Abstract
The DevR/DosR regulator is believed to play a key role in dormancy adaptation mechanisms of Mycobacterium tuberculosis in response to a multitude of gaseous stresses, including hypoxia, which prevails within granulomas. DevR activates transcription by binding to target promoters containing a minimum of two binding sites. The proximal site overlaps with the SigA −35 element, suggesting that DevR-SigA interaction is required for activating transcription. We evaluated the roles of 14 charged residues of DevR in transcriptional activation under hypoxic stress. Seven of the 14 alanine substitution mutants were defective in regulon activation, of which K191A, R197A, and K179A+K168A (designated K179A*) mutants were significantly or completely compromised in DNA binding. Four mutants, namely, E154A, R155A, E178A, and K208A, were activation defective in spite of binding to DNA and were classified as positive-control (pc) mutants. The SigA interaction defect of the E154A and E178A proteins was established by in vitro and in vivo assays and implies that these substitutions lead to an activation defect because they disrupt an interaction(s) with SigA. The relevance of DevR interaction to the transcriptional machinery was further established by the hypoxia survival phenotype displayed by SigA interaction-defective mutants. Our findings demonstrate the role of DevR-SigA interaction in the activation mechanism and in bacterial survival under hypoxia and establish the housekeeping sigma factor SigA as a molecular target of DevR. The interaction of DevR and RNA polymerase suggests a new and novel interceptable molecular interface for future antidormancy strategies for Mycobacterium tuberculosis.
INTRODUCTION
DevR/DosR is a key response regulator of Mycobacterium tuberculosis that induces a robust adaptation program in response to a variety of environmental stresses. Upon exposure to hypoxia, nitric oxide, carbon monoxide, or ascorbic acid, the DevRS two-component system (also called DosRS) induces the expression of ∼48 genes that are referred to as the DevR/DosR regulon (1–5). DevR regulon function is believed to be essential for bacterial survival during dormancy (6–8). A typical two-component system comprises a histidine kinase that is often membrane associated and that in response to an environmental signal transfers an activating phosphosignal to a response regulator, usually a DNA binding protein, which in turn regulates transcription. DevR is a typical two-domain response regulator of the NarL subfamily and contains a conserved aspartic acid phosphorylation site, Asp54, in its N-terminal domain and the DNA binding function in its C-terminal effector domain (9–11). The crystal structure of the DevR protein provides us with insight into the overall structure and details of its interaction with DNA. The elucidation of the structure of inactive DosR/DevR revealed a novel topology and conformation for the protein not seen before in other response regulators of the NarL subfamily (11, 12). We recently showed that the highly conserved residue Thr82 plays a key role in mediating the conformational change in DevR that is essential for cooperative binding to DNA and subsequent gene activation despite an atypical location (13).
We recently assigned the promoters of the DevR regulon to 4 classes based on the number of DevR binding sites (14). The simplest of them, the class I promoters, contain two neighboring Dev boxes. Promoters containing three DevR binding sites are categorized as class II promoters, and class III promoters are those with four tandem Dev boxes. Class IV regulon promoters have the most complex structure, and they not only contain primary and secondary DevR binding sites but also display an extended DNase I-protected region (14). A universal feature of all these promoters is the presence of a minimum of two binding sites in tandem and their helical phase arrangement. DevR interacts first with a primary site and then cooperatively binds to an adjacent site(s) that invariably has low sequence conservation (14). Another conserved feature of target promoters was the juxtaposing of the −35 element and the proximal DevR binding site (15–17), suggesting that DevR interacts with RNA polymerase to activate transcription. This hypothesis was supported by the isolation of a peptide interacting with the C-terminal domain of DevR (DevRC) that inhibited DevR-mediated transcription but not the DNA binding property of DevR (18).
The isolated DNA binding DevRC can activate transcription, albeit weakly, which suggests that one or more activation regions lie within this domain (19). However, the full-length protein, which supports cooperative interactions with DNA, is required for robust activation (19). Toward defining the amino acids necessary for gene activation function, we undertook a partial mutational analysis of residues located in the DNA binding domain of DevR. We report the identification of four M. tuberculosis “positive-control” mutants (pc mutants) that were defective in transcription activation but whose phosphorylation and DNA binding properties were largely unaffected. pc mutants have been defined earlier as mutants that bind to DNA but, unlike the wild-type (WT) protein, fail to activate transcription (20). Two pc mutants, E154A and E178A, were characterized further. The interaction between WT DevR protein and the principal sigma factor SigA was demonstrated by various protein-protein interaction assays, and a mutation in either E154 or E178 disrupted these interactions. The combined defects in DevR-SigA interactions, M. tuberculosis regulon activation, and hypoxic survival support the argument that the reason these substitutions are defective is because they disrupt DevR interaction(s) with SigA. The pc mutants characterized in this study reveal a surface(s) on DevR that interacts with RNA polymerase to activate transcription. Based on our findings, we propose that disrupting the molecular interaction(s) between DevR and RNA polymerase is a novel and additional step in the DevR signaling pathway for developing inhibitors of hypoxia-induced dormancy in M. tuberculosis.
MATERIALS AND METHODS
Plasmids, bacterial strains, and culture conditions.
M. tuberculosis strains were cultured at 37°C in Dubos medium containing 0.05% Tween 80 plus 0.5% albumin, 0.75% dextrose, and 0.085% NaCl (DTA medium). Escherichia coli strains and culture conditions were as described earlier (21). Antibiotics were used at the following concentrations: hygromycin (Hyg), 50 μg/ml for M. tuberculosis and 200 μg/ml for E. coli; kanamycin (Kan), 20 μg/ml for M. tuberculosis; and trimethoprim (Trim), 30, 40, or 50 μg/ml for Mycobacterium smegmatis as indicated. All plasmids and bacterial strains used in this study are described in Tables 1 and 2, respectively.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pFPV27 | E. coli-Mycobacterium shuttle plasmid with promoterless GFP gene, Kanr | 37 |
| p1738 | GFP reporter plasmid containing Rv1738 promoter, Kanr | 16 |
| pJFR19 | E. coli-Mycobacterium integrating shuttle plasmid with 3-kb acetamidase promoter, Hygr | 22 |
| pUAB100 | E. coli-Mycobacterium shuttle plasmid expressing GCN4-mDHFR F[1,2] under hsp60 promoter, Hygr | 23 |
| pUAB200 | E. coli-Mycobacterium shuttle plasmid expressing GCN4-mDHFR F[3] under hsp60 promoter, Kanr | 23 |
| pUS Phsp60 | pJFR19 containing constitutive hsp60 promoter cloned in NdeI and BstBI sites | This study |
| pUS Phsp60 devRwt | pUS Phsp60 containing devR wild type (cloned in NdeI sites); DevR is expressed from the constitutive hsp60 promoter | This study |
| pUS Phsp60 devRmut | pUS Phsp60 containing devR mutant variant generated by site-directed mutagenesis; individual mutant DevR proteins (D152A, E154A, etc.) are expressed from the constitutive hsp60 promoter | This study |
| pUS-mut DevR | pGEX4T1 overexpressing DevR E154A or other mutant DevR proteinsa | This study |
| pAV-FLdevR100 | pUAB100 plasmid containing devR (cloned in BamHI and ClaI sites); DevR-mDHFR F[1,2] protein is expressed from the constitutive hsp60 promoter, Hygr | This study |
| pAV-E154A | pAV-FLdevR100 derivative expressing DevR with E154A mutation | This study |
| pAV-E178A | pAV-FLdevR100 derivative expressing DevR with E178A mutation | This study |
| pVS-SigA200 | pUAB200 plasmid containing SigA (cloned in MfeI and ClaI sites); SigA-mDHFR F[3] is expressed from the constitutive hsp60 promoter, Kanr | This study |
WT and all DevR mutant proteins are expressed with an N-terminal GST tag.
TABLE 2.
