Abstract
Probiotics have been demonstrated to promote growth, stimulate immune responses, and improve food safety of poultry. While widely used, their effectiveness is mixed, and the mechanisms through which they contribute to poultry production are not well understood. Microbial phytases are increasingly supplemented in feed to improve digestibility and reduce antinutritive effects of phytate. The microbial origin of these exogenous enzymes suggests a potentially important mechanism of probiotic functionality. We investigated phytate degradation as a novel probiotic mechanism using recombinant Lactobacillus cultures expressing Bacillus subtilis phytase. B. subtilis phyA was codon optimized for expression in Lactobacillus and cloned into the expression vector pTRK882. The resulting plasmid, pTD003, was transformed into Lactobacillus acidophilus, Lactobacillus gallinarum, and Lactobacillus gasseri. SDS-PAGE revealed a protein in the culture supernatants of Lactobacillus pTD003 transformants with a molecular weight similar to that of the B. subtilis phytase. Expression of B. subtilis phytase increased phytate degradation of L. acidophilus, L. gasseri, and L. gallinarum approximately 4-, 10-, and 18-fold over the background activity of empty-vector transformants, respectively. Phytase-expressing L. gallinarum and L. gasseri were administered to broiler chicks fed a phosphorus-deficient diet. Phytase-expressing L. gasseri improved weight gain of broiler chickens to a level comparable to that for chickens fed a control diet adequate in phosphorus, demonstrating proof of principle that administration of phytate-degrading probiotic cultures can improve performance of livestock animals. This will inform future studies investigating whether probiotic cultures are able to provide both the performance benefits of feed enzymes and the animal health and food safety benefits traditionally associated with probiotics.
INTRODUCTION
Lactobacillus species are important inhabitants of the gastrointestinal tracts of humans and animals and are increasingly being used as probiotic microorganisms due to their health-promoting properties (1, 2). Probiotics, sometimes called direct-fed microbials (DFM) when used in animals (3), are live microorganisms administered to confer a health benefit upon the host (4). Administration of probiotic Lactobacillus to poultry has been demonstrated to promote growth at levels similar to antibiotics (5, 6) and to reduce gastrointestinal colonization of human food-borne pathogens, including Campylobacter (7, 8), Clostridium (9), and Salmonella (10, 11). Because of concern over antibiotic-resistant pathogens and pressure from both consumers and regulatory agencies, probiotics have received increased interest as potential alternatives to antibiotic growth promoters (12). While probiotics are used widely in livestock production (13), their effectiveness is varied, and the mechanisms responsible for their benefits are not well understood.
Phosphorus is an essential nutrient in poultry production (14), with dietary deficiencies leading to excessive financial losses due to increased mortality (15, 16). Phytic acid (myo-inositol hexaphosphate) is an important plant phosphorus storage form and accounts for 50 to 80% of total phosphorus present in cereal grains and legumes commonly used in livestock animal feeds (17, 18). However, phytate phosphorus has low bioavailability and is underutilized due to the lack of endogenous phytate-degrading enzymes in nonruminant livestock, including poultry (19, 20) and swine (21). Additionally, phytic acid exerts antinutritive effects (15), sequestering essential cations, including calcium, magnesium, iron, and zinc, and reducing their bioavailability (22).
Phytases are phosphatases which catalyze the hydrolysis of phytic acid to myo-inositol and inorganic phosphate (23). In-feed administration of microbial phytases to improve digestibility of phytic acid is widely used in the production of poultry and other livestock (24, 25). The resulting increases in phytate phosphorus bioavailability (15, 26, 27) and reduction in the antinutritive effects (28, 29) of phytic acid are well documented. The microbial origin of phytases used in livestock production suggests that degradation of phytic acid may be a potentially important mechanism of probiotic functionality. Combining the nutritional and growth performance benefits of phytase with the food safety and animal health benefits traditionally associated with probiotics is of great interest to livestock producers. In this study, we investigated phytate degradation as a novel mechanism of probiotic functionality. Recombinant Lactobacillus cultures expressing Bacillus subtilis phytase were constructed, and the effect of their administration on growth performance was evaluated in broiler chicks fed a phosphorus-deficient diet. We demonstrate proof of principle that administration of a phytate-degrading probiotic culture can improve the performance of livestock animals.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used or constructed in this study are listed in Table 1. Lactobacillus strains were cultured using de Man, Rogosa, and Sharpe (MRS) medium (Difco, Franklin Lakes, NJ) and incubated in 10% CO2 at 37°C with 5 μg/ml erythromycin (Erm) (EMD Chemicals, Inc., San Diego, CA) added when appropriate. Escherichia coli strains were cultured using Luria-Bertani (LB) medium (Difco, Franklin Lakes, NJ) aerobically at 37°C with 150 μg/ml Erm, when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmida | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| L. acidophilus | ||
| NCFM | Human intestinal isolate | 66 |
| TDCC 60 | NCFM with pTRK882 | This study |
| TDCC 61 | rPhyA+, NCFM with pTD003 | This study |
| L. gallinarum | ||
| ATCC 33319T | Chicken crop isolate, type strain | ATCCa |
| TDCC 62 | ATCC 33319 with pTRK882 | This study |
| TDCC 63 | rPhyA+, ATCC 33319 with pTD003 | This study |
| L. gasseri | ||
| ATCC 33323T | Human isolate, type strain | 62 |
| TDCC 64 | ATCC 33323 with pTRK882 | This study |
| TDCC 65 | rPhyA+, ATCC 33323 with pTD003 | This study |
| E. coli | ||
| MC1061 | Strr, E. coli transformation host | 29 |
| TOP10 | Strr, E. coli transformation host | Invitrogen |
| NCK1814 | MC1061 with pTRK882 | 37 |
| TDCC 33 | TOP10 with pTD001 | This study |
| TDCC 66 | MC1061 with pTD003 | This study |
| Plasmids | ||
| pTRK882 | 4.4 kb, Ermr, constitutive expression vector, Ppgm | 37 |
| pTD001 | 3.5 kb, Ampr, pMAT::phyA | This study |
| pTD003 | 5.6 kb, Ermr, pTRK882::phyA | This study |
ATCC, American Type Culture Collection.
DNA isolation, manipulation, and transformation.
E. coli plasmid DNA was isolated using the QIAprep Spin Miniprep kit (Qiagen Inc., Valencia, CA), while DNA was isolated from Lactobacillus according to the method of Walker and Klaenhammer (30). DNA restriction fragments were purified from agarose gels using the QIAquick gel extraction kit (Qiagen, Germantown, MD). All DNA manipulations were performed using standard molecular cloning techniques (31). Restriction enzymes, T4 ligase, and Taq DNA polymerase were used according to the manufacturer's instructions (NEB, Ipswich, MA). PCR primers are listed in Table 2. Electrocompetent E. coli MC1061 and TOP10 were prepared and transformed according to standard methods (32). Lactobacillus acidophilus and Lactobacillus gasseri were transformed using the method of Luchansky et al. (33), while Lactobacillus gallinarum was transformed using the method of Beasley et al. (34).
