Abstract
Oxidative stress-induced damage, including 8-oxo-guanine and apurinic/apyrimidinic (AP) DNA lesions, were detected in dormant and outgrowing Bacillus subtilis spores lacking the AP endonucleases Nfo and ExoA. Spores of the Δnfo exoA strain exhibited slightly slowed germination and greatly slowed outgrowth that drastically slowed the spores' return to vegetative growth. A null mutation in the disA gene, encoding a DNA integrity scanning protein (DisA), suppressed this phenotype, as spores lacking Nfo, ExoA, and DisA exhibited germination and outgrowth kinetics very similar to those of wild-type spores. Overexpression of DisA also restored the slow germination and outgrowth phenotype to nfo exoA disA spores. A disA-lacZ fusion was expressed during sporulation but not in the forespore compartment. However, disA-lacZ was expressed during spore germination/outgrowth, as was a DisA-green fluorescent protein (GFP) fusion protein. Fluorescence microscopy revealed that, as previously shown in sporulating cells, DisA-GFP formed discrete globular foci that colocalized with the nucleoid of germinating and outgrowing spores and remained located primarily in a single cell during early vegetative growth. Finally, the slow-outgrowth phenotype of nfo exoA spores was accompanied by a delay in DNA synthesis to repair AP and 8-oxo-guanine lesions, and these effects were suppressed following disA disruption. We postulate that a DisA-dependent checkpoint arrests DNA replication during B. subtilis spore outgrowth until the germinating spore's genome is free of damage.
INTRODUCTION
Due to their ability to survive during long periods of metabolic dormancy, spores of Bacillus subtilis represent an excellent model system in which to study the consequences of long-tem exposure to environmental factors that damage DNA. Physical and chemical agents, including UV-A and -B from sunlight, high temperatures, desiccation, and oxidizing chemicals such as hydrogen peroxide, have the potential to cause damage to dormant spore DNA (reviewed in references 1–3). However, spores of the genus Bacillus counter these potential DNA-damaging effects with a number of factors to maintain the integrity of the spore genome. These factors include (i) the spore coats, (ii) the low water content and accumulation of pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) in the spore core, (iii) the low permeability of the spore's inner membrane to hydrophilic small molecules, (iv) the saturation of spore DNA with α/β-type small acid-soluble spore proteins (SASPs), and (v) DNA repair systems (2, 3).
DNA repair cannot take place in metabolically dormant spores. Therefore, DNA lesions generated by chemical and physical factors accumulate during the variable periods of spore dormancy (1, 4). However, accumulated DNA damage can be eliminated during the “return to life” of spores, which occurs thorough a developmental program that can be separated into two stages, germination followed by outgrowth (5, 6). Germination is triggered when specific germinants, generally amino acids or sugars, are sensed by receptors in the spore's inner membrane (5, 6). Several events are followed by this receptor-germinant interaction, including the release of dipicolinic acid and divalent cations from the spore core, hydrolysis of the spore cortex peptidoglycan, and uptake of water into the spore core to levels comparable to those in growing cells. The subsequent full hydration of the spore core during germination allows resumption of enzyme action and the initiation of spore outgrowth that eventually converts the germinated spore into a growing cell (6). Degradation of α/β-type SASP during spore outgrowth frees up spore DNA for transcription, probably also for DNA repair, and eventually for replication (4, 7), and free amino acids produced in this proteolysis support much of the energy metabolism early in spore outgrowth (6).
During long periods of dormancy, spores' DNA can accumulate a variety of lesions (1, 3). Such DNA damage may interfere with transcription and replication during the return of spores to vegetative growth. Therefore, it has been proposed that DNA repair is necessary for efficient spore outgrowth (8). In agreement with this, genes belonging to various DNA repair pathways are expressed during spore formation, and their products are stored in the developing spore (3). In addition, global analysis of gene expression during spore germination and outgrowth has revealed that transcription of DNA repair genes is turned on in the two stages of this developmental process (9). Treatment of spores deficient in these DNA repair proteins with appropriate DNA-damaging agents has confirmed the importance of these repair proteins in allowing spores' efficient return to life (1–3, 10).
Entry of cells into sporulation is a delicately regulated process that can be modulated by DNA damage and conditions that interfere with chromosomal replication. Thus, in post-exponential-phase cultures of B. subtilis, Sda, a protein with checkpoint functions, plays a critical role in preventing starved cells from initiating sporulation under conditions that interfere with DNA replication (11, 12). In addition, when DNA damage is sensed, DisA, a scanning checkpoint protein that operates early in sporulation, delays sporulation until DNA damage has been eliminated (13). Recent studies have established that the sporulation-specific DisA-dependent checkpoint is modulated by the second messenger, cyclic diadenosine monophosphate (14).
DNA lesions can activate specific mechanisms that interfere with DNA replication, thus affecting cell growth or sporulation. However, to our knowledge, no checkpoint mechanisms have been described during spores' return to vegetative growth. In this report, we provide evidence that a DisA-dependent checkpoint that responds to oxidative DNA damage operates during spore germination/outgrowth to ensure the successful return of B. subtilis spores to vegetative growth.
MATERIALS AND METHODS
Bacterial strains and spore preparation.
All B. subtilis strains used in this work were derived from strain PS832, a prototrophic wild-type derivative of strain 168, and are listed in Table 1, along with the plasmids used (15–18). The growth medium used routinely was Luria-Bertani (LB) medium (19). When appropriate, ampicillin (100 μg/ml), erythromycin (1 μg/ml), chloramphenicol (5 μg/ml), neomycin (10 μg/ml), or tetracycline (10 μg/ml) was added to the medium. Liquid cultures were incubated at 37°C with vigorous aeration. Cultures on solid medium were also grown at 37°C. Spores of all strains were prepared at 37°C on 2× Schaeffer's glucose (SG) medium agar plates without antibiotics, and spores were harvested, cleaned, and stored as described previously (20). All dormant spore preparations used in this work were free (≥98%) of growing cells, germinated spores, and cell debris, as determined by phase-contrast microscopy. Where indicated, spores were used at optical density at 600 nm (OD600) values of 0.5 and 1.0, corresponding to ∼0.75 × 108 and 1.5 × 108 viable spores/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or descriptionc | Source or reference |
|---|---|---|
| Bacillus subtilis strains | ||
| 168 | Wild type; trpC2 | Laboratory stock |
| PS832 | Wild-type trpC2 revertant of strain 168 | Laboratory stock |
| PERM454a | ΔexoA::tet Δnfo::neo Tetr Neor | 15 |
| PERM733a | ΔdisA::lacZ Eryr | pPERM732→PS832b |
| PERM758a | ΔexoA::tet Δnfo::neo ΔdisA Tetr Neor Eryr | pPERM732→PERM454b |
| PERM850a | ΔexoA::tet Δnfo::neo ΔdisA amyE::PsspB-disA Tetr Neor Eryr Cmr | pPERM842→PERM758b |
| PERM1008a | disA-gfp Eryr | pPERM1007→PS832b |
| PERM1197a | amyE::PsspB-disA Cmr | pPERM842→PS832b |
| Plasmids | ||
| pMUTIN4 | Integrational B. subtilis vector; Ampr Eryr | 16 |
| pMUTIN-GFP | Integrational B. subtilis vector; Ampr Eryr | 17 |
| pDG364 | Integrational B. subtilis vector (integrates into the amyE locus of B. subtilis); Ampr Cmr | Wayne Nicholson |
| pPERM615 | 674-bp fragment containing the B. subtilis sspB promoter cloned between the EcoRI-BamHI sites of pDG364; Ampr Cmr | 18 |
| pPERM732 | 307-bp internal fragment of the disA ORF inserted between the EcoRI-BamHI sites of pMUTIN4; Ampr Eryr | This study |
| pPERM842 | 1,293-bp DNA fragment of the B. subtilis disA ORF cloned into the BamHI site of pPERM615, generating a PsspB-disA construct; Ampr Cmr | This study |
| pPERM1007 | 1,108-bp DNA fragment encompassing 28 bp upstream from the start codon to the last codon of the disA ORF cloned between the KpnI-ClaI sites of pMUTIN-GFP; Ampr Eryr | This study |
The background for this strain is PS832.
DNA of the plasmid to the left of the arrow was used to transform the strain to the right of the arrow.
Amp, ampicillin; Cm, chloramphenicol; Ery, erythromycin; Neo, neomycin; Tet, tetracycline.
