Abstract
Short-chain fatty acid (SCFA) biosynthesis is pertinent to production of biofuels, industrial compounds, and pharmaceuticals from renewable resources. To expand on Escherichia coli SCFA products, we previously implemented a coenzyme A (CoA)-dependent pathway that condenses acetyl-CoA to a diverse group of short-chain fatty acyl-CoAs. To increase product titers and reduce premature pathway termination products, we conducted in vivo and in vitro analyses to understand and improve the specificity of the acyl-CoA thioesterase enzyme, which releases fatty acids from CoA. A total of 62 putative bacterial thioesterases, including 23 from the cow rumen microbiome, were inserted into a pathway that condenses acetyl-CoA to an acyl-CoA molecule derived from exogenously provided propionic or isobutyric acid. Functional screening revealed thioesterases that increase production of saturated (valerate), unsaturated (trans-2-pentenoate), and branched (4-methylvalerate) SCFAs compared to overexpression of E. coli thioesterase tesB or native expression of endogenous thioesterases. To determine if altered thioesterase acyl-CoA substrate specificity caused the increase in product titers, six of the most promising enzymes were analyzed in vitro. Biochemical assays revealed that the most productive thioesterases rely on promiscuous activity but have greater specificity for product-associated acyl-CoAs than for precursor acyl-CoAs. In this study, we introduce novel thioesterases with improved specificity for saturated, branched, and unsaturated short-chain acyl-CoAs, thereby expanding the diversity of potential fatty acid products while increasing titers of current products. The growing uncertainty associated with protein database annotations denotes this study as a model for isolating functional biochemical pathway enzymes in situations where experimental evidence of enzyme function is absent.
INTRODUCTION
The potential for producing biofuels, industrial compounds, and pharmaceuticals from renewable resources has led to an increased interest in biosynthesis of short-chain (C2 to C7) fatty acids (SCFAs). Developing recombinant strains to produce these molecules could lead to production of polymers such as polyhydroxyalkanoates and pharmaceuticals such as statins from fossil fuel alternatives (1, 2). Supporting the development of microbially synthesized products are the reduced cost and increased prevalence of genomic sequencing. The resulting profusion of metabolic diversity provides a wealth of potential enzymes with known genetic sequences for improving biosynthetic pathways. For example, recent genomic sequencing has unveiled the metabolic diversity of important members from the cow rumen microbiome, an environment rich in SCFAs (3, 4). These genome sequences provide an opportunity to find enzymes that improve production and specificity in SCFA biosynthesis pathways.
One such pathway that would benefit from improved enzyme specificity is coenzyme A (CoA)-dependent biosynthesis of SCFAs. In the CoA-dependent pathway, a thiolase enzyme condenses an acyl-CoA molecule with acetyl-CoA (Fig. 1). The resulting 3-ketoacyl-CoA molecule is then sequentially reduced by reductase, crotonase, and enoyl-reductase enzymes before the 3-hydroxy, unsaturated, and saturated fatty acids, respectively, are cleaved from CoA using a thioesterase enzyme. Previous work from our laboratory has used this pathway with acetyl-CoA and glycolyl-, propionyl-, or isobutyryl-CoA as the condensed substrates to produce a variety of SCFAs using Escherichia coli TesB (EcTesB), including 3-hydroxyvalerate (3-hydroxypentanoate) (1), dihydroxybutyrate (5), 3-hydroxy-4-methylvalerate (5), and a variety of alcohols (6). One benefit of CoA-dependent biosynthesis of SCFAs is the diversity of potential products (7); however, with this diversity comes a need for selective enzymes that increase final product titers by minimizing substrate flux to undesired by-products. The selectivity of the final enzyme in the pathway, the thioesterase, is of particular importance because it influences the product profile by catalyzing fatty acid release from CoA at each step of the pathway (Fig. 1) and is important for secretion of fatty acid products (8). While many acyl-ACP (acyl carrier protein) thioesterases have been investigated for improved fatty acid production, acyl-CoA thioesterases are not as well explored (8–10). Despite its preference for acyl-CoAs in the C14 to C18 range (11), the E. coli acyl-CoA thioesterase TesB produces diverse SCFAs (12). However, locating more selective thioesterases may reduce by-product formation and increase final product titers.
FIG 1.
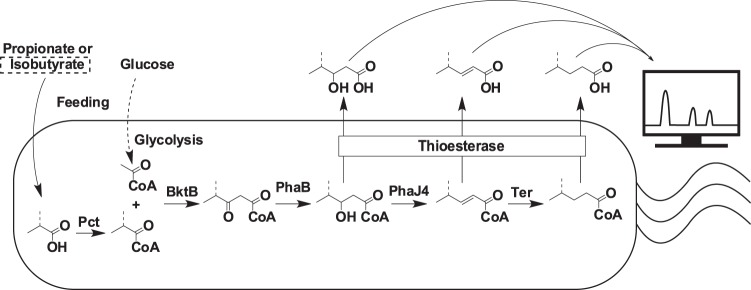
Biochemical pathway and LC-based in vivo screen for thioesterases with improved substrate specificity. The pathway in E. coli for the biosynthesis of SCFAs contains enzymes Pct (Megaphaera elsdenii), BktB (Cupriavidus necator), PhaB (Cupriavidus necator), PhaJ4 (Cupriavidus necator), and Ter (Treponema denticola) with varied thioesterases. The dashed bond indicates the additional carbon incorporated into the fatty acids with feeding of isobutyrate in place of propionate.
Selecting individual thioesterase enzymes for functional screening against short-chain fatty acyl-CoAs is challenging because much of their vast phylogenetic and functional diversity is poorly understood (13). While many thioesterases have been explored for long-chain fatty acid production (14–17), few studies have focused on those that prefer short-chain acyl-CoAs. Several broad-specificity acyl-CoA thioesterases, including E. coli TesB and Saccharomyces cerevisiae Pte1p, can be used for SCFA production but lack the specificity necessary for optimizing biosynthetic pathways (11, 18). One approach to selecting thioesterases for functional screening that improves the likelihood of finding enzymes with the desired specificity is to investigate those proteins with similarity to commonly used and effective enzymes. However, the absence of known selective short-chain acyl-CoA thioesterases restricts this approach. Sampling candidates more broadly increases opportunities for finding enzymes with new substrate specificities but also increases the number of thioesterases with undesired activities. Some combination of these routes can be used to screen sufficient phylogenetic breadth while also increasing the sample size of TesB-like thioesterases to reveal enzymes with greater specificity for short-chain acyl-CoAs.
