Abstract
The rickettsial protein RickA activates host cell factors associated with the eukaryotic actin cytoskeleton and is likely involved with rickettsial host cell binding and infection and the actin-based motility of spotted fever group rickettsiae. The rickA gene sequence and protein vary substantially between Rickettsia species, as do observed motility-associated phenotypes. To help elucidate the function of RickA and determine the effects of species-specific RickA variations, we compared extracellular binding, intracellular motility, and intercellular spread phenotypes of three Rickettsia bellii variants. These included two shuttle vector-transformed R. bellii strains and the wild-type isolate from which they were derived, R. bellii RML 369C. Both plasmid shuttle vectors carried spectinomycin resistance and a GFPuv reporter; one contained Rickettsia monacensis-derived rickA, and the other lacked the rickA gene. Rickettsia bellii transformed to express R. monacensis rickA highly overexpressed this transcript in comparison to its native rickA. These rickettsiae also moved at higher velocities and followed a more curved path than the negative-control transformants. A lower proportion of R. monacensis rickA-expressing bacteria ever became motile, however, and they formed smaller plaques.
INTRODUCTION
Rickettsiae are obligate intracellular Alphaproteobacteria in the order Rickettsiales. Most, but not all, species of rickettsiae co-opt host cell-produced actin to induce bacterial movement in and between host cells, a behavior termed actin-based motility. Rickettsia bellii 369C is one such motile species; it was isolated from ticks of the genera Amblyomma and Dermacentor and is considered nonpathogenic in that it does not obviously negatively affect its host ticks and has never been associated with disease in vertebrates. Most previous studies examining rickettsial motility have done so by using pathogenic species, such as Rickettsia rickettsii (1, 2), or the milder pathogen, Rickettsia parkeri (3, 4). As a nonpathogen also capable of actin-based motility and rapid cell invasion, R. bellii provides a contrast that may help illuminate virulence factors of the pathogenic species. Additionally, because of its capability of infecting both tick and mammal-derived cell lines and its tractability to genetic manipulation, R. bellii was a preferred species for this study.
In Listeria and Shigella, two facultatively intracellular bacterial genera that also utilize actin-based motility, branched actin tails are polymerized by activating the host cell ARP2/3 protein complex (5, 6). The ARP2/3 complex is activated by nucleation-promoting factors, proteins that are naturally produced within eukaryotic cells, including the Wiskott-Aldrich syndrome protein (WASP) family (7), but can be mimicked by intracellular bacteria to hijack the actin polymerization process for the manufacture of a branched actin tail. In Listeria, the nucleation-promoting factor mimic is the protein ActA (8, 9). A similar rickettsial WASP-like protein, RickA, has been considered a candidate for inducing actin-based motility as a nucleation-promoting factor with its associated ability to interact with the ARP2/3 protein complex (10) and to polymerize actin into branched, tail-like structures on microscopic beads (11).
Unlike the branched actin tail produced by the host cell Arp2/3 complex, the actin tails of motile Rickettsia species are unbranched, suggesting the likelihood that proteins other than RickA mediate the process of actin polymerization. Further testing indicated that Sca2 polymerizes an unbranched actin tail by mimicking host cell formin proteins, independent of the presence of RickA, making it a better candidate for the type of actin-based motility observed (12). Still, RickA is considered to be involved in rickettsial motility based on a comparative analysis of the genomes of R. rickettsii and the closely related, but nonmotile, Rickettsia peacockii. The genome of R. rickettsii codes for a functional RickA protein, while in R. peacockii most of the gene has been disrupted by naturally occurring transposon recombination (13, 14). Actin-based motility is notably absent in R. peacockii but fully functional in the active R. rickettsii.
In this study, we describe how transforming R. bellii to overexpress Rickettsia monacensis-derived RickA substantially alters bacterial motility, adherence, and early infiltration into Vero cells. Like R. bellii, R. monacensis replicates in Vero cells and forms actin tails (15). The phenotypic differences highlighted in this study suggest a potential association of the WASP-like RickA with rickettsial actin-based motility, despite the apparent absence of a branched actin tail.
MATERIALS AND METHODS
Cell culture and transformation of rickettsiae.
All experiments were performed using R. bellii strain 369C. Rickettsiae were maintained in the ISE6 Ixodes scapularis tick cell line cultured in L15C medium and incubated at 32°C (16). Infected cells were passaged every 4 or 5 days onto an uninfected cell layer. Cell-free rickettsiae sufficient for two electroporations were obtained from one 25-cm2 flask maintained in 5 ml of medium. A 3.5-ml volume of medium was removed from the flask, and the cell layer containing rickettsiae was resuspended in the remaining 1.5 ml of medium. Resuspended cells were transferred to a 2-ml sterile tube containing 0.2 to 0.25 ml of silicon carbide abrasive 60/90 grit (Lortone, Inc., Mukilteo, WA), vortexed vigorously for 30 s to lyse the cells, transferred to a 5-ml Luer Lock syringe by using a sterile Pasteur pipette, and passed through a sterile 2-μm-pore-size syringe filter to remove most cellular debris. Rickettsiae were pelleted by centrifugation at 4°C, 13,600 × g, for 5 min, washed twice with 1.5 ml 4°C 300 mM sucrose, resuspended in 100 μl ice-cold 300 mM sucrose (50 μl per reaction mixture), and held on ice. Three micrograms of endotoxin-free plasmid DNA was added to each 50-μl aliquot of cell-free rickettsiae, transferred to a chilled 0.1-cm gap cuvette (Bio-Rad, Hercules, CA), incubated on ice for 15 min, and pulsed once at 1.8 kV, 200 Ω, and 25 μF in a Gene Pulser II electroporation system (Bio-Rad). ISE6 cells from a nearly confluent 25-cm2 flask in 1.5 ml medium were used to recover the electroporation mix into a sterile 2-ml tube. The ISE6-rickettsiae mix was centrifuged at room temperature at 700 × g for 2 min to pellet the ISE6 cells and then at 13,600 × g for 1 min to sediment the rickettsiae onto the cells. Rickettsiae were left in contact with the cells at room temperature for 15 min, transferred to a 25-cm2 flask containing 3.5 ml medium, and incubated at 32°C for 24 h before adding 100 μg/ml spectinomycin. If needed, antibiotic concentrations were increased to 200 to 500 μg/ml spectinomycin and streptomycin to control nonspecifically resistant wild-type rickettsiae, and transformants were maintained at 100 μg/ml spectinomycin and streptomycin. Nonspecific antibiotic resistance induced under selection in vitro can develop with the genus Rickettsia (17, 18). To help reduce this in the bacteria examined here, we used double selection (streptomycin and spectinomycin), and transformed rickettsiae were passaged no more than eight times, at which point a new culture was regenerated from stock vials preserved in liquid nitrogen.
