Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) in combination with associated sequences (cas) constitute the CRISPR-Cas immune system, which uptakes DNA from invasive genetic elements as novel “spacers” that provide a genetic record of immunization events. We investigated the potential of CRISPR-based genotyping of Lactobacillus buchneri, a species relevant for commercial silage, bioethanol, and vegetable fermentations. Upon investigating the occurrence and diversity of CRISPR-Cas systems in Lactobacillus buchneri genomes, we observed a ubiquitous occurrence of CRISPR arrays containing a 36-nucleotide (nt) type II-A CRISPR locus adjacent to four cas genes, including the universal cas1 and cas2 genes and the type II signature gene cas9. Comparative analysis of CRISPR spacer content in 26 L. buchneri pickle fermentation isolates associated with spoilage revealed 10 unique locus genotypes that contained between 9 and 29 variable spacers. We observed a set of conserved spacers at the ancestral end, reflecting a common origin, as well as leader-end polymorphisms, reflecting recent divergence. Some of these spacers showed perfect identity with phage sequences, and many spacers showed homology to Lactobacillus plasmid sequences. Following a comparative analysis of sequences immediately flanking protospacers that matched CRISPR spacers, we identified a novel putative protospacer-adjacent motif (PAM), 5′-AAAA-3′. Overall, these findings suggest that type II-A CRISPR-Cas systems are valuable for genotyping of L. buchneri.
INTRODUCTION
The pickle industry relies on the use of naturally occurring bacteria for the fermentation of cucumbers in large industrial tanks (1). To control the diverse microbiota naturally associated with pickles and to preclude spoilage by undesirable microorganisms, salting and brining are implemented in industrial settings. Unfortunately, acid- and halotolerant lactic acid bacteria often contaminate the pickling process, resulting in a secondary fermentation, which spoils the product by generating undesirable attributes (1). Among commonly encountered bacterial contaminants, Lactobacillus buchneri has repeatedly been associated with spoilage of fermenting pickles (1, 2). Recent advances in genome sequencing of this species have shed light on the molecular underpinnings that allow L. buchneri to withstand the pickling process. In particular, determining the complete genome sequences of strains NRRL B-30929 and CD034 (3–5) established several genetic loci for substrate utilization pathways (notably, those for lactate and carbohydrates), including the ability to convert lactic acid into acetic acid (5) and 1,2-propanediol (2). Conversely, the biochemical properties of this robust bacterium have been exploited for silage inoculation to control yeast and mold growth under anaerobic conditions during the fermentation of corn, barley, wheat, and other grains into animal fodder (5–7).
Bacteria used in industrial settings for fermentation purposes are often challenged by ubiquitous bacteriophages, which occasionally interfere with manufacturing processes and product quality (8). Although phage resistance has historically relied on diversifying starter cultures and formulations based on the occurrence of phage defense systems such as restriction-modification and abortive infection (8), the recently discovered clustered regularly interspaced short palindromic repeats (CRISPR) and associated sequences (cas) have shown promise for phage resistance. CRISPR-Cas systems consist of arrays of short DNA repeats interspaced by hypervariable sequences, flanked by cas genes, that provide adaptive immunity against invasive genetic elements, such as phages and plasmids, through sequence-specific targeting and interference (9–16). Typically, invasive DNA sequences are acquired as novel “spacers” (9), each paired with a CRISPR repeat and inserted as a novel repeat-spacer unit in the CRISPR locus. Subsequently, following locus transcription, CRISPR RNAs (crRNAs) guide nucleases for sequence-specific recognition and cleavage (10, 17–23). These systems are widespread in bacteria (∼46%) and archaea (∼90%) and are classified into three main CRISPR-Cas system types (24, 25), based on the cas gene content and biochemistry. Early applications of CRISPR loci exploited the hypervariable nature of CRISPR spacers to type bacterial strains (8, 26). Thereafter, the DNA interference features were readily harnessed to enhance phage resistance in industrial starter cultures used in the dairy industry, by selecting naturally resistant strains with enhanced CRISPR immunity (8, 9, 17, 19, 22). More recently, the ability to direct sequence-specific DNA cleavage has been exploited for precise genome editing in many model organisms (27–29).
Several studies have shown that hypervariable CRISPR loci can readily be exploited for the genotyping of bacterial strains in which these immune systems were once active (8, 26, 30–35). Because novel spacers are integrated in a polarized manner at one extremity of the CRISPR array (9, 17, 26, 36), the spacer organization and content provide phylogenetic insights into the origin of a strain as reflected by a shared ancestral spacer(s); historical events, such as environmental phage exposure; and strain relatedness (through common spacer content) and divergence (inferred from unique spacer content). We aimed to investigate the occurrence of CRISPR-Cas systems in L. buchneri to establish whether these genetic loci can be used for the detection and typing of strains that spoil cucumber fermentation, as there are currently no established genotyping methods for these organisms. Previous studies have shown that genotyping of lactobacilli can be performed successfully and accurately by using the unique DNA sequences that constitute the CRISPR-Cas repeat-spacer array (8, 30, 37).
Diverse phages have been identified in vegetable fermentations, including pickle and sauerkraut fermentations (38–41). Accordingly, a secondary objective was to gain insights into the functional role that CRISPR-Cas systems may play in this species, notably with regard to interplay with invasive genetic elements, to assess activity and the potential for phage resistance.
