Abstract
In this study, we present the discovery and characterization of a highly thermostable endolysin from bacteriophage Ph2119 infecting Thermus strain MAT2119 isolated from geothermal areas in Iceland. Nucleotide sequence analysis of the 16S rRNA gene affiliated the strain with the species Thermus scotoductus. Bioinformatics analysis has allowed identification in the genome of phage 2119 of an open reading frame (468 bp in length) coding for a 155-amino-acid basic protein with an Mr of 17,555. Ph2119 endolysin does not resemble any known thermophilic phage lytic enzymes. Instead, it has conserved amino acid residues (His30, Tyr58, His132, and Cys140) that form a Zn2+ binding site characteristic of T3 and T7 lysozymes, as well as eukaryotic peptidoglycan recognition proteins, which directly bind to, but also may destroy, bacterial peptidoglycan. The purified enzyme shows high lytic activity toward thermophiles, i.e., T. scotoductus (100%), Thermus thermophilus (100%), and Thermus flavus (99%), and also, to a lesser extent, toward mesophilic Gram-negative bacteria, i.e., Escherichia coli (34%), Serratia marcescens (28%), Pseudomonas fluorescens (13%), and Salmonella enterica serovar Panama (10%). The enzyme has shown no activity against a number of Gram-positive bacteria analyzed, with the exception of Deinococcus radiodurans (25%) and Bacillus cereus (15%). Ph2119 endolysin was found to be highly thermostable: it retains approximately 87% of its lytic activity after 6 h of incubation at 95°C. The optimum temperature range for the enzyme activity is 50°C to 78°C. The enzyme exhibits lytic activity in the pH range of 6 to 10 (maximum at pH 7.5 to 8.0) and is also active in the presence of up to 500 mM NaCl.
INTRODUCTION
Bacteriophages, with the exception of the few known filamentous phages, are viruses that specifically infect and multiply in bacteria and, at the final stage of a lytic cycle, destroy the bacterial cell wall to release their progeny (1–3). To achieve this, phages use endolysins that dismantle the cell wall by targeting the peptidoglycan (PG) layer functioning as the bacterial exoskeleton. This extremely robust and complex mesh-like structure is made of glycan strands trans-connected by short peptides (4). The glycan portion of PG is a heteropolymer built of units, each consisting of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), linked together by β-1,4-glycosidic bonds. Short peptides used to cross-link glycan strands are attached by a peptide bond to the MurNAc. Adjacent peptides may be joined by a transpeptide bond or a transpeptide bridge (2, 5, 6). Based on their enzyme specificities, phage endolysins can be divided into five classes: (i) 1,4-β-N-acetylmuramidases, (ii) endo-β-N-acetylglucosaminidases, (iii) lytic transglycosylases that target the glycan moiety, (iv) endopeptidases, and (v) N-acetylmuramoyl-l-alanine amidases that target the peptide moiety (2, 5). Phage endolysins evolved into an enormously diverse group of proteins with respect to their primary structures and enzymatic properties. In general, endolysins infecting Gram-positive bacteria contain both an enzymatic catalytic domain (ECD) and a cell binding domain (CBD), whereas most endolysins from a Gram-negative background possess only an enzymatic catalytic domain.
To date, 723 diverse putative endolysins with 24 different ECDs and 13 CBDs have been identified and can be classified into 89 distinct types based on their structural organization (7).
Phage N-acetylmuramoyl-l-alanine amidases contain a catalytic domain (AMI-2, AMI-3, AMI-5, or AMI02-C) allowing them to cleave PG between the N-acetylmuramoyl residue and the l-amino acid residue (7). The presence of one of these domains has been described in several well-known phage endolysins, including the best-studied example, Escherichia coli phage T7 lysozyme. Despite the extensive studies on lytic enzymes of phages with different host specificities, little is known about endolysins of phages of thermophilic bacteria (8–10). There are only a few records concerning endolysins/lysozymes of Thermus phages in the GenBank database. Among them, only one endolysin of bacteriophage ϕIN93 infecting Thermus aquaticus TZ2 has been characterized (9), yet no described ECDs were detected. Comparative analysis of other Thermus phage endolysins revealed in their protein sequences the presence of a catalytic domain responsible for their specific enzymatic activities (either AMI-3, VANY, or PET-M23) (7). The putative endolysin from Thermus phage P23-45 (P23p108) possesses a PET-M23 domain that is characteristic of peptidases that cleave short peptides that cross-link PG glycan strands (11). Another hypothetical enzyme, N-acetylmuramoyl-l-alanine amidase of Thermus phage P23-77 (P23-77gp12), contains an AMI-3 domain; however, the enzyme's lytic activity has not been tested experimentally (12). Thermus scotoductus MAT2119 phage 2119 (Ph2119) enzyme, described here, is the first known Thermus phage endolysin that contains an AMI-2 domain that has a prominent cleft with a Zn2+ binding site. This domain is also present in T7 lysozyme, where Zn2+ is firmly anchored to the protein by the side chains of His17, His122, and Cys130 and through a water molecule to the negatively charged Tyr46 (13). It is noteworthy that neither T7 lysozyme nor any known Thermus phage endolysin has been found to possess a CBD (7).
Similar structural Zn2+ binding motifs found in T7 lysozyme and its close relative T3 lysozyme are also shared by a group of eukaryotic antibacterial proteins called peptidoglycan recognition proteins (PGRPs), which are involved in innate immunity and are highly conserved from insects to mammals (14). Those invertebrate and vertebrate PGRPs possess a so-called PGRP domain, but only amidase-active PGRPs are characterized by a conserved Zn2+ binding site in the peptidoglycan binding cleft (15). Similar to that of T7 lysozyme, the PGRP Zn2+ binding site consists of two histidines, one cysteine, and one tyrosine. In nonamidase PGRPs, which can only recognize (bind) bacterial PG but do not have the ability to cleave it, the cysteine is replaced with serine. This property can be used for PGRP amidase activity prediction based on amino acid sequence analysis (14).
The objective of the present study was to clone, purify, and characterize the properties of a novel thermostable endolysin from T. scotoductus MAT2119 bacteriophage Ph2119 that has similarity to the eukaryotic peptidoglycan recognition proteins.
MATERIALS AND METHODS
Bacterial strains and media.
The bacteria used in this study were purchased either from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Germany (Thermus thermophilus HB8 DSM 579 and Pseudomonas fluorescens DSM 50090) or the American Type Culture Collection (Bacillus cereus ATCC 13061, Bacillus subtilis ATCC 6633, Deinococcus radiodurans ATCC 13939, Staphylococcus aureus ATCC 25923, and Staphylococcus epidermidis ATCC 12228). The other bacteria used were taken either from the Collection of Plasmids and Microorganisms, University of Gdansk, Gdansk, Poland (E. coli MG1655 [16], Salmonella enterica serovar Panama, Serratia marcescens, Enterococcus faecalis, Staphylococcus intermedius, Sarcina lutea, Streptococcus pyogenes, and Lactococcus lactis subsp. cremoris W15 [17]), or the MATIS collection of microorganisms (T. scotoductus MAT2119 and Thermus flavus MAT1087). The aforementioned bacteria were used as substrates for the Ph2119 endolysin. E. coli strains DH5α (18) and Tuner(DE3) (Invitrogen) were used for molecular cloning and protein overproduction, respectively. The bacteria were cultivated in Luria broth (LB) or Luria agar (LA) medium (18). When necessary, the media were supplemented with 100 μg/ml ampicillin (Ap). T. scotoductus MAT2119 was isolated on R2A medium (containing, per liter, 0.5 g proteose peptone, 0.5 g yeast extract, 0.5 g Casamino acids, 0.5 g glucose, 0.5 g soluble starch, 0.3 g sodium pyruvate, 0.3 g K2PO4, 0.05 g MgSO4 · 7H2O, 15 g agar, pH 7.2 ± 0.2, at 25°C; Difco Laboratories). Bacteria belonging to the genus Thermus were also cultivated at 60°C in TM medium (containing, per liter, 4 g peptone, 2 g yeast extract, pH 7.2 [both from Difco Laboratories], 1 g NaCl, and 10 ml Castenholtz basal salts solution, pH 7.2). Castenholtz basal salts solution contained, per liter, 1 g nitrilotriacetic acid, 0.6 g CaSO4 · 2H2O, 1 g MgSO4 · 7H2O, 0.08 g NaCl, 1.03 g KNO3, 6.89 g NaNO3, 1.11 g Na2HPO4, 10 ml of 0.03% FeCl3, and 10 ml of Nitsch's trace elements (containing, per liter, 5 ml H2SO4, 2.2 g MgSO4 · 5H2O, 0.5 g of ZnSO4 · 7H2O, 0.5 g of H3BO3, 0.016 g of CuSO4, 0.025 g of Na2MoO4 · 2H2O, and 0.046 g of CoCl2 · 6H2O, pH 8.2). S. pyogenes was grown in Todd-Hewitt bullion (THB) (Graso Biotech) and L. lactis in TY medium (18). Vector pET15b (Novagen) was used for the construction of a plasmid overexpressing the gene coding for Ph2119 endolysin. Plasmids pLiz_Ph2119_pMA-T and pMP20, constructed in the present study, were deposited in the Collection of Plasmids and Microorganisms, University of Gdansk, Gdansk, Poland.