TABLE 2.
PCR primers
| Target Gene | Primer | Sequence (5′→3′) |
|---|---|---|
| ermC | pGK12_ermF | ATTCTCTTGGAACCATAC |
| pGK12_ermR | ACTGCCATTGAAATAGAC | |
| phyA | phy_1258F | ATTATCAACTGCTGCTGGTT |
| phy_1976R | ATCAACAACTTGACCCTTTG |
Recombinant phytase expression in Lactobacillus.
The phyA gene from B. subtilis (35) was codon optimized for expression in L. acidophilus using the OPTIMIZER web server (36) and commercially synthesized with EcoRI and NotI restriction sites to facilitate cloning. The synthetic DNA sequence was provided by the manufacturer (Life Technologies, Inc., Carlsbad, CA) in a plasmid (pTD001). The synthetic phyA gene was isolated from pTD001 and ligated into pTRK882 (37) for constitutive high-level expression in Lactobacillus. The resulting plasmid, pTD003, was transformed into and subsequently propagated in E. coli MC1061. The plasmids pTD003 and pTRK882 were introduced into Lactobacillus species by electrotransformation. Transformations were confirmed by PCR using gene specific primers (Table 2).
SDS-PAGE.
Supernatants from overnight Lactobacillus cultures were concentrated and purified by dialysis using Microsep advanced centrifugal devices (Pall Corporation, Ann Arbor, MI). Total protein was precipitated using 100% (wt/vol) trichloroacetic acid (TCA) (Sigma-Aldrich) and pelleted by centrifugation. Protein pellets were washed 3 times using 80% (wt/vol) acetone and resuspended in phosphate-buffered saline (PBS). Protein concentrations were determined using the Bradford method (38). Protein was separated by SDS-PAGE using Any kD Mini-PROTEAN TGX Precast protein gels (Bio-Rad Laboratories, Hercules, CA) in Tris-glycine-SDS buffer (Bio-Rad) with a low-range protein standard (Bio-Rad). Wells were loaded with 3.5 μg of protein in Laemmli buffer (39). Gels were stained with GelCode Blue Safe protein stain (Thermo Scientific, Waltham, MA) for visualization of protein.
Phytate hydrolysis.
Phytate hydrolysis by Lactobacillus transformants was observed using a modification of the method of Bae et al. (40). Lactobacillus colonies were selected, aseptically transferred onto the surfaces of MRS agar plates (5 μg/ml Erm), and incubated for 36 h. Plates were then overlaid with modified MRS (41), in which 0.5% (wt/vol) sodium phytate (Pfaltz & Bauer, Waterbury, CT) was the sole phosphorus source, and incubated for an additional 24 h. Plates were stained with cobalt chloride solution and counterstained with an ammonium molybdovanadate solution. Phytate hydrolysis was indicated by zones of clearing.
Phytase enzyme activity assays.
Phytase activity from cell extracts of recombinant Lactobacillus cultures was assayed by determining the amount of inorganic phosphate released from sodium phytate in phytase reaction buffer (6.4 mM sodium phytate, 2 mM CaCl2, 100 mM Tris-HCl, pH 7.0) at 55°C. Enzyme reactions were terminated by the addition of an equal volume of 5% (wt/vol) TCA, and free phosphate was determined colorimetrically (620 nm) using the ammonium molybdate method (42) with a sodium phosphate standard. Cell extracts were prepared by bead beating (37) as described previously in phytase extract buffer (2 mM CaCl2, 100 mM Tris-HCl, pH 7.0). Protein concentrations were determined using the Bradford method (38). Phytase specific activity was reported as U mg−1 total protein (μmol PO43− released min−1 mg−1). Data were analyzed using analysis of variance (ANOVA), and significant differences between strains were determined using Duncan's multiple-range test.
Broiler chickens.
On the day of hatch, male broiler chicks (Ross × Ross) were obtained from a commercial hatchery, weighed individually, wing banded, and assigned to pens based on body weight to ensure that all treatment groups began with statistically similar weights. Broiler chicks were housed in battery brooders and given access to water and experimental rations ad libitum. All experimental procedures were performed in accordance with protocols approved by the Texas A&M University Institutional Animal Care and Use Committee (IACUC).
Broiler performance trial.
A total of 144 broiler chicks were separated into 6 treatment groups of 24 birds each. Four experimental treatment groups were fed a phosphorus-deficient diet (0.25% available phosphate [aP]) and administered recombinant Lactobacillus cultures (108 CFU) in 1 ml maximum recovery diluent (MRD) (Difco) by oral gavage daily. Chicks were administered L. gallinarum TDCC 63 (rPhyA+), L. gallinarum TDCC 62 (empty vector), L. gasseri TDCC 65 (rPhyA+), and L. gasseri TDCC 64 (empty vector). Control groups were fed a diet adequate in phosphorus (0.40% aP) (positive control) or the phosphorus-deficient diet (0.25% aP) (negative control) and administered a mock inoculation of 1 ml sterile MRD by oral gavage daily. Broiler chicks were weighed individually at days 0, 7, 14, and 21 posthatch. Data were analyzed using ANOVA, and significant differences between treatment groups were determined using Duncan's multiple-range test with individual birds as the experimental unit.
Experimental diets.
A phosphorus-deficient basal starter diet was formulated with 0.25% available phosphate (aP) and all other nutrients meeting or exceeding industry-type broiler diet requirements for market broilers for days 0 to 21 posthatch (Table 3). The positive-control diet adequate in phosphorus was formulated by increasing the aP to 0.40% with the addition of KH2PO4 to the basal diet. Feed samples were analyzed by an independent commercial laboratory for total phosphorus content (43).
TABLE 3.
Ingredient profile and nutrient concentrations for the basal starter diet
| Parameter | % |
|---|---|
| Ingredients | |
| Corn | 60.03 |
| Soybean meal (48% crude protein) | 34.14 |
| Limestone | 1.70 |
| Sodium chloride | 0.46 |
| Fat (animal-vegetable blend) | 2.24 |
| l-Lysine HCl | 0.17 |
| dl-Methionine (99%) | 0.26 |
| Vitaminsa | 0.25 |
| Mineralsb | 0.05 |
| Monocalcium PO4 | 0.60 |
| l-Threonine | 0.03 |
| Calculated nutrient concn | |
| Crude protein | 22.00 |
| Metabolizable energy (kcal/kg) | 3,050 |
| Methionine | 0.58 |
| Total sulfur amino acids | 0.95 |
| Lysine | 1.30 |
| Threonine | 0.85 |
| Tryptophan | 0.26 |
| Calcium | 0.85 |
| Sodium | 0.20 |
| Total phosphorusc | 0.50 |
| Available phosphorus | 0.25 |
Vitamin premix added at this rate yields 11,023 IU vitamin A, 3,858 IU vitamin D3, 46 IU vitamin E, 0.0165 mg B12, 5.845 mg riboflavin, 45.93 mg niacin, 20.21 mg d-pantothenic acid, 477.67 mg choline, 1.47 mg menadione, 1.75 mg folic acid, 7.17 mg pyroxidine, 2.94 mg thiamine, and 0.55 mg biotin per kg diet. The carrier is ground rice hulls.