To address this need, we functionally screened 62 putative thioesterases in a pathway for production of the SCFAs 3-hydroxyvalerate, trans-2-pentenoate, valerate, and 4-methylvalerate in E. coli. Six enzymes were homologous to variants active on short-chain acyl-CoAs, while the remaining 56 represent all of the annotated thioesterases from five bacterial strains of interest. Based upon in vivo fatty acid titers of those recombinant strains, six enzymes were chosen for in vitro analysis on a broad range of acyl-CoAs to determine their substrate preferences. The combination of in vivo and in vitro data indicates that we have uncovered thioesterases with greater specificity for and production of unsaturated, saturated, and branched SCFAs in E. coli relative to EcTesB and endogenous thioesterase activity.
MATERIALS AND METHODS
Bacterial strains.
Rumen isolate Prevotella ruminicola 23 was obtained from Roderick Mackie of the University of Illinois, Urbana-Champaign, IL. Genomic DNA from rumen isolates Fibrobacter succinogenes S85 and Ruminococcus albus 7 were obtained from Paul Weimer of the U.S. Dairy Forage Research Center, USDA-Agricultural Research Service, Madison, WI. Alcanivorax borkumensis SK2 (ATCC 700651) and genomic DNA from Pseudomonas aeruginosa PAO1 (ATCC 47085) were purchased from the ATCC. Genomic DNA of Rhodopseudomonas palustris CGA009 was obtained from Caroline Harwood of the University of Washington, Seattle, WA. Rhodococcus opacus PD630 was obtained from Anthony Sinskey of the Massachusetts Institute of Technology, Cambridge, MA. Pseudomonas syringae pv. maculicola ES4326 was obtained from Fred Ausubel at Massachusetts General Hospital (Boston, MA). E. coli MG1655(DE3) ΔendA ΔrecA was previously constructed in our laboratory (1).
Plasmid and strain construction.
Plasmid pET/ter/bktB/pct was constructed previously (6), and plasmid pCDF/phaB/phaJ4 was constructed by subcloning phaB from pET/bktB/phaB (5) into multiple-cloning site I by NdeI/XhoI restriction digestion and phaJ4 into multiple-cloning site II of pCDFDuet-1 (EMD Millipore) using primers listed in Table S1 in the supplemental material. Plasmid pET/bktB/pct was constructed from pET/ter/bktB/pct by BamHI/NotI restriction digestion, followed by treatment with mung bean nuclease (New England BioLabs) and blunt ligation of the 8-kb fragment.
Genomic DNA was isolated from bacterial strains using the Wizard genomic DNA purification kit (Promega). Custom oligonucleotides were purchased for PCR amplification of all individual thioesterases and CoA ligases from purified genomic DNA (Integrated DNA Technologies). Primers used for amplification are listed in Table S1 in the supplemental material. Following amplification, individual genes were inserted into the expression vector pACYCDuet-1 (EMD Millipore) using polymerase incomplete primer extension (PIPE)-based cloning (19). The genes encoding R. palustris CoA ligases FcsA (Rpa4267) and VcsA (Rpa3299) (20) and the genes encoding the six thioesterases chosen for further analysis were inserted into vector pTEV5 for protein purification using PIPE-based methods (21). The pTEV5 construct produced an enzyme with an N-terminal hexahistidine tag removable by TEV protease. Due to solubility problems in pTEV5, the genes encoding thioesterases P. ruminicola 1687 (Pr1687) and F. succinogenes 2108 (Fs2108) were amplified from genomic DNA and cloned using restriction enzymes NdeI and EcoRI into pTYB22 (New England BioLabs), which produced an enzyme with an N-terminal chitin-binding domain removable by intein self-cleavage. Plasmid sequences were confirmed using PCR amplification and DNA sequencing (GENEWIZ).
Gene deletions of yciA, yigI, and tesB in E. coli MG1655(DE3) ΔendA ΔrecA were made using P1 transduction with strains JW1245-1, JW5588-1, and JW0442-1, respectively, from the Keio collection as donor cells (22). The kanamycin resistance gene was removed using FLP-mediated recombination as previously described (23).
Culture conditions.
Recombinant strains of E. coli MG1655(DE3) ΔendA ΔrecA were grown at 30°C in Luria-Bertani (LB) medium overnight in a shaking incubator at 250 rpm. Fifty microliters of the overnight culture was used to inoculate a 50-ml LB culture supplemented with 10 g/liter of glucose containing 100 mg/liter of ampicillin, 50 mg/liter of streptomycin, and, when pACYCDuet-1 was present, 16 mg/liter of chloramphenicol. Cultures were grown at 30°C until an optical density at 600 nm (OD600) of 0.8 was reached, at which point isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM with either propionate or isobutyrate to a final concentration of 15 mM. Cultures were incubated at 30°C for 48 h postinduction prior to fatty acid titer determination.
Fatty acid analysis.
Culture samples were centrifuged to pellet cells, and culture supernatant was removed for high-performance liquid chromatography (HPLC) analysis. A 5-μl sample of culture supernatant was injected into an Agilent 1100 series instrument equipped with refractive index detection (RID). Samples were processed through an Aminex HPX-87H anion-exchange column (Bio-Rad Laboratories) with an isocratic flow of a 5 mM H2SO4 mobile phase at a rate of 0.6 ml/min and column and detector temperatures set to 35°C. Concentrations of valerate, 3-hydroxyvalerate, trans-2-pentenoate, and 4-methylvalerate were determined using linear regression of external standards.
Protein purification.