Bioinformatics analysis.
To search for a putative endolysin gene, the sequence of T. scotoductus phage Ph2119 was subjected to in silico 6-frame translation using a bacterial genetic code (NCBI translation table 11 [http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi]). Putative open reading frames (ORFs) were identified based on the presence of stop codons. Then, for each putative ORF longer than 60 amino acid residues, a hidden Markov model (HMM) was constructed using the HHblits package (19). The resulting HMMs were used to search (with HHblits) the databases of Protein Data Bank (PDB)- and Pfam-derived HMMs obtained from the website ftp://toolkit.lmb.uni-muenchen.de/pub/HH-suite/databases/. All significant matches (with E values of <1e−3) were extracted and screened for the presence of Pfam and PDB codes characteristic of endolysins: ECDs and CBDs. The relationship between Ph2119 endolysin, phage endolysins, and peptidoglycan recognition proteins was visualized with Circoletto software (20), available through the Bioinformatics Analysis Team server (http://tools.bat.infspire.org). Protein sequences were aligned by use of the CLUSTAL W program (21), available through the European Bioinformatics Institute server (http://www.ebi.ac.uk). The three-dimensional structure of Ph2119 endolysin was predicted using homology modeling. Related proteins with experimentally determined structures were identified using HHpred (via the MPI Bioinformatics Toolkit [19, 22]) against the PDB database. A homology model was built with Modeler (23) based on the crystal structure of Drosophila melanogaster peptidoglycan recognition protein LB (PDB ID 1OHT) (15). The codon adaptation index (CAI) was calculated using the JCat computer program (24). The gene encoding Ph2119 endolysin was analyzed with DNASIS software (Hitachi Software Engineering), and the isoelectric point (pI) was predicted with Isoelectric Point Calculator (http://isoelectric.ovh.org).
DNA manipulations.
Standard protocols were used for molecular cloning (18). The gene coding for phage Ph2119 endolysin was synthesized by GeneArt Gene Synthesis Service (Life Technologies). The nucleotide sequence was optimized for efficient protein overproduction in E. coli using GeneOptimizer technology by adjusting the codon quality distribution and G+C content of the gene. The synthetic gene (468 bp in length) was assembled from synthetic oligonucleotides. The DNA fragment obtained was cloned into the pMA-T vector (Life Technologies) linearized with SfiI enzyme, resulting in plasmid pLiz_Ph2119_pMA-T. The nucleotide sequence of the synthetic gene was verified by automated DNA sequencing. The overexpression plasmid, pMP20, was constructed by cloning into a pET15b vector (Novagen), previously linearized with NdeI and BamHI, a 0.5-kb DNA fragment carrying a synthetic gene coding for Ph2119 endolysin that was obtained by PCR followed by double digestion with NdeI and BamHI. The restriction sites used in molecular cloning were provided by a pair of primers (5′-GAGCTCCATATGCGTATTCTGGAACCG-3′ and 5′-CCAGGATCCTTATTTATTTACCACCTTTTTC-3′ [the underlining indicates NdeI and BamHI sites, respectively]). Plasmid pLiz_Ph2119_pMA-T was used as a template in the PCR. The cloning procedure presented enabled overproduction of Ph2119 endolysin possessing at the N terminus a hexahistidine (His tag) sequence. Recombinant plasmids were transformed into an appropriate E. coli strain and verified by restriction analysis and automated DNA sequencing. Restriction enzymes and DNA-modifying enzymes were purchased from New England BioLabs or Thermo Scientific. Enzymatic reactions were carried out under conditions suggested by the suppliers. PCRs were performed with 2× PCR Master Mix (A&A Biotechnology).
Purification of Ph2119 endolysin from recombinant E. coli.
The Ph2119 endolysin enzyme was prepared from E. coli Tuner(DE3) transformed with pMP20. Bacteria carrying the overexpression plasmid were cultivated at 30°C in LB medium (1 liter) supplemented with Ap and 0.4% glucose to repress the basal expression from the T7 promoter to an optical density at 600 nm (OD600) of 0.5. At that point, overproduction of Ph2119 endolysin was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium to a final concentration of 1 mM. Induction proceeded for 4 h at 37°C. Cells were harvested by centrifugation and suspended in 35 ml of NPI buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.1% Triton X-100, 10% [vol/vol] glycerol, 2 mM 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The bacteria were disrupted by sonication (30 bursts of 10 s at an amplitude of 12 μm). Purification steps were carried out at room temperature, except for sonication, when the sample was kept cold on ice. Cellular debris was removed by centrifugation (10,000 × g; 20 min), and the clarified lysate was treated for 15 min at 75°C, followed by centrifugation (10,000 × g; 20 min). The supernatant obtained was loaded into a 1- by 2-cm Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen) equilibrated with NPI buffer. The column was washed with NPI buffer (15 ml) and then with 20 mM imidazole in NPI buffer. Ph2119 endolysin bound to the column was eluted with 250 mM imidazole in NPI buffer. Fractions with the Ph2119 endolysin, more than 95% pure as verified by SDS-PAGE, were pooled and dialyzed against buffer D (25 mM potassium phosphate [KPO4] buffer, pH 8.0, 50 mM KCl, 0.1% Triton X-100, 10 mM 2-mercaptoethanol, 50% glycerol, and 0.2 mM ZnSO4) and stored at −80°C. The protein concentration was measured using the Bradford assay (25).
Determination of Ph2119 endolysin molecular weight.
A purified preparation of Ph2119 endolysin was analyzed by SDS-polyacrylamide gel electrophoresis (26) in order to estimate its purity and Mr under denaturing conditions. The protein position after electrophoresis was visualized by Coomassie brilliant blue R-250 staining. The Mr value of Ph2119 endolysin was determined using a calibration curve obtained with standard proteins as a reference (PageRuler Prestained Protein Ladder [Mr, 130,000 to 10,000]; Thermo Scientific).
Measurement of Ph2119 lytic activity.