Trace mineral premix added at this rate yields 149.6 mg manganese, 125.1 mg zinc, 16.5 mg iron, 1.7 mg copper, 1.05 mg iodine, 0.25 mg selenium, a minimum of 6.27 mg calcium, and a maximum of 8.69 mg calcium per kg of diet. The carrier is calcium carbonate, and the premix contains less than 1% mineral oil.
Analyzed total phosphorus was 0.67% for the phosphorus-deficient basal diet and 0.81% in the positive-control diet with adequate phosphorus.
RESULTS
Recombinant phytase expression in Lactobacillus.
The 1,149-bp phyA (BSU19800) gene, encoding a phytase (44) from B. subtilis (35), was selected for recombinant expression in Lactobacillus. Protein domain analysis of the 382-amino-acid sequence predicted the presence of a Gram-positive signal peptide (amino acids 1 to 26), suggesting that the protein would likely be secreted via the sec pathway (45). B. subtilis phyA was codon optimized for expression in Lactobacillus using OPTIMIZER (36). The codon adaptation index of the native phyA sequence was 0.27, and this improved to 1.00 after optimization. The optimized sequence was commercially synthesized and subcloned into pTRK882. The resulting plasmid, pTD003 (Fig. 1), and the empty vector, pTRK882, were transformed into L. acidophilus NCFM, L. gallinarum ATCC 33319T, and L. gasseri ATCC 33323T. Transformations were confirmed by PCR to detect ermC and recombinant phyA (rphyA) (data not shown). Amplification of both phyA and ermC indicated successful transformation by pTD003, and amplification of ermC alone indicated successful transformation by pTRK882.
FIG 1.
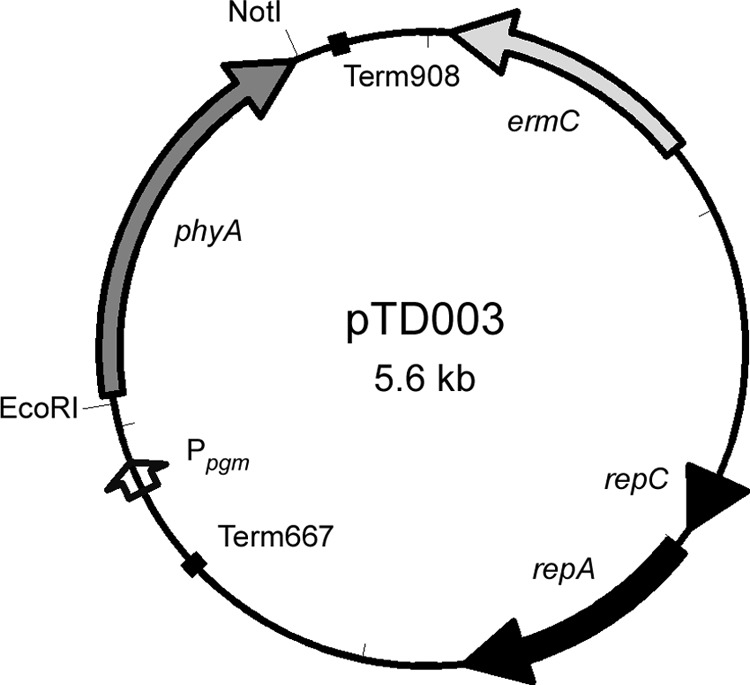
Plasmid map of pTD003. Black arrows, replication determinants; light gray arrow, erythromycin resistance marker, ermC; black boxes, transcriptional terminators; white arrow, Ppgm promoter; dark gray arrow, codon-optimized phytase gene, phyA.
SDS-PAGE.
Total protein in culture supernatants from Lactobacillus cultures was separated using SDS-PAGE (Fig. 2). A protein with a molecular mass of approximately 44 kDa was present in supernatants of L. acidophilus TDCC 61, L. gallinarum TDCC 63, and L. gasseri TDCC 65. While a faint protein band of similar molecular mass did appear in the supernatant of L. gasseri TDCC 64, this protein was not detected in supernatants of the empty-vector controls, L. acidophilus TDCC 60, and L. gallinarum TDCC 62. The molecular mass of the secreted mature phytase from B. subtilis is 44 kDa (44). These data suggest that recombinant PhyA phytase (rPhyA) is expressed and secreted by Lactobacillus cultures transformed with pTD003.
FIG 2.
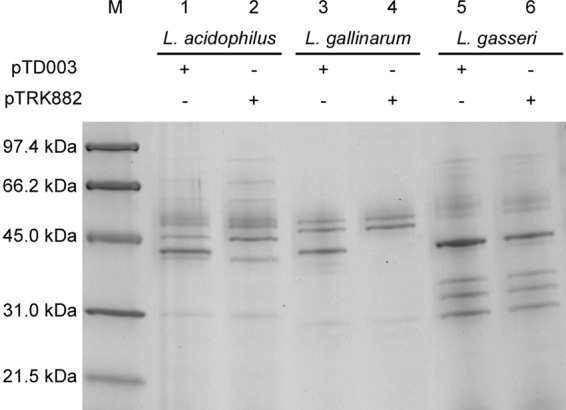
SDS-PAGE. Supernatants from Lactobacillus cultures were analyzed using SDS-PAGE. Lane 1, L. acidophilus TDCC 61; lane 2, L. acidophilus TDCC 60; lane 3, L. gallinarum TDCC 63; lane 4, L. gallinarum TDCC 62; lane 5, L. gasseri TDCC 65; lane 6, L. gasseri TDCC 64; lane M, molecular weight marker.
Phytate hydrolysis.
Phytate hydrolysis by Lactobacillus cultures was evaluated qualitatively (Fig. 3). Zones of clearing appeared around colonies of pTD003-transformed cultures, L. acidophilus TDCC 61, L. gallinarum TDCC 63, and L. gasseri TDCC 65. However, little to no clearing appeared around colonies of the empty-vector control cultures, L. acidophilus TD 60, L. gallinarum TDCC 62, and L. gasseri TDCC 64.
FIG 3.
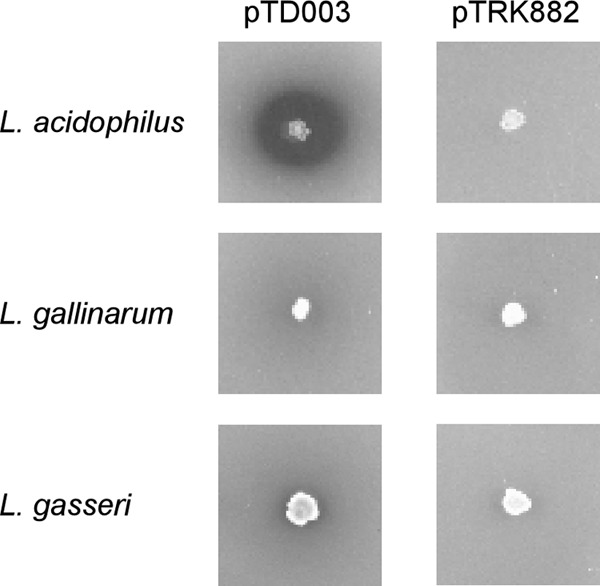
Phytate hydrolysis. Lactobacillus cultures were spotted onto MRS agar and incubated for 36 h. Plates were overlaid with modified MRS agar containing 0.5% sodium phytate, incubated for 24 h, and stained with cobalt chloride and ammonium molybdovanadate solutions. Zones of clearing indicate phytate hydrolysis.