Overall, we provide a comparative analysis of the occurrence of CRISPR-Cas systems in L. buchneri genomes and subsequently investigate the diversity of these loci in select strains isolated from industrial settings. We also provide an in silico analysis of important genetic elements that characterize these systems. In addition, we provide insights into the genetic diversity of these spoilage organisms in space and time, unraveling their phylogenetic origin and establishing their divergent evolutionary paths. Finally, we investigate the origin of CRISPR spacers and reveal events that are consistent with immunization against lytic bacteriophages, which often occur in these environments.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Lactobacillus buchneri strains were obtained from the USDA-ARS Food Science Research Unit, Raleigh, NC (Table 1). All samples originated from an industrial manufacturing environment where cucumbers are fermented into pickles. Mixed populations of lactic acid bacteria were first grown on filter-sterilized fermented cucumber slurry (FCS) and then selected for their persistence under a diverse range of pH and salt conditions and their ability to metabolize lactic acid. Colonies were isolated on de Man-Rogosa-Sharpe (MRS) agar plates and subsequently identified using morphology and 16S rRNA gene sequencing (1, 2). All strains identified at the species level as Lactobacillus buchneri were then used in our study. Strains originated from various sources, including commercial tank of origin and isolation time (Table 1). Isolates were designated by an identification number, suspended in glycerol, and stored at −80°C until the start of the experiment.
TABLE 1.
Lactobacillus buchneri strains used in this study
| Isolate ID | GenBank accession no. |
Notes | Reference | |
|---|---|---|---|---|
| 16S rRNA genea | CRISPR | |||
| LA1147 | JQ249035 | KF624608 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1173 | JQ249034 | KF624603 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1175 | JQ249037 | KF624603 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1178 | JQ249040 | KF624604 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1181 | JQ249043 | KF624602 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1184 | JQ249046 | KF624611 | Reduced-NaCl (2%) spontaneous fermented cucumber spoilage, day 7 | 2 |
| LA1151 | JQ249047 | KF624602 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1152 | JQ249048 | KF624602 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1154 | JQ249052 | KF624611 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1155 | JQ249053 | KF624608 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1156 | JQ249054 | KF624609 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1157 | JQ249055 | KF624607 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1158 | JQ249056 | KF624607 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1159 | JQ249057 | KF624608 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1160 | JQ249058 | KF624607 | Anaerobic spoilage in FCS with 4% NaCl and pH 3.8 source, days 4–6 | 2 |
| LA1161 | JQ249060 | KF624605 | Commercial anaerobic spoilage with 4% NaCl and pH 3.8 FCS source, days 4–6 | 2 |
| LA1163 | JQ249062 | KF624608 | Commercial anaerobic spoilage with 4% NaCl and pH 3.8 FCS source, days 4–6 | 2 |
| LA1164 | JQ249063 | KF624608 | Commercial anaerobic spoilage with 4% NaCl and pH 3.8 FCS source, days 4–6 | 2 |
| LA1166 | JQ249064 | KF624608 | Commercial anaerobic spoilage with 4% NaCl and pH 3.8 FCS source, days 4–6 | 2 |
| LA1167 | JQ249065 | KF624605 | Commercial anaerobic spoilage with 4% NaCl and pH 3.8 FCS source, days 4–6 | 2 |
| LA1149 | JQ086334 | KF624608 | Commercial cucumber fermentation spoilage isolate from 2010 | 1 |
| LA1185 | NA | KF624606 | Unpublished isolate from study of Johanningsmeier et al. (2) | Unpublished data |
| LA1187 | NA | KF624602 | Unpublished isolate from study of Johanningsmeier et al. (2) | Unpublished data |
| LA1188 | NA | KF624606 | Unpublished isolate from study of Johanningsmeier et al. (2) | Unpublished data |
| LA0030 | NA | KF624610 | Unpublished isolate from study of Johanningsmeier et al. (2) | Unpublished data |
| LA0251 | NA | KF624610 | Unpublished isolate from study of Johanningsmeier et al. (2) | Unpublished data |
NA, not applicable.
In silico analyses.
Two complete Lactobacillus buchneri genome sequences, for strains CD034 (5) and NRRL B-30929 (3), and an additional draft genome, for ATCC 11577, were obtained from GenBank (accession no. NC_018610, NC_015428, and NZ_ACGH01000000) via the National Center for Biotechnology Information's website (http://www.ncbi.nlm.nih.gov/) (42). The CRISPR database CRISPRdb (43) and CRISPRFinder were used to identify putative CRISPR loci in the published L. buchneri genomes and new CRISPR loci in draft genomes, respectively. After identifying several putative CRISPR loci in L. buchneri genomes, the basic local alignment sequence tool (BLAST) (44) was used to compare and contrast the sequences of cas genes, CRISPR repeats, and CRISPR spacers to those of closely related systems found in Lactobacillus salivarius UCC118, Lactobacillus brevis subsp. gravensis ATCC 27305, Lactobacillus pentosus KCA1, and Pediococcus acidilactici DSM 20284. Additionally, BLASTp analyses were used to characterize the cas genes in L. buchneri (24), establish the CRISPR-Cas system type and subtype, and align and determine the identity and similarity of conserved cas genes between the different bacterial species most closely related to L. buchneri. The putative trans-encoded CRISPR RNA (tracrRNA) sequence and structure were predicted by homology to characterized homologous sequences and predicted secondary structures (23, 45). Additionally, the repeat and spacer sequences were analyzed for homology to known sequences in the GenBank database. Repeat sequences showing homology to L. buchneri were identified using BLASTn, a nonredundant nucleotide search tool. Sequences showing >80% similarity over the entire 36-bp repeat in the type II-A locus and over the entire 32-bp repeat in the type I-E locus were used for the comparative analysis of L. buchneri. The unique spacer sequences were compared to known foreign genetic elements, such as viruses and plasmids, in the following databases: nonredundant nucleotide collection (nr/nt), genomic sequence surveys (gss), high-throughput genomic sequences (HTGS), and whole shotgun sequences (wgs). Spacer sequences are depicted in an overview by unique color combinations, as previously described (26). A protospacer hit was considered reliable if it showed at least 80% identity over the entire spacer sequence. Once a reliable protospacer was determined, the flanking sequences (∼10 nucleotides [nt]) on both sides were subjected to a comparative analysis to determine whether conserved nucleotides derived from a protospacer-adjacent motif (PAM) were present (26, 36, 46). WebLogo (47) was used to generate a frequency table allowing the identification of a novel PAM.