Acyl-CoA ligases FcsA and VcsA and thioesterases EcTesB, P. putida TesB (PpTesB), E. coli YdiI (EcYdiI), and P. ruminicola 655 (Pr655) were overproduced using pTEV5 constructs in E. coli BL21Star(DE3) (Invitrogen). One liter of cells was grown at 30°C in LB medium containing 100 mg/liter of ampicillin until an OD600 of 0.5 was reached, at which point IPTG was added to the cultures at a concentration of 100 mg/liter. Postinduction, the cells were grown for 15 h at 30°C and then harvested by centrifugation and resuspended in 2.5× vol/wt buffer A (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10% [vol/vol] glycerol). Protein purification followed previously described protocols for nickel chelate chromatography followed by cleavage with TEV protease (24). Proteins were flash frozen in liquid nitrogen, and the concentration was determined using the Bradford assay with bovine serum albumin as a standard (25) (Bio-Rad).
P. ruminicola 1687 (Pr1687) and F. succinogenes 2108 (Fs2108) proteins were purified using the intein-mediated purification with an affinity chitin-binding tag (IMPACT) expression vector pTYB22 (New England BioLabs). Cells were grown and induced using the same conditions as described for pTEV5 constructs, and proteins were purified with chitin affinity chromatography followed by intein cleavage mediated by dithiothreitol (DTT) using previously described conditions (26). Proteins were stored and quantified as described for nickel chelate chromatography-purified proteins.
Enzymatic synthesis of acyl-CoAs.
Acyl-CoA synthesis reactions were carried out in 50 mM HEPES (pH 7.5), 1 mM DTT, 5 mM MgCl2, 5 mM ATP, and 2 mM CoA. The fatty acid substrate was added at 7.5 mM for butyrate, 3-hydroxyvalerate, trans-2-pentenoate, valerate, 4-methylvalerate, hexanoate, octanoate, decanoate, and dodecanoate, while 2 mM fatty acid substrate was used for tetradecanoate. To increase solubility, 1% and 3% (wt/vol) Triton X-100 was added to reaction mixtures with dodecanoate and tetradecanoate, respectively. CoA ligase enzymes were added at 500 nM for all reactions. VcsA was added to butyrate, 3-hydroxyvalerate, valerate, 4-methylvalerate, and hexanoate reaction mixtures, while FcsA was added to octanoate, decanoate, dodecanoate, and tetradecanoate reaction mixtures. Reactions were run overnight at room temperature for all substrates except trans-2-pentenoate, which was run at 30°C for 6 h because these conditions reduced the appearance of degradation products. Acyl-CoA ligases were precipitated from reactions at 95°C for 5 min and then removed by centrifugation.
Acyl-CoA products were purified from substrates using an Agilent 1200 series HPLC with diode array detection (DAD). A 100-μl reaction volume was injected onto an Agilent Eclipse XDB-C18 column, and separation was achieved using a mobile phase of 50 mM ammonium acetate and 0.1% (wt/vol) acetic acid (solvent A)–50 mM ammonium acetate, 0.1% (wt/vol) acetic acid, and 70% (vol/vol) acetonitrile (solvent B) gradient. The method began at 100% solvent A from 0 to 5 min, followed by a 0 to 100% gradient of solvent B from 5 to 50 min, followed by an isocratic step of 100% solvent B from 50 to 55 min. The gradient was run at a flow rate of 1 ml/min, and CoA was monitored by measuring absorbance at 258 nm. Fractions containing peaks corresponding to acyl-CoAs were collected, flash frozen in liquid nitrogen, and lyophilized. Dried acyl-CoAs were then resuspended in water, and the concentration was determined by the absorbance at 258 nm using the molar extinction coefficient of CoA (14,328 M−1 cm−1) within the linear range of detection (27).
Thioesterase activity assays.
Thioesterase activity was measured using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), which reacts with free CoA after it is released through thioesterase-mediated bond cleavage. Reactions were carried out in 1 mM DTNB, 100 mM HEPES (pH 8.0), and 20 μM acyl-CoA and were run in the linear range of each thioesterase. Substrate concentrations matched those used previously for EcTesB analysis (11). Reaction progress was monitored through the change in absorbance at 412 nm using the molar extinction coefficient of 5-thio-2-nitrobenzoate (14,150 M−1 cm−1), which is formed when DTNB reacts with free CoA (28).
RESULTS
In vivo functional screening for thioesterases that alter fatty acid production.
Acyl-CoA thioesterase substrate specificity plays an important role in deciding the SCFA profile produced using a CoA-based pathway. For example, if the target product is a saturated fatty acid, a thioesterase with broad substrate specificity will release fatty acids at each step in the biosynthetic pathway, depleting substrate pools and reducing the final product titer (Fig. 1). To find thioesterases that increase specificity for and production of 3-hydroxy, unsaturated, saturated, and branched fatty acids in the pathway, 62 putative thioesterases were chosen for screening. A group of six thioesterases, three TesB and three TesB2 enzymes, were chosen that had 38 to 50% amino acid identity to E. coli MG1655 TesB (EcTesB), which makes 3-hydroxy and saturated fatty acids in vivo (1, 6, 29), and 40 to 41% amino acid identity to A. borkumensis TesB2 enzyme, which was previously described as having specificity for 3-hydroxy acyl-CoAs (30) (Table 1).
TABLE 1.