Phytase activity of recombinant Lactobacillus cultures.
Phytase activity from cell pellets of recombinant Lactobacillus cultures was evaluated (Table 4). The phytase activities of L. acidophilus TDCC 61, L. gallinarum TDCC 63, and L. gasseri TDCC 65 were approximately 4-, 18-, and 10-fold greater than those of the respective empty-vector control cultures, respectively. Lactobacillus empty-vector (pTRK882) transformants are wild type for phytase activity and account for background phytate degradation by nonspecific phosphatases. The phytase activities of L. gallinarum TDCC 63 and L. gasseri TDCC 65 were approximately 3- and 2-fold greater, respectively, than that of L. acidophilus TDCC 61.
TABLE 4.
Phytase activities of recombinant Lactobacillus cultures
| Culture | Sp act (U/mg)a |
Activity increaseb | |
|---|---|---|---|
| pTD003 | pTRK882 | ||
| L. acidophilus | 0.168 ± 0.019 c | 0.046 ± 0.029 | 4.04 ± 2.46 c |
| L. gallinarum | 0.556 ± 0.077 a | 0.034 ± 0.011 | 18.61 ± 5.80 a |
| L. gasseri | 0.387 ± 0.041 b | 0.038 ± 0.003 | 10.68 ± 0.33 b |
International units, μmol PO43− released min−1 mg−1 total protein. Data are means ± SEMs for replicate reactions from three independent assays. Different letters within columns indicate that the means differ significantly (P < 0.05).
Fold increase between pTD003 (rPhyA+)- and pTD882 (empty-vector)-transformed cultures. Data are means ± SEMs for replicate reactions from three independent assays. Different letters within columns indicate that the means differ significantly (P < 0.05).
Broiler performance trial.
The effects of rPhyA-producing Lactobacillus cultures on the performance of broiler chicks were evaluated (Fig. 4). There were no differences in body weight between the treatment groups at days 0 and day 7 posthatch. For mock-inoculated control groups, the body weight of chicks fed a diet adequate in phosphorus (positive control) was greater than that of those fed a phosphorus-deficient diet (negative control) at days 14 and 21 posthatch (P < 0.05). The body weight of chicks administered rPhyA-producing L. gallinarum (TDCC 63) and L. gasseri (TDCC 65) was not significantly different from that of those administered the respective empty-vector control cultures, L. gallinarum TDCC 62, and L. gasseri TDCC 64 or the negative-control group. However, the body weight of chicks administered L. gasseri TDCC 65 was not significantly different from that of the positive-control group (P > 0.05). While performance was not significantly increased compared to that with the negative control or relevant empty-vector control, the administration of rPhyA-producing L. gasseri improved weight gain of broiler chickens to a level statistically comparable to that of chicks fed a diet adequate in phosphorus.
FIG 4.
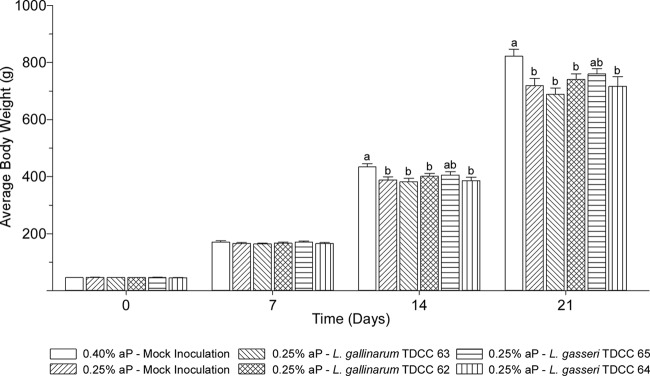
Average body weights of broiler chicks. Male broiler chicks were divided between six treatment groups (n = 24 broiler chickens in each treatment) and either fed a control diet adequate in phosphorus (0.40% aP) and administered a mock inoculation (MRD) or fed a phosphorus-deficient diet (0.25% aP) and administered either a mock inoculation (MRD) or cultures of L. gallinarum TDCC 63 (rPhyA+), L. gallinarum TDCC 62 (empty vector), L. gasseri TDCC 65 (rPhyA+), or L. gasseri TDCC 64 (empty vector) by oral gavage daily. Broiler chicks were weighed individually at days 0, 7, 14, and 21 posthatch. Data shown are the mean body weight for each treatment group, and error bars represent the standard error of the mean (SEM). Different letters indicate that means are significantly different (P < 0.05).
DISCUSSION
The objective of this study was to investigate phytate degradation as a novel mechanism of probiotic functionality. An important role of the gastrointestinal microbiota is to indirectly augment host metabolism by utilizing undigested food and producing short-chain fatty acids and micronutrients which can be utilized by the host (46). The microbial origin of exogenous enzymes used in livestock production, including phytases, suggests that direct augmentation of host metabolism through the in situ production and delivery of these enzymes by microorganisms in the gastrointestinal tract may potentially be an important mechanism of probiotic functionality. While these enzyme activities have been suggested as selection criteria for probiotic cultures (41, 47), biocatalysis by probiotics in the gastrointestinal tract has not been explored.
Phytate-degrading activity has been reported in Lactobacillus species and has been suggested to improve the nutritional quality of fermented cereal grains (48–50). De Angelis et al. (51) reported the purification of a phytase from Lactobacillus sanfranciscensis. However, the significantly greater substrate specificity of this enzyme toward p-nitrophenyl phosphate over phytate suggests that this enzyme would more appropriately be classified as a non-phytate-specific acid phosphatase. Phytate degradation has been attributed to nonspecific acid phosphatases in other lactobacilli (52, 53). Additionally, a phytase gene has not yet been identified in a Lactobacillus species.
Because true phytase-producing Lactobacillus cultures have not yet been identified, recombinant cultures were used to model phytate degradation by probiotic microbes. The phyA gene from B. subtilis (35) encodes a β-propeller phytase with high specificity for phytic acid and activity over broad pH and temperature ranges (44). Analysis of the amino acid sequence using SignalP (45) predicted the presence of a Gram-positive secretion signal, suggesting that heterologous expression of this protein in Lactobacillus would result in production of a secreted protein. Thus, we selected the B. subtilis phyA for expression in Lactobacillus. Interestingly, the popularity of probiotic and DFM products containing spore-forming bacteria, including B. subtilis, has increased (54–57). Bacillus species are workhorse bacteria in microbial fermentations and are highly prized as producers of industrially important enzymes (58). Heterologous expression of B. subtilis phytase using Lactobacillus in this study not only demonstrates biocatalytic phytate degradation as a mechanism of probiotic functionality but will guide future studies investigating this specific mechanism in Bacillus species, further supporting their use in probiotic and DFM products.