Strains used in this study
| Mycobacterium strain | Relevant features | Source |
|---|---|---|
| M. tuberculosis WT | ΔdevR/pUS Phsp60 devRwt, expresses WT DevR protein | This study |
| M. tuberculosis mutant | ΔdevR/pUS Phsp60 devRmut, expresses individual DevR mutant protein (D152A, E154A, etc.) | This study |
| Comp WT | ΔdevR electroporated with pUS Phsp60 devRwt and p1738 expressing WT DevR protein | This study |
| Comp XnA | ΔdevR electroporated with pUS Phsp60 devRD152A (or other mutants) and p1738; X and n represent amino acid residue and position, respectively, in WT DevR protein with alanine (A) substitution | This study |
| C-1738 | ΔdevR electroporated with pUS Phsp60 and p1738 (no source of DevR) | This study |
| M. smegmatis GCN4/GCN4 (positive) | Mycobacterium smegmatis mc2155 cotransformed with pUAB100 and pUAB200 | This study |
| M. smegmatis DevR/GCN4 (negative) | Mycobacterium smegmatis mc2155 cotransformed with pAV-FLdevR100 and pUAB200 | This study |
| M. smegmatis DevR/SigA | Mycobacterium smegmatis mc2155 cotransformed with pAV-FLdevR100 and pVS-SigA200 | This study |
| M. smegmatis E154A/SigA | Mycobacterium smegmatis mc2155 cotransformed with pAV-E154A and pVS-SigA200 | This study |
| M. smegmatis E178A/SigA | Mycobacterium smegmatis mc2155 cotransformed with pAV-E178A and pVS-SigA200 | This study |
Generation of PCR plasmid template for site-directed mutagenesis.
The hsp60 promoter and wild-type devR coding sequences were cloned in the pJFR19 integrative plasmid (22). For this, the hsp60 promoter (23) was amplified by PCR from M. tuberculosis H37Rv genomic DNA using Pfu Turbo DNA polymerase (Invitrogen, USA) and primers hsp60BstBI f and hsp60NdeI r (see Table S1 in the supplemental material) to generate pUS Phsp60. Next, the devR sequence was amplified by PCR as described above using primers FLNdeI f and FLNdeI r (see Table S1 in the supplemental material) and cloned into pUS Phsp60 to generate pUS Phsp60devRwt.
Mutagenesis of DevR.
Alanine substitution mutants of DevR were generated by PCR using plasmid pUS Phsp60devRwt as a DNA template (mut variants in Table 1). Individual codons were changed to GCA (residues R173 and E178), GCT (residue D215), GCG (residues E154, R155, E163, K168, K179, K191, R197, E206, and K208), or GCC (residues D152 and R209). Site-directed mutagenesis was performed by PCR using specific oligonucleotides primers (see Table S1 in the supplemental material) and Pfu Turbo DNA polymerase in a reaction mix of 50 μl containing 8% dimethyl sulfoxide (DMSO). Reaction conditions were 95°C (2 min) followed by 30 cycles of 95°C (30 s), 65°C (30 s), and 72°C (14 min). The 50-μl reaction mix was subjected to DpnI digestion (10 U) for 1 h at 37°C to remove template DNA. Four microliters of the DpnI digestion mix was used to transform E. coli XL-1 Blue, and transformants were selected on LB agar plates containing hygromycin (200 μg/ml). The presence of the desired mutation was confirmed by DNA sequencing. DNA sequencing revealed the presence of a K168A substitution introduced inadvertently during the generation of the K179A mutation that resulted in generation of double mutant K179A* (K168A+K179A). Thus, DevR mutant K168A was used in parallel in all the experiments to rule out an effect of the K168A mutation in double mutant K179A*.
Construction of M. tuberculosis strains constitutively expressing DevR WT or individual DevR mutant proteins.
To analyze the effects of the mutations in vivo, pUS Phsp60devRwt or pUS Phsp60 devRmut recombinant plasmids were electroporated into the M. tuberculosis ΔdevR strain (24). M. tuberculosis transformants were selected on 7H11 agar containing hygromycin (50 μg/ml). Each M. tuberculosis transformant (denoted M. tuberculosis WT, D152A, E154A, R155A, etc.) expresses the respective DevR mutant and is isogenic to M. tuberculosis H37Rv.
Construction of DevR mutant protein-overexpressing plasmids and purification of the mutant proteins from Escherichia coli.
devR mutant coding sequences were amplified from individual integrative pUS Phsp60devRmut plasmids (Table 1) by PCR using UGSTdevR f and UGSTdevR r (for primer sequences, see Table S1 in the supplemental material) and cloned into the E. coli expression plasmid pGEX4T1 (21) to generate the pUS devRmut plasmid series. Plasmid pUS devRmut represents each individual DevR substitution and expresses N-terminally glutathione S-transferase (GST)-tagged DevR mutant protein (Table 1). GST-DevR mutant proteins were overexpressed in E. coli C43(DE3), and the recombinant proteins were purified as described earlier (21) and used in phosphorylation assays, electrophoretic mobility shift assays (EMSAs), GST pulldown assay, and enzyme-linked immunosorbent assay (ELISA). All plasmid constructs generated in this study were verified by DNA sequencing.
Western blotting of M. tuberculosis lysates.
Immunoblot analysis of M. tuberculosis lysates was performed to determine the levels of DevR protein expression in the individual strains. For this, frozen M. tuberculosis stocks were revived in DTA medium, subcultured twice, grown in a shaker incubator at 220 rpm (50 ml in a 250-ml screw-cap airtight Teflon flask) to an A595 of ∼0.2 to 0.3, and subsequently processed for immunoblotting. Briefly, a 20-ml aliquot was chilled on ice and centrifuged immediately at 2,800 × g for 10 min at 4°C, and the pellet was stored at −20°C (“aerobic”). The remaining culture (30 ml) was distributed (10-ml aliquots in 50-ml tubes that were tightly closed) and kept standing for 5 days to generate “hypoxic” cultures as described previously (7). The cells were harvested from dedicated culture tubes after appropriate incubation, and whole-cell lysates were prepared as described previously (38). DevR and SigA proteins were detected in the lysates (containing ∼15 μg protein) by Western blotting using anti-DevR and anti-SigA as described earlier (21).
RT-qPCR analysis.
To assess the effects of the mutations on various target promoters, reverse transcriptase quantitative PCR (RT-qPCR) analysis for various genes of the DevR regulon was carried out. RNA was isolated from M. tuberculosis cultures expressing DevR and individual mutant proteins as described previously (21). Briefly, a 20-ml aliquot was snap-chilled on ice and centrifuged immediately as described above (“aerobic”), and the remaining culture was kept standing for 5 days as described above (“hypoxic”). The harvested cell pellets were each resuspended in 1 ml of TRI reagent (Molecular Research Center, USA) and lysed in a mini-bead beater using 0.1-mm zirconium-silica beads (Biospec, USA). RNA was purified as described earlier (15). For RT-qPCR, cDNA was synthesized using 200 ng of total RNA by reverse transcription with 50 U of MultiScribe reverse transcriptase and random hexamer primers as per the manufacturer's instructions (Applied Biosystems, USA). Two microliters of cDNA was subjected to qPCR using gene-specific primers (see Table S1 in the supplemental material) and Power SYBR green PCR master mix in a 25-μl reaction mixture in a MyiQ thermal cycler (Bio-Rad, USA). The reaction conditions were 94°C (10 min) followed by 40 cycles of 94°C (30 s), 56 to 65°C (45 s), and 72°C (30 s). An RT-negative (without reverse transcriptase) reaction was used to account for residual DNA, and the transcript numbers were normalized to that of 16S rRNA. These normalized values were used to determine the relative fold change in expression (hypoxic versus aerobic [H5/Aer]) for various gene targets in each strain with respect to the WT strain (100%). The experiments were performed in triplicate, and the results are expressed as mean ± standard deviation (SD).
Measurement of GFP fluorescence.
Green fluorescent protein (GFP) reporter assays were conducted as described previously (16). Briefly, M. tuberculosis cultures harboring the M. tuberculosis p1738 promoter-driven GFP plasmid (16) and expressing either WT or individual mutant DevR proteins from a chromosomally integrated gene (Comp strains in Table 2) were grown under aerobic and hypoxic conditions in DTA medium at 37°C (as described above). Relative fluorescence units per unit of optical density (RFU/OD) was calculated from hypoxic cultures grown in 96-well plates and aerobic cultures grown by shaking in parallel for 120 h. The RFU values of the appropriate vector control were subtracted. The promoter activity is expressed in relative fluorescence units per unit of OD at 595 nm (RFU/OD ± SD) of GFP.
In vitro phosphorylation of DevR.