List of thioesterases screened for activity in a biosynthetic pathway for the production of short-chain fatty acids
| TesB or TesB2 enzyme (reference) | E. coli MG1655 | P. putida KT2440 | P. ruminicola 23 | R. albus 7 | F. succinogenes S85 |
|---|---|---|---|---|---|
| E. coli TesB (11) | EcTesA | Pp244 | Pr655 | Ra865 | Fs266 |
| P. putida TesB | EcPaaI | Pp254 | Pr1075 | Ra874 | Fs270 |
| R. opacus TesB | EcEntH | Pp262 | Pr1498 | Ra880 | Fs368 |
| P. syringae TesB2 | EcFadM | Pp301 | Pr1510 | Ra1843 | Fs803 |
| A. borkumensis TesB2 (30) | EcYbfF | Pp580 | Pr1668 | Ra1929 | Fs944 |
| P. aeruginosa TesB2 | EcYbgC | Pp1218 | Pr1687 | Ra1938 | Fs1747 |
| EcYbhC | Pp1466 | Pr2385 | Ra2059 | Fs2108 | |
| EcYciA | Pp1980 | Ra2801 | |||
| EcYiiD | Pp2050 | Ra3109 | |||
| EcYigI | Pp2308 | ||||
| EcYdiI | Pp2318 | ||||
| Pp3281 | |||||
| Pp3807 | |||||
| Pp4105 | |||||
| Pp4180 | |||||
| Pp4181 | |||||
| Pp4975 | |||||
| Pp5198 | |||||
| Pp5331 | |||||
| Pp5356 | |||||
| PpPhaJ1 |
To incorporate a greater phylogenetic and functional diversity of thioesterases, the remaining 56 proteins encompassed all of the annotated thioesterases from five bacterial strains (Table 1). E. coli MG1655 (GenBank accession number NC_000913.2) thioesterases were chosen with the dual purpose of identifying those that increase product titers of desired SCFAs for use in heterologous pathways and those that reduce product titers and should be targeted for deletion from our host strain. The second source of thioesterases, Pseudomonas putida KT2440 (GenBank accession number NC_002947.3), was chosen for its phylogenetic similarity to E. coli and because it is a known producer of polyhydroxyalkanoates, which may indicate the presence of thioesterases with specificity to 3-hydroxy acyl-CoAs (31). The remaining three organisms, Prevotella ruminicola 23 (GenBank accession number NC_014033.1), Fibrobacter succinogenes S85 (GenBank accession number NC_017448.1), and Ruminococcus albus 7 (GenBank accession number NC_014833.1), were chosen because they are prevalent in the cow rumen microbiome and contribute to the high concentrations of SCFAs found there (3, 4, 32).
Each of the 62 putative thioesterases was individually overexpressed in E. coli containing all the necessary genes for CoA-dependent biosynthesis of valerate (Fig. 1). Previous work from our laboratory identified Treponema denticola Ter, Megaphaera elsdenii Pct, and Cupriavidus necator (formerly R. eutropha H16) BktB, PhaB, and PhaJ4 as suitable upstream pathway enzymes for valerate production (reference 6 and unpublished data). Cells were grown in LB medium containing glucose until log phase, then pathway genes were induced, and cultures were supplemented with either propionate or isobutyrate for synthesis of straight or branched SCFAs, respectively (Fig. 1). Liquid chromatography (LC)-based analysis of the culture supernatant was used to screen for enzymes that altered the fatty acid product profiles. Specifically, the resulting chromatograms were examined for increases and decreases of 3-hydroxyvalerate, trans-2-pentenoate, valerate, and 4-methylvalerate. Profiles produced by recombinant strains were compared to controls with either no recombinant thioesterase or with overproduced EcTesB, a thioesterase used previously by our group (1, 5, 6).
Overproduction of more than 20% of the thioesterases functionally screened in this study resulted in observable changes in substrate, intermediate, and product titers relative to the control strains (see Table S2 in the supplemental material). Four thioesterases, E. coli YciA (EcYciA), P. putida 1466 (Pp1466), P. putida 3807 (Pp3807), and P. putida 4975 (Pp4975), were associated with significant reduction in valerate and 4-methylvalerate production combined with increased acetate and propionate titers (see Table S2). Two thioesterases, P. ruminicola 1510 (Pr1510) and F. succinogenes 368 (Fs368), were associated with a production phenotype involving reduced titers of measured substrates, intermediates, and final products combined with increased glucose consumption (see Table S2). The most logical explanation for this phenotype is channeling of substrates to long-chain fatty acid biosynthesis. Because long-chain fatty acids could not be quantified with our HPLC system, additional experiments outside the scope of this work are required to determine the activity profile of these enzymes.
Host strain development and identification of thioesterases for in vitro analysis.
From the 12 annotated E. coli thioesterases screened during this study, three were chosen for deletion from the host strain. The tesB gene was deleted because its overexpression resulted in increased titers of 3-hydroxyvalerate, which could be a final product but also acts as a shunt product in the formation of trans-2-pentenoate or valerate; yciA was deleted because its overexpression increased final titers of the precursor-derived and exogenously supplied acids acetate and propionate, which correlates with previous work (33), while decreasing final product titers of valerate and 4-methylvalerate; and yigI was deleted because its overexpression resulted in decreased production of 4-methylvalerate and increased final acetate and isobutyrate titers. The resulting triple thioesterase deletion strain, E. coli MG1655(DE3) ΔendA ΔrecA ΔtesB ΔyciA ΔyigI, was used for further in vivo analysis of several active thioesterases.