B. subtilis phyA was codon optimized and cloned into pTRK882, under the control of the constitutive high-expressing Ppgm promoter from L. acidophilus NCFM (37), in order to maximize expression in Lactobacillus species. This expression system has been previously demonstrated to be effective in enzyme expression (37), the production and delivery of immune modulating cytokines (59), and an anthrax vaccine (60), and its wide host range allowed the transformation of L. acidophilus, L. gallinarum, and L. gasseri. L. acidophilus NCFM and L. gasseri ATCC 33323 were originally isolated from the human gastrointestinal tract (61, 62). These cultures are commonly used as model organisms in research investigating mechanisms of probiotic functionality because they are readily transformed (63, 64) and genetically tractable (37, 65) and because a complete genome sequences is available for these microorganisms (62, 66). L. gallinarum was originally isolated from the crop of a chicken (67) and has been demonstrated to reduce gastrointestinal colonization of Campylobacter jejuni in experimentally challenged broiler chickens (8). Plasmid transformation and heterologous protein expression in L. gallinarum ATCC 33319 have not been reported previously.
SDS-PAGE revealed the presence of a protein with a molecular mass similar to that of B. subtilis phytase (44) in the supernatants of L. acidophilus TDCC 61, L. gallinarum TDCC 63, and L. gasseri TDCC 65, which was likely to be recombinant rPhyA expressed using pTD003. Additionally, a protein of similar molecular mass was also present in the supernatant of the empty-vector control culture of L. gasseri TDCC 64. The LAB-Secretome database (68) predicted three secreted proteins expressed by L. gasseri ATCC 33323 with molecular masses between 39 kDa and 51 kDa, which may be the protein present.
Differential media containing phytate are commonly used for detection and qualitative evaluation of phytase activity (40, 41, 69). Phytase activity is indicated by zones of clearing around colonies cultured using phytate-containing media. However, reduced pH around colonies of acid-producing bacteria may also cause the appearance of zones of clearing. False-positive detection of phytase activity can be reduced by staining with aqueous cobalt chloride and ammonium molybdovanadate solutions (40). Staining of differential-screening plates requires colonies to be washed from the plate surface prior to detection of enzymatic activity (40, 41). In this study, an overlay medium (70) containing phytate was used to remove the need to wash colonies from the plate surface. This modification is expected to facilitate future screening for phytate-degrading Lactobacillus cultures by allowing isolates to be picked through the overlay agar for subculture.
Recombinant expression of phytase in Lactobacillus has been demonstrated previously (71, 72). However, comparison with these studies was impossible because activity was not evaluated quantitatively (72) or because specific activity was not reported (71). Comparison with published studies of wild-type Lactobacillus cultures was also complicated because specific activity was not reported (41) or was reported in nonstandard units (51, 53, 73). Nonetheless, we have determined that our recombinant cultures produce 10- to 50-fold-greater activity than previously reported for wild-type lactobacilli (41, 51, 53, 73).
L. gallinarum TDCC 63 and L. gasseri TDCC 65 were selected for administration to broiler chicks because they produced greater phytase activity than L. acidophilus TDCC 61 (Table 4). Broiler chicks were inoculated daily with 108 CFU Lactobacillus by oral gavage. Broiler chicks have been administered 108 CFU Lactobacillus by oral gavage in studies investigating their administration in poultry (8, 74, 75) in order to maximize detection of any potential beneficial effects. Because colonization by allochthonous lactobacilli is transient, the probiotic cultures were administered daily (76) in order to maximize the presence of administered lactobacilli in the gastrointestinal tracts of the experimental animals. While the probiotic potential of phytate-degrading Lactobacillus cultures has been explored previously (41, 47, 71), this is the first study to evaluate the effect of their administration in vivo.
Nutritional models using phosphorus-deficient corn soybean meal rations are widely used to investigate phytate phosphorus metabolism in poultry (77, 78). Body weight gain is depressed in broiler chicks fed rations deficient in aP relative to those fed a diet adequate in aP. A decrease in the growth depression caused by aP deficiency is an effective and commonly used measure of the ability of exogenous phytase and other feed additives to improve bioavailability of phytate phosphorus (79–81).
Yi et al. (82) demonstrated that supplementation with commercial exogenous phytase improved the 3-week weight gain of broiler chicks fed a phosphorus-deficient diet (0.27% aP) to a level similar to that for those fed a diet adequate in phosphorus (0.47% aP). It is generally accepted that the aP content of broiler chicken rations supplemented with commercial phytases can be reduced by 0.1% or more without a significant decrease in weight gain (83, 84). The body weight gain of chicks administered L. gasseri TDCC 65 (rPhyA+) was not significantly greater than those of other groups fed a phosphorus-deficient diet (Fig. 4). However, weight gain was improved to a level statistically comparable to that for the control group fed a diet adequate in phosphorus. Similar results were seen in early studies investigating supplementation with crude exogenous phytase preparations (78). Additionally, weight gain was improved only in chicks administered rPhyA+ L. gasseri (TDCC 65) and not in those administered the empty-vector L. gasseri (TDCC 66). These L. gasseri ATCC 33323-derived cultures are isogenic strains which are either phyA+ or wild type for phytase expression, indicating that that improved weight gain was due to increased bioavailability of phytate phosphorus mediated by phytase expression in L. gasseri TDCC 65 (rPhyA+).
While the in vitro phytase activity of L. gallinarum TDCC 63 was greater than that of L. gasseri TDCC 65, the in vivo effectiveness of probiotic cultures is multifactorial. Other factors potentially affecting the behavior of these cultures when administered to poultry include the ability of these organisms to adhere and persist in various locations in the gastrointestinal tract (85, 86), their ability to survive or tolerate acid and bile (86), and the efficiency with which they are able to produce and secrete these enzymes in the gastrointestinal tract.
Recombinant expression of B. subtilis phytase in Lactobacillus has allowed us to demonstrate that administration of phytate-degrading probiotic cultures can increase the bioavailability of phytate phosphorus and improve the performance of nonruminant livestock animals fed a phosphorus-deficient diet. While phytate degradation by Lactobacillus reported previously was attributed to nonspecific phosphatases, a sufficiently large screen may identify Lactobacillus cultures expressing this desired activity. Alternatively, true specific phytase activity may not be critical if sufficient phytate degradation can be produced from nonspecific phosphatases. While it is unlikely that regulatory agencies would approve the use of recombinant microorganisms in commercial livestock production, their use has allowed us to investigate this novel mechanism and inform future studies which will identify and investigate the potential of wild-type probiotic microorganisms able to improve utilization of phytate and other indigestible feed constituents. We have demonstrated proof of principle of in situ enzyme production and degradation of indigestible feed constituents by microorganisms in the gastrointestinal tract as a novel mechanism of probiotic functionality. Although administration of exogenous enzymes is currently relatively inexpensive, the identification of probiotic cultures able to increase the bioavailability of phytate phosphorus at levels similar to those with exogenous enzymes may reduce the need for isolation and purification of enzymes from industrial fermentations. Alternatively, rather than being a replacement for exogenous enzymes, the identification of probiotic organisms producing phytase or other important enzymes may offer a value-added benefit in addition to the food safety and animal health benefits traditionally associated with probiotic administration.