Autophosphorylation assays were carried out as described previously (13) using WT and mutant DevR proteins purified from E. coli. Briefly, 2 units of acetate kinase (catalog no. A6781; Sigma-Aldrich, India) was incubated with 5 μCi [γ-32P]ATP (3,500 Ci/mmol; Brit, Hyderabad, India) in a 10-μl reaction mix containing 25 mM Tris-Cl (pH 7.5), 60 mM potassium acetate, and 10 mM MgCl2 at 25°C for 20 min. The purified mutant or wild-type protein (final concentration, ∼3 μM) was then added to this reaction mix, and the mixture was added to a buffer containing 40 mM Tris-Cl (pH 8.0), 20 mM NaCl, 0.2 mM EDTA, and 0.2 mM dithiothreitol (DTT) and incubated at room temperature for 15 to 30 min. The reaction was terminated with 4 μl of stop solution containing 300 mM Tris-Cl (pH 6.8), 60% glycerol, 12% SDS, 7.5% β-mercaptoethanol, and 0.6% bromophenol blue and subsequently analyzed by electrophoresis on a 15% SDS-polyacrylamide gel and phosphorimaging. DevR phosphorylation by phosphotransfer from the sensor kinase DevS201 (cytoplasmic C-terminal fragment of DevS containing 201 amino acids) was carried out as described previously (25). Briefly, DevS201 (final concentration, 7.5 μM) was incubated in buffer containing 50 mM Tris-Cl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 500 μM ATP, and 0.1 μCi [γ-32P]ATP at 25°C for 60 min. DevR protein (WT or mutant E154A or E178A, 5 μM) was added to the reaction mix and incubated for 0.5 and 1 min. The reaction was terminated with 4 μl of stop buffer and analyzed by SDS-PAGE and phosphorimaging. Each experiment was performed at least thrice, and representative results are shown.
DNA binding assay.
Electromobility shift assays (EMSAs) were performed with purified WT/mutant DevR proteins and a 40-bp double-stranded DNA fragment corresponding to the P+S binding sites in the tgs1-Rv3131 promoter region. This EMSA has been reliably used to establish sequence-specific interactions of DevR with DNA (13, 14, 17, 19). Prior to EMSA, DevR (final concentration range, 1 μM to 6 μM) was autophosphorylated by incubation with 50 mM acetyl phosphate for 20 min at 25°C in 40 mM Tris-Cl (pH 8.0) and 5 mM MgCl2 as described previously (25). The percentage of DNA bound to DevR∼P was determined by densitometric analysis of the fraction of free DNA in each lane and expressed as a percentage of the binding efficiency of the WT protein, which was taken as 100. The assay was performed twice, and the mean values are plotted.
GST pulldown assay.
GST-DevR, His6-SigA, and GST (only tag) proteins were purified from E. coli cultures harboring expression plasmids pSC DevR (21), pET23b SigA (26), and pGEX4T1 (21), respectively, using standard procedures. Purified GST-DevR or GST tag alone (200 pmol) was allowed to bind to glutathione-Sepharose in binding buffer (20 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 0.5% Triton X-100, 5% glycerol) at 4°C with end-over-end mixing for 1 h. The unbound protein was removed by washing with 30 bed volumes of binding buffer, and subsequently 80 pmol of SigA protein was incubated with immobilized GST-DevR/GST for 2 h at 4°C. After rigorous washing, resin-bound GST-DevR/GST along with any bound protein was eluted from the beads with 2× SDS-PAGE buffer and heating at 100°C for 10 min. Load and elution fractions were resolved by SDS-PAGE, followed by detection of bound proteins in elution fractions by Western blotting using anti-GST and anti-SigA antibodies.
ELISA.
DevR-SigA interaction was also assessed in an ELISA format in which 100 pmol of purified GST-DevR or GST tag alone was applied overnight at 4°C to a 96-well ELISA plate in coating buffer (0.5 M carbonate-bicarbonate buffer, pH 9.6). Control wells were coated with bovine serum albumin (BSA) or left uncoated. Blocking was carried out for 2 h at 37°C with 3% BSA made in Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl). The wells were then washed thrice with TBS, and 40 pmol of purified His6-SigA was added. After incubation at room temperature for 2 h, the plate was washed rigorously twice each with TBS containing 0.1% Tween 20 and TBS. Monoclonal anti-His6 horseradish peroxidase (HRP)-conjugated antibody (1: 2000, Sigma) was added and incubated at room temperature for 1 h, followed by washing as described above. o-Phenylenediamine was used as a substrate for color detection, and the absorbance was measured at 490 nm.
MPFC assay.
The mycobacterial protein fragment complementation (MPFC) assay was used to assess in vivo protein-protein interaction between M. tuberculosis proteins SigA and DevR in the surrogate host M. smegmatis as described previously (23). Briefly, WT DevR- and SigA-coding sequences were cloned in pUAB100 and pUAB200, respectively, as described previously (23) to generate plasmids pAV-FLdevR100 and pVS-SigA200 (Table 1). E154A and E178A substitutions were generated in pAV-FLdevR100 by site-directed mutagenesis using specific primers (see Table S1 in the supplemental material) as described by the manufacturer (Accuprime Pfx DNA polymerase and GeneArt SDM system; Invitrogen, USA). The DevR- and SigA-expressing plasmids were coelectroporated into M. smegmatis mc2155 to generate SigA and WT DevR/E154A/178A protein expression pairs. The cotransformants were screened on 7H11 agar medium supplemented with 0.5% glycerol, 0.5% glucose, and 0.2% Tween 80 (7H11) with kanamycin (Kan) (25 μg/ml) and hygromycin (Hyg) (50 μg/ml) for the presence of both expression plasmids. The transformants were then subcultured onto 7H11 with Kan plus Hyg and trimethoprim (Trim) at 30, 40, and 50 μg/ml to detect growth indicative of protein-protein interaction. The protein expression pairs GCN4/GCN4 and DevR/GCN4 were taken as positive and negative controls, respectively.
Viability of M. tuberculosis mutant strains.
Frozen stocks of M. tuberculosis WT and the E154A and E178A mutants were subcultured thrice in DTA medium (A595, ∼0.4). The cultures were diluted to an A595 of 0.005, and 10-ml aliquots were dispensed in 50-ml tubes and grown either with shaking at 220 rpm (aerobic setup) or standing in tight screw-cap tubes with a headspace ratio of 0.6 (hypoxic setup). The cultures were sampled once only from separate tubes dedicated for each time point of the hypoxia setup. Bacterial CFU at defined time points were estimated by plating serial dilutions in duplicate on MB 7H11 agar containing ADC and incubating the plates at 37°C for 5 weeks. The experiment was performed in triplicate.
RESULTS
Experimental design and rationale.
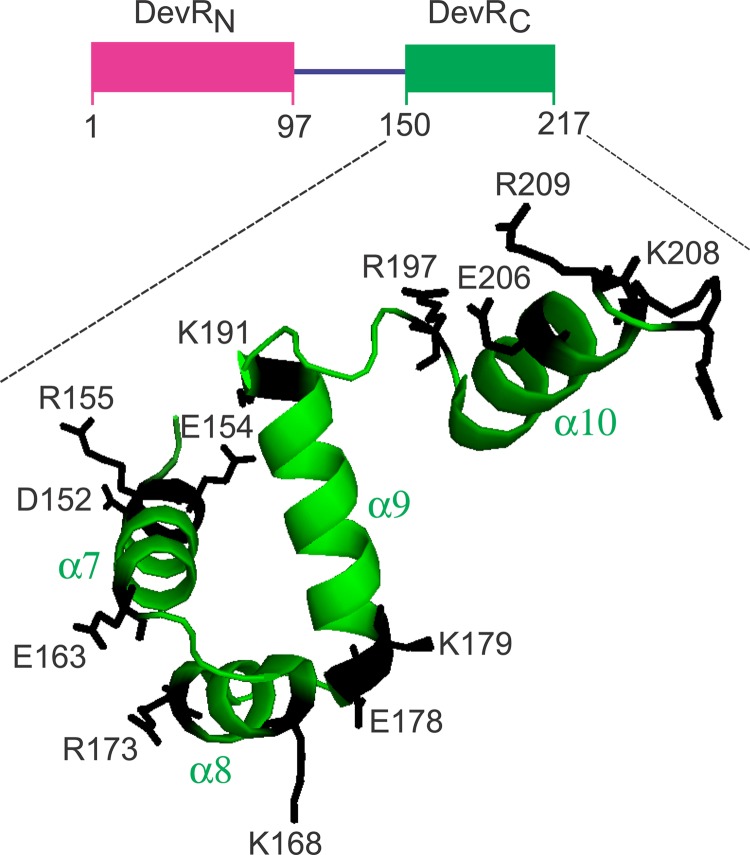
We set out to evaluate the involvement of some residues of the C-terminal domain in the transcriptional activation mechanism of DevR. We assumed that the residues on the protein surface are likely to interact with RNA polymerase at the target promoter(s), and the present study was restricted to charged residues only. The C-terminal domain of DevR (PDB file name 3C3W) (11) contains 20 charged residues (Arg [R], Lys [K], Asp [D], and Glu [E]), of which the role of the K182 residue in DNA binding and regulon activation was demonstrated recently (27). DevRC activates DevR regulon expression, albeit weakly (19), implying that this domain interacts with the transcriptional machinery. Therefore, in the present study, 14 amino acids (R, K, D, or E) were each singly mutated to alanine in the full-length DevR protein, and the effects of individual mutations on the biochemical properties, gene activation, and hypoxic survival of Mycobacterium tuberculosis were analyzed (Fig. 1).