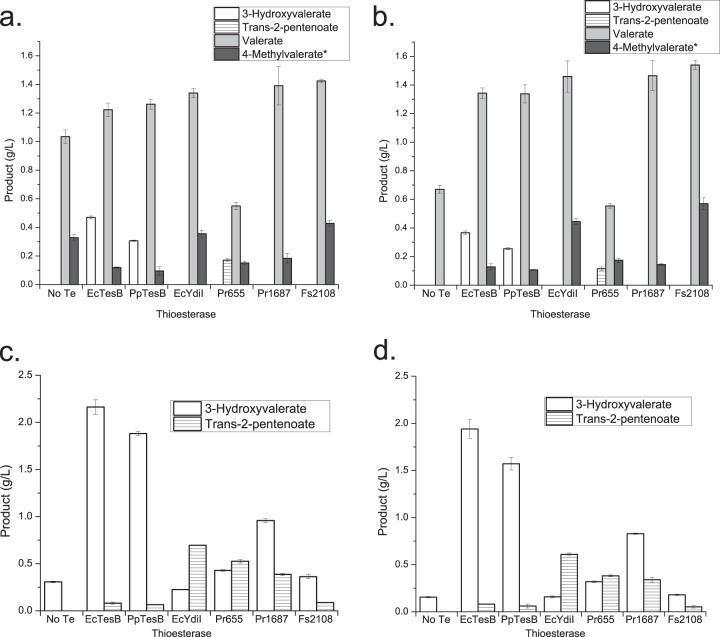
From the full set of 62 functionally screened thioesterases, EcTesB, PpTesB, EcYdiI, Fs2108, Pr655, and Pr1687 were chosen for more detailed in vivo and in vitro analyses because their overproduction resulted in increased titers of 3-hydroxyvalerate, trans-2-pentenoate, valerate, or 4-methylvalerate. Propionate and isobutyrate feeding experiments were performed in triplicate with these six thioesterases in both E. coli MG1655(DE3) ΔendA ΔrecA and the triple-thioesterase deletion strain containing pET/ter/bktB/pct and pCDF/phaB/phaJ4 in an effort to distinguish recombinant thioesterase activity from the background and potentially increase final product titers (Fig. 2a and b). The greatest 3-hydroxyvalerate titers were produced in recombinant strains overproducing TesB enzymes from E. coli and P. putida compared with the other 60 putative thioesterases (Fig. 2). The maximum titer (2.163 g/liter) resulted from overproduction of EcTesB (Table 2) and is similar to previously published titers from our laboratory (1). 3-Hydroxyvalerate titers were 20% lower in the triple-deletion strain overproducing EcTesB than in E. coli with native thioesterases.
FIG 2.
In vivo fatty acid production. Shown are titers of fatty acids produced from E. coli MG1655(DE3) ΔendA ΔrecA (a and c) and E. coli MG1655(DE3) ΔendA ΔrecA ΔtesB ΔyciA ΔyigI (b and d) expressing CoA-based pathways for the biosynthesis of valerate or 4-methylvalerate (a and b) or 3-hydroxyvalerate or trans-2-pentenoate (c and d) with six different thioesterases. The product titers shown were obtained from three separate shake flasks for each thioesterase, with error bars representing standard deviations. Each strain contained plasmid pCDF/phaB/phaJ4 with either pET/ter/bktB/pct (a and b) or pET/bktB/pct (c and d). Each strain also contained plasmid pACYCDuet-1 with the thioesterases displayed on the x axis. An absence of product bars for a thioesterase indicates that titers were below the limit of detection for the HPLC RID detector. The asterisk indicates that 4-methylvalerate titers were determined using three separate cultures for each thioesterase fed 15 mM isobutyrate in the place of propionate.
TABLE 2.
Maximum fatty acid titers observed for each product of the CoA-dependent pathway
| Strain | Maximum titer, g/liter (mean ± SD) |
|||
|---|---|---|---|---|
| 3-Hydroxyvaleratea | trans-2-Pentenoateb | Valeratec | 4-Methylvaleratec | |
| MG1655 ΔendA ΔrecA | 2.163 ± 0.078 | 0.695 ± 0.002 | 1.425 ± 0.011 | 0.428 ± 0.019 |
| MG1655 ΔendA ΔrecA ΔtesB ΔyciA ΔyigI | 1.940 ± 0.099 | 0.609 ± 0.015 | 1.540 ± 0.034 | 0.570 ± 0.041 |
Thioesterase EcTesB.
Thioesterase EcYdiI.
Thioesterase Fs2108.
Observable titers of unsaturated SCFAs were uncommon among the recombinant strains, with only Pr655 overproduction resulting in detectable trans-2-pentenoate during initial screening (Fig. 2a and b). To determine whether strains containing the other five thioesterases selected for further analysis could produce detectable unsaturated SCFAs in vivo, each was introduced into a strain lacking the downstream enoyl reductase gene ter, which does not produce the favored substrate valeryl-CoA (Fig. 2c and d). In these recombinant strains, product titers of trans-2-pentenoate were greatest for EcYdiI, Pr655, and Pr1687, with EcYdiI overproduction resulting in the greatest trans-2-pentenoate titer, 695 mg/liter (Table 2). To our knowledge, this is the largest published titer of an unsaturated SCFA from an engineered pathway in E. coli.
Three thioesterases, Fs2108, Pr1687, and EcYdiI, were chosen for further analysis because their overproduction increased titers of the saturated acids valerate and 4-methylvalerate while decreasing shunt product titers (Fig. 2a and b). Overproduction of thioesterase Fs2108 achieves the greatest final titers for both the straight SCFA valerate and the branched SCFA 4-methylvalerate (Table 2), with 3-hydroxy and unsaturated fatty acid intermediate titers below the limit of detection. While overproduction of Fs2108 resulted in a modest (15%) improvement in valerate titer over the EcTesB control, an improvement of more than 200% was observed for 4-methylvalerate titer over the same control. This improvement was possible in part because 4-methylvalerate titers were significantly lower than valerate titers for all 62 thioesterases tested, leaving more room for improvement. Overproduction of Fs2108 in the deletion strain resulted in approximately 10% more valerate and 30% more 4-methylvalerate than in the host containing all native thioesterases, which likely results from reduced hydrolysis of 3-hydroxyacyl-CoA and trans-2-acyl-CoA precursors. No 4-methylvalerate production from the triple-deletion strain was observed when isobutyrate was supplied without thioesterase overexpression, which indicates that the remaining native thioesterases have poor activity on 4-methylvaleryl-CoA (Fig. 2a and b).
Determination of in vitro SCFA substrate specificity for active thioesterases.
