∼60 W) with a UD-201 Ultrasonic Disruptor (Tomy, Tokyo, Japan). The lysates were centrifuged (15,000 × g, 4°C, 15 min), and the respective supernatants were subjected to heat treatment (15 min at 85°C). After removing heat-labile proteins by centrifugation (15,000 × g, 4°C, 30 min), the supernatants were applied to a Resource Q anion exchange column (GE Healthcare, Little Chalfont, United Kingdom) (6 ml) equilibrated with buffer A (50 mM Tris-HCl buffer, pH 7.5). Recombinant proteins were eluted with a linear gradient of 0 to 1 M NaCl in buffer A. Fractions containing recombinant TK0944/TK0943 or TK2127/TK0943 were concentrated and applied to a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) with a mobile phase of 50 mM Tris-HCl buffer (pH 7.5) containing 150 mM NaCl at a flow rate of 0.5 ml min−1. Protein concentrations were determined with a protein assay kit (Bio-Rad) with bovine serum albumin as a standard. The approximate sizes and molecular weights of the proteins were examined by gel filtration with a Superdex 200 HR 10/30 column using the standard Blue Dextran 2000, a gel filtration HMW calibration kit, and a gel filtration LMW calibration kit (GE Healthcare).
Activity measurements of recombinant TK0944/TK0943 and TK2127/TK0943 proteins.
For kinetic analyses and the examination of activity toward various acid compounds, ADP formation was coupled with pyruvate kinase and lactate dehydrogenase (PK/LDH), and the oxidation of NADH was monitored spectrophotometrically at 340 nm. Reactions were performed at 55°C. The reaction mixture contained the acid substrate (10 mM), ATP (1 mM), CoA (1.5 mM), MgCl2 (5 mM), phosphoenolpyruvate (1.5 mM), NADH (0.15 mM), recombinant protein (2 μg ml−1), and pyruvate kinase/lactate dehydrogenase from rabbit muscle (Sigma, St. Louis, MO) (PK/LDH, 20 U/14 U) in 50 mM MES (2-morpholineethanesulfonic acid)-NaOH buffer (pH 6.5). The equation v = Vmax{[S]/Ks1 + [S]2/(Ks1Ks2)}/{1 + 2[S]/Ks1 + [S]2/(Ks1Ks2)} was used to calculate the kinetic parameters with respect to acid substrates, where v is the reaction velocity, Vmax is the maximum velocity, [S] is the substrate concentration, and Ks1 and Ks2 are the dissociation constants of the first and second acid substrates, respectively. When measuring activity directed at CoA thioester cleavage, the formation of free CoASH was determined using 5,5′-dithiobis(2-nitrobenzoic acid) with methods described in a previous study (19). The reaction mixture contained 0.5 mM acetyl-CoA (Sigma), isovaleryl-CoA (Sigma), or isobutyryl-CoA (Sigma) or 1 mM succinyl-CoA (Sigma), 0.4 mM ADP (Oriental Yeast, Tokyo, Japan), 20 mM sodium phosphate, 0.1 mM 5,5′-dithiobis(2-nitrobenzoic acid), 10 mM MgCl2, and recombinant protein (2 μg ml−1) in 50 mM MES-NaOH (pH 6.5). Reactions were performed at 55°C, and the increase in absorbance at 412 nm was monitored continuously.
Effects of exogenous histidine on 2-(imidazol-4-yl)acetyl-CoA synthetase activity in T. kodakarensis cells.
2-(Imidazol-4-yl)acetyl-CoA synthetase activity was measured in the extracts of cells grown in the absence or presence (0.95, 2.39, or 4.77 mM) of exogenous histidine in ASW-YT-S0 medium. Activity was measured according to the methods described above using 34.5 μg of cell extract instead of the recombinant protein. Background levels observed in the absence of 2-(imidazol-4-yl)acetate were subtracted to calculate activity levels.
Detection of 2-(imidazol-4-yl)acetate in the growth medium.
T. kodakarensis KOD1 cells were cultivated in ASW-YT-Pyr medium (2) supplemented with 0.2 g liter−1 of His at 85°C. After 10 h, aliquots of the culture were taken, cooled on ice, and centrifuged (20,000 × g, 4°C, 5 min). The supernatant was filtered with USY-1 disposable ultrafilter units (Advantec, Tokyo, Japan). A 10-μl volume of aliquot was applied to a VP-ODS Shim-pack (Shimadzu, Kyoto, Japan) (150 mm by 4.6 mm) at 40°C. The mobile phase was a gradient of acetonitrile in 50 mM sodium phosphate buffer (pH 2.3) containing 5 mM sodium hexanesulfonate at a flow rate of 0.8 ml min−1. 2-(Imidazol-4-yl)acetate was detected with a SPD-M20A photodiode array detector (Shimadzu) with a wavelength of 230 nm.