Spore germination and outgrowth.
Spore germination and outgrowth were performed at 37°C with 25 ml of 2× SG medium supplemented with 10 mM l-alanine, except for the fluorescence-activated cell sorting (FACS) assay, in which spore germination was carried out with 25 mM Tris-HCl (pH 8) plus 10 mM l-alanine. Spores in water were first heat shocked for 30 min at 70°C, cooled on ice, and inoculated into germination medium at 37°C at an initial OD600 of ∼0.5. For the FACS assays, spore germination began with an initial OD600 of 1. The OD600 of cultures was monitored with a Pharmacia Ultrospec 2000 spectrophotometer, and where noted, the values were plotted as a fraction of the initial OD600 (OD600 at time t/initial OD600) versus time.
Genetic and molecular biology techniques.
To isolate chromosomal DNA from dormant spores, the following procedure was followed. Six milliliters of wild-type, nfo exoA, and nfo exoA disA spores at an OD600 of 1.0 was pelleted by centrifugation and washed two times with phosphate-buffered saline (PBS) (0.7% Na2HPO4, 0.3% KH2PO4, 0.4% NaCl [pH 7.5]). The spore pellets were processed for spore coat removal as previously described (20), collected by centrifugation (13,500 × g for 2 min), and washed five times with STE buffer (10 mM Tris-HCl [pH 8], 10 mM EDTA, 150 mM NaCl). After suspension in 0.3 ml of lysis buffer (50 mM EDTA, 100 mM NaCl [pH 7.5]), decoated spores were first treated for 15 min with lysozyme (7.5-mg/ml final concentration) and then treated for 5 min with SDS at a final concentration of 1.5% (vol/vol) at 37°C (21). The cell lysates were extracted 3 times with equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol) and once with chloroform-isoamyl alcohol (24:1; vol/vol). Chromosomal DNA was precipitated from the inorganic phase by adding a 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 volumes of cold ethanol. The DNA pellet collected by centrifugation (13,500 × g for 25 min at 4°C) was washed two times with cold ethanol, dried, suspended in 10 mM Tris-HCl (pH 7.8)–1 mM EDTA, and treated with RNase to obtain RNA-free DNA preparations. Chromosomal DNA purity and integrity were assessed by alkaline gel electrophoresis (AGE) and subsequent image analysis, as described below. DNA concentrations were determined by UV spectrophotometry as previously described (22).
Chromosomal DNA from outgrowing spores was isolated as follows. Aliquots (3 ml; 1.5 × 108 viable spores/ml) of wild-type, nfo exoA, and nfo exoA disA dormant spores that had germinated and outgrown for 90 min in 2× SG medium were collected by centrifugation (13,500 × g for 2 min), washed two times with 1 ml of lysis buffer, and suspended in 0.3 ml of the same buffer, and RNA-free chromosomal DNA was isolated as described above. To quantify genomic DNA from spores germinated and outgrown for 30 and 60 min in 2× SG medium, DNA was first isolated from outgrown spores that were directly susceptible to lysozyme degradation, and the fraction of cells that resisted lysozyme was pelleted by centrifugation. This pellet, which consisted of lysozyme-resistant spores likely containing intact spore coats, was subjected to spore coat removal (20), washed five times with STE buffer, and processed for chromosomal DNA isolation as described above. The chromosomal DNA isolated from both fractions was quantified as described above.
Procedures for transformation and isolation of plasmid DNA were described previously (22, 23). For medium-scale preparation and purification of plasmid DNA, commercial ion-exchange columns were used according to the instructions of the supplier (Qiagen, Inc., Valencia, CA).
Strain construction.
To generate pPERM732, a plasmid that integrates by Campbell-type recombination into the disA locus of B. subtilis, interrupting disA and generating a transcriptional fusion with the lacZ gene, the following strategy was used. A 307-bp DNA fragment from nucleotides (nt) 275 to 582 of the disA open reading frame (ORF) was PCR amplified from chromosomal DNA of B. subtilis PS832 by using the oligonucleotide primers 5′-CGGAATTCCGAATACTCAGCTGATGC-3′ (forward) and 5′-GCGGATCCGACAGACAAGACATCACTG-3′ (reverse). The primers were designed to insert EcoRI and BamHI sites, respectively (underlined). Amplification was performed with Vent DNA polymerase (New England BioLabs, Beverly, MA), and the PCR product was cloned into the pMUTIN4 vector (16) to obtain pPERM732 (Table 1). This plasmid was used to transform B. subtilis strains PS832 (wild-type) and PERM454 (nfo exoA), generating B. subtilis strains PERM733 (disA Eryr) and PERM758 (disA nfo exoA), respectively.
To overexpress disA from the strong forespore-specific promoter of the sspB gene (PsspB), the following strategy was implemented. The complete ORF of disA plus 153 bp downstream of the disA stop codon were PCR amplified by using chromosomal DNA from B. subtilis PS832 and the oligonucleotide primers 5′-CGGGATCCATGGAAAAAGAGAAAAAA-3′ (forward) and 5′-CCGGATCCTGTATATTTAACAGTACAA-3′ (reverse). The primers were designed to insert BamHI cloning sites (underlined). Amplification was performed with Vent DNA polymerase (New England BioLabs), and the PCR product (1,293 bp) was cloned into the pCR-BluntII-TOPO vector (Invitrogen, Eugene, OR), giving pPERM827. The 1,293-bp disA ORF was released from this plasmid with BamHI, purified from a low-melting-point agarose gel, and cloned into the BamHI site of pPERM615 (18), placing disA immediately downstream of PsspB. After ensuring that the orientation of the PsspB-disA cassette was correct, the resulting construct, pPERM842 (Table 1), was introduced by transformation into competent cells of B. subtilis PS832 and PERM758, generating strains PERM1197 (amyE::PsspB-disA) and PERM850 (nfo exoA disA amyE::PsspB-disA), respectively. The recombination events leading to inactivation of disA or insertion of the PsspB-disA cassette into the amyE locus were confirmed by PCR using specific oligonucleotide primers (data not shown).
To generate a B. subtilis strain producing a DisA-green fluorescent protein (GFP) fusion protein, the following strategy was implemented. Using chromosomal DNA of B. subtilis PS832 as a template, a 1,108-bp DNA fragment located between 28 bp upstream of the start codon and the last codon of disA was PCR amplified with the oligonucleotide primers 5′-GCGGTACCGCTTCGTACTTCATTAGGAG-3′ (forward) and 5′-GCATCGATCAGTTGCTGTCTAAATAATGC-3′ (reverse). The primers were designed to insert KpnI and ClaI sites (underlined) into the resulting PCR product. The 1,108-bp PCR fragment was cloned into pCR-Bunt-II-TOPO (Invitrogen), giving pPERM975. Plasmid pPERM975 was treated with KpnI and ClaI to release the disA fragment, which was then cloned between the KpnI and ClaI sites of the integrative vector pMUTIN-GFP, giving pPERM1007 (Table 1). Plasmid pPERM1007 was used to transform B. subtilis strain PS832 to Eryr. This process generated strain PERM1008, harboring a transcriptional disA-gfp fusion in the disA locus (Table 1). The single-crossover event leading to integration of the disA-gfp fusion into the disA locus was verified by PCR with specific oligonucleotide primers (data not shown).
Flow cytometry.
Dormant and germinating spore samples at an OD600 of 1 were stained in 25 mM Tris-HCl buffer (pH 8) with 0.1 μM SYTO16 (Invitrogen) for ∼5 min prior to flow cytometry, according to the instructions of the manufacturer. Flow cytometry was carried out on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) with excitation at 488 nm. Fluorescence signals were collected through a 530-nm-band-pass filter, and at least 104 spores were analyzed in each run.
Outgrowth kinetics of germinated B. subtilis spores.
Spores in water were heat activated (70°C for 30 min), cooled on ice, and inoculated to an OD600 of ∼1 in prewarmed (37°C) 25 mM Tris-HCl buffer (pH 7.4) with 10 mM l-alanine. Spore germination was monitored by determining the OD600 of cultures, which falls ∼55% upon complete spore germination, and after 2 h, the extent of spore germination was determined by examination of several hundred spores by phase-contrast microscopy.