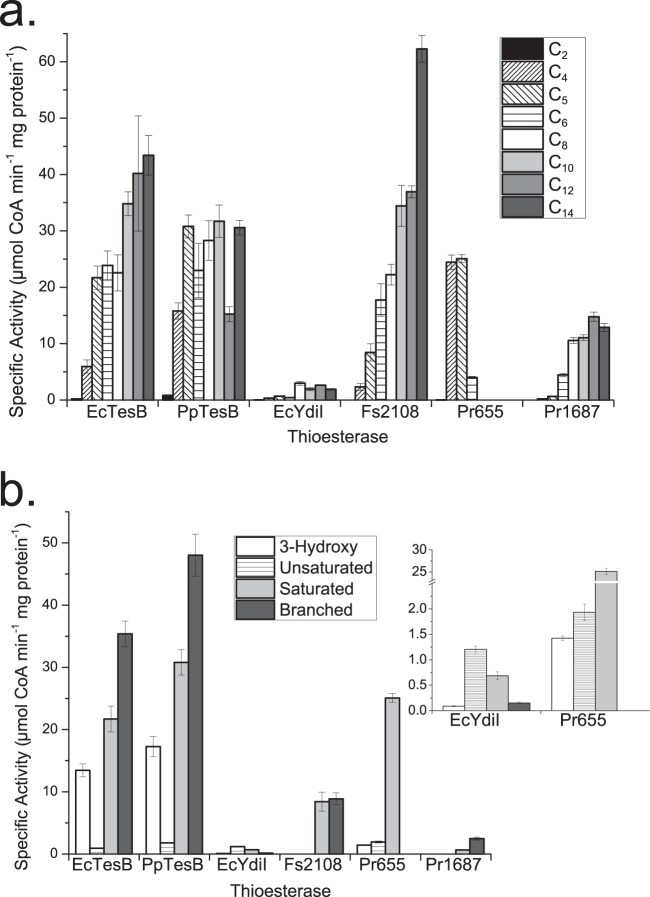
To determine the substrate preferences for the selected thioesterases, all six were overexpressed in E. coli BL21Star(DE3) and purified. Overexpression of Pr1687 and Fs2108 required the use of a vector that inserted an N-terminal chitin binding domain to obtain soluble protein. EcTesB, PpTesB, EcYdiI, and Pr655 were soluble with an N-terminal hexahistidine tag. Thioesterases were then tested for in vitro acyl-CoA hydrolysis activity on 20 μM acetyl-CoA, butyryl-CoA, valeryl-CoA, hexanoyl-CoA, octanoyl-CoA, decanoyl-CoA, dodecanoyl-CoA, and tetradecanoyl-CoA using an Ellman's reagent-based assay described previously for determining specific activity (11) (Fig. 3a). EcTesB, Fs2108, and Pr1687 showed a preference for longer-chain acyl-CoAs. PpTesB also showed a preference for longer acyl-CoAs, but the relationship was less linear than for EcTesB, Fs2108, and Pr1687. Pr655 showed a strong preference for butyryl- and valeryl-CoAs, and activity on acyl-CoAs longer than hexanoyl-CoA was below the limit of detection for this assay. EcYdiI had low activity for all substrates tested. A recent publication showing that EcYdiI has strong activity on the aromatic compound 1,4-dihydroxy-2-naphthoyl-CoA provides justification for the weak activity of this enzyme on the substrates provided in this study (34).
FIG 3.
Acyl-CoA specificity of six active thioesterases. (a) In vitro specific activity of six thioesterases on C2 to C14 acyl-CoAs. Specific activities represent the averages from three enzymatic reactions, with error bars representing standard deviations. They were determined in the linear range for each thioesterase with acyl-CoA concentrations of 20 μM for acetyl-CoA (C2), butyryl-CoA (C4), valeryl-CoA (C5), hexanoyl-CoA (C6), octanoyl-CoA (C8), decanoyl-CoA (C10), dodecanoyl-CoA (C12), and tetradecanoyl-CoA (C14). (b) In vitro specific activities of six thioesterases on 3-hydroxyvaleryl CoA (3-hydroxy), trans-2-pentenoyl-CoA (unsaturated), valeryl-CoA (saturated), and 4-methylvaleryl-CoA (branched). The inset shows enlarged activities of EcYdiI and Pr655 for comparison of low-level activities. Specific activities were determined using the same conditions as described for panel a. The absence of a bar for a given substrate or thioesterase specific activity indicates that the activity was below the limit of detection at absorbance of 412 nm.
Specific activities were also measured for all six thioesterases on 3-hydroxy, unsaturated, saturated, and branched valeryl-CoAs (Fig. 3b). For 3-hydroxyvaleryl-CoA, EcTesB and PpTesB had 6-fold-greater specific activity than the next best thioesterase. Both EcYdiI and Pr655 showed low activity for 3-hydroxyvaleryl-CoA, while specific activity of Fs2108 and Pr1687 for the same substrate was below the limit of detection. No thioesterase had specific activity greater than 2 μM CoA min−1 mg of protein−1 on trans-2-pentenoyl-CoA. Pr655, EcTesB, PpTesB, and EcYdiI displayed similar specific activities for this substrate; however, the small differences in specific acitivity translated into significant deviations in trans-2-pentenoate product titers when the thioesterases were overproduced in recombinant strains lacking the downstream enzyme Ter (Fig. 3b). Fs2108 and Pr1687 specific activities on trans-2-pentenoyl-CoA were below the limit of detection, but in vivo product profiles of strains lacking ter reflect a slight preference for 3-hydroxyvaleryl-CoA over trans-2-pentenoyl-CoA (Fig. 2d).
All six enzymes selected for further analysis had detectable activity on valeryl-CoA; however, EcTesB, PpTesB, Pr655, and Fs2108 had much greater activity than did EcYdiI and Pr1687 (Fig. 3b). Activity against the 4-methylvaleryl-CoA (branched) substrate was greater than or equal to activity on straight valeryl-CoA for EcTesB, PpTesB, Fs2108, and Pr1687. EcYdiI and Pr655 both showed weak activities against 4-methylvaleryl-CoA. In vitro analysis of the enzymes associated with the greatest titers of valerate revealed that Fs2108 had greater specific activity for both valeryl-CoA and 4-methylvaleryl-CoA than either Pr1687 or EcYdiI (Fig. 3b). Further, Fs2108 specific activity for both 3-hydroxyvaleryl-CoA and trans-2-pentenyl-CoA precursors was below the limit of detection for in vitro assays. The greater specific activity of Fs2108 on 4-methylvaleryl-CoA and valeryl-CoA combined with reduced specific activity on precursor acyl-CoAs likely accounts for the increased 4-methylvalerate and valerate titers found for Fs2108.