RESULTS
Phylogenetic examination of the α subunits of NDP-forming acyl-CoA synthetase in the Thermococcales.
As mentioned above, almost all Thermococcales members harbor five ACS α subunit paralogs on their genomes, along with two β subunit paralogs. A phylogenetic examination was performed on the ACS α subunits from all members of the Thermococcales whose genome sequences were available (Fig. 1). The analysis revealed that the proteins clustered into five groups, which we designated groups 1 to 5. The structural similarities among members of a single group are high. In group 1, the protein most distantly related to TK0665 is the protein from Pyrococcus sp. NA2, which is 84.7% identical to the TK0665 protein. On the other hand, the protein most closely related to TK0665 from the other groups is the PAB1230 protein from “Pyrococcus abyssi,” which is only 45.2% identical. Likewise, the proteins most distantly related to TK0139 (group 2), TK1880 (group 3), TK0944 (group 4), and TK2127 (group 5) but belonging to the same group displayed identities of 77.9%, 76.1%, 80.2%, and 77.5%, respectively. The proteins from other groups that were most closely related to these proteins displayed 47.4%, 50.7%, 50.8%, and 47.0% identity, respectively. This suggests that each α subunit from one organism corresponds to a single α subunit in most of the other organisms. This raises the possibility that members of a particular group share similar substrate specificities with one another. Supporting this, it has been shown that ACS IIPf (α: PF0532) and ACS IITk (α: TK0139), both with α subunit members of group 2, display similar substrate preferences (12, 19).
Expression of the TK0944/TK0943 and TK2127/TK0943 genes in E. coli.
Among the five groups of NDP-forming acyl-CoA synthetase α subunits of the Thermococcales (Fig. 1), at least one member of groups 1 to 3 has been biochemically examined with enzymes from either P. furiosus or T. kodakarensis (2, 9–15, 19). In this study, we set out to determine the biochemical properties of enzymes that are composed of the group 4 and group 5 α subunits from T. kodakarensis. It has previously been shown that β subunits can assemble with multiple α subunits in vivo. Furthermore, the recombinant TK0943 protein (β subunit) assembles with the TK1880 protein (α subunit) to form an active and thermostable succinyl-CoA synthetase, as well as with the TK0139 protein (α subunit), resulting in an active acetyl-CoA synthetase II in vitro (19). The results indicated that the substrate specificities of these enzymes are dependent on the α subunit, most likely through substrate recognition and binding by domain 5 of the α subunit (19). Here we expressed the TK0944 or TK2127 gene with the TK0943 gene in E. coli. Recombinant proteins with the expected molecular masses (for TK0944, 51.9 kDa; for TK2127, 47.6 kDa; and for TK0943, 27.1 kDa) were recovered in a soluble form, and the cell extracts were subjected to heat treatment, anion exchange, and gel filtration chromatography. SDS-PAGE analysis showed that the recombinant proteins were purified to apparent homogeneity (Fig. 2). The two proteins, although to different extents, displayed acyl-CoA synthetase activity with acetate as the acid substrate.
Fig 2.
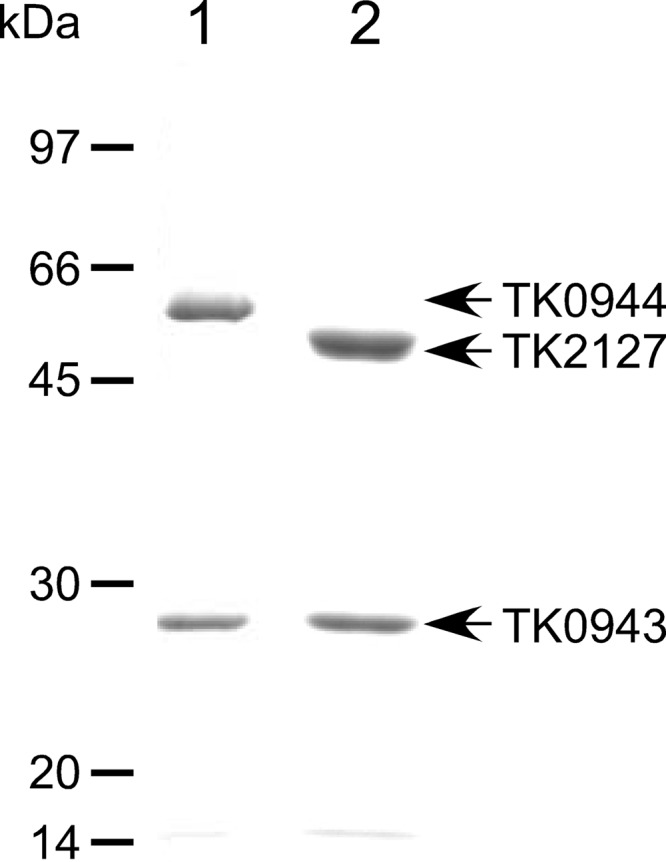
SDS-PAGE analysis of the purified, recombinant TK0944/TK0943 and TK2127/TK0943 proteins. A 3-μg volume of TK0944/TK0943 (lane 1) and TK2127/TK0943 (lane 2) was applied, and the gels were stained with Coomassie brilliant blue.