ACKNOWLEDGMENTS
We thank Sadie L. Dunn-Horrocks, Dale Hyatt, and Joseph M. Sturino (Texas A&M University) for their assistance in conducting this study. We also thank Todd R. Klaenhammer (North Carolina State University) for providing L. acidophilus NCFM and pTRK882.
This research was supported by Texas A&M AgriLife Research and USDA-NIFA Hatch project number TEX09405. Tyler E. Askelson and Ashley Campasino were supported by graduate assistantships from the Texas A&M University Poultry Science Department.
Footnotes
Published ahead of print 22 November 2013
REFERENCES
- 1.Klaenhammer TR, Altermann E, Pfeiler E, Buck BL, Goh YJ, O'Flaherty S, Barrangou R, Duong T. 2008. Functional genomics of probiotic lactobacilli. J. Clin. Gastroenterol. 42(Suppl 3):S160–S162. 10.1097/MCG.0b013e31817da140 [DOI] [PubMed] [Google Scholar]
- 2.Gibson GR, Probert HM, Van Loo J, Rastall RA, Roberfroid MB. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259–275. 10.1079/NRR200479 [DOI] [PubMed] [Google Scholar]
- 3.Sanders ME. 2008. Probiotics: definition, sources, selection, and uses. Clin. Infect. Dis. 46:S58–S61. 10.1086/523341 [DOI] [PubMed] [Google Scholar]
- 4.Klein M, Sanders ME, Duong T, Young HA. 2010. Probiotics: from bench to market. Ann. N. Y. Acad. Sci. 1212(Suppl 1):E1–E14. 10.1111/j.1749-6632.2010.05839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loh TC, Thanh NT, Foo HL, Hair-Bejo M, Azhar BK. 2010. Feeding of different levels of metabolite combinations produced by Lactobacillus plantarum on growth performance, fecal microflora, volatile fatty acids and villi height in broilers. Anim. Sci. J. 81:205–214. 10.1111/j.1740-0929.2009.00701.x [DOI] [PubMed] [Google Scholar]
- 6.Mountzouris KC, Tsirtsikos P, Kalamara E, Nitsch S, Schatzmayr G, Fegeros K. 2007. Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poult. Sci. 86:309–317 [DOI] [PubMed] [Google Scholar]
- 7.Ghareeb K, Awad WA, Mohnl M, Porta R, Biarnes M, Bohm J, Schatzmayr G. 2012. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poult. Sci. 91:1825–1832. 10.3382/ps.2012-02168 [DOI] [PubMed] [Google Scholar]
- 8.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, Konkel ME. 2012. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One 7:e43928. 10.1371/journal.pone.0043928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Ragione R, Narbad A, Gasson M, Woodward M. 2004. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 38:197–205. 10.1111/j.1472-765X.2004.01474.x [DOI] [PubMed] [Google Scholar]
- 10.Pascual M, Hugas M, Badiola JI, Monfort JM, Garriga M. 1999. Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl. Environ. Microbiol. 65:4981–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kizerwetter-Swida M, Binek M. 2009. Protective effect of potentially probiotic Lactobacillus strain on infection with pathogenic bacteria in chickens. Pol. J. Vet. Sci. 12:15–20 [PubMed] [Google Scholar]
- 12.Kabir SML. 2009. The role of probiotics in the poultry industry. Int. J. Mol. Sci. 10:3531–3546. 10.3390/ijms10083531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huyghebaert G, Ducatelle R, Van Immerseel F. 2011. An update on alternatives to antimicrobial growth promoters for broilers. Vet. J. 187:182–188. 10.1016/j.tvjl.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 14.National Research Council Subcommittee on Poultry Nutrition 1994. Nutrient requirements of poultry, 9th ed. National Academy Press, Washington, DC [Google Scholar]
- 15.Adeola O, Cowieson AJ. 2011. Opportunities and challenges in using exogenous enzymes to improve nonruminant animal production. J. Anim. Sci. 89:3189–3218. 10.2527/jas.2010-3715 [DOI] [PubMed] [Google Scholar]
- 16.Cowieson AJ, Hruby M, Pierson EEM. 2006. Evolving enzyme technology: impact on commercial poultry nutrition. Nutr. Res. Rev. 19:90–103. 10.1079/NRR2006121 [DOI] [PubMed] [Google Scholar]
- 17.Reddy NR, Sathe SK, Salunkhe DK. 1982. Phytates in legumes and cereals. Adv. Food Res. 28:1–92 [DOI] [PubMed] [Google Scholar]
- 18.Dalal RC. 1977. Soil organic phosphorus. Adv. Agron. 29:83–113 [Google Scholar]
- 19.Selle PH, Ravindran V, Bryden WL, Scott T. 2006. Influence of dietary phytate and exogenous phytase on amino acid digestibility in poultry: a review. J. Poult. Sci. 43:89–103. 10.2141/jpsa.43.89 [DOI] [Google Scholar]
- 20.Nelson TS. 1967. The utilization of phytate phosphorus by poultry—a review. Poult. Sci. 46:862–871. 10.3382/ps.0460862 [DOI] [PubMed] [Google Scholar]
- 21.Jongbloed AW, Mroz Z, Kemme PA. 1992. The effect of supplementary Aspergillus-Niger phytase in diets for pigs on concentration and apparent digestibility of dry-matter, total phosphorus, and phytic acid in different sections of the alimentary tract. J. Anim. Sci. 70:1159–1168 [DOI] [PubMed] [Google Scholar]
- 22.Graf E, Eaton JW. 1990. Antioxidant functions of phytic acid. Free Radic. Biol. Med. 8:61–69. 10.1016/0891-5849(90)90146-A [DOI] [PubMed] [Google Scholar]
- 23.Mitchell DB, Vogel K, Weimann BJ, Pasamontes L, van Loon APGM. 1997. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology 143:245–252. 10.1099/00221287-143-1-245 [DOI] [PubMed] [Google Scholar]
- 24.Selle PH, Ravindran V, Ravindran C, Bryden WL. 2007. Effects of dietary lysine and microbial phytase on growth performance and nutrient utilisation of broiler chickens. Asian Austral. J. Anim. 20:1100–1107 [Google Scholar]
- 25.Simons PC, Versteegh HA, Jongbloed AW, Kemme PA, Slump P, Bos KD, Wolters MG, Beudeker RF, Verschoor GJ. 1990. Improvement of phosphorus availability by microbial phytase in broilers and pigs. Br. J. Nutr. 64:525–540. 10.1079/BJN19900052 [DOI] [PubMed] [Google Scholar]
- 26.Denbow DM, Grabau EA, Lacy GH, Kornegay ET, Russell DR, Umbeck PF. 1998. Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poult. Sci. 77:878–881 [DOI] [PubMed] [Google Scholar]
- 27.Coppedge J, Klein J, Brown B, Ratliff B, Ruch F, Lee JT. 2011. Effects of co-adminstration of phytase and NSPase on broiler performance and bone ash. Int. J. Poult. Sci. 10:933–939. 10.3923/ijps.2011.933.939 [DOI] [Google Scholar]