Ph2119 endolysin activity was assayed spectrophotometrically by measuring the decrease in optical density of a bacterial suspension at 600 nm. The substrate bacteria (T. thermophilus HB8) were cultivated in TM medium (300 ml) until the late expotential-early stationary phase of bacterial growth was reached. Then, cells were harvested by centrifugation (4,000 × g for 15 min) and suspended in 20 ml of chloroform saturated with 50 mM Tris-HCl (pH 7.7). After incubation at room temperature (45 min), cells were collected by centrifugation (4,000 × g; 15 min), washed with 50 mM Tris-HCl (pH 7.7), and lyophilized (27). The reaction mixture contained 190 μl of chloroform-treated cells of T. thermophilus HB8 in 10 mM KPO4 buffer (pH 8.0) (optical density, 0.8 to 1.0; 1-cm path length) and 10 μl of Ph2119 endolysin at a concentration range of 0.25 to 25 μg/ml. The enzyme was diluted in 10 mM KPO4 buffer (pH 8.0), the mixture was incubated at 60°C in the standard 96-well titration plate (Corning), and a decrease in the OD600 was measured over time in an EnSpire multimode plate reader (PerkinElmer). In the control samples (negative control), 10 μl of 10 mM KPO4 buffer (pH 8.0) instead of the Ph2119 endolysin was added to the cell suspension. The experiments were repeated in triplicate. For the enzyme activity calculations, a standardized calculation method to quantify the lytic activity of endolysins, described by Briers et al. (28), was adopted. The specific lytic activity of Ph2119 endolysin was calculated by use of ActivityCalculator (http://www.biw.kuleuven.be/logt/ActivityCalculator.htm) (28). The results of the negative control were subtracted from the sample values.
Substrate specificity.
The substrate specificity of the Ph2119 endolysin was calculated as the ratio of the lytic activity against T. thermophilus HB8 (the reference substrate) to the enzyme activity against the following bacteria: T. scotoductus, T. flavus, E. coli MG1655, Salmonella serovar Panama, P. fluorescens, S. marcescens, E. faecalis, B. cereus, B. subtilis, D. radiodurans, S. intermedius, S. aureus, S. epidermidis, S. lutea, S. pyogenes, and L. lactis subsp. cremoris W15. Gram-negative bacteria were prepared as the T. thermophilus substrate. Gram-positive bacteria were harvested by centrifugation (4,000 × g; 15 min; 4°C) when they reached the late exponential-early stationary phase of growth, washed, and suspended in reaction buffer (10 mM KPO4, pH 8.0).
Testing the Ph2119 endolysin optimum.
The activity of Ph2119 endolysin (25 μg/ml) assayed in reaction mixtures was tested under various conditions. The variable factors were (i) temperature (10 to 99°C), (ii) pH range (3 to 12), and (iii) NaCl concentration (0 to 500 mM). Chloroform-treated T. thermophilus HB8 cells were used for a lysis assay essentially as described above. In all experiments except pH testing, 10 mM KPO4 buffer, pH 8.0, was used. Each experiment was conducted in triplicate and repeated at least twice. The results obtained were normalized against samples without endolysin.
Thermal stability.
Aliquots of Ph2119 endolysin and hen egg white lysozyme (HEWL) (25 μg/ml; Sigma-Aldrich) were heated at 95°C and submitted to increasing heating times in a Mastercycler ep gradient S thermocycler (Eppendorf) or autoclaved (121°C; 30 min). Samples were subsequently placed on ice before measuring their activity at 60°C for Ph2119 endolysin or at 37°C for HEWL in 10 mM KPO4 buffer (pH 8.0).
Nucleotide sequence accession numbers.
The accession numbers for the nucleotide sequences of the Ph2119 endolysin, gPGRP-LE, PGRP-LB, PGPP-Sa, PGRP1 HOLDI, T3 lysozyme, and T7 lysozyme deposited in the GenBank database are KF408298, AF313391, AF207537, AF207540, AB115774, KC960671 and S75616, respectively.
RESULTS
In silico analysis of the Thermus phage 2119 genome in search of lytic enzymes.
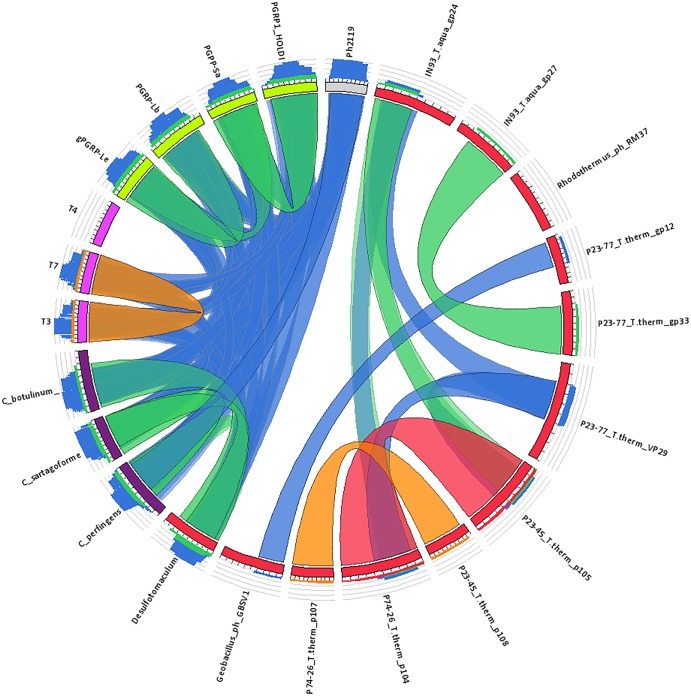
A number of bacteriophages infecting Thermus species were isolated and characterized in a study on thermophilic viruses from geothermal areas in Iceland (J. K. Kristjansson, personal communication). The Thermus strain used in this study, designated MAT2119, was isolated from a sample taken at Hrafntinnusker, which is located in the highlands of Iceland north of the glacier Mýrdalsjökull, where the Katla volcano is positioned. The strain was isolated on R2A medium (Difco Laboratories). Its optimal growth temperature is 65°C. Analysis of the 16S rRNA gene sequence enabled its identification as T. scotoductus (data not shown). The genome of Ph2119 derived from this strain was sequenced (data not shown). The resulting 92 contigs were subjected to extensive bioinformatics analysis aimed at identifying genes coding for lytic enzymes. This approach resulted in identification of a 468-bp ORF encoding a protein that showed some identity (approximately 32 to 34% when the proteins were compared in pairs) to a number of hypothetical N-acetylmuramoyl-l-alanine amidases encoded in the genomes of several Clostridium species—Clostridium perfringens (WP_003465616), Clostridium sartagoforme (WP_016208077), and Clostridium botulinum (YP_001921482)—and the thermophilic bacterium Desulfotomaculum kuznetsovii DSM 6115 (YP_004516476). It is noteworthy that our novel Ph2119 enzyme does not resemble any known thermophilic phage lytic enzymes. Neither a partially characterized lytic enzyme from T. aquaticus TZ2 phage ϕIN93 (9); lytic enzymes of other Thermus phages, T. thermophilus phage P23-77 (VP29) (12) and two hypothetical proteins of Thermus phage P74-26 and phage P23-45 (11); nor hypothetical protein Rm378p070 (NP_835657) from Rhodothermus RM378 show any similarity in amino acid sequence (Fig. 1). Instead, bioinformatics analysis revealed similarity of Ph2119 endolysin to T7 and T3 phage lysozymes and eukaryotic PGRPs, as depicted in Fig. 1. PGRPs are innate immunity proteins that are conserved from insects to mammals; all recognize bacterial peptidoglycan, and many possess the ability to directly destroy bacterial peptidoglycan by hydrolyzing the peptidoglycan amide bond (14). Hence, a candidate Ph2119 endolysin-encoding gene predicted to have an N-acetylmuramoyl-l-alanine amidase catalytic domain, AMI-2 (PF01510/IPR002502), was chosen for further experiments.
FIG 1.