Assembly of the TK0944/TK0943 and TK2127/TK0943 proteins.
Gel-filtration chromatography and SDS-PAGE revealed that the TK0944 or TK2127 protein coeluted with the TK0943 protein, indicating that each α subunit assembled with the TK0943 β subunit. Estimated molecular weights were consistent with an α2β2 subunit assembly and were the same as those observed for previously characterized NDP-forming acyl-CoA synthetases of the Thermococcales. In order to examine the stability of these complexes, we measured the thermostability of TK0944/TK0943 and TK2127/TK0943. The half-lives at 80 and 90°C were 380 and 170 min for TK0944/TK0943 and 140 and 120 min for TK2127/TK0943, respectively. The results of gel-filtration chromatography and the high thermostability of the two proteins indicated that the recombinant TK0944/TK0943 and TK2127/TK0943 proteins were properly assembled and thus were subjected to further enzymatic analyses.
Activity toward various acid substrates.
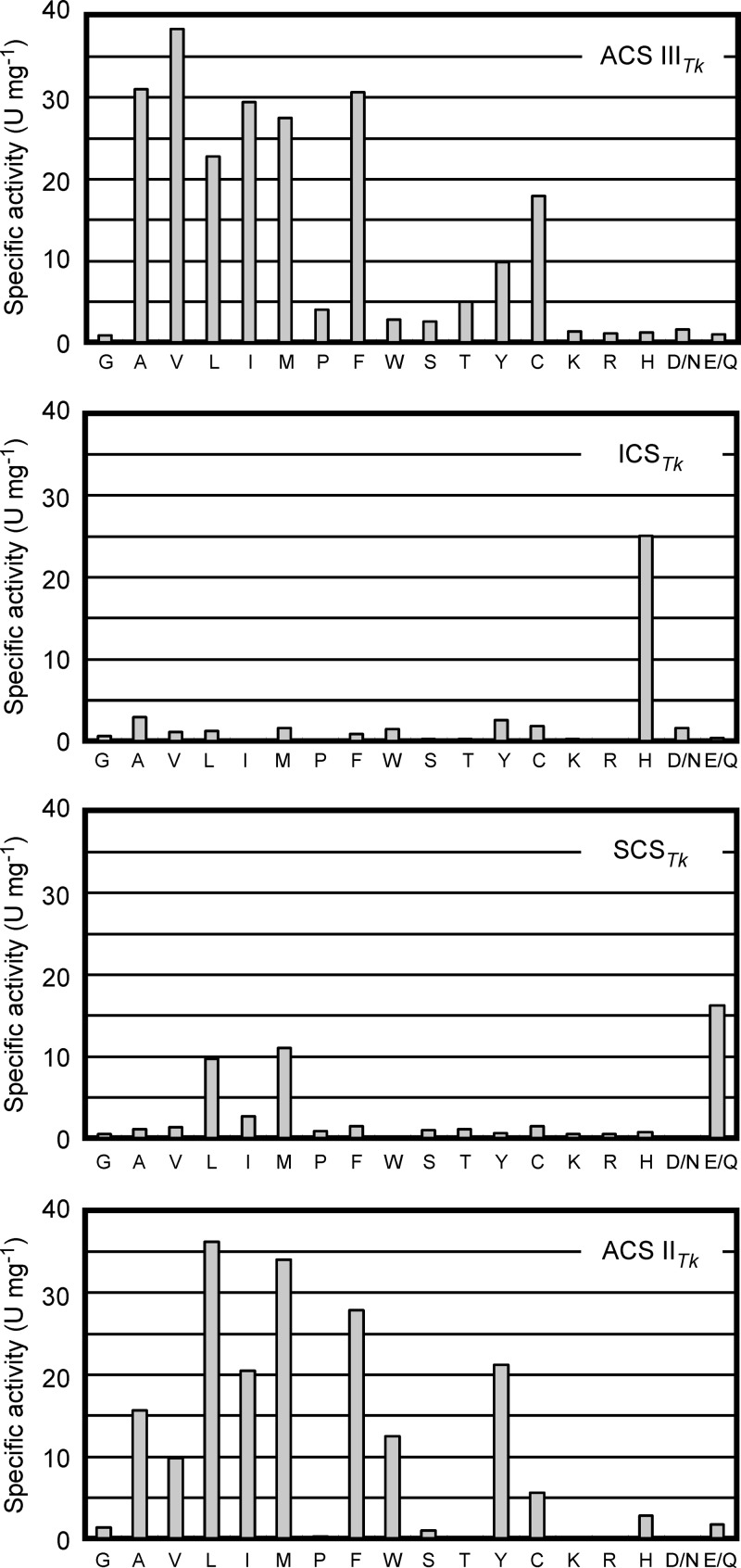
ADP-forming acyl-CoA synthetase activities of the TK0944/TK0943 and TK2127/TK0943 proteins were examined in the acid-utilizing direction with 18 different acid compounds, corresponding to metabolites that are expected to be formed via oxidative degradation of amino acids in the Thermococcales. Each acid substrate was present at a concentration of 10 mM, and ATP was used as the nucleoside 5′-triphosphate. As shown in Fig. 3, the TK0944/TK0943 protein displayed significant levels of acetyl-CoA synthetase activity that were higher than those seen for ACS IITk. Activity was also observed toward a broad range of other substrates, with a preference for those derived from the oxidation of hydrophobic amino acids. As these properties are similar to those observed for ACS IITk (19) and ACS IIPf (11, 12), we gave the TK0944/TK0943 enzyme the designation ACS IIITk. ACS IIITk displayed activities with