To visualize rickettsial actin tails in live cell cultures, Vero cells transfected to express mCherry-fluorescent Lifeact were used (19). They were grown in RPMI medium supplemented with 10% fetal bovine serum and 4 mM l-glutamine at 37°C in a humidified atmosphere of air with 5% CO2 and were subcultured weekly.
L15C cell culture medium formulation.
L15C medium is a modification of L15B medium, previously described in detail (20). Alterations were made to the following reagent concentrations: l-aspartic acid at 0.4485 g/liter, l-glutamine at 0.5 g/liter, l-proline at 0.45 g/liter, l-glutamic acid at 0.25 g/liter, α-ketoglutaric acid at 0.4485 g/liter, d-glucose at 18.018 g/liter, 5 mM NaOH. Prior to use in supplemented medium, L15C was diluted by the addition of 33% water by volume (21). Two supplemented L15C media were used, one for uninfected cells containing 5% fetal bovine serum (FBS; Benchmark), 5% tryptose phosphate broth (TPB; Difco), and 0.1% lipoprotein concentrate (LPC; MP Biomedicals), the other for Rickettsia-infected cells with 10% FBS, TPB, and LPC as above, plus 25 mM HEPES buffer and 0.25% NaHCO3, as previously described, except L15C replaced L15B (21, 22). The pH was adjusted to pH 7.5 by using 1 M NaOH.
Construction of rickettsial shuttle vector.
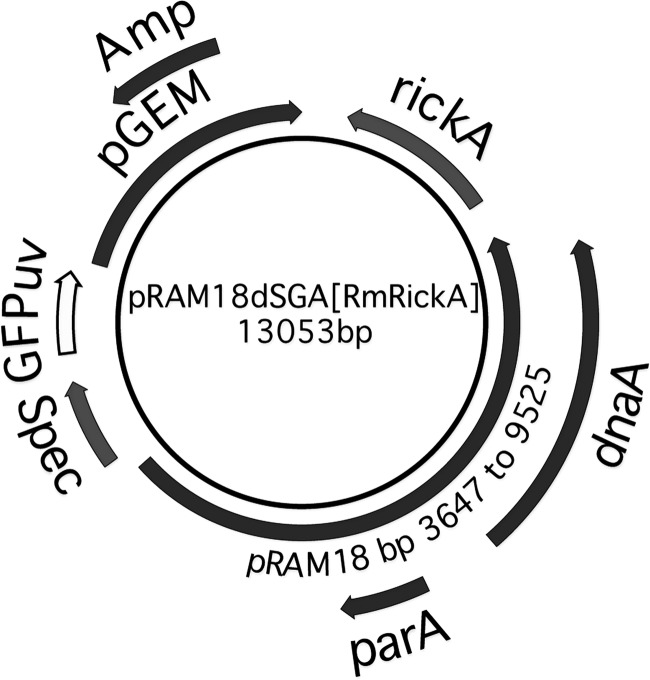
We used the Rickettsia amblyommii AaR/Sc pRAM18-based shuttle vector pRAM18dRGA[MCS], which carries a multiple-cloning site (MCS) (23), and replaced the rifampin resistance marker with a gene carrying genes for spectinomycin and streptomycin resistance, aadA. The functional R. monacensis rickA gene was amplified using wild-type R. monacensis genomic DNA as the template with primers RmonaRickAF2 (5′-ATACGAGCTAGCCATTTTGAAAAACACGGCTGG-3′) and RmonaRickAR2 (5′-GTATACGCTAGCATTGTATGATTTGGGTTGGGCG-3′). The resulting PCR product was digested with the NheI restriction enzyme and inserted into the MCS at the NheI restriction site, creating pRAM18dSGA[RmRickA] (Fig. 1).
FIG 1.
Map of the pRAM18dSGA[RmRickA] shuttle vector. The shuttle vector pRAM18dSGA[RmRIckA] was constructed using the pGEM vector backbone (Promega, Madison, WI), which is selectable by ampicillin (Amp) and thus enables production of pRAM18dSGA[RmRickA] shuttle vectors in Escherichia coli. It includes the region containing the parA and dnaA genes of the R. amblyommii plasmid pRAM18 and the intervening sequence (23), and it carries the genes for spectinomycin and streptomycin resistance (labeled Spec) through insertion of the aadA gene and a GFPuv reporter, both driven by ompA promoters. The rickA gene from R. monacensis, along with the region 5′ of the gene, were PCR amplified and inserted into the multiple-cloning site at the Nhel restriction site.
Preparation of rickettsial genomic DNA and RNA.