FIG 1.
Locations of amino acids on the structure of DevRC/DosRC. Amino acids that were mutated to alanine in this study are indicated. The residues (extending up to 210) at the C-terminal end of DevR (DosR) are shown as per the crystal structure from PDB file 3C3W (11) using program PyMol, and hence the location of D215 is not shown.
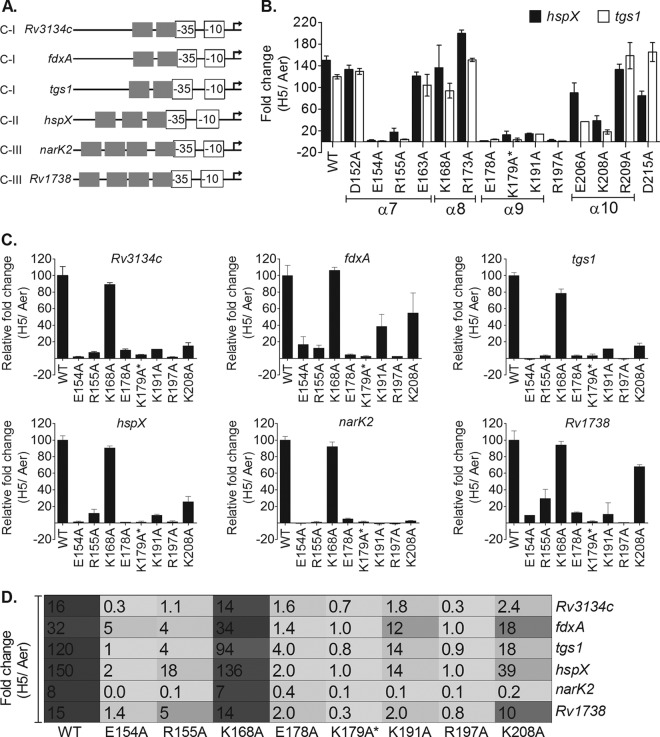
The genes of the DevR regulon are grouped into 4 classes based on the number of DevR binding sites in their promoter regions (14). However, temporal analysis of regulon expression revealed the lack of a direct relationship between the number of binding sites and the timing and magnitude of the activation response. Furthermore, the temporal induction response did not correlate strictly with the affinity of DevR for the various target promoters and suggested that additional factors such as intrinsic promoter strength and other cis elements/trans-acting regulatory proteins may be involved (14). An overlap of the −35 promoter element with the transcription start point (TSP)-proximal DevR binding site is a conserved feature of the target promoters across all promoter classes (Fig. 2A), making it very likely that DevR would interact with the RNA polymerase machinery to activate transcription. Therefore, the question arose whether individual amino acids of DevR are selectively involved in the activation of different promoter classes. Accordingly, promoters representative of class I (Rv3134c, fdxA, and tgs1), class II (hspX), and class III (narK2 and Rv1738) were examined for the effects of individual amino acid mutations in DevR on gene activation.
FIG 2.
Identification of transcription activation-defective devR mutants. (A) Conserved promoter architecture of regulon promoters. The overlap of the −35 element with the TSP-proximal Dev box is shown (15–17, 27). Experimentally determined hypoxic transcription start points (TSPs) are indicated with bent arrows. C-I, C-II, and C-III refer to class I, class II, and class III promoters, respectively. (B) M. tuberculosis mutant strains were screened for tgs1 and hspX transcripts by RT-qPCR analysis. The fold change in transcripts under 5-day “hypoxic” versus “aerobic” (H5/Aer) conditions is shown. (C) Selected mutant strains (from panel B) were further tested for their activation properties on class I (Rv3134c, fdxA, and tgs1), class II (hspX), and class III (narK2 and Rv1738) promoters by RT-qPCR. Relative fold change values (H5/Aer) in mutants with respect to WT DevR (taken as 100) are shown. (D) Comparison of fold activation (H5/Aer) of various regulon promoters in WT and mutant M. tuberculosis strains as measured by RT-qPCR analysis.
Identification and characterization of DevR mutants.
Fourteen M. tuberculosis strains, each expressing a DevR mutant protein harboring an alanine substitution at a single amino acid (Fig. 1), were constructed and screened for DevR regulon gene activation by RT-qPCR analysis of hspX and tgs1 transcripts and the Rv1738 promoter-based GFP reporter assay.
Four of the mutations, namely, in E206, K208, R209, and D215, were generated in amino acids located in the α10 helix and the unstructured tail of DevR. The α10 dimerization helix is uniquely placed in DevR/DosR compared to the other members of the NarL family; it may closely interact with the N-terminal domain, and extensive structural rearrangements are probably required for acquiring the active DNA binding conformation (11). Three of these mutations, namely, in R209, D215, and E206, either did not influence gene activation (R209A), displayed an inconsistent activation defect (D215A), or caused only a moderate activation defect (E206) (Fig. 2). In contrast, the K208A mutation was associated with a consistently severe transcription activation defect (Fig. 2B). The K208A mutant was further analyzed for the activation of various promoter classes. Surprisingly, this mutant behaved differently at the same class of promoters; for example, at class I promoters, it showed a severe defect in tgs1 and Rv3134c activation (85%) but a partial defect in fdxA activation (45%). Similarly, at class III promoters, the K208 mutation severely affected narK2 promoter activity (97% defect) but only moderately affected the divergently transcribed Rv1738 promoter (33% defect) (Fig. 2). On the basis of the combined results of RT-qPCR and the reporter assays, the E206, R209, and D215 residues were considered to be not uniformly essential for DevR-mediated activation and were not analyzed further in the present study.
Among the remaining 10 strains that were analyzed, M. tuberculosis K168A displayed only a mild tgs1-specific defect, the E163A mutant displayed a mild to moderate defect, the D152A strain was not activation defective, and the R173A mutant yielded a hyperactivation phenotype (Fig. 2C). The remaining 6 strains expressing DevR proteins mutated at E154, R155, E178, K179* (*, DevR double mutant K179A+K168A), K191, and R197 showed a consistent phenotype and were severely defective in activation (>89 to 100% defect) (Fig. 2). Analysis of the effects of these mutations on the activation of promoters belonging to different classes revealed a complete defect for the K179A* and R197A mutants, a 90 to 100% defect in the E154A and E178A mutants and an ∼60 to 99% defect in case of the R155A and K191A mutants (Fig. 2C). These mutants also showed an activation defect in the GFP reporter assay (range of 327 to 5,167 RFU/OD in the various mutants compared to 16,498 RFU/OD in the WT). Since the magnitude of the regulon induction response depends on the level of DevR expression during hypoxia (28), DevR levels in selected activation-defective M. tuberculosis strains were compared next by immunoblot analysis.
Level of DevR expression.