DISCUSSION
In this study, 62 putative thioesterases were screened for increased product titers of 3-hydroxy, unsaturated, saturated, and branched products of interest. From these, six were chosen for more thorough in vivo and in vitro analyses. Relating the specific activity profiles of EcTesB, PpTesB, EcYdiI, Fs2108, Pr655, and Pr1687 with their in vivo product profiles of 3-hydroxyvalerate, trans-2-pentenoate, valerate, or 4-methylvalerate leads to several important conclusions. First, the enzymes with the greatest in vivo product titers always had lower specific activity for the associated acyl-CoA than for alternative CoA substrates. For example, Fs2108 has a 6-fold-higher specific activity for tetradecanyl-CoA than for valeryl-CoA, indicating that valeryl-CoA is not the enzyme's preferred substrate. The disparity between the specific activities of Fs2108 for these two substrates indicates that our pathway relies on the promiscuous thioesterase activity of Fs2108. This is particularly evident for two thioesterases, EcYdiI and Pr1697, that were associated with increased valerate production despite having much higher specific activity for long-chain acyl-CoAs in the case of Pr1687 or aromatic acyl-CoAs for EcYdiI (34). The fact that all six thioesterases that were investigated in vitro prefer alternate substrates over those provided in our pathways suggests that both protein engineering and future bioprospecting efforts could further improve on the short-chain fatty acyl-CoA thioesterases discovered here.
Comparison of the in vivo product titers with in vitro substrate specificities of the six chosen thioesterases also suggests that after a specific activity level of 1 to 5 μM CoA min−1 mg of protein−1 is reached for a given acyl-CoA, the enzyme's activity for the pathway precursor acyl-CoAs becomes an important factor influencing final product titer, illustrating the importance of selecting pathway thioesterases with reduced activity on precursor acyl-CoAs. For example, strains overproducing thioesterases Pr1687 and EcYdiI produce more valerate than those overproducing EcTesB despite EcTesB having 30-fold-greater specific activity for valeryl-CoA than Pr1687 and EcYdiI (Fig. 2b). Accounting for this difference is the observation that EcTesB also has strong specific activity on the precursor 3-hydroxyvaleryl-CoA that translates into 3-hydroxyvalerate production, while Pr1687 does not have detectable activity on 3-hydroxyvaleryl-CoA. Further supporting the importance of reduced specific activity on pathway precursors is the observation that recombinant strains overproducing Pr655 produce much less valerate than strains overproducing EcYdiI and Pr1687 (Fig. 2a and b), even though Pr655 maintains greater specific activity for valeryl-CoA than thioesterases EcYdiI and Pr1687 (Fig. 3b). The increased specific activity of Pr655 on 3-hydroxyvaleryl-CoA and trans-2-pentenoyl-CoA precursors relative to those of EcYdiI and Pr1687 suggests that the reduced valerate titer results from increased precursor acyl-CoAs hydrolysis.
Confounding our conclusion that thioesterases with low specific activity on precursor acyl-CoAs have improved final product titers is the observation that recombinant strains overexpressing ydiI produce high valerate titers, with trans-2-pentenoate titers below the limit of detection despite EcYdiI exhibiting higher in vitro specific activity for 20 μM trans-2-pentenoyl-CoA than for 20 μM valeryl-CoA (Fig. 2a and 3b). In our recombinant strains, EcYdiI and the other screened thioesterases compete with the downstream pathway enzyme Ter for the substrate trans-2-pentenoyl-CoA. One plausible explanation for the absence of detectable trans-2-pentenoate from this recombinant strain is that EcYdiI has a Km, a measure of substrate affinity, for both trans-2-pentenoyl-CoA and valeryl-CoA greater than the 20 μM concentration used in our in vitro assay, which is reasonable to assume because EcYdiI has evolved for specificity toward aromatic acids (34). If Ter has a lower Km for trans-2-pentenoyl-CoA than EcYdiI, then it could reduce the intracellular concentration of trans-2-pentenoyl-CoA below 20 μM, causing EcYdiI activity for this compound to become physiologically irrelevant. In this situation, the intracellular valeryl-CoA concentration could increase to a point where low-level specific thioesterase activity would result in significant valerate production. Pr655, on the other hand, could have a lower Km for both trans-2-pentenoyl-CoA and valeryl-CoA, which would compete more effectively for the trans-2-pentenoyl-CoA with downstream enzyme Ter and allow continued low-level production of trans-2-pentenoate (Fig. 2a). Determination of kinetic parameters for these noncognate substrates is limited by the low concentrations of acyl-CoAs obtained from enzymatic synthesis and LC-based purification.
The thioesterase substrate preferences found by our in vitro experiments indicate the breadth of thioesterase functional diversity screened in this study and underline the importance of sampling in the selection of pathway enzymes (Fig. 3a and b). The TesB enzymes of E. coli and P. putida represent the most phylogenetically (50% amino acid identity) and functionally (Fig. 3a and b) similar enzymes, while the remaining four thioesterases appear both phylogenetically and functionally disparate. The TesB thioesterases showed both similar in vivo product profiles and similar in vitro specific activity profiles, which suggests that future screens aimed at acquiring more diverse TesB function should sample enzymes with lower similarity at the amino acid level than the E. coli and P. putida homologs. The diversity of the remaining 60 thioesterase phenotypes is an indication of the functional diversity inherent to bacterial thioesterases and suggests that many opportunities remain for isolating enzymes with improved specificity over those currently used in CoA-dependent biosynthetic pathways.
Our study also highlights the challenges associated with enzyme selection for metabolic pathways. A common route to selecting pathway enzymes is to rely on the proposed function of known enzymes in databases and the literature or to choose enzymes homologous to those with proposed functions. Unfortunately, enzyme annotation in public databases has degraded, as functional analysis has not kept up with the rate of sequence deposition (35). The absence of credible studies on short-chain acyl-CoA thioesterases prompted us to implement a broad functional screen of diverse candidates. Our results demonstrate the power of a well-designed screen for isolating uncharacterized or poorly characterized enzymes that improve product titers. Our methods for screening and in vitro characterization of substrate specificity serve as a template for investigating other poorly characterized enzyme functions for pathway development. The E. coli thioesterase EcYdiI serves as an example of an enzyme whose documented function may eliminate it from contention (34), but once included in our functional screen, it maintains the appropriate levels of promiscuous activity under intracellular conditions to provide the highest specificity for unsaturated SCFAs discovered so far. It is unlikely that selecting enzymes based solely upon annotated function would have identified this level of activity because short-chain acyl-coA thioesterase activity remains poorly characterized.