Equivalent ISE6 cell layers were infected with a 100-fold dilution of a suspension of wild-type R. bellii (WT), R. bellii pRAM18dSGA[MCS] (R. belliiMCS), or R. bellii pRAM18dSGA[RmRickA] (R. belliiRmRickA), creating six replicates for each R. bellii type. Rickettsiae were propagated in ISE6 tick cells and used when cultures were 100% infected and the majority of cells had lysed. Cultures were harvested simultaneously after 3 days, when cell layers were lifting off the flask, indicating maximal infection. Purified bacterial pellets were prepared as described above, except pellets were washed once in SPG medium (24) and then resuspended in either 600 μl cell lysis solution at room temperature or in 300 μl SPG and frozen at −80°C. Rickettsial genomic DNA was prepared with the Puregene Core A kit (Qiagen, Valencia, CA) as per the manufacturer's Gram-negative bacteria protocol. For RNA preparation, pelleted rickettsiae were resuspended in 1 ml TRI reagent (Sigma, St. Louis, MO), transferred to Phase Lock gel (PLG) Heavy 2-ml tubes (5′ Prime, Gaithersburg, MD), and isolated using the manufacturer's protocol for suspension cells and RNA isolation. RNA pellets were dissolved in 50 μl RNase-free elution buffer (Stratagene, La Jolla, CA). RNA was treated with DNase I twice using the concentrated RNA protocol for the Turbo DNA kit (Ambion, Grand Island, NY) and purified with the RNA Clean and Concentrator kit (Zymo Research, Irvine, CA). RNA and DNA were quantified by spectrophotometry with a BioPhotometer (Eppendorf, Hamburg, Germany).
Determination of shuttle vector copy number and expression of native and shuttle vector-conferred rickA.
Rickettsia monacensis or R. bellii rickA-specific primers were tested for specificity at an annealing temperature of 58°C and used to create plasmids for generation of species-specific standard curves and for quantitative PCR (qPCR) and quantitative reverse transcription-PCR (qRT-PCR). Primer sequences were as follows: qRmRickAF2 (5′-CTACTATGGCTCCGGTTCAG-3′), qRmRickAR2 (5′-CATCTCCATAGCAACTCTTCG-3′), qRbRickAF2 (5′-TACGCCACTCCCTGTGTCA-3′), and qRbRickAR (5′-GATGTAACGGTATTATCACCAACAG-3′). Plasmid copy number was determined by qPCR, by comparing R. monacensis or R. bellii rickA to chromosomal gltA as previously described (25) using Brilliant II SYBR green QPCR master mix (Stratagene) with 200 nM each primer and 1 ng of genomic DNA. The relative expression of R. monacensis versus R. bellii rickA was determined by qRT-PCR using 1-step Brilliant II SYBR green QRT-PCR master mix (Stratagene, La Jolla, CA) with 150 nM each primer and 100 ng of RNA. The equation 2−(CT of target gene − CT of reference gene) (CT is the threshold cycle) was used to determine the expression levels of the rickA gene in comparison to gltA.
Binding of rickettsiae to host cells.
R. belliiMCS and R. belliiRmRickA were microscopically examined for binding to Lifeact mCherry-expressing Vero host cells at 1, 2, 4, and 8 h postinoculation (h p.i.). Transformed cells produced fluorescent Lifeact protein, which labels filamentous actin, allowing it to be directly visualized without affecting normal actin function (26). Twenty-four hours prior to inoculation with rickettsiae, 35-mm glass-bottom dishes (P35G-1.5-14-C; Mattek Corporation, Ashland, MA) were seeded with Vero cells. Cells were quantified by counting cells in 15 visual fields and extrapolating cell numbers to the seeded areas of the dishes. Rickettsiae were quantified using a Petroff-Hausser counting chamber. Two hundred bacteria per Vero cell were added to 100 μl of medium placed in the internal well of the glass-bottom dish. Dishes were rocked gently for 2 min, then incubated at 34°C, 5% CO2 until observation.
At the time points of interest, the samples were rinsed with medium to remove unbound bacteria, and whole cells were imaged as a series of confocal slices at 0.5-μm increments by using the equipment described in “Microscopy and imaging,” below. Attached rickettsiae were counted from maximum intensity z-projections of the stacked cell images. The proportion of bacteria bound of the initial 200 bacteria inoculated per cell was calculated. The number of cells observed in this manner ranged from 11 to 22, depending on the number of cell images captured within a 10-min imaging window.
Proportion of motile rickettsiae.
The proportion of bacteria exhibiting actin-based motility in each individual Vero cell was calculated for both R. bellii transformants at 1, 2, 4, 8, 12, 16, and 24 h p.i. in a manner similar to that for the binding assay, described above. Samples were inoculated, rocked, incubated, rinsed, and imaged as for the binding assays. Confocal microscope-generated image stacks and z-projections were examined to determine the total number of rickettsiae present and the number of those rickettsiae exhibiting fluorescent actin tails. At 1, 2, 4, and 8 h p.i., total bacterial numbers were small enough that all bacteria on and in each observed cell were counted. At 12, 16, and 24 h p.i., rickettsial reproduction rendered whole-cell bacteria numbers too large to count, so bacteria on the focal planes associated with the uppermost and lowermost membranes of the host cell were excluded.
Rickettsial velocity and path curvature.
Mean velocity and path curvature were calculated for mobile R. belliiMCS and R. belliiRmRickA at 23°C and 34°C. Videos of motile rickettsiae were captured, and segments illustrating 10 frames of movement were overlaid as a four-dimensional (4D) projection by using the materials described in “Microscopy and imaging,” below. The total distance traveled by a bacterium within this projected segment was measured and divided by the time elapsed to determine velocity (Fig. 2E). Examples of videos analyzed to determine rickettsial velocity and path curvature are included in the supplemental material.
FIG 2.