isobutyrate and thioglycolate, products of Val and Cys degradation, respectively, that were higher than those seen with ACS IITk. In contrast to the broad specificity of ACS IIITk, the TK2127/TK0943 protein exhibited a strict preference for 2-(imidazol-4-yl)acetate, the metabolite expected in the degradation of histidine (Fig. 3). The enzyme was therefore designated 2-(imidazol-4-yl)acetyl-CoA synthetase (ICSTk). To our knowledge, this is the first report identifying an enzyme with this activity.
Fig 3.
Activity levels of TK0944/TK0943 and TK2127/TK0943 proteins for various acid compounds. Amino acids, represented by single-letter codes, and the corresponding acid compounds are as follows: G, formate; A, acetate; V, isobutyrate; L, isovalerate; I, 2-methylbutyrate; M, 3-methylthiopropionate; P, 4-aminobutyrate; F, phenylacetate; W, 2-(indol-3-yl)acetate; S, glycolate; T, lactate; Y, 2-(4-hydroxyphenyl)acetate; C, thioglycolate; K, 5-aminovalerate; R, 4-guanidinobutyrate; H, 2-(imidazol-4-yl)acetate; D/N, malonate; and E/Q, succinate. Activities of TK0944/TK0943 (top [first] panel) and TK2127/TK0943 (second panel) proteins are shown, along with previously reported results for SCSTk (third panel) and ACS IITk (bottom panel) (19).
Enzyme activities were also examined in the ATP-forming (acid-generating) direction with several commercially available substrates at a concentration of 0.5 mM. In the case of ACS IIITk, ATP-forming activity was observed with acetyl-CoA, isobutyryl-CoA, and isovaleryl-CoA but not with succinyl-CoA (see Table S1 in the supplemental material). In the case of ICSTk, clear levels of activity could not be detected, but low levels of activity were observed with acetyl-CoA at a concentration of 1 mM. The substrate specificities are in good agreement with the results obtained for these enzymes in the ADP-forming direction.
Kinetic analyses of ACS IIITk and ICSTk.