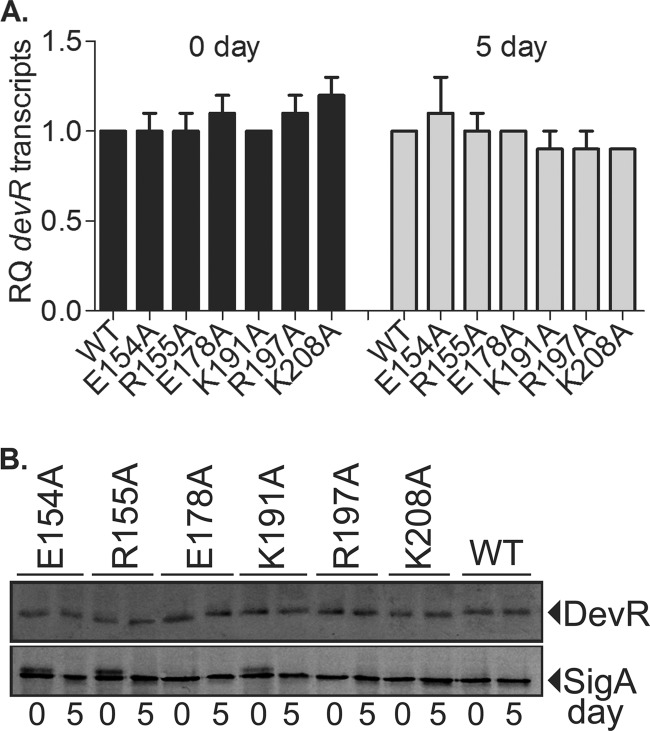
Several classes of loss-of-function mutations in devR, such as mutations resulting in protein instability or leading to a DNA binding defect or compromise in phosphorylation, could in principle lead to an activation-defective phenotype. In M. tuberculosis, DevR expression from the Rv3134c-devRS operon promoter is autoregulated in a hypoxia-responsive manner (15). Moreover, the magnitude of the regulon induction response depends on the level of DevR expression (28). Therefore, the constitutive hsp60 promoter was used to express WT or mutant DevR proteins from an integrated copy of the devR gene to enable the equivalent expression of DevR in various M. tuberculosis strains and to ensure that the observed phenotypes of the mutants were due to the mutation alone and not caused by differences in DevR levels or stability. Indeed, the expression of DevR in the WT and various M. tuberculosis mutant strains was confirmed to be equivalent and constitutive at both the RNA and protein levels (Fig. 3). Therefore, differences in mutant protein expression and/or stability were ruled out as an underlying cause of the activation-defective phenotype.
FIG 3.
DevR expression analysis. (A) devR transcript levels in various M. tuberculosis strains relative to WT in aerobic (day 0) and hypoxic (day 5) cultures. The normalized copy number values were then used to determine the relative quantities (RQ) of individual gene transcripts with reference to WT (see Materials and Methods). (B) Lysates (15 μg protein) of aerobic (day 0) and hypoxic (day 5) cultures of different M. tuberculosis strains expressing the wild-type (WT) or mutant DevR protein from the hsp60 promoter were subjected to immunoblotting. SigA protein served as an internal loading control.
Mechanism(s) underlying the activation defect.
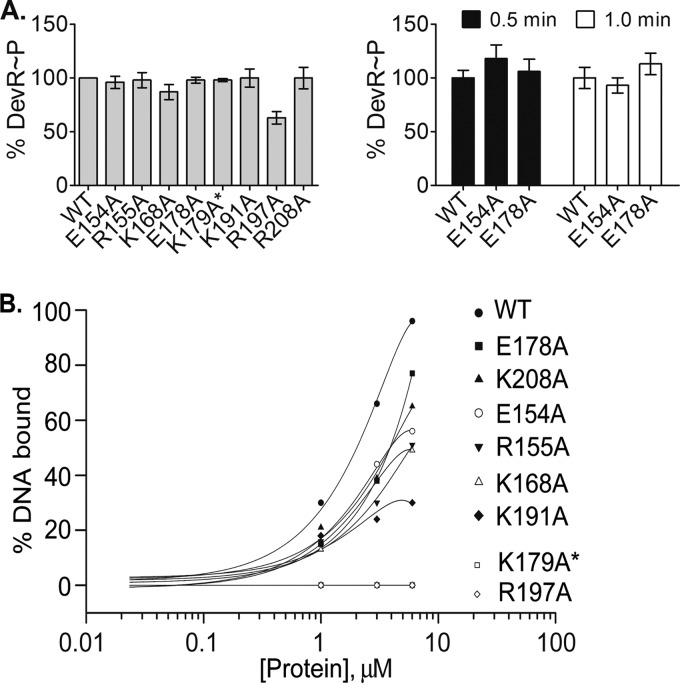
DevR mutant proteins that exhibited an activation defect were overexpressed and purified from E. coli as stable proteins (not shown) and biochemically characterized to decipher the underlying mechanism(s). Since the phosphorylation of DevR is essential for its binding to DNA and for DevR-mediated activation (15), the transcription activation defect exhibited by some mutants could be a consequence of a phosphorylation and/or a DNA binding defect. Accordingly, the in vitro autophosphorylation activity of mutant DevR proteins was evaluated using acetyl-[32P]phosphate, a low-molecular-weight phosphodonor that is frequently used to assess the phosphorylation property of response regulators, including DevR (13, 15, 16, 25). All the mutant proteins were autophosphorylated with an efficiency similar to that for DevR WT (87 to 100%), with the exception of R197A, which exhibited an ∼40% defect (Fig. 4A).
FIG 4.
Phosphorylation and DNA binding properties of mutant proteins. (A) Left panel, autophosphorylation of DevR proteins was carried out using acetyl-[32P]phosphate as described in Materials and Methods. The percent phosphorylation of mutant proteins relative to that of WT protein was derived by phosphorimaging and densitometric analysis. Right panel, DevS201-mediated phosphotransfer to DevR WT and mutant proteins. The percent efficiency of mutant DevR phosphorylation was calculated as described above. DevS201 (∼15 μM) was autophosphorylated in the presence of [γ-32P]ATP prior to addition to the reaction mixture. (B) Interaction of DevR with target promoter DNA. P+S box oligonucleotide sequences of the tgs1-Rv3131 promoter region were incubated with WT or mutant protein. The percentage of DNA bound is plotted versus protein concentration. A representative result from 3 experiments is shown.
The DNA binding properties of various DevR mutant proteins were examined next by a standardized EMSA procedure (13, 14, 17, 19) that is based on the interaction of DevR with a 40-bp double-stranded tgs1-Rv3131 intergenic DNA sequence. Protein binding to the motifs was examined at DevR concentrations that were in the physiological range, estimated as ∼5.4 μM under basal conditions (28) The interaction between DNA and phosphorylated WT DevR and five mutant proteins (E154A, R155A, K168A, E178A, and K208A) generated a single similarly migrating protein-DNA complex even at low protein concentrations, suggesting a similar type of interaction (not shown). Based on the comparative binding efficiencies of these mutant proteins with that of the WT DevR protein, they were not considered to be severely defective in binding to DNA. In contrast, three mutant proteins displayed severe binding defects; the R197A and K179A* mutant proteins were completely defective in binding to DNA, and the K191A mutant protein was ∼ 70% defective in DNA binding at a 6 μM protein concentration (Fig. 4B). The DNA binding defect of the R197A mutant protein may be attributed in part to a phosphorylation defect that disallows formation of the DNA binding active form of DevR. From these results it is concluded that a complete/partial defect in binding to DNA is the likely basis for the observed activation defect of these three mutant proteins. K179A* harbors a double mutation at K179 and K168. Because the K168A mutant exhibits no observable defects in biochemical properties or transcription activation and because the K179 residue was previously shown to directly interact with DNA in the crystal structure (12), the binding defect observed with the K179A* double mutant is the likely underlying cause of its severe activation defect. However, although individually the K168 mutation does not have a phenotype, we cannot exclude the possibility that this substitution could still exacerbate the phenotype of the K179A mutant.
Based on the in vivo and in vitro biochemical analysis described above, four activation-defective mutants, with mutations at E154, R155, E178, and K208, that were not majorly defective in DNA binding were classified as DevR pc mutants.
DevR single mutation E154A or E178A disrupts the interaction of DevR with SigA.
DevR binds in a sequence-specific manner to conserved sites located upstream of its target genes to activate gene expression under inducing conditions (15, 16). The conserved architecture of the regulon promoters suggests that DevR binding to DNA may help recruit and/stabilize RNA polymerase at target promoters. Although it is not known which sigma factor is utilized for transcription, we reasoned that SigA, which is crucial for RNA polymerase recognition of the −35 and −10 promoter elements, could be a likely interacting partner of DevR, as suggested for other bacterial systems (29, 30). This hypothesis was confirmed by protein-protein interaction assays in the present study (Fig. 5).
FIG 5.