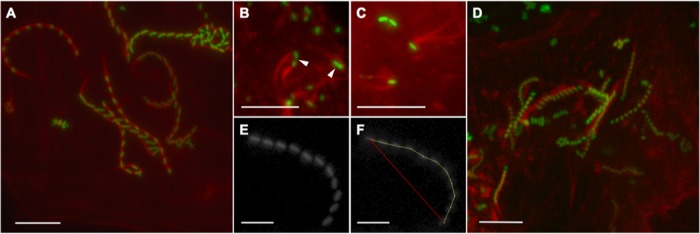
R. bellii transformants undergoing actin-based motility. Time-lapse micrographs in panels A and D to F were compiled as t-projections. (A) R. belliiMCS at 34°C. (B) Dividing rickettsiae may present actin tails on both joined bacteria, resulting in spinning. Measurements from dividing bacteria were not included in mean values of velocity or path curvature. Dividing rickettsiae are indicated by white arrowheads. (C) Twinned tails on single bacteria were occasionally visible, in this case on R. belliiMCS at 34°C. The red channel gamma value of this image was adjusted to accentuate the brightness of the actin tails against the background. (D) The R. belliiRmRickA mean velocity at 23°C is just over half that of the rickettsiae pictured in panel A. (E) Green channel fluorescent 10-movement t-stack of an R. belliiMCS variant at 34°C. (F) The t-stack pictured in panel E was measured to determine the velocity and path curvature. The path length (yellow line) was 43.7 μm and the elapsed time of the 10-movement stack was 118.25 s, yielding a velocity of 0.37 μm/s. The absolute distance traveled over the course of the t-stack was 37.2 μm, as indicated by the red line. Division of the absolute distance traveled (37.2 μm) by the path length (43.7 μm) yielded a path curvature value of 0.8513, which is very close to the mean value for R. belliiMCS at 34°C. Bars, 10 μm.
A path curvature value was calculated from the same 4D projection segments. For each segment, the distance traveled by a bacterium was measured, as was the absolute straight line distance between the first and last frames of the projected image. Path curvature was calculated by dividing the total distance traveled by the absolute distance traveled, to produce a fraction of 1. A path curvature value of 1 therefore indicated a completely straight path, with progressively lower values indicating progressively more curved paths of travel (2).
Rickettsiae impacting the inner face of the host cell membrane most often reflected off it at an angle of reflection roughly corresponding to the angle of incidence. Rickettsiae closely approaching the edge of a host cell were also observed to slow as less vertical space between the upper and lower membranes was available. To avoid measuring path curvature values caused by collision with the cells' outer membrane and reflection off it, as well as slowing velocity due to cell edge approach, bacterial path segments within 2 μm of the cell edge were discarded from the analysis. Actively dividing rickettsiae were also excluded from measurement, because they tended to follow more erratic and spiraling paths as the two actin tails of the dividing bacteria struggled against each other (Fig. 2B; see also Movie S1 in the supplemental material).
Plaque assays.
To evaluate the rate of spread of transformants R. belliiMCS and R. belliiRmRickA through a cell layer, plaque assays were performed (using a method adapted from that described in reference 27). In addition to the two transformant strains, plaque assays were also performed on wild-type R. bellii 369C. Six-well plates were seeded with 90% confluent red fluorescent Lifeact-expressing Vero cells and allowed to grow overnight. The next day, cell-free rickettsiae were prepared and serially diluted from 1 × 10−2 to 1 × 10−7, and 400 μl of each dilution was added to two wells per replication. The 6-well plates were incubated for 2 h at 32°C to allow the rickettsiae to settle and bind to the host cells. Wells were then filled with 1 ml 0.5% agarose dissolved in supplemented RPMI medium to eliminate infection of cells by drifting, extracellular rickettsiae. Over the agarose layer, 2.5 ml of growth medium was added. The plates were then incubated at 32°C and 5% CO2 in air for 10 days to allow plaques to form.
On the 10th day, the medium was removed from the agar overlay, and a second layer consisting of 0.5% agarose with 0.01% neutral red stain was applied. Neutral red is a vital stain, staining only the living cells that endocytose it and leaving the dead cells in the plaque unstained. The plates were then incubated for a further 24 h to allow the living cells to take up the stain.
After staining, the plates were examined, and distinct, nonoverlapping plaques were imaged by using a dissecting microscope. Plaque areas for both transformants and WT were delineated and measured using ImageJ (U.S. National Institutes of Health).
Microscopy and imaging.
Live R. bellii-infected Lifeact mCherry-expressing Vero cells were observed microscopically for a variety of assays as detailed below. Images were obtained using an BX61 DSU spinning disk confocal microscope (Olympus America, Center Valley, PA), a Quantem:512SC electron multiplying charge-coupled-device (EMCCD) camera (Photometrics, Tucson, AZ), and an X-cite Exacte fluorescent light source (Lumen Dynamics, Mississauga, Ontario, Canada). Olympus UPlanSApo 60× (numerical aperture [NA], 1.35) and 100× (NA, 1.40) objectives were used to obtain all images. For the measurements of velocity and path curvature at 34°C, a TU 2000 inverted microscope (Nikon Instruments Inc., Melville, NY) equipped with a Photometrics Cascade EMCCD camera and enclosed in a heat-regulated environmental chamber was used. Images were captured using Metamorph software (Molecular Devices, Sunnyvale, CA), and ImageJ (U.S. National Institutes of Health) was used for still-image z-projections, 4D projections, cropping, adjustment of brightness/contrast, and analytical measurement.
Statistics.
Mean values were assessed by using a two-tailed Student's t test to determine significance. Unless otherwise indicated, the precision of the mean is represented by the standard error (SE).
RESULTS
Shuttle vector copy numbers and expression of native and shuttle vector-conferred rickA.