DNA sequencing of L. buchneri CRISPR-Cas systems.
To prepare for DNA extraction, cells were propagated overnight, resuspended in 10 ml of MRS broth, and grown at 37°C in a Coy Laboratories (Grass Lake, MI) anaerobic chamber. After 48 h, the DNA was extracted using a Zymo Fungal/Bacterial DNA purification kit following the special protocol for Gram-positive bacteria. PCR screening for CRISPR was used to determine which CRISPR-Cas system was present in the isolated strains. To screen for the type II-A repeat found in L. buchneri ATCC 1577, the primers 11577F (5′-GCTTTAGTAGTTCAAAAC-3′) and 11577R (5′-CATCATTGTTTTGAACTACTAC-3′) were used. To screen for the type II-A repeat found in L. buchneri CD034 and NRRL B-30929, the primers CD034F (5′-GGGTTTAACCTTATTGATTTAAC-3′) and CD035R (5′-GAAGGATGTTAAATCAATAAGG-3′) were used. PCR amplification of the cas9 gene was performed using the primer set Cas9.1 (5′-CCTTCAGACTGACGGTTC-3′) and Cas9.Rev (5′-GTCTCGATATTGGGACCTC-3′). PCR amplification of the repeat-spacer array in the 26 strains was performed using the primer set RSA.Fwd (5′-CCAGAATGAATGATCTGTTG-3′) and RSA.Rev (5′-CATCGACGAGAACTTTG-3′). PCR products were purified using a Zymo Research DNA Clean and Concentrator-5 kit and were sent for Sanger sequencing at Eton Biolabs, Raleigh, NC. The previously described in silico analyses were used to visualize the newly obtained repeat-spacer array sequences.
Nucleotide sequence accession numbers.
Sequences for all 10 unique CRISPR genotypes were deposited in GenBank under accession numbers KF624602 to KF624611 (Table 1).
RESULTS
Identification and characterization of CRISPR-Cas systems in L. buchneri genomes.
Multiple putative CRISPR arrays were identified in the L. buchneri complete and draft genomes. Specifically, a type I-E CRISPR-Cas system was identified in the CD034 and ATCC 11577 genomes, while a type II-A CRISPR-Cas system was identified in the CD034, NRRL B-30929, and ATCC 11577 genomes. The type I-E locus was defined by a highly conserved 28-nt CRISPR sequence nearly identical to systems also present in multiple Lactobacillus brevis genomes (see Table S1 in the supplemental material), with variety in the number of spacers across these loci. As anticipated, this CRISPR-Cas system includes the universal cas1 and cas2 genes, together with the type I cas3 signature gene (24) and the previously characterized Cascade and cas6 genes. Note that this system is not ubiquitous in L. buchneri genomes, limiting its potential as a universal target for typing purposes within this species (3, 8).
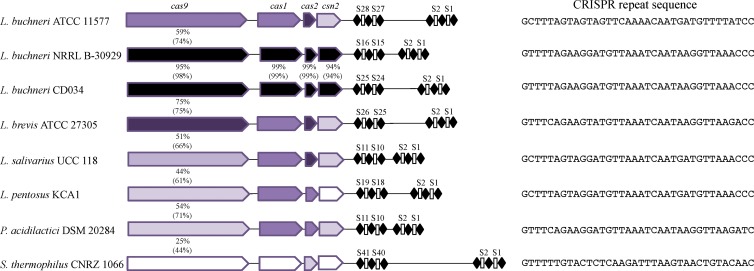
Type II-A CRISPR-Cas systems were identified in all three L. buchneri complete and draft genomes. This locus was defined by a highly conserved 36-nt CRISPR sequence homologous to those found in other Lactobacillus species, including L. salivarius, L. brevis, and L. pentosus (Fig. 1). Likewise, this repeat sequence was somewhat homologous to CRISPR found in more distant genera, including Pediococcus and Streptococcus, notably the Streptococcus thermophilus model type II-A CRISPR-Cas system (Fig. 1). These systems carry the universal cas1 and cas2 genes together with the type II signature cas9 gene, as well as csn2 (24), which is unique to type II-A subtypes (Fig. 1). A comparative analysis of Cas protein sequence conservation among these homologous type II-A systems revealed high similarity between the CD034 and NRRL B-30929 strains and relatively limited homology to the other aforementioned homologous systems, with 25% identity between the CD034 and CNRZ Cas9 protein sequences (Fig. 1). Notwithstanding CRISPR repeat sequence conservation and cas homologies, the hypervariable nature of these CRISPR loci across genera, species, and strains is illustrated by the diversity observed in terms of CRISPR spacer number and sequences, with as few as 11 and up to 28 spacers within lactobacilli (Fig. 1).