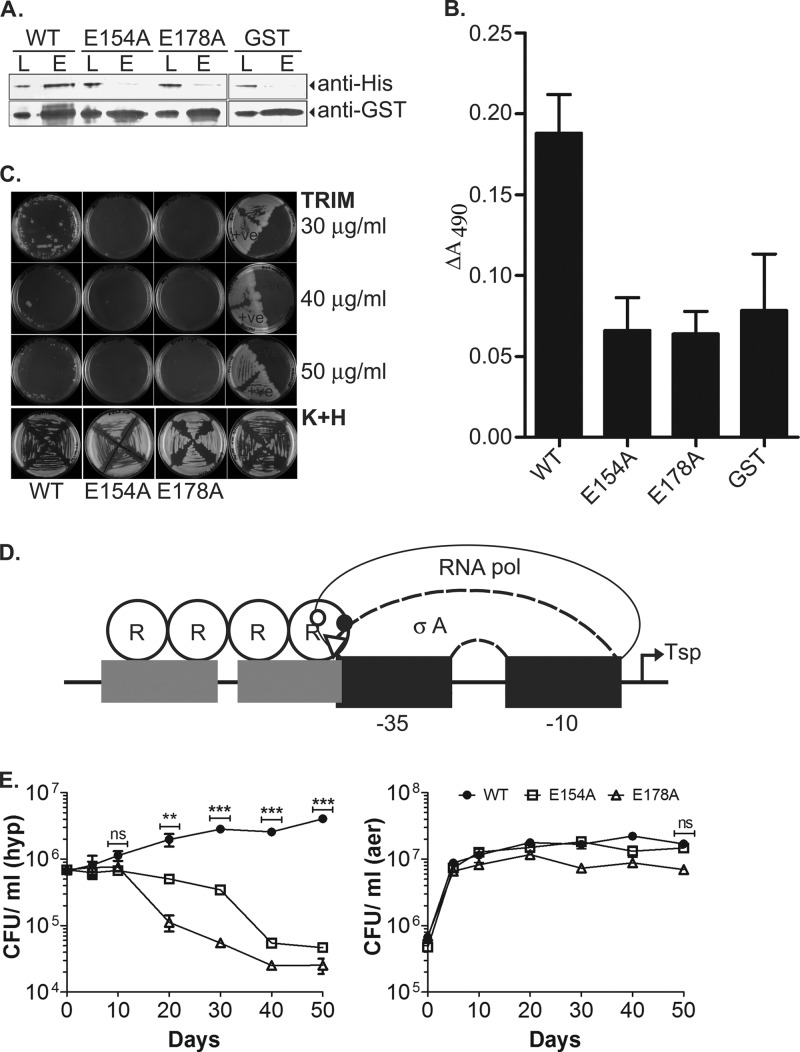
Interaction of DevR with SigA and effect on M. tuberculosis viability. (A) GST pulldown assay using SigA and GST-DevR/GST proteins. L, input load; E, elution. Monoclonal antibodies anti-GST (1:5,000; Sigma) and anti-His6 (1:2,000; Sigma) were used for the detection of DevR/GST and SigA proteins, respectively, by Western blotting. (B) ELISA using proteins DevR, E154A, E178A, GST tag only and SigA. ΔA490 values were obtained after subtracting the absorbance values obtained for control wells coated with BSA protein. (C) MPFC assay to analyze in vivo interaction of M. tuberculosis DevR and SigA. M. smegmatis transformants were subcultured on 7H11 agar containing Kan, Hyg, and Trim and incubated at 37°C for 3 to 6 days. (D) Model of interaction of DevR surface residues E154 (solid circle) and E178 (triangle) with SigA. R155 and K208 residues on DevR that are likely to interact with RNA polymerase (not excluding SigA) are shown by an empty circle. (E) Survival of M. tuberculosis WT and mutant strains. The viability of M. tuberculosis strains was assessed under aerobic and hypoxic conditions over a period of 50 days. The mean CFU ± SD determined from three independent cultures is shown. The difference in hypoxic viability between the WT and the E154A and E178A mutants was statistically significant on days 20 (**, P < 0.005) and 30, 40, and 50 (***, P < 0.0005).
Two DevR pc mutants with the most severe activation defect phenotype, E154A and E178A, were further analyzed for interaction with SigA using GST pulldown assay, ELISA, and mycobacterial protein fragment complementation (MPFC) assay. In the GST pulldown assay, the mutation of either E154 or E178 resulted in the disruption of DevR-SigA interaction. The specificity of DevR interaction was established by the lack of SigA interaction with the GST tag protein (Fig. 5A). The ELISA confirmed the interaction of WT DevR and SigA and indicated a role for the E154 and E178 residues in this interaction (Fig. 5B). The interaction of DevR and SigA was further established by the MPFC in vivo protein-protein interaction assay (Fig. 5C). The MPFC assay is based upon the functional reconstitution of two murine dihydrofolate reductase domains independently fused to the two putative interacting partners (the M. tuberculosis SigA and DevR proteins in this study) within Mycobacterium smegmatis, a surrogate host for M. tuberculosis. The growth of M. smegmatis in medium containing appropriate antibiotics (Kan plus Hyg) confirmed the presence of recombinant plasmids coexpressing SigA and DevR (WT, E154A, or E178A). Protein-protein interaction was screened using selective concentrations of trimethoprim (Trim) that were inhibitory for the endogenous dihydrofolate reductase. M. smegmatis coexpressing WT DevR and SigA grew well at a 30-μg/ml Trim concentration but modestly at a 40- or 50-μg/ml Trim concentration, indicative of an interaction between DevR and SigA. In contrast, the strains coexpressing SigA and either DevR E154A or E178A failed to grow at any of the Trim concentrations (Fig. 5C). Good mycobacterial growth of the positive control (GCN4-GCN4) and no growth of the negative control (DevR-GCN4) in the presence of Trim established assay specificity.
The results of these different interaction assays were indicative of a disruption of DevR-SigA interaction upon the introduction of either the E154A or E178A mutation and hence implicate these residues as being located at the interaction surface of DevR and SigA (Fig. 5D).
Hypoxia survival phenotype of M. tuberculosis strains expressing DevR E154A or E178A.
The M. tuberculosis E154A and E178A strains were next analyzed for their survival fitness properties under hypoxia (Fig. 5E). WT bacteria initially multiplied and maintained their viability at ∼6 times the initial CFU on day 50. However, both the mutant strains were defective in hypoxic adaptation; M. tuberculosis E178A displayed an earlier hypoxic survival defect (∼85% defect on day 20; P = 0.0016), whereas E154A exhibited a >90% defect on day 40 (P = 0.0002). On day 50, ∼ 4% to 7% E178A and E154A mutant bacteria, respectively, were viable relative to the initial CFU on day 0 (P < 0.0001). The aerobic survival of all the strains was similar and was consistent with the defined role for DevR under hypoxia. We conclude that WT DevR activity is essential for regulon induction and hypoxic survival of M. tuberculosis and that the E154 and E178 residues are crucial for DevR function via interaction with the SigA subunit of RNA polymerase.
DISCUSSION
This study aimed to identify residues in M. tuberculosis DevR that are involved in transcription activation. Seven of the 14 residues analyzed in this study were identified as being important for DevR-mediated activation. These amino acids are located on various surfaces of DevR: E154 and R155 in the α7 helix, E178 along with K179 and K191 in the DNA contacting helix α9, R197 in the loop between α helices 9 and 10, and K208 in the α10 dimerization helix (Fig. 1). As we examined the role of only select residues of DevR in transcriptional activation, we do not exclude the role of interactions involving other amino acids in the activation mechanism.
The identification of 4 pc mutants with mutations in the E154, R155, E178, and K208 residues was the highlight of this study. These mutants displayed severe defects in transcription activation that are not majorly attributable to an expression defect or a defect in phosphorylation or DNA binding. The properties of these mutants suggest that the phosphorylation-induced switch between the inactive and active conformations may not differ appreciably in them. Rather, the characterization of the E154A and E178A mutants establishes the essential role of the SigA sigma factor in mediating transcription of the DevR regulon genes. Thus, the hypoxic adaptation and survival phenotype of the mutant strains is attributed to a defect in interaction with RNA polymerase and a failure to express the DevR regulon in a SigA-dependent manner.