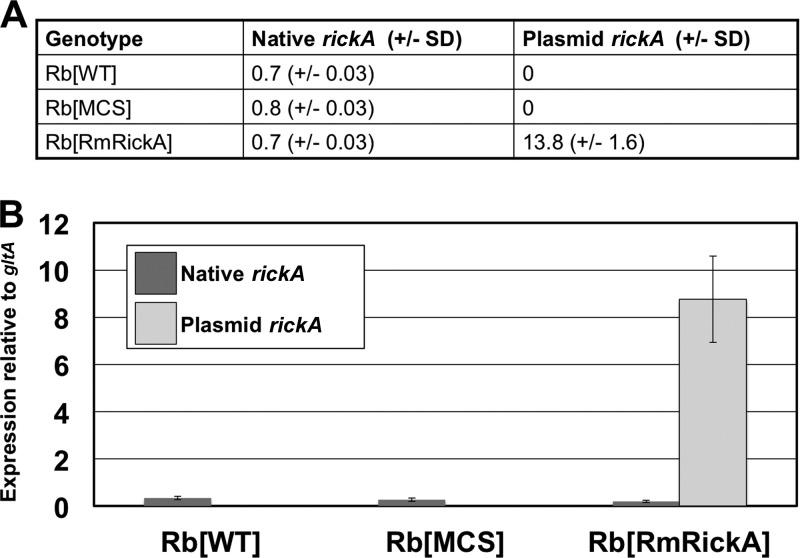
The ratio of R. monacensis or R. bellii rickA to chromosome-encoded gltA was used to estimate the relative number of copies of shuttle vector-carried R. monacensis rickA and native R. bellii rickA. Native R. bellii rickA had approximately a 1-to-1 ratio to gltA, as would be expected; shuttle vector-carried R. monacensis rickA had 13.8 copies per copy of gltA (Fig. 3A). The genome of R. bellii 369C has been sequenced and is known to encode only a single copy of gltA (NCBI gene ID 3996133). Expression of R. bellii rickA was evaluated with respect to the expression of gltA, as well. gltA was selected for this purpose because it is consistently expressed and had been used previously in a similar context for Rickettsia (23, 28, 29). The three R. bellii variants had the following rates of native rickA expression: WT, 0.33; R. belliiMCS, 0.26; R. belliiRmRickA, 0.20. As expected, R. monacensis rickA was highly expressed only in R. belliiRmRickA transformant (8.75 relative to gltA) (Fig. 3B).
FIG 3.
Relative copy number and expression of native and plasmid-associated rickA in transformed and wild-type R. bellii. Plasmid rickA was derived from R. monacensis, which allowed for differentiation of plasmid sequence from the native sequence. (A) Numbers of genetic copies of each type of rickA, normalized to the number of gltA copies. (B) Expression of native and plasmid rickA relative to gltA expression. Plasmid-derived rickA was only present above the threshold of detection in R. belliiRmRickA. Relative gene copy number and expression of native rickA were comparable between all genotypes of R. bellii. Error bars represent standard errors.
Binding.
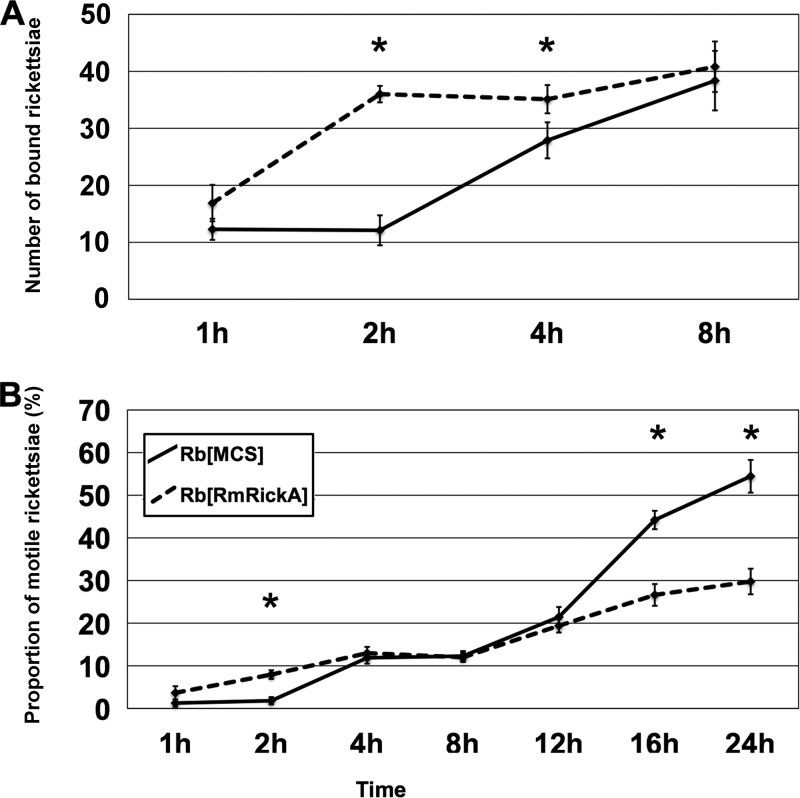
The proportion of bacteria bound per Vero cell was calculated for 1, 2, 4, and 8 h p.i. for both R. belliiMCS and R. belliiRmRickA (Fig. 4). A strong significant difference in the proportion of bound bacteria between the two transformants was apparent at 2 h p.i. and 4 h p.i. (P < 0.00001 and P = 0.03344, respectively). By 8 h p.i., the proportion of bound bacteria was again comparable between groups. These results indicate that most R. belliiRmRickA binding occurred between 1 and 2 h p.i., while binding of R. belliiMCS occurred later, between 2 and 4 h p.i. Relative to R. belliiMCS, R. belliiRmRickA exhibited an accelerated binding rate, but by 8 h p.i. approximately equal proportions of bacteria were bound.
FIG 4.
Change in the proportion of bound and motile transformant rickettsiae over time. Asterisks indicate significant differences between the transformants at the time points marked. (A) At 2 h p.i. the greatest difference between the binding of transformants was observed, as three times more R. belliiRmRickA adhered to the host cells than R. belliiMCS. Rickettsiae were inoculated at a rate of 200 bacteria per Vero cell. (B) A significant difference in the proportion of motile bacteria was also present at 2 h p.i., corresponding to the disparity in binding observed at that time point. The proportion of motile rickettsiae increased through time until, beginning at 16 h p.i., the proportion of motile R. belliiMCS was substantially higher than the proportion of motile R. belliiRmRickA. Error bars show standard errors.
Proportion of motile rickettsiae.