As ACS IIITk displayed significant levels of activity toward several acid substrates, we performed kinetic analyses of the reactions with isobutyrate, acetate, thioglycolate, and phenylacetate at substrate concentrations of 0.2 to 20 mM. Kinetic parameters for CoA and ATP were also determined (Table 1). When various CoA (12 μM to 1,600 μM) and ATP (20 μM to 1,000 μM) concentrations were employed in the presence of 10 mM isobutyrate, the reactions followed Michaelis-Menten kinetics. The Km values of CoA and ATP were 5.6 ± 0.4 and 56 ± 3 μM, respectively. As in the case of SCSTk (19), the ACS IIITk reaction did not follow Michaelis-Menten kinetics when the concentration of acid substrates was adjusted. The data fit well with the assumption that the Ks value of the second acid substrate to the dimeric (αβ)2 catalytic unit (Ks2) differs from that of the first (Ks1) by a factor of a (= Ks2/Ks1). The parameters are shown in Table 1. Among the four acid substrates, acetate displayed a relatively high Ks1 value and low kcat/Ks1, indicating that acetate is not the preferred substrate of ACS IIITk. However, the Ks1 value (1.5 mM) is not high compared to those of ACS IPf (1.1 mM) and ACS IIPf (10.7 mM) toward acetate (11, 12) and thus does not necessarily rule out the involvement of ACS IIITk in the conversion of acetyl-CoA to acetate. Kinetic analyses were also performed on ICSTk with CoA, ATP, and the only acid substrate with relevant levels of activity, 2-(imidazol-4-yl)acetate (Table 1). The Km value toward CoA (62.5 μM) was over 10-fold higher than that observed for ACS IIITk but was approximately equivalent to those observed for SCSTk (60 μM) and ACS IIPf (74 μM). The kcat/Ks1 value with 2-(imidazol-4-yl)acetate (69.7 s−1 mM−1) was higher than that observed for SCSTk with succinate (10 s−1 mM−1) (19), indicating that its catalytic efficiency is comparable to those of previously reported ACS enzymes.
TABLE 1.
Kinetic parameters of ACS IIITk and ICSTka
| Substrate | Kinetic model | Vmax (U mg−1) ± SD | kcat (s−1 active site−1) ± SD | Ks1 or Km (mM) ± SD | Ks2 (mM) ± SD | kcat/Ks1 or kcat/Km (s−1 active site−1 mM−1) |
|---|---|---|---|---|---|---|
| ACS IIITk (TK0944/TK0943) | ||||||
| Isobutyrate | Adair-Pauling | 38.7 ± 0.2 | 50.9 ± 0.3 | 0.132 ± 0.054 | 0.0860 ± 0.0156 | 385 |
| Acetate | Adair-Pauling | 37.4 ± 0.6 | 49.3 ± 0.7 | 1.50 ± 0.07 | 2.06 ± 0.21 | 32.8 |
| Thioglycolate | Adair-Pauling | 19.8 ± 0.3 | 26.1 ± 0.4 | 0.322 ± 0.038 | 1.12 ± 0.14 | 81.0 |
| Phenylacetate | Adair-Pauling | 30.8 ± 0.2 | 40.6 ± 0.2 | 0.329 ± 0.094 | 0.0706 ± 0.0140 | 123 |
| CoA | Michaelis-Menten | 40.4 ± 0.2 | 53.1 ± 0.3 | 0.00560 ± 0.00040 | 9,490 | |
| ATP | Michaelis-Menten | 40.8 ± 0.4 | 53.7 ± 0.5 | 0.0556 ± 0.0026 | 965 | |
| ICSTk (TK2127/TK0943) | ||||||
| 2-(Imidazol-4-yl)-acetate | Adair-Pauling | 30.3 ± 0.5 | 37.8 ± 0.7 | 0.543 ± 0.036 | 2.12 ± 0.26 | 69.7 |
| CoA | Michaelis-Menten | 31.7 ± 0.2 | 39.6 ± 0.3 | 0.0625 ± 0.0025 | 633 | |
| ATP | Michaelis-Menten | 30.4 ± 1.0 | 37.9 ± 1.2 | 0.0300 ± 0.0011 | 1,260 |
Kinetic analyses on acid substrates were performed in the presence of 1 mM ATP and 1.5 mM CoA. Kinetic analyses on CoA were performed in the presence of 10 mM isobutyrate (ACS IIITk) or 2-(imidazol-4-yl)acetate (ICSTk) and 1 mM ATP. Kinetic analyses on ATP were performed in the presence of 10 mM isobutyrate (ACS IIITk) or 2-(imidazol-4-yl)acetate (ICSTk) and 1.5 mM CoA.
Detection of ICS activity in T. kodakarensis cells.