Relationship between Ph2119 and phage endolysins and peptidoglycan recognition proteins visualized by Circoletto software (20). The ribbons represent the local alignments produced by BLAST (with an E value of 0.1), and their colors, blue, green, orange, and red, correspond to the alignment bit scores in the four quartiles. Blue indicates the lowest values of the maximum bit score (i.e., a bit score below 25%), green stands for the next 25%, orange for the third quartile, and, finally, red ribbons illustrate the best relative bit scores of 75% to 100% of the maximum bit score. Ph2119 phage endolysin is shown here as a short gray rectangle at the top of the circle, while endolysins from other thermophilic organisms are shown in red. Ph2119 endolysin is not related to any known Thermus or Rhodothermus phage endolysins (no interconnecting ribbons). This endolysin, however, shows some sequence similarity to AMI-2 domain-containing proteins, namely, endolysins of phages T3 and T7 (E. coli phages are shown in pink), several hypothetical N-acetylmuramoyl-l-alanine amidases encoded in the genome of Clostridium species (in purple), the thermophilic bacterium D. kuznetsovii DSM 6115 (in red), and several insect PGRPs (in lime green). The blue ribbons reflect the level of 22 to 34% amino acid identity between the Ph2119 endolysin and an interconnected protein sequence (BLAST results are available in File S1 in the supplemental material).
Sequence analysis of the gene coding for Ph2119 endolysin.
A 468-bp putative ORF coding for Ph2119 endolysin starts from an initiation codon, AUG, and ends at the UAG termination codon. The overall G+C content of the gene is 55.98%, which is significantly lower than the average G+C content of T. scotoductus SA-O1 genomic DNA (64.89%) (29). The Ph2119 endolysin gene codes for a 155-amino-acid protein with a predicted molecular mass of 17,555 Da. Its pI, calculated with Isoelectric Point Calculator software, is 9.5, which is higher than the pIs of T7 lysozyme (8.7) and T3 lysozyme (9.1). Analysis of the codon usage of the gene coding for Ph2119 endolysin revealed the presence of a significant number of rare codons for E. coli (DNASIS software) (see Fig. S2 in the supplemental material).
In order to determine the differences in the codon usage patterns of the Ph2119 gene and that of bacteria belonging to the genus Thermus, we calculated the value of the CAI using the JCat computer program (28). The CAI enables evaluation of synonymous codon usage bias in relation to the codon usage of highly expressed genes. According to this notion, open reading frames with a high CAI (near 1) belong to the class of highly expressed housekeeping genes. The calculated CAI value obtained for the Ph2119 endolysin gene (0.20) suggests its low rate of expression in T. scotoductus. A similar CAI value (0.156) was obtained when E. coli was considered a host for the Ph2119 endolysin gene.
Structure and sequence analysis of the Ph2119 endolysin.
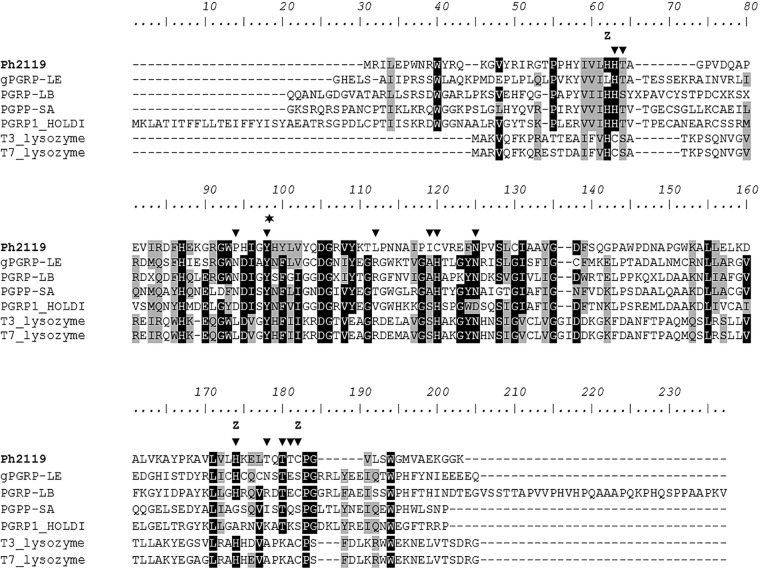
Figure 2 illustrates a multiple-amino-acid sequence alignment of Ph2119 endolysin, T7 and T3 lysozymes, and insect PGRPs that have all been studied experimentally: the fruit fly D. melanogaster gPGRP-LE (30), PGRP-LB (15), and PGRP-SA (31) and the large beetle Holotrichia diomphalia PGRP1 HOLDI (32). The predicted amino acid sequence of Ph2119 endolysin in a pairwise comparison showed only 22% identity to the T7 lysozyme sequence and 23% identity to T3 lysozyme. Both lysozymes have served for a long time as excellent models to study the enzymology of phage lytic enzymes (33, 34). A tyrosine residue that has been shown to be critical for phage T7 lysozyme lytic activity (Tyr46) (13) is also conserved in the Ph2119 endolysin (Tyr58) amino acid sequence (Fig. 2, asterisk). Four highly conserved amino acids that form the T7 lysozyme Zn2+ binding site (His17, Tyr46, His122, and Cys130) are also present in the amino acid sequence of Ph2119 (His30, Tyr58, His132, and Cys140) (Fig. 2, Z).
FIG 2.
Multiple-sequence alignment of amino acid sequences of Ph2119 endolysin and related lytic enzymes. The alignment was performed using the ClustalW computer program. The arrowheads indicate residues of the D. melanogaster PGRP-LE protein involved in substrate binding, determined by the model of PGRP-LE binding monomeric diaminopimelic acid-type peptidoglycan (tracheal cytotoxin) (30). Z denotes residues involved in Zn2+ binding, which is necessary for amidase activity. In phage T7, lysozyme Zn2+ is coordinated by two histidines (His17 and His122) and a cysteine residue (Cys130) (13). The asterisk indicates a tyrosine residue (Tyr46) that has been shown experimentally to be critical for phage T7 lysozyme amidase activity and that is highly conserved across all the sequences. Gray shading reflects amino acid conservation at 80% consensus, whereas the black boxes represent 100% amino acid sequence identity.
Ph2119 endolysin also shares similarity with PGRPs. All these eukaryotic proteins that recognize bacterial peptidoglycan have similar structure designs of the PGRP domain, with three peripheral α-helices and several central β-sheet strands and a substrate binding groove on the front of the molecule (14), which is consistent with the molecular model of Ph2119 endolysin shown in Fig. 3.
FIG 3.
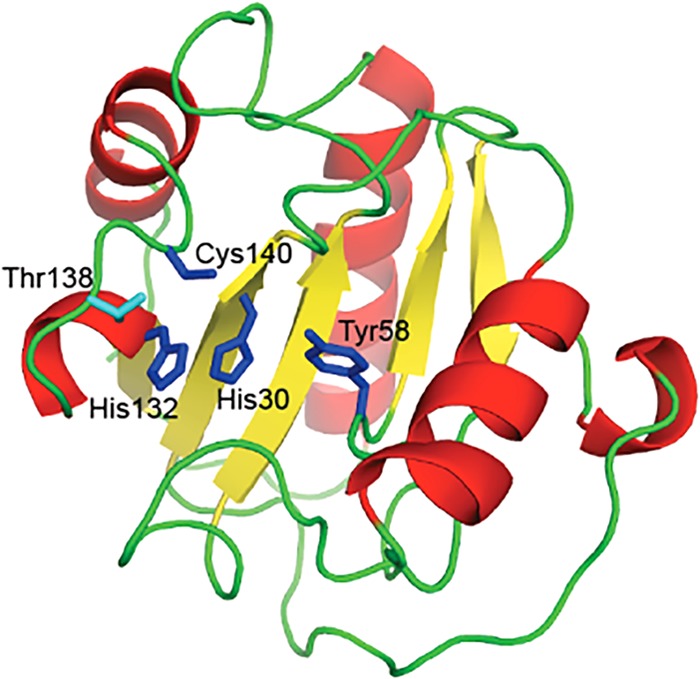
Structural model of Ph2119 endolysin. The model was built based on the crystal structure of D. melanogaster peptidoglycan recognition protein LB (PDB ID 1SK4). His30, Tyr58, His132, and Cys140, represented as blue sticks, are involved in Zn2+ binding. The secondary-structure elements alpha-helices, beta-strands, and loops are shown in red, yellow, and green, respectively.