The proportions of motile bacteria were determined for both R. bellii transformants at 1, 2, 4, 8, 12, 16, and 24 h p.i. (Fig. 4). A significant difference was present earliest between the transformants at 2 h p.i. (P < 0.00001), when R. belliiRmRickA showed an increase in the proportion of motile bacteria relative to R. belliiMCS, likely due to the substantially earlier binding of R. belliiRmRickA, as previously noted. After the proportion of bound R. belliiMCS converged with that of R. belliiRmRickA at around 4 h p.i., a concomitant increase in the proportion of motile R. belliiMCS was also observed. From 4 h p.i. to 12 h p.i., no difference in the proportion of motile rickettsiae was notable. Beginning at 16 h p.i., however, and continuing thereafter, the proportion of R. belliiMCS greatly increased relative to R. belliiRmRickA. At the final time point observed, 24 h p.i., 54% of R. belliiMCS within the host cells were motile, compared with a maximum motility proportion of only 29.8% for R. belliiRmRickA, demonstrating a strong statistical difference (P = 0.00094).
Rickettsial velocity and path curvature.
Considerable qualitative differences were notable between the movement patterns of R. belliiMCS and R. belliiRmRickA. R. belliiMCS demonstrated a higher degree of homogeneity in movement velocity and path curvature than the movement patterns of R. belliiRmRickA, which were more erratic and varied, particularly at 34°C, as indicated by the higher standard error values (described below).
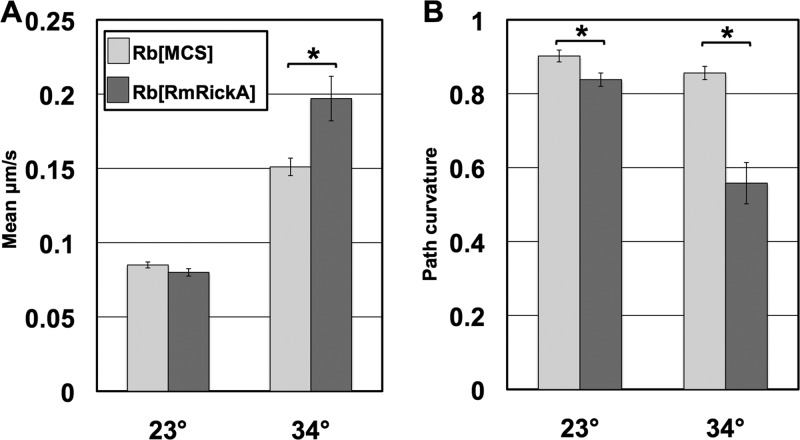
Motility characteristics of individual rickettsiae were measured and compared between R. belliiMCS and R. belliiRmRickA at 23°C and 34°C (Fig. 5). At 23°C, differences in motility between the transformants were relatively muted. Statistical comparison showed no significant difference in velocity at this temperature (P = 0.1236). Mean path curvature varied significantly between the transformants at 23°C, with R. belliiRmRickA following a more curved path than R. belliiMCS (P = 0.0114). At 34°C, observed differences in both mean velocity and mean path curvature were amplified. The mean velocity of R. belliiRmRickA was significantly higher than that of R. belliiMCS (P < 0.00001). The increased variability of observed motility behavior was reflected in the markedly higher standard error value for R. belliiRmRickA measurements. Path curvature also varied more substantially between the two R. bellii transformants at 34°C. The path traveled by R. belliiMCS was relatively straight, with a path curvature value of 0.856 (SE, 0.018), while the path curvature value of R. belliiRmRickA was 0.558 (SE, 0.056). Again, the qualitative variability observed in R. belliiRmRickA was reflected in the markedly higher standard error of its mean path curvature. A statistically significant difference was also notable in the mean path curvatures of the two transformants (P < 0.00001).
FIG 5.
Comparison of mean velocity and mean path curvature values of the rickettsial transformants. Asterisks indicate significant differences between transformants for the same temperature and phenotype observed. (A) Comparisons of mean velocities of R. bellii transformants. Results showed a substantial increase in the mean velocity of both groups at 34°C. (B) Mean path curvature values of transformants. As a path curvature value approaches 1, the straighter the path the bacteria have followed. At 23°C, the mean path of R. belliiRmRickA was marginally, though significantly, more curved than that of R. belliiMCS. At 34°C, however, the path curvature value was much lower in R. belliiRmRickA, corresponding to more curved and convoluted paths. The exacerbation of phenotypic differences at 34°C relative to 23°C indicates the temperature dependence of the response. Error bars show standard errors.
Prior to 4 h p.i., rickettsiae may still be adhering to, and invading, the host cell, as described above for binding. After 4 h p.i., the rickettsial infection of Vero cells progressed to a stage demonstrating typical motility patterns, during which measurements of velocity and path curvature were taken. With either of the R. bellii transformant variants, there were no significant differences in velocity or path curvature between replications at any time points from 4 h p.i. to 28 h p.i. Once normal rickettsial motility had begun at 4 h p.i., no changes in these characteristics occurred through 28 h p.i., the latest time point examined.
Plaque assays.
Plaque areas were examined and measured for R. belliiMCS, R. belliiRmRickA, and WT. Both transformants produced relatively clear plaques in which dead Vero cells failed to endocytose the neutral red stain. Unlike the two transformants, the WT did not produce distinct plaques, although regions of infected cells were visible as areas of reduced neutral red absorption. The WT-infected cell regions were substantially larger than the plaques of R. belliiMCS and R. belliiRmRickA and showed little cellular mortality. Figure 6 shows representative examples of plaques formed by all three types of R. bellii examined. Because the WT did not produce distinct plaques, direct statistical comparison was inappropriate, but the data describing the mean plaque areas were still included in the analysis.
FIG 6.
Representative plaques produced by transformant and wild-type R. bellii. The area of infection formed by the WT is visible as a region with less stain uptake into cells, rather than as a plaque. Because plaques were visualized with the vital stain neutral red, dead cells were unable to endocytose stain. The lighter region associated with the WT demonstrated that the infection did not kill the host cells within the 10-day incubation period, though the lighter color may indicate reduced cell viability.