It is generally accepted that the NDP-forming acyl-CoA synthetases of the Thermococcales play a catabolic role in amino acid metabolism. As ACS IITk, ACS IIITk, and SCSTk do not exhibit relevant levels of acyl-CoA synthetase activity toward 2-(imidazol-4-yl)acetate, we anticipated that it would be possible to examine the intracellular levels of ICSTk by measuring ICS activity. This would not be possible with the individual ACS enzymes, as ACS activity would be a sum of the activities of ACS I to IIITk. Although we have not examined the substrate preference of ACS ITk, we assumed it unlikely that the enzyme displays ICS activity, considering the properties of ACS IPf (11, 12). T. kodakarensis cells were grown in ASW-YT-S0 medium in the absence or presence (0.95, 2.39, or 4.77 mM) of exogenous histidine. While activity levels were 0.020 μmol min−1 (U) mg extract−1 in the absence of histidine, levels of 0.045 to 0.053 μmol min−1 mg extract−1 were observed with the addition of exogenous histidine. The levels of acetyl-CoA synthetase activity did not change under these conditions, with 0.169 μmol min−1 mg extract−1 in the absence of histidine and 0.150 to 0.171 μmol min−1 mg extract−1 in the presence of histidine, supporting the assumption that the ICS activity was due to the function of ICSTk and not to the function of ACS ITk. Furthermore, by examining the culture supernatant after 10 h of growth in ASW-YT-Pyr medium containing 0.95 mM His, we were able to detect the production of 5.3 mg liter−1 of 2-(imidazol-4-yl)acetate, the expected end product of His degradation. The results suggest that ICSTk plays a role in histidine catabolism in T. kodakarensis.
DISCUSSION
Taken together with the results from previous studies, the data presented here provide the first overall indications as to what types of NDP-forming acyl-CoA synthetases are present in the Thermococcales (summarized in Table 2). We cannot rule out the possibility that members of a single group from different organisms exhibit entirely different substrate specificities. However, the striking relatedness among the members within a single group (>76% identical to T. kodakarensis homologs), compared to those between members of different groups (<51% identical to T. kodakarensis homologs), suggests otherwise (Fig. 1).
TABLE 2.
The substrate specificities of the five ADP-forming acyl-CoA synthetases in Thermococcales
| Enzyme | Abbreviation | Activity detected with: | Involved in the metabolism of: | References or source |
|---|---|---|---|---|
| Acetyl-CoA synthetase I | ACS I | Acetate | Ala | 10, 12, 13 |
| Isobutyrate | Val | |||
| Isovaleratea | Leua | |||
| Acetyl-CoA synthetase II | ACS II | Acetate | Ala | 12, 19 |
| Isobutyrate | Val | |||
| Isovalerate | Leu | |||
| 2-Methylbutyrate | Ile | |||
| 3-Methylthiopropionate | Met | |||
| Phenylacetate | Phe | |||
| 2-(4-Hydroxyphenyl)acetate | Tyr | |||
| 2-(Indol-3-yl)acetate | Trp | |||
| Acetyl-CoA synthetase III | ACS III | Acetate | Ala | This study |
| Isobutyrate | Val | |||
| Isovalerate | Leu | |||
| 2-Methylbutyrate | Ile | |||
| 3-Methylthiopropionate | Met | |||
| Phenylacetate | Phe | |||
| Thioglycolate | Cys | |||
| 2-(4-Hydroxyphenyl)acetatea | Tyra | |||
| Succinyl-CoA synthetase | SCS | Succinate | Glu | 2, 19 |
| Isovalerate | Leu | |||
| 3-Methylthiopropionate | Met | |||
| 2-(Imidazol-4-yl)acetyl-CoA synthetase | ICS | 2-(Imidazol-4-yl)acetate | His | This study |
Relatively low activity levels.