In addition, PGRPs with amidase activity directly destroy bacterial peptidoglycan by hydrolyzing the peptidoglycan amide bond. An example is D. melanogaster PGRP-LB, which possesses a Zn2+ binding site in the peptidoglycan binding groove characteristic of amidase-active PGRPs (14). In the PGRP-LB structure, a zinc ion is coordinated by the side chains of His42, His152, and Cys160 and most likely through a water molecule by the conserved Tyr78 (15). Similarly, in the case of Ph2119 endolysin, the potential Zn2+ binding site is probably formed by His30, Tyr58, His132, and Cys140 (Fig. 2 and 3, blue sticks).
Nonlytic PGRPs, such as PGRP-LE, PGRP-SA, and PGRP1 HOLDI, lack amino acid residues that form a Zn2+ binding site. Instead, they have a serine residue in place of the conserved cysteine of PGRP-LB (Cys160 [Fig. 2, Z]). In PGRPs exerting lytic activity (e.g., PGRP-LB), the Thr158 residue present on the surface of the protein corresponds to Lys128, critical to the amidase activity of the T7 lysozyme (15). It was shown that replacing Lys128 with Thr greatly reduced the lytic activity of the T7 lysozyme (13). The involvement of Thr158 in substrate binding was proven for PGRP-LB (15). In contrast to the T7 lysozyme, but in accordance with PGRP-LB, Thr138 is also conserved in the amino acid sequence of Ph2119 endolysin (Fig. 2 and 3, light blue stick).
Overproduction and purification of the Ph2119 endolysin.
In order to establish an efficient system for Ph2119 endolysin overproduction in E. coli cells, the corresponding gene sequence was optimized with respect to the host codon usage. The resulting optimized gene sequence with a higher CAI value (0.6) and a G+C content lowered to 46.8% was then synthesized by the GeneArt Gene Synthesis Service (Life Technologies) (the optimized sequence is shown in Fig. S2 in the supplemental material).
When we tried to overexpress a synthetic gene coding for Ph2119 endolysin in a heterologous system, we found that the T7 expression system used was leaking, most probably due to the background transcription of the lacUV5 promoter that controls expression of the T7 RNA polymerase gene. This problem was overcome by adding glucose to a final concentration of 0.4% and lowering the culture temperature to 30°C. The overproduction of Ph2119 endolysin in E. coli Tuner(DE3)(pMP20) is shown in Fig. 4A. Four hours after induction (1 mM IPTG), a distinct band of Ph2119 endolysin was observed on SDS-PAGE (Fig. 4A, lane 2). The expression of the Ph2119 endolysin gene was not toxic to the E. coli host, as we did not observe any inhibition of bacterial growth.
FIG 4.
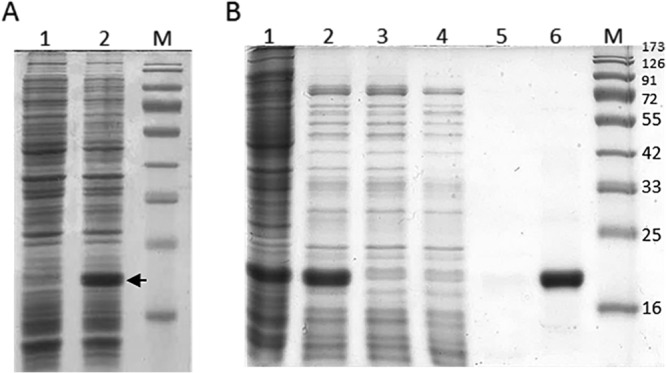
Overproduction of Ph2119 endolysin in E. coli Tuner(DE3) cells. Shown is SDS-PAGE (12.5%) of the cellular proteins from 100-μl cultures of bacteria. (A) E. coli Tuner(DE3)(pMP20) before induction (lane 1) and 4 h after induction (1 mM IPTG) (lane 2). The position of Ph2119 endolysin is indicated by an arrow. (B) Successive steps in the purification of Ph2119 endolysin on a column with Ni-NTA agarose. Lanes: 1, total protein; 2, soluble protein after heat treatment (15 min at 75°C); 3, flowthrough; 4, 1st wash with 10 mM imidazole in buffer NPI; 5, 2nd wash with 20 mM imidazole in buffer NPI; 6, Ph2119 endolysin eluted with 250 mM imidazole in buffer NPI; M, molecular masses of reference proteins (Thermo Scientific) (in kilodaltons).
The standard purification protocol involved affinity chromatography on a column with Ni-NTA agarose. We found that this procedure could be further optimized by heat treatment of the cell lysates (75°C; 15 min), which excluded a significant portion of the E. coli proteins (Fig. 4B, lane 2). The final preparation of purified Ph2119 endolysin was dialyzed against buffer D (see Materials and Methods) supplemented with 0.2 mM ZnSO4, since many N-acetylmuramoyl-l-alanine amidases require divalent cations for their activity (13). From 1 liter of E. coli Tuner(DE3)(pMP20) culture, we were able to obtain 24 mg of a homogeneous enzyme preparation with a specific activity of 3,079 U/mg (R2 = 0.968). On Coomassie brilliant blue-stained gels, the enzyme was found to be at least 95% pure (Fig. 4B, lane 6). The relative Mr of the recombinant Ph2119 endolysin (with His-tag moiety) was estimated to be 19,600 ± 500 in relation to the protein molecular weight markers with known molecular weight. This value is greater than the molecular weight calculated from the predicted amino acid sequence of Ph2119 endolysin (Mr, 17,555) (see Fig. S3, lane 1, in the supplemental material).
Characterization of the Ph2119 endolysin optimum.
The Ph2119 endolysin activity was measured using buffers (pH 3.0 to 12.0) with a constant ionic strength of 0.01 M. The enzyme exhibits lytic activity against reference T. thermophilus HB8 cells in the pH range of 6 to 10 (with the maximum level at pH 7.5 to 8), as shown in Fig. 5A. Lytic activity at various pH values was also measured in biological buffers, and the lysis percentages measured in relation to the optimal 10 mM KPO4, pH 8.0, were as follows: MES (morpholineethanesulfonic acid), pH 6.0, 33% ± 0.7%; MOPS (morpholinepropanesulfonic acid), pH 7.0, 42% ± 4.9%; Tricine, pH 8.0, 92% ± 0.7%; Tris-HCl, pH 8.0, 17% ± 3.3%; and Tris-HCl, pH 8.5, 40% ± 4.0% (Tris buffer pH measured at 25°C) (data not shown). The effect of NaCl on the lytic activity of Ph2119 endolysin was tested in reaction mixtures containing a suspension of chloroform-treated T. thermophilus cells in 10 mM KPO4 buffer (pH 8.0) and an NaCl concentration ranging from 0 to 500 mM. At 25 mM NaCl, the lytic activity was reduced to 71.6%, and with increasing NaCl concentrations, the activity level remained between 68 and 62% (Fig. 5B). Enzyme activities at various temperatures from 10°C to 99°C were determined in 10 mM KPO4, pH 8.0. Ph2119 endolysin was functional over a temperature range from 10°C to 99°C, with activity over 94% at 50°C to 78°C (Fig. 5C).
FIG 5.