The mean plaque area for R. belliiMCS was 1.8 mm2 (SE, 0.102), and for R. belliiRmRickA it was 0.84 mm2 (SE, 0.086). The strongly significant difference (P < 0.0001) supports a slower rate of spread for R. belliiRmRickA through the cell layer than was observed for R. belliiMCS. The visible mean area of non-plaque-forming infection of the WT was 2.7 mm2 (SE, 0.299), demonstrating a large infection area but one with a high degree of variability.
DISCUSSION
We examined phenotypic differences between R. bellii modified to overexpress R. monacensis-derived RickA (R. belliiRmRickA), a multiple-cloning-site-modified R. bellii control (R. belliiMCS) and, where feasible, a nonfluorescent wild-type R. bellii isolate. These differences were investigated to help elucidate the function of RickA in relation to rickettsial host cell binding, internalization, and intracellular motility. As described in the results above, numerous notable and statistically significant phenotypic differences were observed.
It has been demonstrated previously that alteration of suspected motility-associated proteins may affect plaque formation in Rickettsia. Plaque assays of R. rickettsii Sca2-defective mutants demonstrated reduced plaque areas and a lack of actin tails, although cell-to-cell movement of bacteria must have occurred for visible plaques to have formed (30). In our experiments, plaques formed by R. belliiRmRickA were significantly smaller than those produced by R. belliiMCS, indicating that overexpression of rickA reduced the rate at which the rickettsiae traveled through the cell layer. The mechanism of cell-to-cell movement of Rickettsia has not been resolved, and further study is necessary to ascertain what, if any, role RickA has in the process. Possibly, expression of the heterologous RickA resulted in enhanced adhesion of R. belliiRmRickA to the host cell membrane. In the absence of regulatory sequences needed to downregulate RickA later on, host cell entry could have been delayed, resulting in reduced plaque size. Sequences that regulate expression of rickA have not been identified in Rickettsiaceae, and the exact mechanisms whereby RickA influences infection of new host cells (i.e., cell-to-cell spread) remain to be elucidated. Plaque formation by wild-type R. bellii seemed categorically different than that observed in the transformant strains. The transformants produced relatively well-defined plaques that were comparable in form and easily measurable. The wild-type bacteria did not produce cleared areas in which the dead cells were entirely unable to take up the vital neural red stain. Instead, the extent of bacterial spread through the cell layer was visible as a region in which less stain was endocytosed but in which cells still clearly survived. One explanation is that the transformant R. bellii varieties demonstrated greater toxicity to the Vero cells due to either the genetic transformation itself or to a difference in growth density within the host cells. In any case, experimental data regarding the wild-type bacteria have been included for comparison.
The substantially more rapid binding of R. belliiRmRickA to host cells compared to R. belliiMCS (Fig. 4) indicated a possible role of RickA in cell membrane adhesion. Interestingly, a nucleation-promoting factor produced by mouse embryonic fibroblasts, WASH (a subclass of the WASP family), was recently demonstrated to affect cell adhesion and migration by regulating endocytic trafficking of α5-integrin (31). This finding, together with the accelerated adhesion of R. belliiRmRickA, indicates that RickA may be important for infiltration through the host cell membrane.
The proportions of both transformants that developed an actin tail and demonstrated actin-based motility increased over time as the infection progressed. The much lower proportion of motile R. belliiMCS versus motile R. belliiRmRickA at 16 h p.i. and 24 h p.i. suggests that overexpression of RickA negatively impacts normal motility. A potential mechanism for this disruption may be distilled from the literature. Like the cell-produced WASH protein and other related nucleation-promoting factors, RickA has a WCA domain (which includes the WASP homology 2 domain, central domain, and acidic domain, hence, WCA) that is responsible for binding to, and activating, the cellular Arp2/3 complex (11). In PC12 cells transfected to overexpress the WASH-WCA domain, the normal structure of the actin cytoskeleton is disrupted and actin colocalizes with the nucleation-promoting factor (32). Similar disruptions of the normal actin cytoskeleton were also observed in cells overexpressing the WCA regions of other nucleation-promoting factors, such as WASP and Scar1, inhibiting lamellipodia formation and stress fiber polymerization (33). Although the quantities of RickA produced by R. belliiRmRickA are certainly smaller than the quantities of these proteins endogenously overexpressed by the transfected cells, it is possible that hyperphysiologic quantities of RickA may impact normal Arp2/3 function, at least in the immediate vicinity of the transformed rickettsiae, disrupting actin tail formation.
Previous examination of rickettsial velocity demonstrated substantial differences in the velocities R. rickettsii and R. typhi (2). Our results indicated that phenotypic differences in motility were more apparent in transformants of a single species that expressed heterologous RickA (R. belliiRmRickA) or only marker proteins (R. belliiMCS) when infecting Vero cells at 34°C rather than 23°C. The velocity of R. bellii at 23°C was comparable with values previously published for R. rickettsii at room temperature (2) but it was higher at 34°C, indicating a significant temperature-dependent response. An unexpected result of these experiments was that the mean velocity of motile R. belliiRmRickA was actually higher than that of R. belliiMCS, providing further indication that RickA plays a direct role in actin-based motility. The increased curvature and erratic nature of the path followed by R. belliiRmRickA could mean that overexpressed RickA, while providing increased velocity, interferes with the directionality of the actin polymerization process of R. bellii as portrayed by R. belliiMCS.
In conclusion, R. bellii transformed to overexpress R. monacensis RickA demonstrated substantial phenotypic differences from the control transformant bearing an empty multiple-cloning site. These differences suggested that RickA may mediate enhanced rickettsial adhesion and invasion of host cells and is possibly also involved in orderly assembly of actin tails in wild-type rickettsiae. This is supported by our observation that cell-free rickettsiae overexpressing RickA adhered more rapidly and were internalized, or initiated motility, earlier than controls but traveled more erratically within cells. Taken together, these results indicate that an overabundance of RickA accelerates binding to host cells while interfering with normal actin-based motility.
Supplementary Material
ACKNOWLEDGMENTS
Funding for these experiments was provided by National Institutes of Health grants R01 AI049424 and R01 AI081690.