Group 1 enzymes, represented by ACS IPf, exhibit relatively high levels of activity toward acetate, propionate, butyrate, isobutyrate, and, to a lesser extent, isovalerate in the acid-utilizing direction. 2-(Indol-3-yl)acetate, phenylacetate, and succinate could not be utilized. Acid-forming activity has also been observed for acetyl-CoA, propionyl-CoA, butyryl-CoA, and isobutyryl-CoA but not for phenylacetyl-CoA (10, 12). The results suggest that group 1 enzymes are involved in the metabolism of Ala, Val, and possibly Leu. Members of group 2, represented by ACS IIPf and ACS IITk, accommodate a broader range of substrates, and besides the substrates recognized by ACS IPf, the enzymes also display activity toward aromatic acids such as phenylacetate, 2-(indol-3-yl)acetate, and 2-(4-hydroxyphenyl)acetate, as well as 2-methylbutyrate and 3-methylthiopropionate (12, 19). This implies the involvement of group 2 enzymes in the metabolism of Ala, Val, Leu, Ile, Met, Phe, Trp, and Tyr. It should be noted that ACS IPf and ACS IIPf display comparable Km values for acetyl-CoA and isobutyryl-CoA, making it difficult to estimate which enzyme plays a larger role in the metabolism of Ala and Val. Group 3, represented by SCSTk, recognizes only a few acid compounds, namely, succinate, 3-methylthiopropionate, and isovalerate, corresponding to acid compounds implicated in Glu/Gln, Met, and Leu metabolism, respectively (19). SCSTk seems to be the only enzyme involved in succinate formation in T. kodakarensis, as formation of succinate could no longer be detected in the disruption strain of the α subunit gene (19). Group 4, represented by ACS IIITk, displays a broad substrate preference similar to that seen with group 2 enzymes. Relatively high levels of activity were also observed for thioglycolate and isobutyrate, suggesting the involvement of this group in the degradation of a wide range of amino acids which may include Cys and Val. ICSTk of group 5 displays strict substrate specificity, and relevant levels of activity were observed only for 2-(imidazol-4-yl)acetate, strongly suggesting an exclusive role in the metabolism of His. Judging from these results (Table 2), NDP-forming acyl-CoA synthetases are not present for the breakdown of Gly, Asp, Asn, Arg, and Lys, and this may also be the case for Thr, Ser, and Pro.
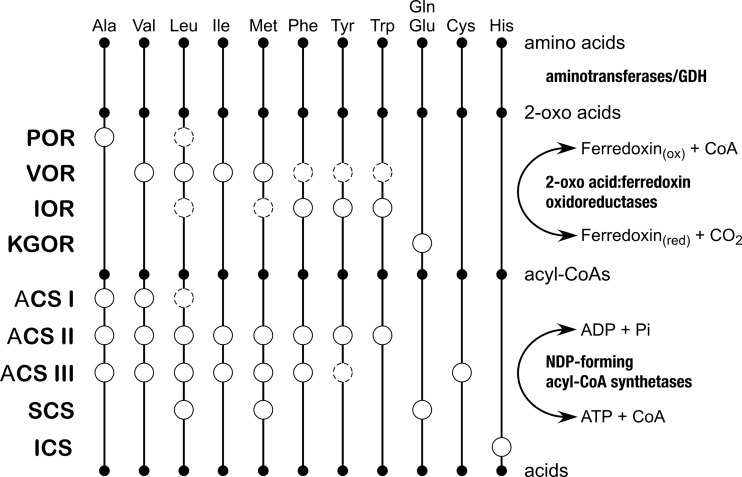
Four enzymes from the Thermococcales that are responsible for the oxidation of 2-oxo acids, POR, IOR, VOR, and KGOR, have been examined. POR from P. furiosus has been found to be relatively specific for pyruvate, indicating a role of the enzyme in Ala metabolism (3, 7). Analysis of the IOR from P. furiosus indicates that the enzyme is involved in the oxidation of aromatic amino acids Trp, Phe, and Tyr and possibly Met and Leu (6, 7). VOR from various Pyrococcus and Thermococcus strains displays activity toward 2-oxo acid derivatives of branch-chained Val, Leu, Ile, and Met (5, 7). KGOR from Thermococcus litoralis has been shown to be specific to 2-oxoglutarate, indicating its role in the metabolism of Glu/Gln (7). KGOR from T. kodakarensis has been genetically shown to be the major 2-oxo acid:ferredoxin oxidoreductase in Glu/Gln metabolism, as succinate generation was no longer observed in its disruption strain (2). As a whole, the amino acids for which a 2-oxo acid:ferredoxin oxidoreductase is present correspond well to those suggested to be metabolized by the five groups of NDP-forming acyl-CoA synthetases. Enzymes catalyzing both steps are clearly present for Ala, Val, Leu, Ile, Met, Phe, Tyr, Trp, and Glu/Gln. In contrast, it is most likely that Gly, Arg, Lys, Pro, Ser, Thr, and Asp/Asn are not degraded through these pathways, suggesting either that other catabolic mechanisms for these amino acids are present or that these amino acids are utilized for anabolic purposes. As we have identified an acyl-CoA synthetase apparently specific for His degradation and, moreover, detected 2-(imidazol-4-yl)acetate in the culture medium, it is most likely that a 2-oxo acid:ferredoxin oxidoreductase(s) responsible for the degradation of His is present. The enzyme may well be one of the four 2-oxo acid:ferredoxin oxidoreductases described above, as activity of these enzymes toward the corresponding substrate 3-(imidazol-4-yl)pyruvate has not been examined. It remains to be determined whether Cys is also degraded by the 2-oxo acid:ferredoxin oxidoreductases, followed by ACS III. Based on our present knowledge, we can now summarize which enzymes among the four 2-oxo acid:ferredoxin oxidoreductases and the five NDP-forming acyl-CoA synthetases are responsible for the degradation of individual amino acids in the Thermococcales (Fig. 4).