The culture containing germinated and nongerminated spores was harvested by centrifugation (15,000 × g [20°C]), and the pellet was washed with 250 μl of sterile PBS and suspended in 100 μl of 20% Nycodenz (Sigma, St. Louis, MO). Aliquots of the suspension (∼100 μl) were layered onto a solution of 50% Nycodenz in 2-ml Eppendorf tubes, which were centrifuged for 45 min at 15,000 × g at 20°C. Under these conditions, dormant spores are pelleted, and germinated spores float (24). The germinated spores were separated from spore pellets, washed with water to remove Nycodenz, suspended in PBS, and inoculated into 15 ml of 2× SG medium at 37°C and at an initial OD600 of ∼0.01. The OD600 of cultures was monitored as described above, and the values were plotted versus time.
Fluorescence microscopy.
Cell morphology, nucleoid structure, and GFP foci were analyzed by fluorescence microscopy. Samples (1 ml) of cultures from germinated spores were removed at various times, centrifuged briefly (15,000 × g [20°C]), and mixed with 0.2 ml of fixative solution (3% [vol/vol] paraformaldehyde and 5% glutaraldehyde in HEPES-buffered saline [273 mM NaCl, 9.9 mM KCl, 1.27 mM Na2HPO4 · 2H2O, 11.1 mM dextrose, 42 mM HEPES {pH 7}]). After 30 min at room temperature, fixation was continued on ice for 50 min. The samples were washed twice by centrifugation with PBS and suspended in 100 μl of GTE (5 mM glucose, 25 mM Tris-HCl, 10 mM EDTA [pH 8.0]). Aliquots (10 μl) of this suspension were mixed with 10 μl of 2 μg/ml 4′,6′-diamino-2-phenylindole (DAPI) in water to stain DNA and were kept at room temperature for 15 min. Where indicated, cells were also suspended in 100 μl of 10 μg/ml FM4-64 (Invitrogen) and kept at room temperature for 15 min. To fix cells for microscopic analysis, glass slides were prewashed with 0.01% (vol/vol) poly-l-lysine (Sigma-Aldrich). Samples spotted onto glass slides were covered with Vectashield (Vector Laboratories, Inc., Burlingame, CA) and stored at 4°C. Fluorescence microscopy was performed with a Zeiss Axioscope A1 microscope equipped with an AxioCam ICc1 camera. Fluorescence and phase-contrast images were acquired by using AxioVision V 4.8.2 software and adjusted only for brightness and contrast. Excitation and emission wavelengths employed were 475 and 508 nm for GFP, 350 and 470 nm for DAPI, and 506 and 750 nm for FM4-64, respectively.
Quantitation of fluorescence emission.
The fluorescence emitted by spores of B. subtilis strain PERM1008 (disA-gfp) during germination and outgrowth was quantitated by fluorescence spectrometry. Briefly, dormant spores of B. subtilis PERM1008 in water were heat shocked (30 min at 70°C), cooled on ice, and inoculated into 2× SG germination medium at 37°C at an OD600 of ∼0.5. Cell samples (1.5 ml) were harvested by centrifugation (13,500 × g for 2 min at 4°C) at various times during germination and outgrowth, and cell pellets were washed and suspended in 1.5 ml of PBS. Samples of the cell suspensions (0.1 ml) were collected, serially diluted into PBS, and plated onto solid LB medium to determine viable counts. Finally, the fluorescence emitted by each sample was quantified with an LS-55 Perkin-Elmer fluorescence spectrometer (Perkin-Elmer, Waltham, MA) set at excitation and emission wavelengths of 488 nm and 515 nm, respectively. Emission values were reported as fluorescence units/106 viable cells. The basal values of fluorescence emitted by spores of the wild-type strain during germination and outgrowth were subtracted from the fluorescence values of the disA-gfp strain at each time point; these corrections were always <10%.
Determination of β-galactosidase activity.
B. subtilis strain PERM733 carrying the disA-lacZ fusion was grown and sporulated in liquid Difco sporulation medium (DSM) (25). Samples (1.5 ml) were collected during vegetative growth and throughout sporulation, pellets were washed with cold 0.1 M Tris-HCl (pH 7.5), and the cell pellets were stored at −20°C until determination of β-galactosidase activity, as described previously (20, 26). Briefly, washed cell samples were disrupted with lysozyme and centrifuged (13,500 × g for 2 min), and the level of β-galactosidase in the supernatant was measured by using o-nitrophenyl-β-d-galactoside (ONPG) as the substrate and assigned to the mother cell fraction (this actually consisted of mother cells plus lysozyme-sensitive forespores). The pellet fraction, which consisted of lysozyme-resistant forespores containing spore coats, was subjected to spore coat removal (20) and washed five times with 50 mM Tris-HCl (pH 7.5), and lysozyme treatment and β-galactosidase assays were repeated to determine the β-galactosidase activity in forespores that had lost spore coats (20, 27). β-Galactosidase specific activities were expressed in Miller units relative to the OD600 of the initial sporulating culture (19).
The levels of β-galactosidase expressed by spores carrying the disA-lacZ fusion during germination and outgrowth were determined as follows. Dormant spores of strain PERM733 were germinated in 2× SG medium as described above. Triplicate samples (3 ml each) were collected during germination and outgrowth and washed with 0.1 M Tris-HCl (pH 7.5), and cell pellets were stored at −20°C. Two cell samples were processed for determination of β-galactosidase with ONPG as a substrate, as described above, and the other sample was subjected to lyophilization and then processed for protein quantification, as previously described (18). β-Galactosidase specific activities were calculated with the formula (OD420)(1,000)/(volume)(min)(mg of protein). Values of the β-galactosidase specific activity in the wild-type strain during growth, sporulation, and germination were subtracted from values for the disA-lacZ strain at each time point. These corrections were always ≤10%.
Detection of oxidative DNA damage.
Samples of chromosomal DNA isolated from germinated or dormant spores of each strain were incubated with 18 units of endonuclease IV (EndoIV), which cleaves DNA at apurinic/apyrimidinic (AP) sites, or 14 units of formamidopyrimidine-DNA glycosylase (Fpg), which cleaves DNA at 8-oxo-guanine (8-oxo-G) residues, respectively, according to the instructions of the supplier (New England BioLabs). Enzyme reactions were done for 3 h, and reaction mixtures containing 3 or 5 μg of DNA were then electrophoresed on a 1% alkaline agarose gel, which was then stained with ethidium bromide, as described previously (22). The method for quantitative image analysis of these gels is described in Table 2.
TABLE 2.
Quantification of EndoIV and Fpg degradation of chromosomal DNA isolated from dormant or outgrowing spores of different B. subtilis strainsa
| Enzyme | Avg % band intensity ± SD remaining after enzyme treatment of strainb: |
|||||
|---|---|---|---|---|---|---|
| WT |
nfo exoA |
nfo exoA disA |
||||
| DS | OG | DS | OG | DS | OG | |
| EndoIV | 87 ± 9.8 | 94 ± 8.8 | 86 ± 7.6 | 75 ± 8.6 | 47 ± 5.1 | 12 ± 0.7 |
| Fpg | 18 ± 2.1 | 97 ± 9.2 | 9 ± 1.1 | 67 ± 7.3 | 10.5 ± 1.2 | 8.5 ± 0.94 |
Samples of chromosomal DNA isolated from outgrowing spores (OG) or dormant spores (DS) of each strain were treated with EndoIV or Fpg, respectively, and reaction mixes were electrophoresed on a 1% alkaline agarose gel and stained with ethidium bromide, all as described in Materials and Methods and the legend to Fig. 1A to C.
For each reaction, the intensity of the chromosomal DNA band remaining in the gel well after EndoIV or Fpg treatment was quantified by densitometry using Quantity One 1-D software (Bio-Rad Laboratories, Hercules, CA), using as a reference the chromosomal DNA band in the gel well from an identical amount of total chromosomal DNA in an untreated control reaction that was run in an adjacent lane to the same amount of total EndoIV or Fpg-treated chromosomal DNA, as shown in Fig. 1A to C. Values are averages ± standard deviations for duplicate determinations in two separate experiments (with different lots of spores).
RESULTS
Oxidative damage to dormant spore DNA delays germination and outgrowth of B. subtilis spores lacking AP endonucleases.