We thank Mike Herron for helping to develop the live imaging microscopy techniques and providing technical advice.
Published ahead of print 2 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03352-13.
REFERENCES
- 1.Van Kirk LS, Hayes SF, Heinzen RA. 2000. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect. Immun. 68:4706–4713. 10.1086/IAI.68.8.4706-4716.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinzen RA. 2003. Rickettsial actin-based motility: behavior and involvement of cytoskeletal regulators. Ann. N. Y. Acad. Sci. 990:535–547. 10.1111/1749-6632.2003.tb07424.x [DOI] [PubMed] [Google Scholar]
- 3.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. 2010. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe 7:388–398. 10.1016/j.chom.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch MD, Reed SCO, Lamason RL, Serio AW. 2012. Expression of an epitope-tagged virulence protein in Rickettsia parkeri using transposon insertion. PLoS One 7(5):e37310. 10.1371/journal.pone.0037310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouin E, Welch MD, Cossart P. 2005. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8:35–45 http://dx.doi.org/10.1016/j.mib.2004.12.013 [DOI] [PubMed] [Google Scholar]
- 6.Ireton K. 2013. Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol. 3:130079. 10.1098/rsob.130079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snapper SB, Rosen FS. 1999. The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annu. Rev. Immunol. 17:905–929. 10.1146/annurev.immunol.17.1.905 [DOI] [PubMed] [Google Scholar]
- 8.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521–531. 10.1016/0092-8674(92)90188-I [DOI] [PubMed] [Google Scholar]
- 9.Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wächter M, Wuenscher M, Chakraborty T. 1992. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11:1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, Li R, Cossart P. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457–461. 10.1038/nature02318 [DOI] [PubMed] [Google Scholar]
- 11.Jeng RL, Goley ED, D'Alessio JA, Chaga OY, Svitkina TM, Borisy GG, Heinzen RA, Welch MD. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell. Microbiol. 6:761–769. 10.1111/j.1462-5822.2004.00402.x [DOI] [PubMed] [Google Scholar]
- 12.Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. 2010. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12:1057–1063. 10.1038/ncb2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simser JA, Rahman MS, Dreher-Lesnick SM, Azad AF. 2005. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol. Microbiol. 58:71–79. 10.1111/j.1365-2958.2005.04806.x [DOI] [PubMed] [Google Scholar]
- 14.Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4(12):e8361. 10.1371/journal.pone.0008361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simser JA, Palmer AT, Fingerle V, Wilske B, Kurtti TJ, Munderloh UG. 2002. Rickettsia monacensis sp. nov., a spotted fever group Rickettsia, from ticks (Ixodes ricinus) collected in a European city park. Appl. Environ. Microbiol. 68:4559–4566. 10.1128/AEM.68.9.4559-4566.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 80:533–543. 10.2307/3283188 [DOI] [PubMed] [Google Scholar]
- 17.Rachek LI, Tucker AM, Winkler HH, Wood DO. 1998. Transformation of Rickettsia prowazekii to rifampin resistance. J. Bacteriol. 180:2118–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drancourt M, Raoult D. 1999. Characterization of mutations in the rpoB gene in naturally rifampin-resistant Rickettsia species. Antimicrob. Agents Chemother. 43:2400–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtti TJ, Mattila JT, Herron MJ, Felsheim RF, Baldridge GD, Burkhardt NY, Blazar BR, Hackett PB, Meyer JM, Munderloh UG. 2008. Transgene expression and silencing in a tick cell line: a model system for functional tick genomics. Insect Biochem. Mol. Biol. 38:963–968 http://dx.doi.org/10.1016/j.ibmb.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munderloh UG, Kurtti TJ. 1989. Formulation of medium for tick cell culture. Exp. Appl. Acarol. 7:219–229 [DOI] [PubMed] [Google Scholar]
- 21.Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37:2518–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattila JT, Burkhardt NY, Hutcheson HJ, Munderloh UG, Kurtti TJ. 2007. Isolation of cell lines and a rickettsial endosymbiont from the soft tick Carios capensis (Acari: Argasidae: Ornithodorinae). J. Med. Entomol. 44:1091–1101. 10.1603/0022-2585(2007)44[1091:IOCLAA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 23.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG. 2011. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One 6(12):e29511. 10.1371/journal.pone.0029511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin A, Tucker AM, Hines A, Wood DO. 2004. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 70:2816–2822. 10.1128/AEM.70.5.2816-2822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldridge GD, Burkhardt NY, Labruna MB, Pacheco RC, Paddock CD, Williamson PC, Billingsley PM, Felsheim RF, Kurtti TJ, Munderloh UG. 2010. Wide dispersal and possible multiple origins of low-copy-number plasmids in Rickettsia species associated with blood-feeding arthropods. Appl. Environ. Microbiol. 76:1718–1731. 10.1128/AEM.02988-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ammerman NC, Beier-Sexton M, Azad AF. 2008. Laboratory maintenance of Rickettsia rickettsii. Curr. Protoc. Microbiol. Chapter 3:Unit 3A.5. 10.1002/9780471729259.mc03a05s11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai J, Winkler HH. 1996. Transcriptional regulation in the obligate intracytoplasmic bacterium Rickettsia prowazekii. J. Bacteriol. 178:5543–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai J, Winkler HH. 1997. Transcriptional regulation of the gltA and TLC genes in Rickettsia prowazekii growing in a respiration-deficient host cell. Acta Virol. 41:285–288 [PubMed] [Google Scholar]
- 30.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 78:2240–2247. 10.1128/IAI.00100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duleh SN, Welch MD. 2012. Regulation of integrin trafficking, cell adhesion, and cell migration by WASH and the Arp2/3 complex. Cytoskeleton (Hoboken) 69:1047–1058. 10.1002/cm.21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monfregola J, Napolitano G, D'Urso M, Lappalainen P, Ursini MV. 2010. Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J. Biol. Chem. 285:16951–16957. 10.1047/jbc.109.078501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machesky LM, Insall RH. 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347–1356 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.