FIG 1.
Type II-A CRISPR-Cas systems. (Left) Architecture of the type II-A CRISPR-Cas systems in select lactic acid bacteria containing the type II signature cas9 gene together with the universal cas1 and cas2 genes as well as csn2, which is uniquely found in type II-A systems. The color scale reflects sequence similarity to the CD034 reference sequences, with amino acid identity (top number) and isofunctional conservation (bottom number), ranging from lowest (light) to highest (dark). (Right) Repeat sequences of the type II-A CRISPR in select lactic acid bacteria.
Diversity of type II-A CRISPR loci.
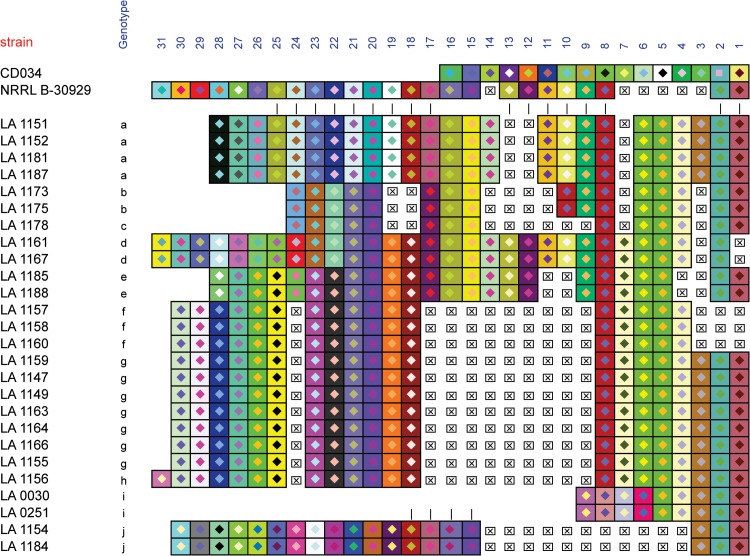
In order to determine the occurrence of type II-A CRISPR-Cas systems in diverse L. buchneri strains, we carried out PCR analyses to ascertain the presence of the signature cas9 gene (using internal primers) and the size of the CRISPR array (using primers flanking the repeat-spacer array) in a series of industrial isolates (Table 1). The results consistently showed that this CRISPR-Cas system is ubiquitous and hypervariable in L. buchneri (Fig. 2). Indeed, we further determined the repeat-spacer variability by sequencing the PCR amplicons and reconstructing the spacer content of these strains (Fig. 3). Comparative analysis of CRISPR spacer contents and sequences across 26 strains revealed 10 different CRISPR genotypes containing between 9 and 29 spacers (Fig. 3). We observed conservation of ancestral CRISPR spacers, revealing a common origin, including that for the NRRL B-30929 strain, namely, spacers 1 and 2, reflecting trailer-end conservation (45). Furthermore, the first block of spacers (positions 1 through 8) was widely conserved throughout our strain collection. Conversely, distinct sets of consecutive spacers were shared only between certain sets of strains, revealing divergent evolutionary paths. One such set of shared consecutive spacers can be seen for genotypes e, f, g, and h, where spacers at position 18 through 23 are strictly conserved in all 13 strains across these four genotypes. This contrasts with spacers at positions 17 through 25 that are shared only between genotype a and the NRRL B-30929 strain. The same applies to spacers at positions 15 and 16, which are shared “only” between genotypes a through e. Overall, internal deletions and leader-end spacer diversity revealed hypervariability between even closely related strains. Consistent with previous reports indicating a preference for internal deletions at the trailer end (26, 48), we observed 13 distinct spacer loss events (Fig. 3), the large majority of which (11/13 events) occur within the trailer half of the loci. Interestingly, LA 1156 exhibits an additional spacer at the leader end, which reflects novel spacer integration and suggests that this locus is active.
FIG 2.
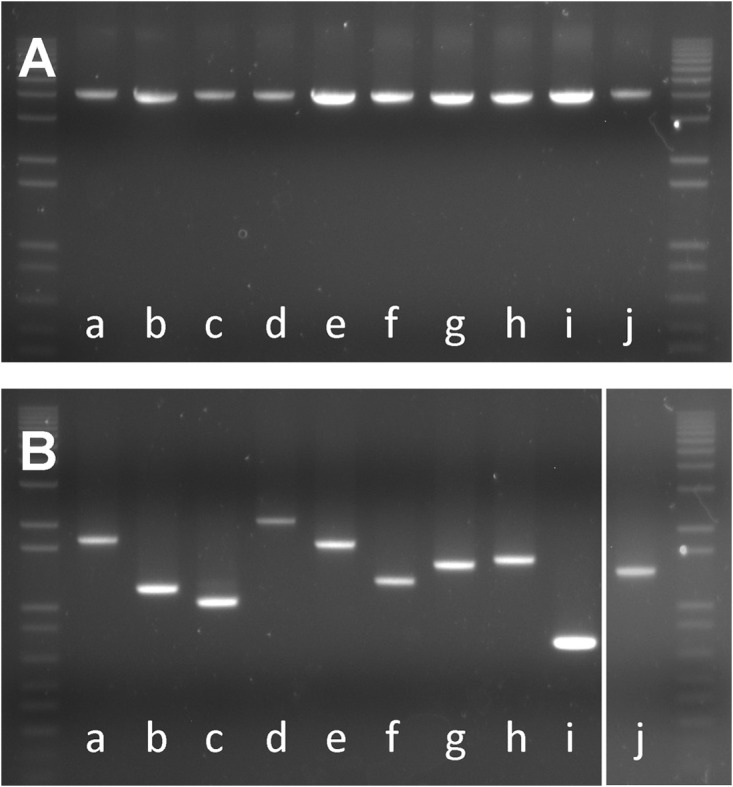
PCR-based detection of the CRISPR-Cas elements in L. buchneri isolates. (A) Visualization of conserved cas9 amplicons in 10 representative isolates. (B) Visualization of the hypervariable type II-A repeat-spacer CRISPR array in 10 diverse and representative isolates. A 1-kb DNA ladder is shown on the sides of both gels.