Fig 4.
A diagram illustrating the possible involvement of different 2-oxo acid:ferredoxin oxidoreductases and NDP-forming acyl-CoA synthetases of the Thermococcales in the catabolism of individual amino acids. Open circles with solid lines indicate that the enzyme has been shown to exhibit relatively high levels of activity toward the corresponding substrate. Open circles with dotted lines indicate the presence of lower degrees of activity.
Supplementary Material
Footnotes
Published ahead of print 25 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00877-13.
REFERENCES
- 1.Itoh T. 2003. Taxonomy of nonmethanogenic hyperthermophilic and related thermophilic archaea. J. Biosci. Bioeng. 96:203–212. 10.1263/jbb.96.203, [DOI] [PubMed] [Google Scholar]
- 2.Yokooji Y, Sato T, Fujiwara S, Imanaka T, Atomi H. 2013. Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195:1940–1948. 10.1128/JB.01979-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blamey JM, Adams MWW. 1993. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161:19–27. 10.1016/0167-4838(93)90190-3 [DOI] [PubMed] [Google Scholar]
- 4.Smith ET, Blamey JM, Adams MWW. 1994. Pyruvate ferredoxin oxidoreductases of the hyperthermophilic archaeon, Pyrococcus furiosus, and the hyperthermophilic bacterium, Thermotoga maritima, have different catalytic mechanisms. Biochemistry 33:1008–1016. 10.1021/bi00170a020 [DOI] [PubMed] [Google Scholar]
- 5.Heider J, Mai X, Adams MWW. 1996. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178:780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mai X, Adams MWW. 1994. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 269:16726–16732 [PubMed] [Google Scholar]
- 7.Mai X, Adams MWW. 1996. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178:5890–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363. 10.1101/gr.3003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bräsen C, Schmidt M, Grotzinger J, Schönheit P. 2008. Reaction mechanism and structural model of ADP-forming acetyl-CoA synthetase from the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a second active site histidine residue. J. Biol. Chem. 283:15409–15418. 10.1074/jbc.M710218200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glasemacher J, Bock AK, Schmid R, Schönheit P. 1997. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 244:561–567. 10.1111/j.1432-1033.1997.00561.x [DOI] [PubMed] [Google Scholar]
- 11.Hutchins AM, Mai X, Adams MWW. 2001. Acetyl-CoA synthetases I and II from Pyrococcus furiosus. Methods Enzymol. 331:158–167. 10.1016/S0076-6879(01)31054-6 [DOI] [PubMed] [Google Scholar]
- 12.Mai X, Adams MWW. 1996. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:5897–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musfeldt M, Selig M, Schönheit P. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schäfer T, Schönheit P. 1991. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus—acetate formation from acetyl-CoA and ATP synthesis are catalyzed by an acetyl-CoA synthetase (ADP forming). Arch. Microbiol. 155:366–377 [Google Scholar]
- 15.Schäfer T, Selig M, Schönheit P. 1993. Acetyl-CoA synthetase (ADP forming) in archaea, a novel enzyme involved in acetate formation and ATP synthesis. Arch. Microbiol. 159:72–83. 10.1007/BF00244267 [DOI] [Google Scholar]
- 16.Labes A, Schönheit P. 2001. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming). Arch. Microbiol. 176:329–338. 10.1007/s002030100330 [DOI] [PubMed] [Google Scholar]
- 17.Musfeldt M, Schönheit P. 2002. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic Archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184:636–644. 10.1128/JB.184.3.636-644.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bräsen C, Schönheit P. 2004. Unusual ADP-forming acetyl-coenzyme A synthetases from the mesophilic halophilic euryarchaeon Haloarcula marismortui and from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Arch. Microbiol. 182:277–287. 10.1007/s00203-004-0702-4 [DOI] [PubMed] [Google Scholar]
- 19.Shikata K, Fukui T, Atomi H, Imanaka T. 2007. A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal NDP-forming acetyl-CoA synthetases. J. Biol. Chem. 282:26963–26970. 10.1074/jbc.M702694200 [DOI] [PubMed] [Google Scholar]
- 20.Sánchez LB, Galperin MY, Müller M. 2000. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J. Biol. Chem. 275:5794–5803. 10.1074/jbc.275.8.5794 [DOI] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 22.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. 10.1155/2004/204953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.