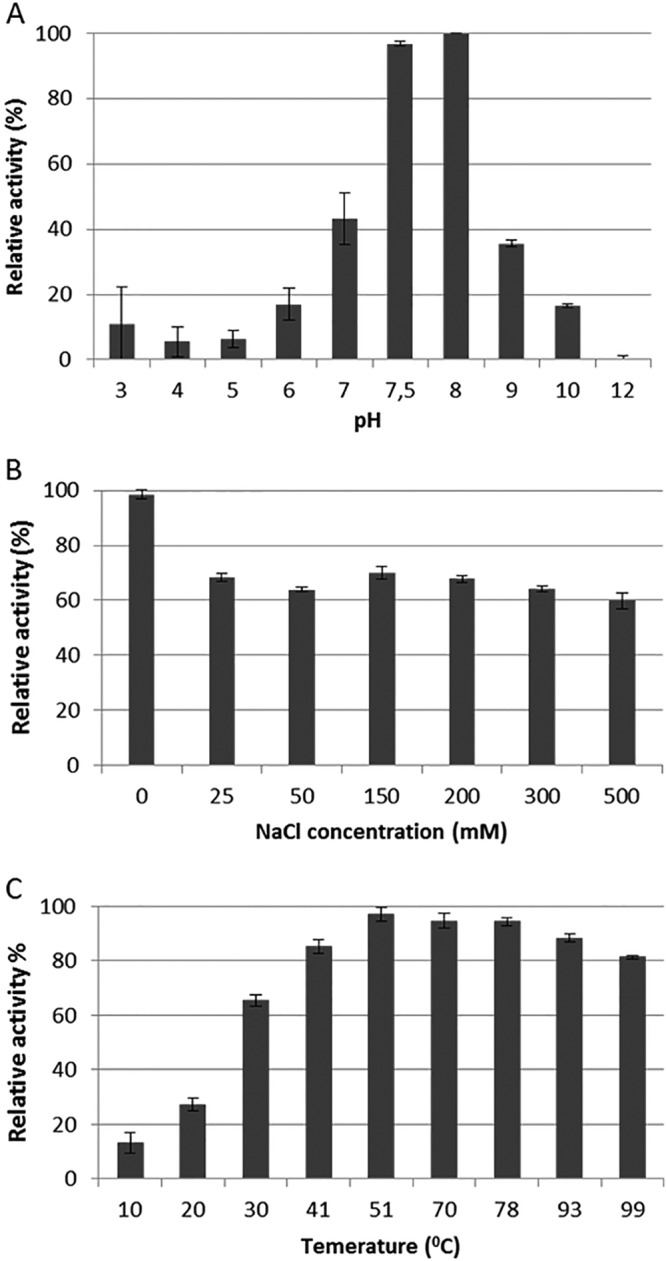
Effects of pH (A), NaCl (B), and temperature (C) on the lytic activity of Ph2119 endolysin. Relative activity against T. thermophilus cells was calculated by comparing the lytic activity under specific conditions with the maximal lytic activity among the data set. Each experiment was repeated in triplicate; the error bars indicate standard deviations.
Determination of thermal stability.
In order to estimate the thermostability of Ph2119 endolysin, samples of the enzyme were incubated for a prolonged time (up to 17 h) at 95°C or autoclaved (121°C; 30 min), followed by an activity assay. The results shown in Fig. 6 indicate that Ph2119 endolysin is highly thermostable. After incubation at 95°C for 6 h, it retained 86.7% of its original activity. Furthermore, after 17 h at 95°C, 17.4% residual activity remained, although autoclaving totally inactivated the enzyme. At the same time, HEWL, which was used as a control, was completely inactivated after 5 min at 95°C.
FIG 6.
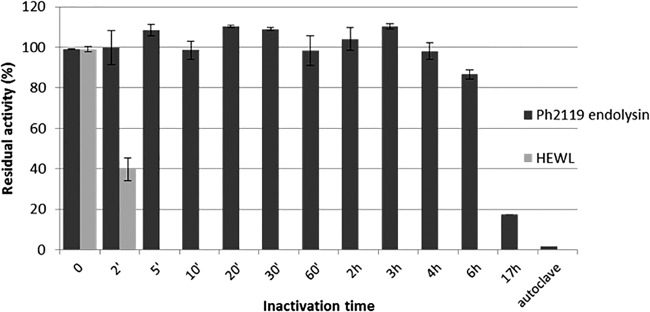
Examination of the thermal stability of the Ph2119 endolysin. Samples of Ph2119 (25 μg/ml) endolysin and HEWL (25 μg/ml) were incubated in 10 mM KPO4, pH 8.0, at 95°C (0 to 17 h) or autoclaved (30 min; 121°C). Then, samples were placed on ice before measuring their activity against T. thermophilus HB8 cells at optimal temperature (60°C for Ph2119 endolysin and 37°C for HEWL). The activities of Ph2119 endolysin and HEWL are indicated as percentages relative to untreated samples. Each experiment was repeated in triplicate; the error bars indicate standard deviations.
Substrate specificity.
Substrate specificity was examined by comparing the relative activities of Ph2119 endolysin and HEWL under optimal conditions and temperatures (37°C for HEWL and 60°C for Ph2119 endolysin). The results obtained are summarized in Table 1. It was shown that while the HEWL lytic activity was directed toward both Gram-negative and Gram-positive bacteria, the Ph2119 endolysin was mainly active against Gram-negative bacteria, including T. thermophilus HB8 (100%), T. scotoductus MAT2119 (100%), T. flavus MAT1087 (99%), E. coli (34%), S. marcescens (28%), P. fluorescens (13%), and Salmonella serovar Panama (10%). Ph2119 endolysin showed no activity against Gram-positive bacteria, with the exception of D. radiodurans (25%) and B. cereus (15%).
TABLE 1.
Substrate specificities of Ph2119 endolysin versus HEWL lysozyme
| Organism | Relative activitya (%) |
|
|---|---|---|
| Ph2119 endolysin | HEWL | |
| T. scotoductus MAT2119 | 100 | 61 |
| Thermus flavus MAT1087 | 99 | 98 |
| T. thermophilus HB8 DSM 579 | 100 | 43 |
| E. coli MG1655 | 34 | 100 |
| Salmonella serovar Panama | 10 | 35 |
| P. fluorescens DSM 50090 | 13 | 40 |
| S. marcescens | 28 | 35 |
| E. faecalis | 4 | 0 |
| B. cereus ATCC 13061 | 15 | 75 |
| B. subtilis ATCC 6633 | 2 | 0 |
| D. radiodurans ATCC 13939 | 25 | 21 |
| S. intermedius | 0 | 5 |
| S. aureus ATCC 12228 | 0 | 0 |
| S. epidermidis | 0 | 0 |
| S. lutea | 2 | 48 |
| S. pyogenes | 0 | 11 |
| L. lactis | 0 | 11 |
Relative activities are expressed as the percentage of activity toward T. thermophilus HB8 DSM 579 for Ph2119 endolysin and toward E. coli MG1655 for HEWL.
DISCUSSION
In this report, we present the cloning, overproduction, and characterization of a highly thermostable endolysin from bacteriophage Ph2119 infecting Thermus strain MAT2119 isolated from geothermal areas in Iceland. Proteins isolated from thermophilic bacteria have proved to be relevant in many applications in biotechnology. Our protocol for isolation of a gene coding for Ph2119 endolysin was based on a bioinformatics approach currently widely applied to this kind of research instead of using metagenomic libraries for lytic activity screening (35). The latter approach may encounter two major problems: (i) clonal toxicity due to the presence next to the gene for endolysin of a gene coding for holin, a protein generally toxic to E. coli, and (ii) proper selection of a substrate to test endolysin activity. A bioinformatics-based approach for selection of a target gene for experimental analysis allowed us to overcome these difficulties and to save time and resources compared to extensive functional screening. Comparative analysis revealed that the Ph2119 endolysin is homologous with N-acetylmuramoyl-l-alanine amidases possessing AMI-2 enzymatic catalytic domains.