FIG 3.
CRISPR spacer overview. Visualization of the type II-A CRISPR spacer content for all 26 isolates and comparative analysis with the loci from the two complete L. buchneri genomes. Only spacers are represented, with all conserved repeats removed. Each square represents a CRISPR spacer, and unique color combinations represent unique spacer sequences. Deletions/missing spacers are represented by “x.” Spacers are numbered in order of predicted acquisition in the locus. Each unique spacer combination was assigned a genotype (letters).
Overall, the diversity found within this set of strains is interesting considering that the samples came from at least three very distinct industrial and laboratory settings.
Origin of CRISPR spacers and locus activity.
In order to determine the likely origin of the CRISPR spacers, we investigated their homology to known sequences. Upon searching for homologous sequences not associated with CRISPR arrays, we identified several matches to foreign genetic elements such as plasmids and bacteriophages (Table 2), as anticipated (9, 17, 49). We did observe several matches to plasmids and phages associated with lactobacilli, including examples where there is perfect identity between a CRISPR spacer and a protospacer from an invasive nucleic acid. This, together with the aforementioned novel spacer insertion event, is consistent with the involvement of these type II-A CRISPR-Cas systems with adaptive immunity in L. buchneri.
TABLE 2.
L. buchneri CRISPR spacer matches
| Strain | Spacer no. | Sequence |
No. of nt matches in spacer (among 30 nt) | Protospacer match (accession no.) | Annotation | ||
|---|---|---|---|---|---|---|---|
| Left flank | Protospacer | Right flank | |||||
| LA 1175 | 15 | AAAATTCAGA | CAACAAAAAAAGCGCTCCGCAACGGCCATT | GCAAAACGCT | 30 | Food metagenome (ASXE01000335) | Putative phage Mu Gam protein |
| LA 1154 | 12 | ATGAAGTTCA | AGCTGTGTCAAACTACGTTGAATCCCAAGG | ACAAAACTTA | 29 | Food metagenome (ASXE01000117) | Putative phage transcriptional activator |
| LA 1152 | 2 | CTGGTTTTAT | AAACGGATATTGCGGCTTATATTAACGAGC | TGAAATGGTT | 30 | Lactobacillus brevis/pLB925A02 | Plasmid mobilization protein |
| LA 1152 | 2 | CTGGTTTTAT | AAACGGATATTGCGGCTTATATTAACGAGC | TGAAATGGTT | 30 | Lactobacillus buchneri CD034/pCD034-1 | Mobilization protein |
| LA 1147 | 10 | AGAATATCGA | CAACGCAGCTAAAGATAATCGTCAGAATTA | CCAGAAATTA | 29 | Food metagenome (ASXE01000848) | Putative phage nucleotide-binding protein |
| NRRL B-30929 | 9 | TAAGCTTGGT | GGAAAAAGGTGGCGGCCGCTTTGTGCAAGG | TCAAGAAATG | 30 | Lactobacillus kefiranofaciens ZW3/pWW2 | Conjugal transfer protein |
| NRRL B-30929 | 3 | TTACGCTTTA | ACCGAGTTTCGTGATCTCAAAAGTAGCTAC | GCAAAAACTA | 30 | Lactobacillus paracasei/pLP5402 | Plasmid replication initiation |
| LA 1188 | 1 | TTCTTAGATG | CCGCTTACTTGCCGTTAAAGCGGGATATCG | TTCAAAAAGA | 28 | Lactobacillus plantarum ZJ316/pLP-ZJ103 | Transposase |
| LA 1161 | 21 | AAAATTCAGA | CAACAAAAAAAGCGCTCCGCAACGGCCATT | GCAAAACGCT | 30 | Food metagenome (ASXE01000848) | Putative phage nucleotide-binding protein |
| LA 1161 | 23 | CATTATGCTA | AAGGTTCAGGTGTCTCACACGCTGAACTAG | ACAAAATTAT | 29 | Lactobacillus kisonensis F0435 | Phage tail tape measure protein |
| LA 1161 | 24 | CTTTATCTAG | GAAATAAGCAGCCTCATTTGAAGCACCATG | CCAAAAATGA | 30 | Food metagenome (ASXE01000470) | Putative phage lysozyme |
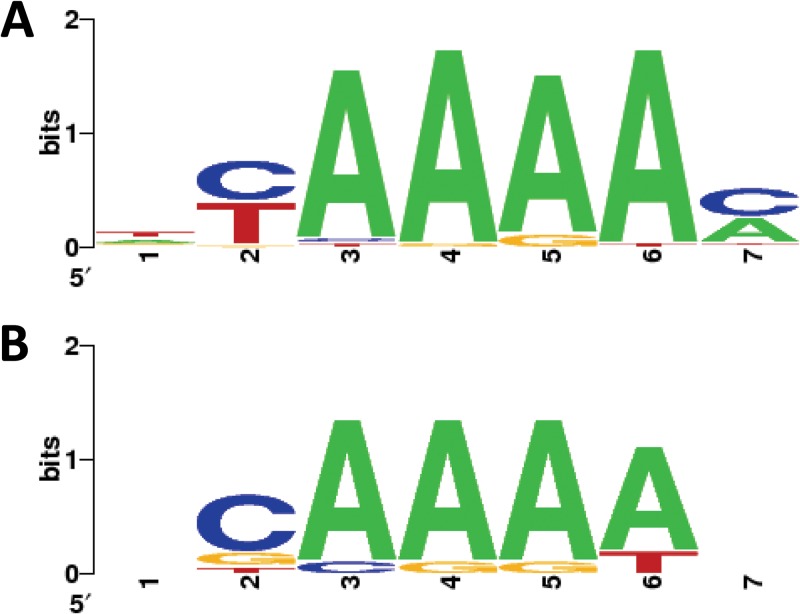