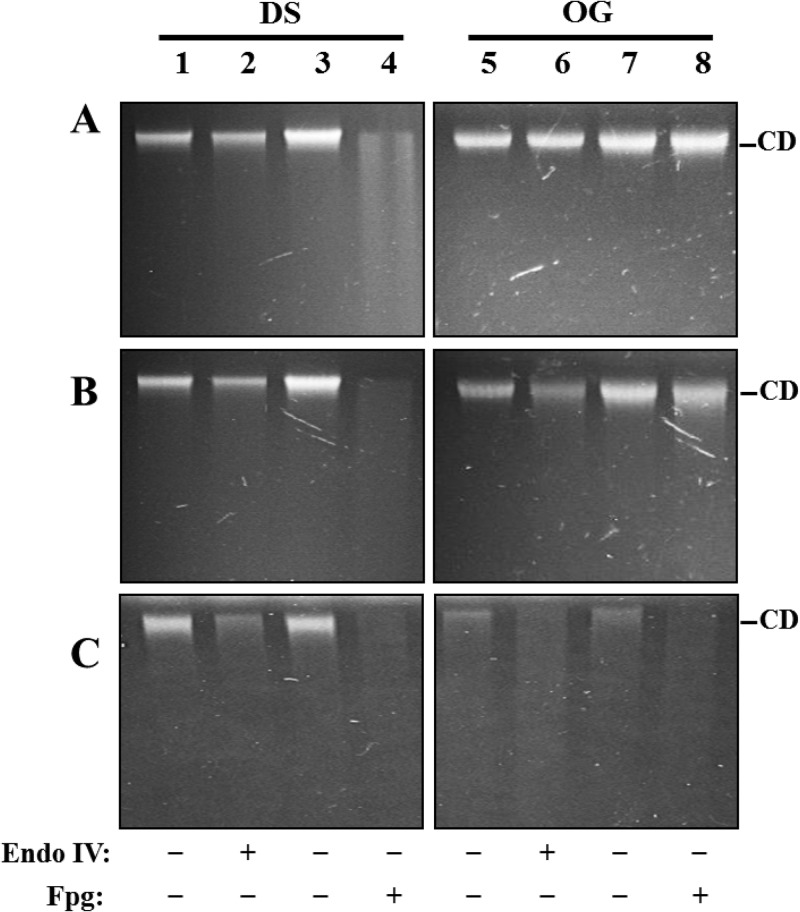
Oxidative stress promotes distinct types of DNA lesions that are processed mainly by the base excision repair (BER) pathway, including AP sites, strand breaks, and oxidized DNA bases such as 8-oxo-G (28, 29). Consequently, we investigated whether this type of DNA damage is present in dormant and outgrown wild-type and nfo exoA spores. To this end, chromosomal DNAs from dormant wild-type and nfo exoA spores as well as from these spores incubated for 90 min in 2× SG medium plus l-alanine were treated with EndoIV and Fpg to detect AP sites and 8-oxo-G, respectively (28). Analysis of dormant spore DNA by AGE revealed the existence of a small amount of AP sites and a larger amount of 8-oxo-G in wild-type spore DNA (Fig. 1A, lanes 1 to 4, and Table 2), with slightly higher levels of these two lesions in nfo exoA spore DNA (Fig. 1B, lanes 1 to 4, and Table 2). Results from the AGE analysis also revealed the presence of AP sites in DNA from outgrown nfo exoA spores (Fig. 1B, lanes 5 and 6, and Table 2). However, AP sites were less abundant in DNA from outgrown wild-type spores than from outgrown nfo exoA spores (Fig. 1A and B, compare lanes 5 and 6, and Table 2). In addition, although the assay with Fpg detected high levels of 8-oxo-G in DNA from the outgrown nfo exoA spores, such lesions were significantly less abundant in DNA from outgrown wild-type spores (Fig. 1A and B, compare lanes 7 and 8, and Table 2). Overall, the results noted above indicated that the great majority of AP sites and 8-oxo-G lesions in wild-type dormant spore DNA were repaired during outgrowth, while most but not all of this damage in dormant nfo exoA spores was repaired during outgrowth (compare Fig. 1A and B for dormant and outgrown wild-type and nfo exoA spores). These results suggest that (i) much of the oxidative DNA lesions such as AP sites and 8-oxo-G detected during outgrowth of wild-type and nfo exoA spores are generated during spore morphogenesis and/or dormancy, (ii) there are repair enzymes other than Nfo and ExoA that repair these lesions during spore outgrowth, and (iii) Nfo and ExoA play a significant role in the processing of these types of DNA damage during outgrowth. In support of the latter notion, B. subtilis spores lacking Nfo and ExoA were slow in returning to vegetative growth (Fig. 2A), indicating that unrepaired DNA lesions normally processed by Nfo and ExoA are involved in the latter defect.
FIG 1.
AGE analysis of DNA isolated from dormant or outgrown spores of different strains and with or without treatment with EndoIV or Fpg. Chromosomal DNAs from dormant spores (DS) or outgrown spores (OG) of the wild-type (A), nfo exoA (B), or nfo exoA disA (C) strain were untreated (lanes 1, 3, 5, and 7) or treated with 18 units of EndoIV (lanes 2 and 6) or 14 units of Fpg (lanes 4 and 8), all as described in Materials and Methods. Reaction mixture samples were electrophoresed on a 1% alkaline agarose gel that was then stained with ethidium bromide as described in Materials and Methods. The results shown are representative of two experiments that yielded essentially similar results. CD, high-molecular-weight chromosomal DNA. The amounts of total chromosomal DNA run in the various lanes were 3 μg (A and B, lanes 1, 2, 5, and 6, and C, lanes 1 to 8) and 5 μg (A and B, lanes 3, 4, 7, and 8).
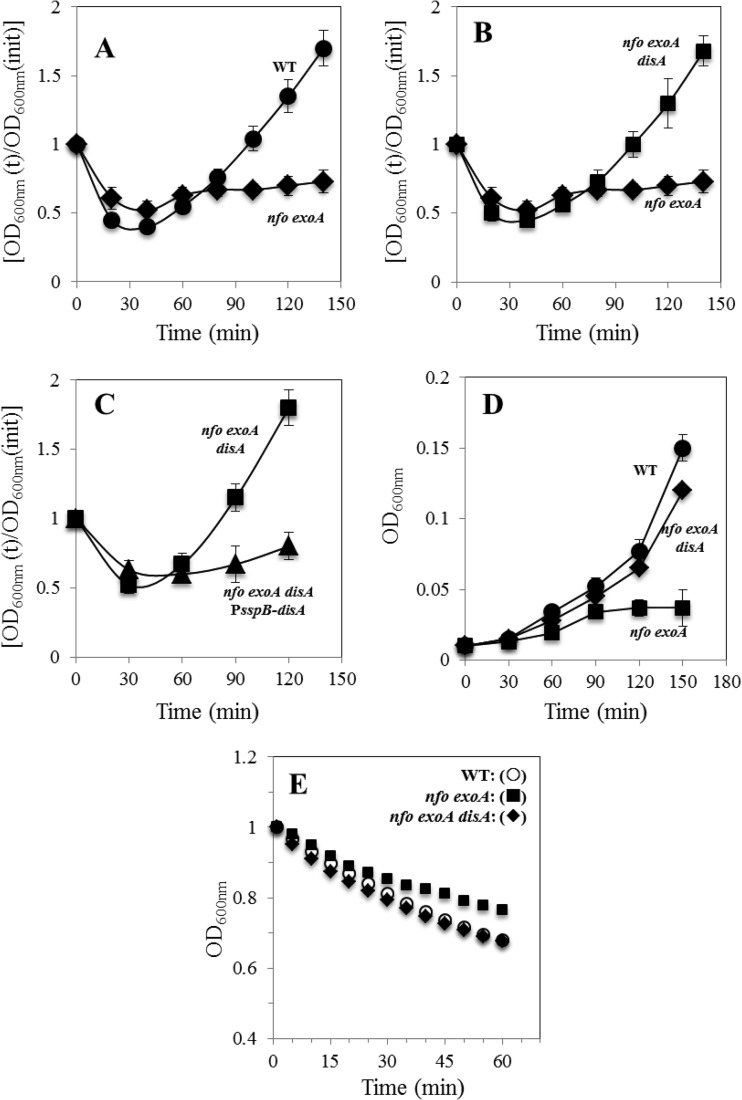
FIG 2.