- 28.Ravindran V, Selle PH, Bryden WL. 1999. Effects of phytase supplementation, individually and in combination, with glycanase, an the nutritive value of wheat and barley. Poult. Sci. 78:1588–1595 [DOI] [PubMed] [Google Scholar]
- 29.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207. 10.1016/0022-2836(80)90283-1 [DOI] [PubMed] [Google Scholar]
- 30.Walker DC, Klaenhammer TR. 1994. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 176:5330–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 2001. Current protocols in molecular biology. John Wiley and Sons, New York, NY [Google Scholar]
- 32.Seidman CE, Struhl K, Sheen J, Jessen T. 2001. Introduction of plasmid DNA into cells. Curr. Protoc. Mol. Biol. unit 1.8. 10.1002/0471142727.mb0108s37 [DOI] [PubMed] [Google Scholar]
- 33.Luchansky JB, Tennant MC, Klaenhammer TR. 1991. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J. Dairy Sci. 74:3293–3302. 10.3168/jds.S0022-0302(91)78515-9 [DOI] [PubMed] [Google Scholar]
- 34.Beasley SS, Takala TM, Reunanen J, Apajalahti J, Saris PE. 2004. Characterization and electrotransformation of Lactobacillus crispatus isolated from chicken crop and intestine. Poult. Sci. 83:45–48 [DOI] [PubMed] [Google Scholar]
- 35.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Cummings NJ, Daniel RA, Denizot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo MF, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. 10.1038/36786 [DOI] [PubMed] [Google Scholar]
- 36.Puigbò P, Guzmán E, Romeu A, Garcia-Vallvé S. 2007. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35:W126–W131. 10.1093/nar/gkm219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong T, Miller MJ, Barrangou R, Azcarate-Peril MA, Klaenhammer TR. 2011. Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microb. Biotechnol. 4:357–367. 10.1111/j.1751-7915.2010.00200.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 39.Gallagher SR. 2001. One-dimensional SDS gel electrophoresis of proteins. Curr. Protoc. Protein Sci., chapter 10, unit 10.1. 10.1002/0471140864.ps1001s00 [DOI] [PubMed] [Google Scholar]
- 40.Bae HD, Yanke LJ, Cheng KJ, Selinger LB. 1999. A novel staining method for detecting phytase activity. J. Microbiol. Methods 39:17–22. 10.1016/S0167-7012(99)00096-2 [DOI] [PubMed] [Google Scholar]
- 41.Raghavendra P, Halami PM. 2009. Screening, selection and characterization of phytic acid degrading lactic acid bacteria from chicken intestine. Int. J. Food Microbiol. 133:129–134. 10.1016/j.ijfoodmicro.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 42.Shimizu M. 1992. Purification and characterization of phytase from Bacillus subtilis (Natto) N-77. Biosci. Biotechnol. Biochem. 56:1266–1269. 10.1271/bbb.56.1266 [DOI] [Google Scholar]
- 43.Parker DS, Armstrong DG. 1987. Antibiotic feed additives and livestock production. Proc. Nutr. Soc. 46:415–421. 10.1079/PNS19870056 [DOI] [PubMed] [Google Scholar]
- 44.Tye AJ, Siu FK, Leung TY, Lim BL. 2002. Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl. Microbiol. Biotechnol. 59:190–197. 10.1007/s00253-002-1033-5 [DOI] [PubMed] [Google Scholar]
- 45.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 46.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota—introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 47.Taheri HR, Moravej H, Tabandeh F, Zaghari M, Shivazad M. 2009. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult. Sci. 88:1586–1593. 10.3382/ps.2009-00041 [DOI] [PubMed] [Google Scholar]
- 48.Anastasio M, Pepe O, Cirillo T, Palomba S, Blaiotta G, Villani F. 2010. Selection and use of phytate-degrading LAB to improve cereal-based products by mineral solubilization during dough fermentation. J. Food Sci. 75:M28–35. 10.1111/j.1750-3841.2009.01402.x [DOI] [PubMed] [Google Scholar]
- 49.Reale A, Konietzny U, Coppola R, Sorrentino E, Greiner R. 2007. The importance of lactic acid bacteria for phytate degradation during cereal dough fermentation. J. Agric. Food Chem. 55:2993–2997. 10.1021/jf063507n [DOI] [PubMed] [Google Scholar]
- 50.Songre-Ouattara LT, Mouquet-Rivier C, Icard-Verniere C, Humblot C, Diawara B, Guyot JP. 2008. Enzyme activities of lactic acid bacteria from a pearl millet fermented gruel (ben-saalga) of functional interest in nutrition. Int. J. Food Microbiol. 128:395–400. 10.1016/j.ijfoodmicro.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 51.De Angelis M, Gallo G, Corbo MR, McSweeney PLH, Faccia M, Giovine M, Gobbetti M. 2003. Phytase activity in sourdough lactic acid bacteria: purification and characterization of a phytase from Lactobacillus sanfranciscensis CB1. Int. J. Food Microbiol. 87:259–270. 10.1016/S0168-1605(03)00072-2 [DOI] [PubMed] [Google Scholar]
- 52.Palacios MC, Haros M, Rosell CM, Sanz Y. 2005. Characterization of an acid phosphatase from Lactobacillus pentosus: regulation and biochemical properties. J. Appl. Microbiol. 98:229–237. 10.1111/j.1365-2672.2004.02447.x [DOI] [PubMed] [Google Scholar]
- 53.Zamudio M, Gonzalez A, Medina JA. 2001. Lactobacillus plantarum phytase activity is due to non-specific acid phosphatase. Lett. Appl. Microbiol. 32:181–184. 10.1046/j.1472-765x.2001.00890.x [DOI] [PubMed] [Google Scholar]
- 54.Cutting SM. 2011. Bacillus probiotics. Food Microbiol. 28:214–220. 10.1016/j.fm.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 55.Hong HA, Duc LH, Cutting SM. 2005. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 29:813–835. 10.1016/j.femsre.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 56.Shivaramaiah S, Pumford NR, Morgan MJ, Wolfenden RE, Wolfenden AD, Torres-Rodriguez A, Hargis BM, Tellez G. 2011. Evaluation of Bacillus species as potential candidates for direct-fed microbials in commercial poultry. Poult. Sci. 90:1574–1580. 10.3382/ps.2010-00745 [DOI] [PubMed] [Google Scholar]
- 57.Tompkins TA, Hagen KE, Wallace TD, Fillion-Forte V. 2008. Safety evaluation of two bacterial strains used in Asian probiotic products. Can. J. Microbiol. 54:391–400. 10.1139/W08-022 [DOI] [PubMed] [Google Scholar]