Several studies have established that PAMs are implicated in novel spacer acquisition (49) and Cas9-mediated sequence-specific cleavage of target nucleic acid (19, 21, 23). Thus, we aligned the sequences flanking the protospacers that showed homology to CRISPR spacer sequences and consistently observed the presence of a 5′-AAAA-3′ PAM two nucleotides downstream of the protospacer (Fig. 4). Aligning the flanking sequences of the 35 hits revealed a 5′-AAAA-3′ tetranucleotide 2 nt downstream of the protospacer, which was confirmed by looking at the top 11 matches (Table 2; Fig. 4). This PAM sequence is homologous to the 5′-AGAA-3′ PAM previously established for the closely related CRISPR1-Cas type II-A system from S. thermophilus (26, 49).
FIG 4.
PAMs. The 10-nt sequences flanking the 3′ ends of the protospacer sequences were aligned to generate a WebLogo (46). (A) PAM inferred from 35 matches (protospacer matches showing >80% similarity); (B) PAM inferred from the top 11 matches (protospacer matches showing >90% similarity) listed in Table 2.
Because Cas9 is the core protein driving spacer-dependent target recognition and cleavage, we further analyzed the Cas9 sequence and investigated the presence of biochemically relevant residues in L. buchneri. We first looked at the N-terminal RuvC motif and observed the presence of the conserved and important Asp residue implicated in the nicking of the positive-strand target DNA, namely, D31 within the IGLDIGT motif (19, 21). Next, using the L. buchneri NRRL Cas9 protein sequence as a template (annotated csn1 [GenBank accession no. AEB74124.1] in the publicly available genome sequence), we investigated the presence of conserved residues implicated in nicking of the negative target strand, namely, H868-X13-N882-X8-N891 (19, 21), and observed a pattern consistent with this exact spacing, as well as conservation of these three biochemically relevant residues, namely, YDIDHI, NNRVL, and INNG.
Further in silico analyses were carried out to characterize elements of type II-A CRISPR-Cas systems. Because the tracrRNA plays a critical role in type II CRISPR-Cas systems, crRNA biogenesis, and interference (21, 50), which relies on partial sequence complementarity between the CRISPR and the tracrRNA (21, 45), we investigated the presence of a tracrRNA in the vicinity of the repeat-spacer array. We used the CRISPR repeat sequence to look for partial matches in intergenic sequences flanking cas9, as previously described. We identified a putative 90-nt tracrRNA which shows complementarity to the CRISPR repeat (Fig. 5) and contains three predicted hairpins at the 3′ end, reminiscent of the idiosyncratic tracrRNA structure of other type II-A systems (23). Consistent with previous reports (45), the putative L. buchneri tracrRNA is located between the cas9 and cas1 genes in both CD034 and NRRL B-30929.
FIG 5.

Sequence and structural details for core CRISPR-Cas system elements. A section of the repeat-spacer array is shown (center) with the corresponding protospacer (top), including flanking sequences (±10 nt) comprising the PAM, and the predicted tracrRNA sequence and structure (bottom), including the complementary anti-CRISPR repeat, as well as three putative hairpins reminiscent of characterized tracrRNAs.
DISCUSSION
We investigated the occurrence and diversity of CRISPR-Cas immune systems in L. buchneri and characterized a type II-A system. Specifically, we showed (i) strict conservation of the 5′-GTTTTAGAAGGATGTTAAATCAATAAGGTTAAACCC-3′ CRISPR sequence; (ii) typical cas gene content and architecture for this particular subtype, which includes the cas9 signature gene; (iii) high spacer diversity between even closely related isolates, reflecting a common origin yet extensive divergence; (iv) leader-end spacer polymorphisms and matches to viral sequences consistent with phage immunity; and (v) core elements necessary for functional exploitation, namely, important residues within Cas9, a novel PAM, and the necessary tracrRNA sequence and structure.