Germination and outgrowth of spores of different B. subtilis strains. (A to C) Dormant spores of the wild-type (WT) (●), nfo exoA (◆), nfo exoA disA (■), and nfo exoA disA amyE::PsspB-disA (▲) strains were heat shocked and germinated, and spore germination and outgrowth were measured by monitoring the OD600 of the cultures, as described in Materials and Methods. Values are averages ± standard deviations for triplicate determinations in three separate experiments (with different batches of spores). (D and E) Outgrowth (D) and germination (E) of spores of different strains. (D) Germinated spores of the wild-type (●), nfo exoA (■), and nfo exoA disA (◆) strains separated from dormant spores on a Nycodenz density gradient were inoculated into 2× SG medium, and outgrowth was monitored by determining the culture's OD600, all as described in Materials and Methods. Values are averages ± standard deviations for duplicate determinations in two separate experiments (with different batches of spores). (E) Dormant spores of wild-type (WT), nfo exoA, and nfo exoA disA strains were heat shocked and then germinated in 25 mM Tris-HCl buffer (pH 7.4) with 10 mM l-alanine, and germination was monitored by determining the culture's OD600, all as described in Materials and Methods. The results are representative, and the experiment was performed at least two times with different batches of spores, all of which gave similar results.
Disruption of disA suppresses the slow germination/outgrowth phenotype of BER-deficient spores.
Among several checkpoint mechanisms involved in controlling patterns of cell differentiation in B. subtilis, DisA is the only protein regulating a checkpoint event that connects DNA damage with spore formation (13). Therefore, we investigated whether DisA is also responsible for delaying the transition of nfo exoA spores into vegetative growth. Strikingly, we found that (i) the outgrowth of nfo exoA disA spores was essentially similar to that of wild-type spores (Fig. 2A and B) and (ii) nfo exoA spores also deficient for DisA retained a significantly higher proportion of unrepaired AP sites and oxidized guanines during spore outgrowth than did spores that were proficient for this checkpoint protein (Fig. 1B and C, compare lanes 5 to 8). These results strongly suggest that DisA is involved in the retardation of events leading ultimately to vegetative growth in the DNA repair-deficient nfo exoA spores. Further support for this hypothesis was that expression of disA from the strong forespore-specific PsspB promoter (18, 30, 31) restored the slow-outgrowth phenotype of nfo exoA spores (Fig. 2C). However, overexpression of disA did not affect the outgrowth of wild-type spores (data not shown). Interestingly, while germinating wild-type and disA spores were equally affected by hydrogen peroxide, a statistically significantly increase in the mutation frequency to Rifr was observed for spores lacking DisA, providing additional support for the checkpoint function of this protein (results not shown).
Role of DisA during spore germination and outgrowth.
Two major stages can be distinguished during the return of B. subtilis spores to vegetative growth: germination and outgrowth (6). As shown in Fig. 2A, nfo exoA spores were slightly slowed in germination but greatly slowed in outgrowth. Therefore, the role played by DisA was separately analyzed in each developmental stage. To investigate the role of DisA during outgrowth, spores of the different strains were first induced to germinate at 37°C in 25 mM Tris-HCl (pH 7.4) plus l-alanine, and germinated spores were separated from dormant spores by density gradient centrifugation (24). Analysis of the kinetics of outgrowth of the isolated germinated spores in 2× SG medium revealed that the absence of DisA suppressed the slow-outgrowth defect of nfo exoA spores (Fig. 2D). Importantly, with wild-type spores, the absence of DisA did not significantly affect spore outgrowth (data not shown).
To investigate the effects of DisA on germination but not outgrowth, heat-shocked wild-type, nfo exoA, and nfo exoA disA spores were germinated at 37°C in 25 mM Tris-HCl (pH 7.4) plus l-alanine, and the OD600 of cultures was monitored (Fig. 2E). Notably the nfo exoA spores germinated slower than did the wild-type spores, and this difference, while small, was seen in at least two different preparations of these spores. In addition, the slower germination of the nfo exoA spores was suppressed by also disrupting disA, as no significant difference was observed in the germinations of the wild-type and nfo exoA disA spores (Fig. 2E). Similar results were obtained when the outgrowth of wild-type, nfo exoA, and nfo exoA disA spores was analyzed by monitoring spore DNA levels by flow cytometry (data not shown). Therefore, although there was a slight slowing of germination by DisA with nfo exoA spores, DisA played a much more significant role in spore outgrowth.
Analysis of disA expression during sporulation and spore germination and outgrowth.
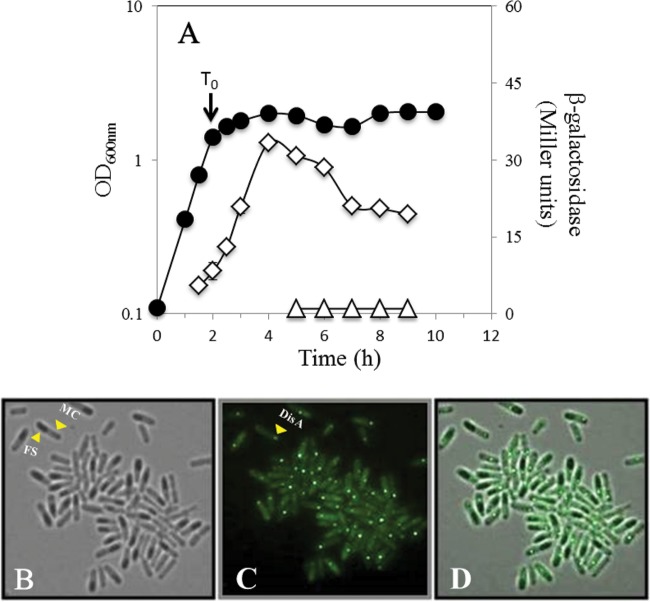
It has been shown that a DisA-deficient strain has no detectable defect in cell division (13). In contrast, we found here that DisA exerts a checkpoint control in spore outgrowth and perhaps to a lesser extent in germination. Therefore, we investigated whether expression of disA occurred in the forespore compartment during sporulation, thus ensuring that DisA would be present in the dormant spore. Analysis of the temporal and spatial expression of a disA-lacZ fusion at the disA locus showed that there was minimal, if any, expression in vegetative cells, with maximum levels of expression being reached 2 to 3 h after the onset of sporulation (Fig. 3A). The disA-driven β-galactosidase activity directly extracted by lysozyme was still detected later in sporulation, but analysis of β-galactosidase in developing spores directly by decoating followed by lysozyme treatment revealed no enzyme in the forespore compartment (Fig. 3A). To further confirm these results, we used fluorescence microscopy to locate a DisA-GFP fusion protein in sporangia collected 7 h after the onset of sporulation. Results confirmed the existence of a discrete DisA-GFP fluorescent signal in the mother cell but not in the forespore compartment (Fig. 3B to D). Of note, a previously reported DisA-GFP protein detected during the first stages of B. subtilis sporulation (13) exhibited the same pattern of fluorescence exhibited by the GFP fusion protein employed here, namely, globular discrete foci that were formed by an octamer of DisA subunits (32). Thus, the DisA-GFP chimera described in this work is also most likely functional. Together, these results indicate that sporulation-associated expression of disA does not result in packaging of DisA in the forespore.
FIG 3.
Levels of β-galactosidase from a B. subtilis strain containing a disA-lacZ fusion (A) and compartmental synthesis of DisA-GFP in B. subtilis sporangia (B to D). (A) B. subtilis strain PERM733 (disA-lacZ) was grown at 37°C in DSM, and the OD600 was measured (●). Cell samples were collected during vegetative growth and at various times during sporulation and treated with lysozyme, and initial extracts were assayed for β-galactosidase (◊), as were extracts from initially lysozyme-resistant forespores (△), all as described in Materials and Methods. Values shown are averages of duplicate independent experiments ± standard deviations of the β-galactosidase specific activity. (B to D) B. subtilis strain PERM1008 (disA-gfp) was grown at 37°C in DSM. Cell samples collected 7 h after time zero (T0) were processed and photographed under fluorescence microscopy as described in Materials and Methods. (B) Bright field; (C) GFP channel; (D) overlay of the two images. T0 is the time when the slopes of the logarithmic and stationary phases of growth intersected. Arrows indicate mother cell (MC) and forespore (FS) compartments (B) and DisA-GFP (C).