- 58.Schallmey M, Singh A, Ward OP. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1–17. 10.1139/w03-076 [DOI] [PubMed] [Google Scholar]
- 59.McFarland AP, Savan R, Wagage S, Addison A, Ramakrishnan K, Karwan M, Duong T, Young HA. 2011. Localized delivery of interferon-beta by Lactobacillus exacerbates experimental colitis. PLoS One 6:e16967. 10.1371/journal.pone.0016967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. 2009. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc. Natl. Acad. Sci. U. S. A. 106:4331–4336. 10.1073/pnas.0900029106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanders ME, Klaenhammer TR. 2001. The scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84:319–331. 10.3168/jds.S0022-0302(01)74481-5 [DOI] [PubMed] [Google Scholar]
- 62.Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610–4625. 10.1128/AEM.00054-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allison G, Klaenhammer T. 1996. Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl. Environ. Microbiol. 62:4450–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luchansky JB, Muriana PM, Klaenhammer TR. 1988. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol. Microbiol. 2:637–646. 10.1111/j.1365-2958.1988.tb00072.x [DOI] [PubMed] [Google Scholar]
- 65.Russell W, Klaenhammer T. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361–4364. 10.1128/AEM.67.9.4361-4364.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906–3912. 10.1073/pnas.0409188102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujisawa T, Benno Y, Yaeshima T, Mitsuoka T. 1992. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int. J. Syst. Bacteriol. 42:487–491. 10.1099/00207713-42-3-487 [DOI] [PubMed] [Google Scholar]
- 68.Zhou MM, Theunissen D, Wels M, Siezen RJ. 2010. LAB-Secretome: a genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of lactic acid bacteria. BMC Genomics 11:651. 10.1186/1471-2164-11-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tseng YH, Fang TJ, Tseng SM. 2000. Isolation and characterization of a novel phytase from Penicillium simplicissimum. Folia Microbiol. (Praha) 45:121–127. 10.1007/BF02817409 [DOI] [PubMed] [Google Scholar]
- 70.Liu JR, Yu B, Liu FH, Cheng KJ, Zhao X. 2005. Expression of rumen microbial fibrolytic enzyme genes in probiotic Lactobacillus reuteri. Appl. Environ. Microbiol. 71:6769–6775. 10.1128/AEM.71.11.6769-6775.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuo R, Chang J, Yin Q, Chen L, Chen Q, Yang X, Zheng Q, Ren G, Feng H. 2010. Phytase gene expression in Lactobacillus and analysis of its biochemical characteristics. Microbiol. Res. 165:329–335. 10.1016/j.micres.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 72.Kerovuo J, Tynkkynen S. 2000. Expression of Bacillus subtilis phytase in Lactobacillus plantarum 755. Lett. Appl. Microbiol. 30:325–329. 10.1046/j.1472-765x.2000.00660.x [DOI] [PubMed] [Google Scholar]
- 73.Tang AL, Wilcox G, Walker KZ, Shah NP, Ashton JF, Stojanovska L. 2010. Phytase activity from Lactobacillus spp. in calcium-fortified soymilk. J. Food Sci. 75:M373–M376. 10.1111/j.1750-3841.2010.01663.x [DOI] [PubMed] [Google Scholar]
- 74.Garriga M, Pascual M, Monfort JM, Hugas M. 1998. Selection of lactobacilli for chicken probiotic adjuncts. J. Appl. Microbiol. 84:125–132. 10.1046/j.1365-2672.1997.00329.x [DOI] [PubMed] [Google Scholar]
- 75.Farnell MB, Donoghue AM, de los Santos FS, Blore PJ, Hargis BM, Tellez G, Donoghue DJ. 2006. Upregulation of oxidative burst and degranulation in chicken heterophils stimulated with probiotic bacteria. Poult. Sci. 85:1900–1906 [DOI] [PubMed] [Google Scholar]
- 76.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74:4985–4996. 10.1128/AEM.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perney KM, Cantor AH, Straw ML, Herkelman KL. 1993. The effect of dietary phytase on growth performance and phosphorus utilization of broiler chicks. Poult. Sci. 72:2106–2114. 10.3382/ps.0722106 [DOI] [PubMed] [Google Scholar]
- 78.Nelson TS, Shieh TR, Wodzinski RJ, Ware JH. 1971. Effect of supplemental phytase on the utilization of phytate phosphorus by chicks. J. Nutr. 101:1289–1293 [DOI] [PubMed] [Google Scholar]
- 79.Viveros A, Brenes A, Arija I, Centeno C. 2002. Effects of microbial phytase supplementation on mineral utilization and serum enzyme activities in broiler chicks fed different levels of phosphorus. Poult. Sci. 81:1172–1183 [DOI] [PubMed] [Google Scholar]
- 80.Sebastian S, Touchburn SP, Chavez ER. 1998. Implications of phytic acid and supplemental microbial phytase in poultry nutrition: a review. World Poultry Sci. J. 54:27–47. 10.1079/WPS19980003 [DOI] [Google Scholar]
- 81.Ravindran V, Cabahug S, Ravindra G, Selle PH, Bryden WL. 2000. Response of broiler chickens to microbial phytase supplementation as influenced by dietary phytic acid and non-phytate phosphorous levels. II. Effects on apparent metabolisable energy, nutrient digestibility and nutrient retention. Br. Poult. Sci. 41:193–200. 10.1080/00071660050022263 [DOI] [PubMed] [Google Scholar]
- 82.Yi Z, Kornegay E, Ravindran V, Denbow D. 1996. Improving phytate phosphorus availability in corn and soybean meal for broilers using microbial phytase and calculation of phosphorus equivalency values for phytase. Poult. Sci. 75:240–249. 10.3382/ps.0750240 [DOI] [PubMed] [Google Scholar]
- 83.Selle PH, Ravindran V. 2007. Microbial phytase in poultry nutrition. Anim. Feed Sci. Technol. 135:1–41. 10.1016/j.anifeedsci.2006.06.010 [DOI] [Google Scholar]
- 84.Slominski BA. 2011. Recent advances in research on enzymes for poultry diets. Poult. Sci. 90:2013–2023. 10.3382/ps.2011-01372 [DOI] [PubMed] [Google Scholar]
- 85.Fuller R. 1973. Ecological studies on Lactobacillus flora associated with crop epithelium of fowl. J. Appl. Bacteriol. 36:131–139. 10.1111/j.1365-2672.1973.tb04080.x [DOI] [Google Scholar]
- 86.Saarela M, Mogensen G, Fonden R, Matto J, Mattila-Sandholm T. 2000. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84:197–215. 10.1016/S0168-1656(00)00375-8 [DOI] [PubMed] [Google Scholar]