Based on the surface topology of DNA-bound DevRC (see Fig. S1 in the supplemental material) and the conserved promoter architecture, the E178 residue seems to be positioned appropriately for contact with RNA polymerase at all target promoters. This hypothesis is supported by the observed interaction of WT DevR and SigA in all the protein-protein interaction assays and the severe activation phenotype of the E178 mutant strain for all promoter classes. Another SigA-interacting surface on DevR was identified through the phenotype of the E154A pc mutant (Fig. 5D; see Fig. S1 in the supplemental material). The R155 and K208 residues, which also display a pc phenotype, are present at the α7/α8 interface in the DevR tetramer structure (12). A comparative analysis of the consequences of mutations in DevR and its orthologs provides useful insights into the activation mechanisms of the NarL subfamily of response regulators (see Fig. S2 in the supplemental material). The E163 residue in DevR is homologous to the Q169 residue in NarL (see Fig. S2 in the supplemental material), and both mutant forms are proficient in transcription activation (reference 31 and this study). Likewise, K179 in DevR and SsrB and R171 in FixJ were found to have similar DNA binding functions (references 32 and 33 and this study), and the mutation of this residue led to an abrogation of DNA binding in all three proteins. Similarly, the residue corresponding to R197 in DevR is V197 in SsrB, and alanine substitution mutations affected the DNA binding activities of both these proteins (reference 32 and this study). In contrast, R173 of DevR is homologous to R179 in NarL, and while an alanine substitution was proficient in the former, it was partially defective in activation in the latter (reference 31 and this study). Another noteworthy observation was the positional conservation of the E154 and R155 residues in DevR and related orthologs, thereby implying a common function for these residues, possibly in interaction with the transcriptional machinery. Interestingly, the SigA-interacting E178 residue of DevR was seen to be equivalent to D192 in PhoB of Escherichia coli in a pairwise alignment (not shown), and the latter residue was important for PhoB interaction with σ70 of RNA polymerase (34).
The defects in activation noted in the K179A*, K191A, and R197A strains are attributed to a defect in DNA binding. The observed defect of K179A* is consistent with DosR/DevR-DNA cocrystal structure data where K179 was shown to contact bases in DNA (12). Interestingly, the K191 residue is completely conserved in DevR and its orthologs (see Fig. S2 in the supplemental material). Although no function is attributed to it so far, this residue is likely important for the binding of DevR to DNA and may play a similar role in other regulators. However, we cannot rule out the possibility that the mutation may have altered the local structure such that protein contact with DNA is affected.
We obtained useful insights from the analysis of target promoters, all of which featured a conserved overlap of the −35 SigA promoter element with a DevR binding motif but varied in the number of binding sites, in whether transcription was divergent or unidirectional, or in the magnitude and timing of the induction response. All the pc mutants were severely defective in the activation of the narK2 promoter, whereas the Rv1738 promoter was the least affected. The variation in the activation defect for this pair of genes is particularly striking as they are transcribed divergently from a common intergenic region (14). The observed range of defects in promoter activation among the various DevR mutants may reflect variations in the binding orientation of DevR, particularly at divergent promoters (which share DevR binding sites), or in DevR-RNA polymerase interactions at different promoters and differences in intrinsic promoter strength. Thus, the disruption of a single interaction due to mutation may have a small or large effect on transcription activation at different target promoters. The diversity in interactions at various regulon promoters could greatly increase the available combinations of potential regulatory interactions for finely tuning gene expression. For all promoter classes, the E154 and E178 pc mutants displayed an unambiguously severe phenotype, while the R155A pc mutant showed a moderate to severe activation defect. In contrast, as described in Results, the K208A mutant behaved differently at the same class of promoters. The pc phenotype of this mutant may be attributed to a role for the K208 residue in stabilizing the α10 dimerization helix in phosphorylated DevR (12). The different responses at similar promoter classes may be attributed to mutation-driven changes at the tetramer interface that influence its interaction with DNA and/transcriptional machinery at the target promoters.
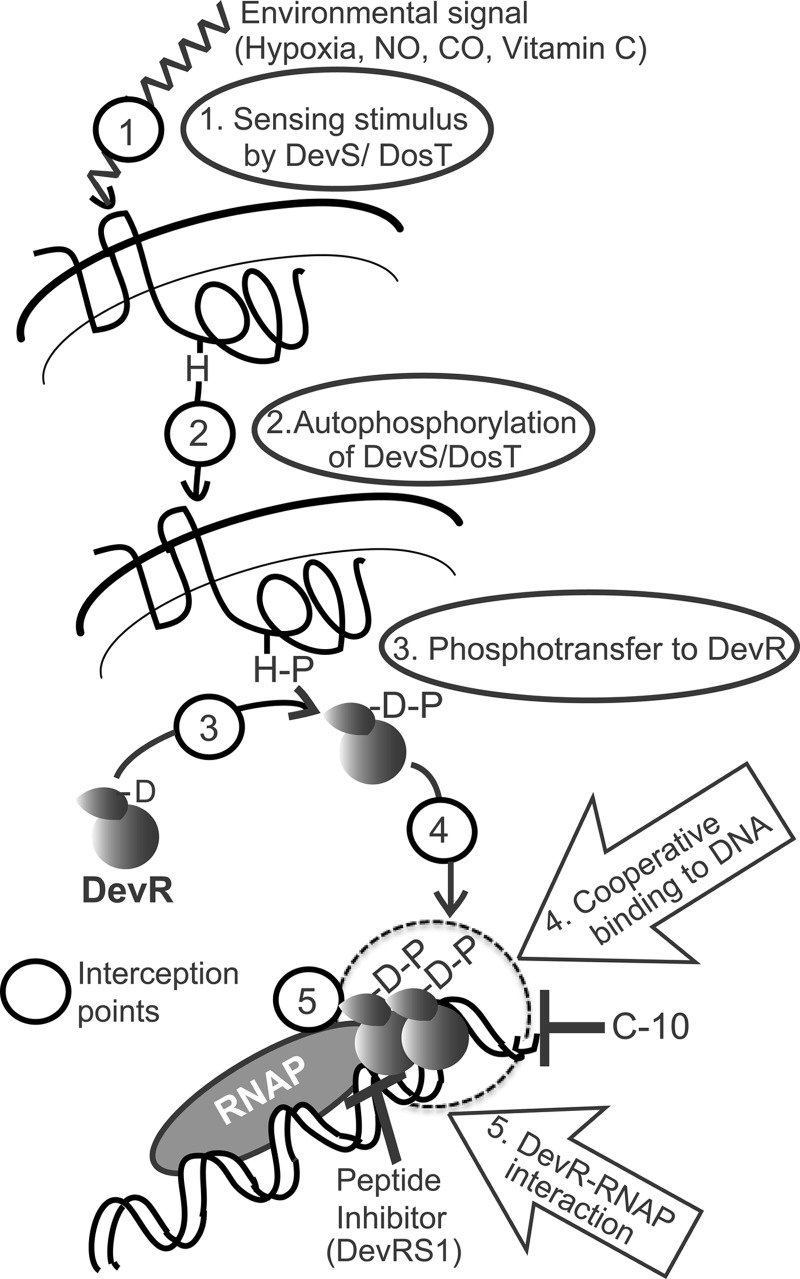
In conclusion, the isolation of pc mutants in this study has revealed an activating function for the C-terminal domain of DevR in addition to its previously known DNA binding activity. Further, the loss-of-function properties of the M. tuberculosis pc mutants establishes that DevR mediates the hypoxia response via interaction with SigA. For the first time, four DevR determinants that interact with RNA polymerase have been discovered. Our findings establish DevR interaction with the transcriptional machinery as an essential step of the DevR signaling cascade in addition to the previously known steps of signal sensing, kinase autophosphorylation, transfer of the phosphosignal to DevR (35), and DevR binding to DNA, including cooperative interactions (13, 27) (Fig. 6). We suggest that targeting this interaction is a new and novel strategy for the blockade of DevR signaling essential for M. tuberculosis adaptation and survival.
FIG 6.