This domain shows similarity to a conserved PGRP domain of PGRPs with peripheral α-helices and several central β-sheet strands (14, 36). Almost 100 PGRP family members known to date have at least one PGRP domain (36, 37). Some of the PGRPs, known as amidase-active PGRPs, not only recognize but also destroy bacterial peptidoglycan. All amidase-active PGRPs have a Zn2+ binding site in the peptidoglycan binding groove consisting of two histidines, one cysteine, and one tyrosine. Four conserved amino acids (His30, His132, Cys140, and Tyr58) are also present in the Ph2119 endolysin primary structure. Surprisingly, there is no similarity at the amino acid sequence level between Ph2119 endolysin and other endolysins from thermophilic bacteria characterized to date: endolysin from Geobacillus sp., infecting bacteriophage GVE2 isolated from a deep-sea hydrothermal vent (8), and lysozyme from T. aquaticus TZ2, infecting bacteriophage ϕIN93 (9).
The antibacterial activity of Ph2119 endolysin is not restricted to the genus Thermus but also includes Gram-negative mesophilic bacteria—E. coli, S. marcescens, P. fluorescens, and Salmonella serovar Panama—and Gram-positive D. radiodurans and B. cereus. This is in agreement with the clear distinction in species specificity between endolysins originating from phages infecting Gram-negative and Gram-positive hosts. The latter have a narrow antibacterial spectrum (38), whereas endolysins from phages infecting Gram-negative bacteria, similar to Ph2119 endolysin, have a broader species range (39–41). This distinction might be due to the lack of a CBD in most endolysins derived from Gram-negative bacteria, while endolysins of Gram-positive bacteria possess a clearly defined CBD that increases the substrate specificity of the enzymes (42, 43).
The substrate specificity of Ph2119 endolysin may also be affected by the composition of the bacterial peptidoglycan. The structure of the peptidoglycan of Gram-positive bacteria is more variable than that of Gram-negative microorganisms (44). The basic structure of the peptidoglycan of the Gram-positive D. radiodurans is similar to that of the closely related T. thermophilus HB8 (6). Also, the Gram-positive B. cereus reveals PG similarities to Gram-negative bacteria possessing diaminopimelic acid (DAP) in place of lysine (Lys) at position 3 of short peptides cross-linking glycan strands in the PG structure. It was shown previously that amidase-active PGRPs may distinguish between DAP-type and Lys-type peptidoglycans (14). In the Ph2119 endolysin amino acid sequence, there are conserved Gly52 and Trp53 residues corresponding to Gly and Trp residues of PGRP-LE (30), suggesting binding of Ph2119 endolysin in preference to DAP-type peptidoglycan.
T. thermophilus, however, represents the rare peptidoglycan A3β chemotype with l-ornithine at position 3 of the peptide subunit. The basic unit of T. thermophilus HB8 peptidoglycan is a muropeptide consisting of N-acetylglucosaminyl-N-acetylmuramyl-l-Ala-d-Glu-(γ)-l-Orn[(δ)-Gly-Gly]-d-Ala-d-Ala (6, 44, 45). There are no reports about interactions between PGRP and l-ornithine peptidoglycan, so the finding that Ph2119 endolysin may recognize bacteria with DAP-type and l-ornithine-type peptidoglycans is extremely interesting.
In this work, the Ph2119 endolysin activity optimum was determined. Ph2119 endolysin works between pH 6.0 and 10, with the maximum at 7.5 to 8.0, which is in agreement with phage lysozyme activity optima between 7.0 and 8.0 (optimal pH, 7.0 for T4 and λ lysozymes and pH 8.0 for P22 lysozyme) (41). Ph2119 endolysin does not work well in Tris-HCl buffer at pH 8.0 and 8.5, providing only 17% and 40% cell lysis, respectively. The reason for that might be the Tris buffer pH instability at different temperatures (46). In the experiment, the appropriate pH of the Tris-HCl buffer was established at room temperature (25°C), while the temperature at which the activity assay was performed was 60°C.
It was shown that Ph2119 tolerated elevated concentrations of NaCl up to 500 mM. Tolerance of phage endolysins for the presence of high salt levels ranges from high resistance (100% activity at 500 mM NaCl of KZ144 endolysin from the Pseudomonas aeruginosa-infecting phage fKZ [42]) through moderate inhibition (40% residual activity at 200 mM NaCl of LysB4 endolysin from the B. cereus-infecting phage B4 [47]) to almost complete inhibition (6% residual activity at 133 mM NaCl of P22 lysozyme from the Salmonella enterica serovar Typhimurium-infecting phage P22 [41]). It was also demonstrated that the Ph2119 endolysin was active over a temperature range from 10°C to 99°C, with activity over 94% at 50°C to 78°C. It also retains approximately 87% of its lytic activity after 6 h of incubation at 95°C. An open question still remains: what makes this protein resistant to high temperature? The thermal stability of proteins seems to be a complex phenomenon that can be affected by many factors. In general, proteins can be stabilized by decreasing their entropy of unfolding (48). The proline residue has the lowest conformational entropy in an unfolded state that consequently leads to protein stabilization (49). Proline represents as much as 9% of the Ph2119 endolysin primary structure (14 residues). At the same time, prolines represent only 2.7% and 3.3% of the residues in the amino acid sequences of mesophilic T7 and T3 lysozymes, respectively. However, in thermostable lysozymes—gp36C from bacteriophage ϕKMV (21) or lysozyme from bacteriophage ϕIN93 (9)—the proline content is also low, only 2.1% and 3%, respectively, which suggests that other factors are responsible for their thermal stability. One of them might be the presence of disulfide bonds formed by cysteine residues that stabilize proteins by decreasing the entropy of the protein's unfolded state (48).
High thermostability of Ph2119 endolysin makes it suitable for applications under extreme temperature conditions, which is important from a practical point of view. It was shown previously that phage endolysins/lysozymes can be successfully applied in food preservation (50, 51, 52) and in agriculture to achieve resistance to phytopathogenic bacteria (53), and as the number of multi-antibiotic-resistant bacteria increases, they are more often considered novel antibacterial agents (1–3, 54, 55). Taking into account their relevance, it is clear that phage endolysins have a great potential for use, not only in medicine, but also in the fields of applied microbiology and biotechnology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the European Union's Seventh Framework Programme managed by the Research Executive Agency (REA) (FP7/2007-2013 and FP7/2007-2011) under grant agreement 286556 to the Exgenome project (Exgenome Molecular Enzymes).
We thank Anna Kloska (Department of Molecular Biology, University of Gdansk) for help with the PerkinElmer multimode plate reader.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03074-13.