Comparative genome analysis of CRISPR content in L. buchneri genomes revealed the occasional presence of a type I-E system and the universal occurrence of a type II-A system (24). The conserved type II-A system provides an attractive single-locus target for investigating the occurrence and diversity of L. buchneri strains and could be broadly useful for genotyping of this species. Many studies have established that CRISPR loci can be targeted for genotyping in multiple species and provide insights into the phylogenetic relationships between organisms, including closely related isolates (30–35, 51). The preliminary results shown here suggest that CRISPR loci could be targeted to investigate population diversity and evolution over space and time. Intriguingly, the most prevalent genotype (genotype g) was detected in multiple strains (LA 1147, LA 1149, LA 1155, LA 1159, LA 1163, LA 1164, and LA 1166) isolated at different times from separate locations. Likewise, genotype a was detected from strains isolated from different sources (Fig. 3; Table 1). This suggests that some genotypes are naturally widespread and relatively robust. Nevertheless, we also repeatedly observed the concurrent presence of multiple genotypes within space and time, suggesting a naturally diversified population. The diverse prevalence of multiple genomes may reflect predation by bacteriophages, as previously suggested (49, 52). Furthermore, shared ancestral spacers could provide a genetic basis to establish phylogenetic relationships between strains and/or clusters of strains. This is of particular industrial interest given the widespread use of L. buchneri in silage inocula as well as their exploitation in bioethanol manufacturing. Likewise, this target could be instrumental in the detection, typing, and monitoring of strains that contaminate industrial vegetable fermentations such as cucumber pickling.
With regard to the potential of CRISPR-based genotyping in general, and within L. buchneri in particular, it is important to point out the many advantages of these loci, which include targeting a hypervariable region flanked by conserved sequences (the former can segregate between sequences, while the latter can be exploited for primer design); providing a time-resolved record of environmental immunization events (which reflects both a common phylogenetic origin between strains and recent divergence); and using loci that are genomically stable, short, easily identifiable, and widespread (8).
In addition to its genotyping potential, given the extensive circumstantial evidence implicating type II CRISPR-Cas immune systems in adaptive immunity against phages, this system has potential for phage defense exploitation in industrial cultures. Perhaps this novel system can be exploited to enhance phage resistance in L. buchneri strains used to inoculate silage, similar to what has been implemented in S. thermophilus starter cultures (8, 9, 14, 17, 22). Studies have shown that resistance can be enhanced naturally by selecting phage-resistant mutants with novel CRISPR spacers or that novel immunity can be engineered using molecular biology (9, 19). In L. buchneri, the observation of the concurrent presence of two CRISPR genotypes that share spacer content, with the exception of a single novel spacer at the leader end, strongly suggests that this locus has the ability to naturally acquire novel spacers in a polarized manner, as previously shown for active CRISPR loci.
Furthermore, Cas9, with its conserved residues, associated PAM, and tracrRNA, could be exploited as a novel nuclease for genome editing. Several recent studies have repeatedly established that Cas9 has tremendous potential for genome editing applications given the ability to reprogram DNA cleavage by this nimble endonuclease (20, 27–29). The identification of a novel PAM (5′-AAAA-3′), together with the accompanying putative tracrRNA, opens new avenues for flexible Cas9-mediated genome editing using native elements or guide RNAs. Further studies will investigate the potential of this type II-A system for genotyping, phage resistance, and genome editing.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kurt Selle, Evelyn Durmaz, Rosemary Sanozky-Dawes, and the entire Todd Klaenhammer lab for providing technical assistance during this project. We also thank Suzanne Johanningsmeier and Janet Hayes at the USDA-ARS Food Science Research Unit, Raleigh, NC, for allowing the use of their strains in this study. Additionally, we thank Philippe Horvath at DuPont for making the CRIPSR visualization tool available to us to generate the CRISPR spacer overview.
This study was supported by start-up funds from North Carolina State University.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03015-13.
REFERENCES
- 1.Franco W, Perez-Diaz IM, Johanningsmeier SD, McFeeters RF. 2012. Characteristics of spoilage-associated secondary cucumber fermentation. Appl. Environ. Microbiol. 78:1273–1284. 10.1128/AEM.06605-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johanningsmeier SD, Franco W, Perez-Diaz I, McFeeters RF. 2012. Influence of sodium chloride, pH, and lactic acid bacteria on anaerobic lactic acid utilization during fermented cucumber spoilage. J. Food Sci. 77:M397–M404. 10.1111/j.1750-3841.2012.02780.x [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Leathers TD, Copeland A, Chertkov O, Goodwin L, Mills DA. 2011. Complete genome sequence of Lactobacillus buchneri NRRL B-30929, a novel strain from a commercial ethanol plant. J. Bacteriol. 193:4019–4020. 10.1128/JB.05180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eikmeyer FG, Köfinger P, Poschenel A, Jünemann S, Zakrzewski M, Heinl S, Mayrhuber E, Grabherr R, Pühler A, Schwab H, Schlüter A. 2013. Metagenome analyses reveal the influence of the inoculant Lactobacillus buchneri CD034 on the microbial community involved in grass silaging. J. Bacteriol. 167:334–343. 10.1016/j.jbiotec.2013.07.021 [DOI] [PubMed] [Google Scholar]
- 5.Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, Linke B, Goesmann A, Grabherr R, Schwab H, Puhler A, Schulter A. 2012. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J. Biotechnol. 161:153–166. 10.1016/j.jbiotec.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 6.Dreihuis F, Elferink SJ, Spoelstra SF. 1999. Anaerobic lactic acid degradation during ensilage of whole crop maize inoculated with Lactobacillus buchneri inhibits yeast growth and improves aerobic stability. J. Appl. Microbiol. 87:583–594. 10.1046/j.1365-2672.1999.00856.x [DOI] [PubMed] [Google Scholar]