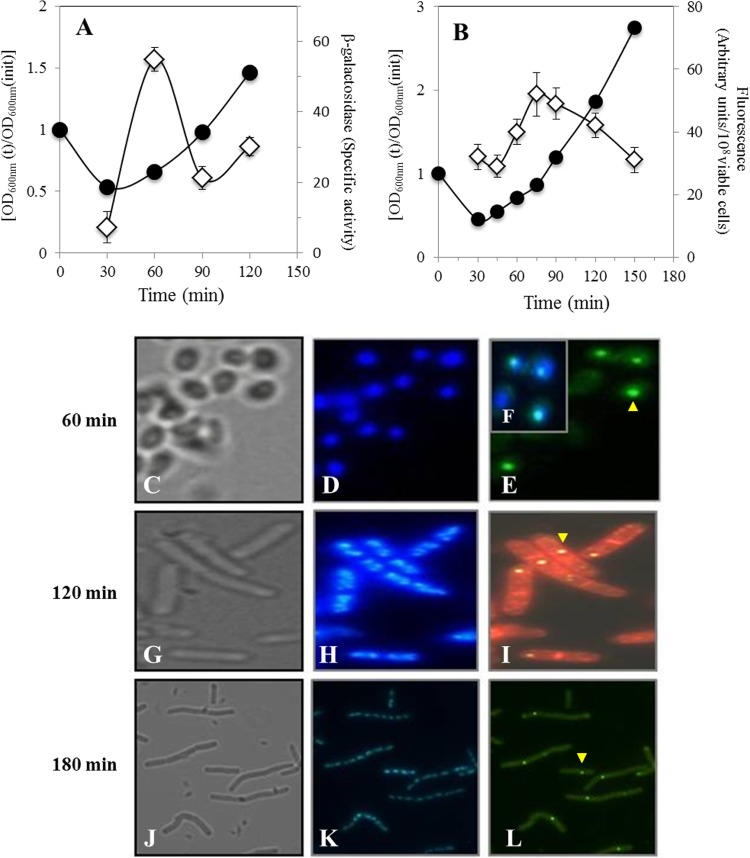
Given the results noted above, we further explored the possibility that disA is also expressed during germination and outgrowth by measuring the expression level of a disA-lacZ fusion and the synthesis of DisA-GFP protein during these developmental stages. The maximum β-galactosidase specific activity in germinating spores carrying the disA-lacZ fusion was reached 60 min after the initiation of germination during the transition between germination and outgrowth, confirming that transcription of disA takes place during outgrowth (Fig. 4A). Strikingly, fluorescence microscopy revealed the presence of discrete DisA-GFP foci in spores/cells collected at different times during outgrowth and vegetative growth (Fig. 4C to L). As shown in Fig. 4C to E, the fluorescent DisA-GFP signal colocalized with the nucleoid of spores that were collected 1 h after the initiation of germination. In fact, inspection of 100 outgrown spores expressing DisA-GFP revealed that all the foci were associated with the nucleoids (data not shown). Importantly, when germinated spores underwent the first division event, the DisA-GFP foci remained associated with the chromosome of only one of these cells (Fig. 4G to I). Analysis of samples collected 3 h after germination initiation showed that after 2 to 3 rounds of cell division, the DisA-GFP foci generally remained associated with the chromosome of only one of the cells (Fig. 4J to L). A careful analysis revealed that ∼85% of chains of 2 to 4 cells (120 min after germination initiation) and 4 to 8 cells (180 min after germination initiation) contained 1 DisA-GFP focus, whereas only ∼15% contained 2 fluorescent DisA-GFP foci (Table 3). These results strongly suggest that disA is expressed and that its product is synthesized during the return of spores to vegetative growth. To better analyze the temporal pattern of DisA synthesis during germination and outgrowth, the fluorescence levels of spores harboring the disA-gfp fusion were quantified by spectrofluorometry. The results of this analysis showed that the fluorescence emission in spores of this strain reached a maximum ∼75 min after germination initiation (Fig. 4B), confirming that significant DisA synthesis takes place during spore outgrowth. The later appearance of DisA-GFP with respect to the maximum value observed for the disA-lacZ fusion may reflect the time that it takes for the GFP to become fluorescent during the fusion protein's maturation. Consequently, these results suggest that DisA is synthesized during spore outgrowth, presumably to scan the chromosome and prevent DNA replication if DNA damage is encountered.
FIG 4.
Levels of β-galactosidase from a B. subtilis strain containing a disA-lacZ fusion (A), fluorescence emitted (B), and analysis of DisA-GFP synthesis (C to L) by a B. subtilis strain harboring a disA-gfp fusion during spore germination and outgrowth. Heat-shocked spores of B. subtilis PERM733 (disA-lacZ) and PERM1008 (disA-gfp) were germinated in 2× SG medium. (A). Samples collected at the indicated times were washed with 0.1 M Tris-HCl (pH 7.5), and cell pellets were stored at −20°C. Cell samples were processed for determination of β-galactosidase and for protein quantification as described in Materials and Methods. ●, OD600; ◊, β-galactosidase. (B) Samples collected at the indicated times were washed and suspended in PBS, and the samples' GFP fluorescence was measured as described in Materials and Methods. ●, OD600; ◊, GFP fluorescence. (C to L) Samples collected at 60 min (C to E), 120 min (G to I), and 180 min (J to L) were processed and photographed as described in Materials and Methods. (C, G, and J) Bright field; (D, H, and K) DAPI staining; (E, I, and L) GFP channel; (F) overlay of images in panels D and E. The sample in panel I (120 min) was stained with FM4-64 (red). The yellow arrowheads point to DisA-GFP.
TABLE 3.
Parameters of accumulation of DisA-GFP during spore germination/outgrowtha
| Time of analysis after germination onset (min) | No. of cell chains analyzed | No. of cells/chain | No. of septa/chain | No. of chains with 1 DisA fluorescent focus | No. of chains with 2 DisA fluorescent foci |
|---|---|---|---|---|---|
| 120 | 200 | 2–4 | 1–3 | 184 | 16 |
| 180 | 200 | 4–8 | 3–7 | 186 | 14 |
Spores of strain PERM1008 (disA-gfp) were germinated and outgrown in 2× SG medium at 37°C, and at various times, cells were examined by microscopy as described in Materials and Methods.
Analysis of DNA replication during germination and outgrowth of nfo exoA and nfo exoA disA spores.
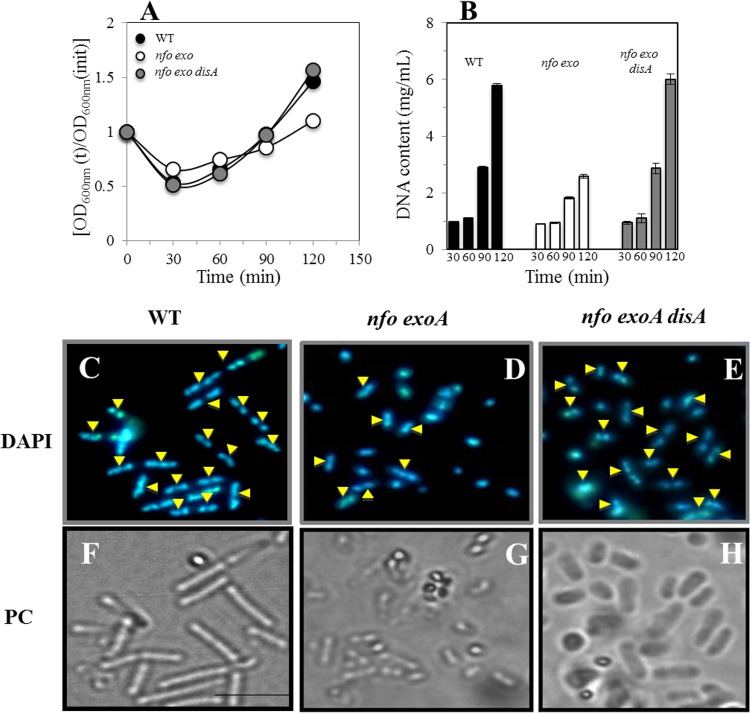
The results described above indicated that the absence of Nfo and ExoA delayed spores' return to vegetative growth, most probably due to a DisA-dependent checkpoint arrest in response to oxidative DNA damage. Therefore, we investigated whether the slow outgrowth of nfo exoA spores was accompanied by a delay in DNA replication and whether DisA is involved in this delay (Fig. 5A and B). Analysis of wild-type, nfo exoA, and nfo exoA disA spores outgrowing in 2× SG medium revealed that the amount of DNA remained relatively constant for ∼60 min (Fig. 5B). However, subsequently, both wild-type and nfo exoA disA spores accumulated DNA similarly and much more rapidly than did nfo exoA spores, consistent with the slower outgrowth of the nfo exoA spores. These results suggest that DisA is involved in regulating DNA replication during the return of spores to vegetative growth. To obtain further evidence for this suggestion, spores of the nfo exoA and nfo exoA disA strains collected 90 min after initiation of germination were stained with DAPI and analyzed by fluorescence microscopy. Strikingly, whereas ∼80% of outgrown nfo exoA disA and wild-type spores had undergone cell division and chromosome segregation after 90 min, only 25% of the nfo exoA spores had done so (Fig. 5C to H). Together, these results support the suggestion that when reactive oxygen species (ROS)-promoted damage is present in spore DNA, DisA is directly or indirectly involved in delaying the DNA replication that accompanies the return of spores to vegetative growth.