REFERENCES
- 1.Fenton M, Ross P, McAuliffe O, O'Mahony J, Coffey A. 2010. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs 1:9–16. 10.4161/bbug.1.1.9818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loessner MJ. 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8:480–487. 10.1016/j.mib.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 3.Fischetti VA. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13:491–496. 10.1016/j.tim.2005.08.007 [DOI] [PubMed] [Google Scholar]
- 4.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778:1714–1734. 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 5.Borysowski J, Weber-Dabrowska B, Gorski A. 2006. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 231:366–377 [DOI] [PubMed] [Google Scholar]
- 6.Quintela JC, Zollner P, Garcia-del Portillo F, Allmaier G, de Pedro MA. 1999. Cell wall structural divergence among Thermus spp. FEMS Microbiol. Lett. 172:223–229. 10.1111/j.1574-6968.1999.tb13472.x [DOI] [PubMed] [Google Scholar]
- 7.Oliveira H, Melo LD, Santos SB, Nobrega FL, Ferreira EC, Cerca N, Azeredo J, Kluskens LD. 2013. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 87:4558–4570. 10.1128/JVI.03277-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye T, Zhang X. 2008. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 78:635–641. 10.1007/s00253-008-1353-1 [DOI] [PubMed] [Google Scholar]
- 9.Matsushita I, Yanase H. 2008. A novel thermophilic lysozyme from bacteriophage phiIN93. Biochem. Biophys. Res. Commun. 377:89–92. 10.1016/j.bbrc.2008.09.101 [DOI] [PubMed] [Google Scholar]
- 10.Jin M, Ye T, Zhang X. 2013. Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology 159:1597–1605. 10.1099/mic.0.067611-0 [DOI] [PubMed] [Google Scholar]
- 11.Minakhin L, Goel M, Berdygulova Z, Ramanculov E, Florens L, Glazko G, Karamychev VN, Slesarev AI, Kozyavkin SA, Khromov I, Ackermann HW, Washburn M, Mushegian A, Severinov K. 2008. Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23-45 and P74-26: siphoviruses with triplex-forming sequences and the longest known tails. J. Mol. Biol. 378:468–480. 10.1016/j.jmb.2008.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalasvuori M, Jaatinen ST, Laurinavicius S, Ahola-Iivarinen E, Kalkkinen N, Bamford DH, Bamford JK. 2009. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J. Virol. 83:9388–9397. 10.1128/JVI.00869-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Zhang X, Pflugrath JW, Studier FW. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 91:4034–4038. 10.1073/pnas.91.9.4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziarski R, Gupta D. 2006. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 7:232. 10.1186/gb-2006-7-8-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MS, Byun M, Oh BH. 2003. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 4:787–793. 10.1038/ni952 [DOI] [PubMed] [Google Scholar]
- 16.Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mruk I, Cichowicz M, Kaczorowski T. 2003. Characterization of the LlaCI methyltransferase from Lactococcus lactis subsp. cremoris W15 provides new insights into the biology of type II restriction-modification systems. Microbiology 149:3331–3341. 10.1099/mic.0.26562-0 [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19.Remmert M, Biegert A, Hauser A, Soding J. 2012. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9:173–175. 10.1038/nmeth.1818 [DOI] [PubMed] [Google Scholar]
- 20.Darzentas N. 2010. Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26:2620–2621. 10.1093/bioinformatics/btq484 [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34:W335–W339. 10.1093/nar/gkl217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 24.Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D. 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33:W526–W531. 10.1093/nar/gki376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 27.Lavigne R, Briers Y, Hertveldt K, Robben J, Volckaert G. 2004. Identification and characterization of a highly thermostable bacteriophage lysozyme. Cell. Mol. Life Sci. 61:2753–2759. 10.1007/s00018-004-4301-y [DOI] [PubMed] [Google Scholar]
- 28.Briers Y, Lavigne R, Volckaert G, Hertveldt K. 2007. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods 70:531–533. 10.1016/j.jbbm.2006.10.009 [DOI] [PubMed] [Google Scholar]
- 29.Gounder K, Brzuszkiewicz E, Liesegang H, Wollherr A, Daniel R, Gottschalk G, Reva O, Kumwenda B, Srivastava M, Bricio C, Berenguer J, van Heerden E, Litthauer D. 2011. Sequence of the hyperplastic genome of the naturally competent Thermus scotoductus SA-01. BMC Genomics 12:577. 10.1186/1471-2164-12-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. 2006. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281:8286–8295. 10.1074/jbc.M513030200 [DOI] [PubMed] [Google Scholar]
- 31.Reiser JB, Teyton L, Wilson IA. 2004. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution. J. Mol. Biol. 340:909–917. 10.1016/j.jmb.2004.04.077 [DOI] [PubMed] [Google Scholar]
- 32.Lee MH, Osaki T, Lee JY, Baek MJ, Zhang R, Park JW, Kawabata S, Soderhall K, Lee BL. 2004. Peptidoglycan recognition proteins involved in 1,3-beta-d-glucan-dependent prophenoloxidase activation system of insect. J. Biol. Chem. 279:3218–3227. 10.1074/jbc.M309821200 [DOI] [PubMed] [Google Scholar]
- 33.Inouye M, Arnheim N, Sternglanz R. 1973. Bacteriophage T7 lysozyme is an N-acetylmuramyl-l-alanine amidase. J. Biol. Chem. 248:7247–7252 [PubMed] [Google Scholar]
- 34.DeMartini M, Halegoua S, Inouye M. 1975. Lysozymes from bacteriophages T3 and T5. J. Virol. 16:459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz JE, Schuch R, Fischetti VA. 2010. Identifying active phage lysins through functional viral metagenomics. Appl. Environ. Microbiol. 76:7181–7187. 10.1128/AEM.00732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurata S. 2014. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 42:36–41. 10.1016/j.dci.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. 1998. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. U. S. A. 95:10078–10082. 10.1073/pnas.95.17.10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loessner MJ, Maier SK, Daubek-Puza H, Wendlinger G, Scherer S. 1997. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J. Bacteriol. 179:2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsugita A, Inouye M. 1968. Purification of bacteriophage T4 lysozyme. J. Biol. Chem. 243:391–397 [PubMed] [Google Scholar]
- 40.Rao GR, Burma DP. 1971. Purification and properties of phage P22-induced lysozyme. J. Biol. Chem. 246:6474–6479 [PubMed] [Google Scholar]
- 41.Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol. Microbiol. 65:1334–1344. 10.1111/j.1365-2958.2007.05870.x [DOI] [PubMed] [Google Scholar]
- 42.Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335–349. 10.1046/j.1365-2958.2002.02889.x [DOI] [PubMed] [Google Scholar]
- 43.Schmelcher M, Donovan DM, Loessner MJ. 2012. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 7:1147–1171. 10.2217/fmb.12.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quintela JC, Pittenauer E, Allmaier G, Aran V, de Pedro MA. 1995. Structure of peptidoglycan from Thermus thermophilus HB8. J. Bacteriol. 177:4947–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durst RA, Staples BR. 1972. Tris/Tris-HCl; a standard buffer for use in the physiologic pH range. Clin. Chem. 18:206–208 [PubMed] [Google Scholar]
- 47.Son B, Yun J, Lim JA, Shin H, Heu S, Ryu S. 2012. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 12:33. 10.1186/1471-2180-12-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews BW, Nicholson H, Becktel WJ. 1987. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. U. S. A. 84:6663–6667. 10.1073/pnas.84.19.6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1–43. 10.1128/MMBR.65.1.1-43.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia P, Martinez B, Obeso JM, Rodriguez A. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479–485. 10.1111/j.1472-765X.2008.02458.x [DOI] [PubMed] [Google Scholar]
- 51.Callewaert L, Walmagh M, Michiels CW, Lavigne R. 2011. Food applications of bacterial cell wall hydrolases. Curr. Opin. Biotechnol. 22:164–171. 10.1016/j.copbio.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 52.Schmelcher M, Waldherr F, Loessner MJ. 2012. Listeria bacteriophage peptidoglycan hydrolases feature high thermoresistance and reveal increased activity after divalent metal cation substitution. Appl. Microbiol. Biotechnol. 93:633–643. 10.1007/s00253-011-3372-6 [DOI] [PubMed] [Google Scholar]
- 53.Kim WS, Salm H, Geider K. 2004. Expression of bacteriophage phiEa1h lysozyme in Escherichia coli and its activity in growth inhibition of Erwinia amylovora. Microbiology 150:2707–2714. 10.1099/mic.0.27224-0 [DOI] [PubMed] [Google Scholar]
- 54.Hermoso JA, Garcia JL, Garcia P. 2007. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 10:461–472. 10.1016/j.mib.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 55.Fischetti VA. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393–400. 10.1016/j.mib.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.