- 7.Schmidt RJ, Kung L., Jr 2010. The effects of Lactobacillus buchneri with or without a homolactic bacterium on the fermentation and aerobic stability of corn silages made at different locations. J. Dairy Sci. 93:1616–1624. 10.3168/jds.2009-2555 [DOI] [PubMed] [Google Scholar]
- 8.Barrangou R, Horvath P. 2012. CRISPR: new horizons in phage resistance and strain identification. Annu. Rev. Food Sci. Technol. 3:143–162. 10.1146/annurev-food-022811-101134 [DOI] [PubMed] [Google Scholar]
- 9.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 10.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaya D, Davidson M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45:273–297. 10.1146/annurev-genet-110410-132430 [DOI] [PubMed] [Google Scholar]
- 14.Barrangou R, Coute-Monvoisin AC, Stahl B, Chavichcily I, Damange F, Romero DA, Boyaval P, Fremaux C, Horvath P. 2013. Genomic impact of CRISPR immunization against bacteriophages. Biochem. Soc. Trans. 41:1383–1391. 10.1042/BST20130160 [DOI] [PubMed] [Google Scholar]
- 15.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46:311–339. 10.1146/annurev-genet-110711-155447 [DOI] [PubMed] [Google Scholar]
- 16.Barrangou R. 2013. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4:267–278. 10.1002/wrna.1159 [DOI] [PubMed] [Google Scholar]
- 17.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- 18.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. 10.1126/science.1192272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39:9275–9282. 10.1093/nar/gkr606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magadan AH, Dupuis ME, Villion M, Moineau S. 2012. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7:e40913. 10.1371/journal.pone.0040913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. 2013. crRNA and tracrRNA guide Cas9-mediated DNA interferences in Streptococcus thermophilus. RNA Biol. 10:841–851. 10.4161/rna.24203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova KS, Wolf YI, Koonin EV. 2013. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41:4360–4377. 10.1093/nar/gkt157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412. 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31:233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath P, Coute-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62–70. 10.1016/j.ijfoodmicro.2008.05.030 [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Kariyawasam S, Jayarao BM, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Subtyping Salmonella enterica serovar Enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl. Environ. Microbiol. 77:4520–4526. 10.1128/AEM.00468-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl. Environ. Microbiol. 77:1946–1956. 10.1128/AEM.02625-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shariat N, Kirchner MK, Sandt CH, Trees E, Barrangou R, Dudley EG. 2013. Subtyping of Salmonella enterica serovar Newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J. Clin. Microbiol. 51:2328–2336. 10.1128/JCM.00608-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shariat N, DiMarzio MJ, Dettinger L, Sandt CH, Lute JR, Barrangou R, Dudley EG. 2013. The combination of CRISPR-MVLST and PFGE provides increased discriminatory power for differentiating human clinical isolates of Salmonella enterica subsp. enterica serovar Enteritidis. Food Microbiol. 34:164–173. 10.1016/j.fm.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 35.Yin S, Jensen MA, Bai J, Debroy C, Barrangou R, Dudley EG. 2013. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl. Environ. Microbiol. 79:5710–5720. 10.1128/AEM.00950-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, Horvath P, Heidenreich J, Perna NT, Barrangou R, Steele JL. 2012. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533. 10.1186/1471-2164-13-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrangou R, Yoon SS, Breidt F, Jr, Fleming HP, Klaenhammer TR. 2002. Identification and characterization of Leuconostoc fallax strains isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 268:2877–2884. 10.1128/AEM.68.6.2877-2884.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon SS, Barrangou-Poueys R, Breidt F, Jr, Klaenhammer TR, Fleming HP. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68:973–976. 10.1128/AEM.68.2.973-976.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon SS, Barrangou-Poueys R, Breidt F, Jr, Fleming HP. 2007. Detection and characterization of a lytic Pediococcus bacteriophage from the fermenting cucumber brine. J. Microbiol. Biotechnol. 17:262–270 [PubMed] [Google Scholar]
- 41.Lu Z, Pérez-Díaz IM, Hayes JS, Breidt F. 2012. Bacteriophage ecology in a commercial cucumber fermentation. Appl. Environ. Microbiol. 78:8571–8578. 10.1128/AEM.01914-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2009. GenBank. Nucleic Acids Res. 37:D26–D31. 10.1093/nar/gkn723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grissa I, Verngnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chylinski K, Rhun AL, Charpentier E. 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10:726–737. 10.4161/rna.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defense system. Microbiology 155:733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinberger AD, Sun CL, Pluciński MM, Denef VJ, Thomas BC, Horvath P, Barrangou R, Gilmore MS, Getz WM, Banfield JF. 2012. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput. Biol. 8:e1002475. 10.1371/journal.pcbi.1002475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun. 4:1430. 10.1038/ncomms2440 [DOI] [PubMed] [Google Scholar]
- 50.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimarzio M, Shariat N, Kariyawasam S, Barrangou R, Dudley EG. 24 June 2013. Antibiotic resistance in Salmonella typhimurium associates with CRISPR sequence type. Antimicrob. Agents Chemother. 10.1128/AAC.00913-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levin BR, Moineau S, Bushman M, Barrangou R. 2013. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 9:e1003312. 10.1371/journal.pgen.1003312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.