Interception steps in the DevR signaling cascade. Hypoxia, NO, CO, and ascorbic acid are sensed by DosT and DevS (interception step 1), resulting in their activation by autophosphorylation (interception step 2). Transfer of phosphosignal from these sensors to DevR leads to its phosphorylation and its activation (interception step 3). Activated DevR binds to target gene promoters and triggers regulon activation (interception step 4). Step 4 is the target of the C-10 molecule, which inhibits DevR function and hypoxic survival of M. tuberculosis (36). Cooperative interaction of DevR with DNA is required for full gene activation (13, 19), and this is a novel interception step (step 4). Peptide inhibitor DevRS1 inhibits DevR-dependent gene expression and hypoxic viability (18) and likely intercepts the signaling cascade at step 5. The dashed circle indicates the DevR-DNA-RNA polymerase interaction interface. Here we describe that targeting DevR-SigA interaction is a new and novel strategy for the blockade of DevR signaling (interception step 5) and is an antibacterial strategy. Steps 4 and 5 are novel points for interception.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Department of Biotechnology, Government of India, to J.S.T. J.S.T. thanks the Department of Biotechnology, Government of India, for a Tata Innovation Fellowship, and U.S.G. thanks the Council for Scientific and Industrial Research (CSIR) for a Senior Research Associateship (Scientist's Pool Scheme). K.S. thanks the CSIR for a Senior Research Fellowship, and A.V. thanks the Indian Council of Medical Research for a Senior Research Fellowship.
We thank Kohinoor Kaur for providing GST protein, Navin Kumar for helpful discussions, and Gurvisha Sandhu of LeadInvent Technologies Pvt Ltd. for her expert assistance in generating DNA-protein complex structures.
Footnotes
Published ahead of print 6 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01270-13.
REFERENCES
- 1.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, Jyothisri K, Tyagi JS. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141–159. 10.1054/tuld.2000.0240 [DOI] [PubMed] [Google Scholar]
- 2.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843. 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713. 10.1084/jem.20030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiloh MU, Manzanillo P, Cox JS. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3:323–330. 10.1016/j.chom.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taneja NK, Dhingra S, Mittal A, Naresh M, Tyagi JS. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. 10.1371/journal.pone.0010860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boon C, Dick T. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760–6767. 10.1128/JB.184.24.6760-6767.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163. 10.1146/annurev.micro.55.1.139 [DOI] [PubMed] [Google Scholar]
- 8.Chaos MC, Rubin MJ. 2010. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu. Rev. Microbiol. 64:293–311. 10.1146/annurev.micro.112408.134043 [DOI] [PubMed] [Google Scholar]
- 9.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150:865–875. 10.1099/mic.0.26218-0 [DOI] [PubMed] [Google Scholar]
- 10.Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082–23087. 10.1074/jbc.M401230200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wisedchaisri G, Wu M, Sherman DR, Hol WG. 2008. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J. Mol. Biol. 378:227–242. 10.1016/j.jmb.2008.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisedchaisri G, Wu M, Rice AE, Roberts DM, Sherman DR, Hol WG. 2005. Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J. Mol. Biol. 354:630–641. 10.1016/j.jmb.2005.09.048 [DOI] [PubMed] [Google Scholar]
- 13.Gautam US, Sikri K, Tyagi JS. 2011. The residue threonine 82 of DevR (DosR) is essential for DevR activation and function in Mycobacterium tuberculosis despite its atypical location. J. Bacteriol. 193:4849–4858. 10.1128/JB.05051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chauhan S, Sharma D, Singh A, Surolia A, Tyagi JS. 2011. Comprehensive insights into Mycobacterium tuberculosis DevR (DosR) regulon activation switch. Nucleic Acids Res. 39:7400–7414. 10.1093/nar/gkr375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan S, Tyagi JS. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190:4301–4312. 10.1128/JB.01308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan S, Tyagi JS. 2008. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J. Bacteriol. 190:5394–5403. 10.1128/JB.00488-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chauhan S, Tyagi JS. 2009. Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. J. Bacteriol. 191:6075–6081. 10.1128/JB.00310-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhingra S, Kaur K, Taneja NK, Tyagi JS. 2012. DevR (DosR) binding peptide inhibits adaptation of Mycobacterium tuberculosis under hypoxia. FEMS Microbiol. Lett. 330:66–71. 10.1111/j.1574-6968.2012.02534.x [DOI] [PubMed] [Google Scholar]
- 19.Gautam US, Chauhan S, Tyagi JS. 2011. Determinants Outside the DevR C-Terminal Domain Are Essential for Cooperativity and Robust Activation of Dormancy Genes in Mycobacterium tuberculosis. PLoS One 6:e16500. 10.1371/journal.pone.0016500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarente L, Nye JS, Hochschild A, Ptashne M. 1982. Mutant lambda phage repressor with a specific defect in its positive control function. Proc. Natl. Acad. Sci. U. S. A. 79:2236–2239. 10.1073/pnas.79.7.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagchi G, Chauhan S, Sharma D, Tyagi JS. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151:4045–4053. 10.1099/mic.0.28333-0 [DOI] [PubMed] [Google Scholar]
- 22.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. 2006. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188:1856–1865. 10.1128/JB.188.5.1856-1865.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A, Mai D, Kumar A, Steyn AJ. 2006. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U. S. A. 103:11346–11351. 10.1073/pnas.0602817103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, Stoker NG. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134–1140. 10.1128/IAI.71.3.1134-1140.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saini DK, Malhotra V, Tyagi JS. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565:75–80. 10.1016/j.febslet.2004.02.092 [DOI] [PubMed] [Google Scholar]
- 26.Agarwal N, Tyagi AK. 2003. Role of 5′-TGN-3′ motif in the interaction of mycobacterial RNA polymerase with a promoter of ‘extended −10′ class. FEMS Microbiol. Lett. 225:75–83. 10.1016/S0378-1097(03)00483-X [DOI] [PubMed] [Google Scholar]
- 27.Gupta RK, Chauhan S, Tyagi JS. 2011. K182G substitution in DevR or C(8)G mutation in the Dev box impairs protein-DNA interaction and abrogates DevR-mediated gene induction in Mycobacterium tuberculosis. FEBS J. 278:2131–2139. 10.1111/j.1742-4658.2011.08130.x [DOI] [PubMed] [Google Scholar]
- 28.Majumdar SD, Vashist A, Dhingra S, Gupta R, Singh A, Challu VK, Ramanathan VD, Kumar P, Tyagi JS. 2012. Appropriate DevR (DosR)-mediated signaling determines transcriptional response, hypoxic viability and virulence of Mycobacterium tuberculosis. PLoS One 7:e35847. 10.1371/journal.pone.0035847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busby S, Ebright RH. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23:853–859. 10.1046/j.1365-2958.1997.2771641.x [DOI] [PubMed] [Google Scholar]
- 30.Kumar A, Buckner Starke C, DeZalia M, Moran CP., Jr 2004. Surfaces of Spo0A and RNA polymerase sigma factor A that interact at the spoIIG promoter in Bacillus subtilis. J. Bacteriol. 186:200–206. 10.1128/JB.186.1.200-206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin AV, Stewart V. 2010. Functional roles for the GerE-family carboxyl-terminal domains of nitrate response regulators NarL and NarP of Escherichia coli K-12. Microbiology 156:2933–2943. 10.1099/mic.0.040469-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll RK, Liao X, Morgan LK, Cicirelli EM, Li Y, Sheng W, Feng X, Kenney LJ. 2009. Structural and functional analysis of the C-terminal DNA binding domain of the Salmonella typhimurium SPI-2 response regulator SsrB. J. Biol. Chem. 284:12008–12019. 10.1074/jbc.M806261200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurashima-Ito K, Kasai Y, Hosono K, Tamura K, Oue S, Isogai M, Ito Y, Nakamura H, Shiro Y. 2005. Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter. Biochemistry 44:14835–14844. 10.1021/bi0509043 [DOI] [PubMed] [Google Scholar]
- 34.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15–26. 10.1006/jmbi.1996.0298 [DOI] [PubMed] [Google Scholar]
- 35.Saini DK, Tyagi JS. 2005. High-throughput microplate phosphorylation assays based on DevR-DevS/Rv2027c 2-component signal transduction pathway to screen for novel antitubercular compounds. J. Biomol. Screen. 10:215–224. 10.1177/1087057104272090 [DOI] [PubMed] [Google Scholar]
- 36.Gupta RK, Thakur TS, Desiraju GR, Tyagi JS. 2009. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J. Med. Chem. 52:6324–6334. 10.1021/jm900358q [DOI] [PubMed] [Google Scholar]
- 37.Valdivia RH, Hromockyj AE, Monack D, Ramakrishnan L, Falkow S. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47–52. 10.1016/0378-1119(95)00706-7 [DOI] [PubMed] [Google Scholar]
- 38.Rodrigue S, Brodeur J, Jacques PE, Gervais AL, Brzezinski R, Gaudreau L. 2007. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505–1513. 10.1128/JB.01371-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.