FIG 5.
Outgrowth, DNA concentration, and microscopic analysis of chromosome segregation in outgrowing spores of different B. subtilis strains. (A) Heat-shocked spores of wild-type (WT), nfo exoA, and nfo exoA disA strains were germinated, and spore germination and outgrowth were measured by monitoring the OD600 of cultures, as described in Materials and Methods. (B) Levels of DNA in samples from the cultures described in panel A, collected at different times during germination and outgrowth, were measured as described in Materials and Methods. (C to E) Outgrown spores collected 90 min after initiation of germination were stained with DAPI, examined, and photographed, as described in Materials and Methods. (C and F) DAPI-stained (C) and phase-contrast (PC) (F) images of wild-type spores. (D and G) DAPI-stained (D) and phase-contrast (G) images of nfo exoA spores. (E and H) DAPI-stained (E) and phase-contrast (H) images of nfo exoA disA spores. Yellow arrowheads show cells that have replicated and segregated their chromosome. For each strain, >200 cells were analyzed in 6 different fields.
DISCUSSION
Nfo and ExoA AP endonucleases that are found in organisms of the three domains of life play prominent roles in the processing of AP sites resulting from the spontaneous depurination and depyrimidination of DNA (15, 28). Moreover, these enzyme operate over 3′-blocking groups such as phosphates, phosphoglycolates, and 3′-α,β-unsaturated aldehydes existing in DNA as products of the attack of reactive oxygen species or generated by the combined action of glycosylase/lyase activities (3, 8, 28). In B. subtilis, nfo and exoA are expressed not only during sporulation but also during spore germination/outgrowth (9, 15, 33). Indeed, both enzymes are important in protecting germinating and dormant B. subtilis spores against oxidative DNA damage (8, 34). The results in this work demonstrated the existence of 8-oxo-G and AP sites in DNA from dormant and outgrowing spores and that these lesions were more abundant in spores, in particular outgrowing spores, that were deficient in the AP endonucleases Nfo and ExoA. However, the amount of 8-oxo-G found in dormant spores decreased significantly during outgrowth of both wild-type and nfo exoA spores, suggesting that repair proteins in addition to Nfo and ExoA process 8-oxo-G in this developmental stage. In agreement with this contention, it has been shown that nth, a gene encoding a DNA glycosylase able to act on 8-oxo-G and AP sites (28), is also expressed during spore outgrowth (9). Notably, results from this and previous work (8) showed that B. subtilis spores lacking Nfo and ExoA are significantly delayed in their return to vegetative growth, presumably because of the oxidative lesions present during spore germination/outgrowth (Fig. 1B). Together, these results strongly suggest that there is oxidative DNA damage in both dormant and germinating/outgrowing spores and that Nfo and ExoA are important in counteracting such damage in this developmental stage.
Comparison of germination and outgrowth curves of wild-type and nfo exoA spores showed that despite their slowed outgrowth, the nfo exoA spores did ultimately resume vegetative growth (data not shown). Since there is no significant difference in the doubling times of wild-type and nfo exoA cells (8), we reasoned that a checkpoint mechanism was involved in the phenotypic defect exhibited by spores lacking Nfo and ExoA. Moreover, since oxidative damage-induced DNA lesions were detected during spore outgrowth, we further speculated that oxidative damage to spore DNA could be involved in activating such a checkpoint. Among candidate proteins with known checkpoint functions in B. subtilis (11–13), DisA was of particular interest, as this protein exerts a checkpoint control associated with DNA damage early in sporulation (13). Essentially, when starved cell DNA is damaged, DisA delays nucleoid segregation into the forespore compartment until the chromosomes are free of damage (13). Our new results indicated that inactivation of disA also suppressed the delayed germination/outgrowth phenotype of spores lacking Nfo and ExoA. This result strongly suggests that DisA is responsible for the germination/outgrowth defect observed for these spores. Further genetic evidence supporting this suggestion was that overexpression of disA in spores reestablished the slow-outgrowth phenotype in nfo exoA disA spores, while overexpression of disA in wild-type spores did not affect spore germination/outgrowth. In addition to this evidence, analysis of DNA lesions in outgrown spores showed higher levels of accumulation of AP sites and 8-oxo-G only when DisA was absent from the nfo exoA spores, strongly suggesting that DisA retards outgrowth until the spore chromosome is free of damage before spores can proceed into vegetative growth. Additional support for this contention was provided by mutagenesis experiments showing that outgrown spores deficient for DisA and treated with hydrogen peroxide showed a higher propensity to generate colonies with a Rifr phenotype than did hydrogen peroxide-treated outgrown wild-type spores. From these results, it can be inferred that besides double-strand breaks (13), 8-oxo-G and/or AP sites may elicit the checkpoint function of DisA. However, as noted above, processing of these oxidative lesions by DNA glycosylase/lyase enzymes produces single- and double-stranded breaks that may interfere with replication and activate DisA (3, 8, 28). Current knowledge indicates that the “return to life” of spores occurs thorough a developmental program that can be separated into two stages: germination and outgrowth (5, 6). An independent analysis of spores' germination or outgrowth kinetics indicated that nfo exoA spores exhibit a delay in both of these developmental stages, although the delay was relatively small for germination. Furthermore, both defects were suppressed by disruption of disA. Based on this evidence, we speculate that DisA could be required early in outgrowth to ensure efficient expression of genes and also later to act as a DNA damage-sensitive checkpoint prior to DNA replication.
To carry out its sporulation-associated checkpoint function, DisA accumulates during the first 3 h of sporulation (13). Our genetic analysis strongly suggests that DisA also has a checkpoint function during spore germination/outgrowth. Two possibilities whereby disA expression leads to the latter function were considered: (i) disA transcription occurs during sporulation, resulting in DisA packaging in spores, and/or (ii) expression of disA takes place during spore germination/outgrowth. However, expression of disA-lacZ and DisA-GFP fusions during sporulation occurred only in the mother cell. Previous results using Northern blot analysis showed that disA is transcribed as part of the ctsR-mcsA-mcsB-clpC-radA-disA hexacistronic operon from alternate σA- and σB-dependent promoters located upstream of ctsR (35). Based on our demonstration of disA expression during sporulation, it is possible that an unknown σE- or σK-dependent sporulation-specific promoter directs the transcription of disA in the mother cell compartment. disA-lacZ and disA-gfp expression also took place during spore germination/outgrowth. In particular, fluorescence microscopy and spectroscopy of germinating/outgrowing spores with a disA-gfp fusion detected a GFP signal possessing the described properties of DisA (13), most notably the formation of a single globular focus associated with the nucleoid of outgrown spores. Interestingly, although our results clearly showed the expression of disA and the presence of its encoded product in outgrowing spores, a study analyzing global transcription of the B. subtilis genome during germination/outgrowth did not detect disA mRNA (9). However, results from a different transcriptomic study with germinating spores subjected to different stresses showed induction of disA expression 60 min after the beginning of germination (36). It remains to be investigated whether expression of disA during this developmental stage takes place from the σA promoter located upstream of cstR or some other promoter. Indeed, it has been reported that σB and σM promoters located upstream of ctsR (37) and in the open reading frame of radA/sms (38), respectively, direct the transcription of disA. In good agreement with that report, we found that NaCl (4%) and vancomycin (0.2 μg/ml) significantly increased the expression level of the disA-lacZ fusion (data not shown). In contrast, neither mitomycin C nor H2O2 induced disA-lacZ expression (data not shown), suggesting that disA is not a member of the PerR and SOS regulons (39, 40). Future experiments will be required to identify the sigma factors involved in directing the expression of disA in the mother cell compartment and during spore outgrowth and also to assess if disA mRNA is part of a polycistronic transcription unit in both differentiation stages.
Comparison of germination and outgrowth curves of nfo exoA and nfo exoA disA spores together with experiments that analyzed the spatiotemporal expression and synthesis of DisA strongly suggest that this protein is a major component of a checkpoint when oxidative DNA damage is present in germinating/outgrowing spore DNA. Several lines of evidence described in this work support this contention: (i) the DisA-GFP detected by fluorescence microscopy during spore outgrowth was in a focus, a structural property inherent in DisA's checkpoint function (13, 32); (ii) DisA-GFP was associated with the chromosome of outgrown spores; (iii) the delayed outgrowth of nfo exoA spores was accompanied by a delay in chromosome segregation as well as in DNA replication; (iv) repair of oxidative lesions in DNA and the slow-germination outgrowth phenotype were suppressed by disA disruption in spores deficient for Nfo and ExoA; and (v) there was increased mutagenesis in hydrogen peroxide-treated outgrown disA spores compared to hydrogen peroxide-treated outgrown wild-type spores. Overall, our results provide novel evidence supporting the previously postulated prediction that DisA may constitute a more general mechanism that provides checkpoint control in different cellular and developmental contexts (13).
Finally, it must be pointed that the checkpoint mechanism in Nfo- and ExoA-deficient outgrowing spores described in this work resembles a similar phenomenon described for cells of Saccharomyces cerevisiae lacking the AP endonucleases Apn1 and Apn2 and Rad1 or Rad10 nucleotide excision repair (NER) proteins (41). Essentially, the accumulation of unrepaired AP sites in this mutant that are normally processed by DNA glycosylases/AP-lyases generates 3′-blocking single-strand breaks. The replication of the DNA strand containing these lesions generates double-strand breaks that elicit a G2/M checkpoint dependent on the RAD9 protein (41). As noted above, B. subtilis spores accumulate AP sites but mostly 8-oxo-G; however, processing of the latter lesion by DNA glycosylase with associated AP-lyase activity may also lead to the production of double-strand breaks when replication is activated during outgrowth, thus activating the DisA-dependent checkpoint mechanism.
ACKNOWLEDGMENTS
This work was supported by the University of Guanajuato (grant DAIP-044-2012) and by the Consejo Nacional de Ciencia y Tecnología (CONACYT) (grant 205744) of México. S.S.C., J.R.I.-R., R.C.B.-O., and F.H.R.-G. were supported by scholarships from CONACYT. The work of P.S. was supported by a Department of Defense Multi-Disciplinary University Research initiative through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286.
Footnotes
Published ahead of print 15 November 2013
REFERENCES
- 1.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572. 10.1128/MMBR.64.3.548-572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- 3.Pedraza-Reyes M, Ramírez-Ramírez N, Vidales-Rodríguez LE, Robleto EA. 2012. Mechanisms of bacterial spores survival, p 73–84 In Abel-Santos E. (ed), Bacterial spores: current research and applications. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 4.Setlow P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172–180. 10.1016/j.tim.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 5.Paidhungat M, Setlow P. 2001. Spore germination and outgrowth, p 537–548 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 6.Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556. 10.1016/j.mib.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 7.Setlow P. 1988. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function and degradation. Annu. Rev. Microbiol. 42:319–338. 10.1146/annurev.mi.42.100188.001535 [DOI] [PubMed] [Google Scholar]
- 8.Ibarra JR, Orozco AD, Rojas JA, López K, Setlow P, Yasbin RE, Pedraza-Reyes M. 2008. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 190:2031–2038. 10.1128/JB.01625-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keijser JF, Ter Beek A, Rauwerda H, Schuren F, Montijn R, van der Spek H, Brul S. 2007. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J. Bacteriol. 189:3624–3634. 10.1128/JB.01736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkholder WF, Kurtser I, Grossman AD. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269–279. 10.1016/S0092-8674(01)00211-2 [DOI] [PubMed] [Google Scholar]
- 12.Veening JW, Murray H, Errington J. 2009. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 23:1959–1970. 10.1101/gad.528209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. 2006. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125:679–690. 10.1016/j.cell.2006.03.039 [DOI] [PubMed] [Google Scholar]
- 14.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12:594–601. 10.1038/embor.2011.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salas-Pacheco JM, Setlow B, Setlow P, Pedraza-Reyes M. 2005. Role of the Nfo (YqfS) and ExoA apurinic/apyrimidinic endonucleases in protecting Bacillus subtilis spores from DNA damage. J. Bacteriol. 187:7374–7381. 10.1128/JB.187.21.7374-7381.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097–3104. 10.1099/00221287-144-11-3097 [DOI] [PubMed] [Google Scholar]
- 17.Kaltwasser M, Wiegert T, Schumann W. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 68:2624–2628. 10.1128/AEM.68.5.2624-2628.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barraza-Salas M, Ibarra-Rodríguez JR, Mellado SJ, Salas-Pacheco JM, Setlow P, Pedraza-Reyes M. 2010. Effects of forespore-specific overexpression of apurinic/apyrimidinic endonuclease Nfo on the DNA-damage resistance properties of Bacillus subtilis spores. FEMS Microbiol. Lett. 302:159–165. 10.1111/j.1574-6968.2009.01845.x [DOI] [PubMed] [Google Scholar]
- 19.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Nicholson WL, Setlow P. 1990. Sporulation, germination, and outgrowth, p 391–450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Sussex, England [Google Scholar]
- 21.Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 27–74 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Sussex, England [Google Scholar]
- 22.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Boylan RJ, Mendelson NH, Brooks D, Young FE. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S, Setlow P. 2009. Isolation and characterization of superdormant spores of Bacillus species. J. Bacteriol. 191:1787–1797. 10.1128/JB.01668-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711. 10.1073/pnas.54.3.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedraza-Reyes M, Gutierrez-Corona F, Nicholson WL. 1994. Temporal regulation and forespore-specific expression of the spore photoproduct lyase gene by sigma-G RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 176:3983–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason JM, Hackett RH, Setlow P. 1988. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd ed. ASM Press; Washington, DC: 2006. [Google Scholar]
- 29.Barzilai A, Yamamoto K. 2004. DNA damage responses to oxidative stress. DNA Repair 3:1109–1115. 10.1016/j.dnarep.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 30.Cabrera-Martínez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457–2464. 10.1128/JB.185.8.2457-2464.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982–3990. 10.1128/JB.183.13.3982-3990.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte G, Hartung S, Büttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30:167–178. 10.1016/j.molcel.2008.02.020 [DOI] [PubMed] [Google Scholar]
- 33.Urtiz-Estrada N, Salas-Pacheco JM, Yasbin RE, Pedraza-Reyes M. 2003. Forespore-specific expression of Bacillus subtilis yqfS, which encodes type IV apurinic/apyrimidinic endonuclease, a component of the base excision repair pathway. J. Bacteriol. 185:340–348. 10.1128/JB.185.1.340-348.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moeller R, Setlow P, Pedraza-Reyes M, Okayasu R, Reitz G, Nicholson WL. 2011. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in radiation resistance and radiation-induced mutagenesis of Bacillus subtilis spores. J. Bacteriol. 193:2875–2879. 10.1128/JB.00134-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hecker M, Pané-Farré J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236. 10.1146/annurev.micro.61.080706.093445 [DOI] [PubMed] [Google Scholar]
- 36.Moeller R. 2008. Characterization of different types of radiation- and pressure-induced DNA damage in Bacillus subtilis spores and their global transcriptional response during spore germination. Ph.D. thesis University of Carolo-Wilhelmina, Braunschweig, Germany [Google Scholar]
- 37.Krüger E, Msadek T, Hecker M. 1996. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol. Microbiol. 20:713–723. 10.1111/j.1365-2958.1996.tb02511.x [DOI] [PubMed] [Google Scholar]
- 38.Eiamphungporn W, Helmann JD. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830–848. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276–3286. 10.1128/JB.184.12.3276-3286.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovett CM, Love PE, Yasbin RE, Robertson JW. 1988. SOS-like induction in Bacillus subtilis: induction of the RecA protein analog and a damage-inducible operon by DNA damage in Rec+ and DNA repair-deficient strains. J. Bacteriol. 170:1467–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillet M, Boiteux S. 2002. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 21:283–2841. 10.1093/emboj/21.11.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]