During this study, we isolated acyl-CoA thioesterases from diverse bacterial sources that increase production of saturated, unsaturated, and branched SCFAs through improvements in acyl-CoA substrate specificity. By comparing in vivo product profiles with in vitro specific activities of thioesterases that produced the greatest 3-hydroxyvalerate, trans-2-pentenoate, valerate, and 4-methylvalerate titers, we discovered that the most productive thioesterases found during functional screening (i) use promiscuous activity to produce the SCFAs monitored in this study and (ii) maintain low specific activity for pathway precursors relative to the preferred acyl-CoA. These findings indicate this study as a model for isolating enzymes for biochemical pathway functions that are poorly characterized in the literature. Further, the thioesterases we identified provide opportunities for increasing titers of desirable products as well as developing new pathways for the production of unsaturated SCFAs. We have also provided important in vivo and in vitro data on the production phenotypes and substrate specificities of poorly characterized acyl-CoA thioesterases for short-chain acyl-CoAs that are valuable for future bioprospecting and engineering studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the U.S. Department of Agriculture Agricultural Research Service for providing genomic DNA from cow rumen strains.
This work was supported by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office. The content of this article does not necessarily reflect the position or the policy of the U.S. government, and no official endorsement should be inferred.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03303-13.
REFERENCES
- 1.Tseng HC, Harwell CL, Martin CH, Prather KL. 2010. Biosynthesis of chiral 3-hydroxyvalerate from single propionate-unrelated carbon sources in metabolically engineered E. coli. Microb. Cell Fact. 9:96. 10.1186/1475-2859-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SH, Park OJ. 2009. Uses and production of chiral 3-hydroxy-gamma-butyrolactones and structurally related chemicals. Appl. Microbiol. Biotechnol. 84:817–828. 10.1007/s00253-009-2143-0 [DOI] [PubMed] [Google Scholar]
- 3.Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, Deneke J, Drinkwater C, Ivanova NN, Mikhailova N, Chertkov O, Goodwin LA, Currie CR, Mead D, Brumm PJ. 2011. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One 6:e18814. 10.1371/journal.pone.0018814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purushe J, Fouts DE, Morrison M, White BA, Mackie RI, Coutinho PM, Henrissat B, Nelson KE. 2010. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb. Ecol. 60:721–729. 10.1007/s00248-010-9692-8 [DOI] [PubMed] [Google Scholar]
- 5.Martin CH, Dhamankar H, Tseng HC, Sheppard MJ, Reisch CR, Prather KL. 2013. A platform pathway for production of 3-hydroxyacids provides a biosynthetic route to 3-hydroxy-gamma-butyrolactone. Nat. Commun. 4:1414. 10.1038/ncomms2418 [DOI] [PubMed] [Google Scholar]
- 6.Tseng HC, Prather KL. 2012. Controlled biosynthesis of odd-chain fuels and chemicals via engineered modular metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 109:17925–17930. 10.1073/pnas.1209002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. 2013. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. U. S. A. 110:11290–11295. 10.1073/pnas.1307129110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handke P, Lynch SA, Gill RT. 2011. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab. Eng. 13:28–37. 10.1016/j.ymben.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 9.Jing F, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ. 2011. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 12:44. 10.1186/1471-2091-12-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X, Vora H, Khosla C. 2008. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10:333–339. 10.1016/j.ymben.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 11.Nie L, Ren Y, Janakiraman A, Smith S, Schulz H. 2008. A novel paradigm of fatty acid beta-oxidation exemplified by the thioesterase-dependent partial degradation of conjugated linoleic acid that fully supports growth of Escherichia coli. Biochemistry 47:9618–9626. 10.1021/bi801074e [DOI] [PubMed] [Google Scholar]
- 12.Naggert J, Narasimhan ML, DeVeaux L, Cho H, Randhawa ZI, Cronan JE, Jr, Green BN, Smith S. 1991. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J. Biol. Chem. 266:11044–11050 [PubMed] [Google Scholar]
- 13.Cantu DC, Chen Y, Reilly PJ. 2010. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 19:1281–1295. 10.1002/pro.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voelker TA, Davies HM. 1994. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 176:7320–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu A, Tan X, Yao L, Lu X. 2013. Fatty alcohol production in engineered E. coli expressing Marinobacter fatty acyl-CoA reductases. Appl. Microbiol. Biotechnol. 97:7061–7071. 10.1007/s00253-013-5027-2 [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Vora H, Khosla C. 2010. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab. Eng. 12:378–386. 10.1016/j.ymben.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 17.Gronenberg LS, Marcheschi RJ, Liao JC. 2013. Next generation biofuel engineering in prokaryotes. Curr. Opin. Chem. Biol. 17:462–471. 10.1016/j.cbpa.2013.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda I, Delessert S, Hasegawa S, Seto Y, Zuber S, Poirier Y. 2006. The peroxisomal Acyl-CoA thioesterase Pte1p from Saccharomyces cerevisiae is required for efficient degradation of short straight chain and branched chain fatty acids. J. Biol. Chem. 281:11729–11735. 10.1074/jbc.M511762200 [DOI] [PubMed] [Google Scholar]
- 19.Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71:982–994. 10.1002/prot.21786 [DOI] [PubMed] [Google Scholar]
- 20.Crosby HA, Pelletier DA, Hurst GB, Escalante-Semerena JC. 2012. System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J. Biol. Chem. 287:15590–15601. 10.1074/jbc.M112.352104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237. 10.1016/j.plasmid.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon MD, Rush JS, Thomas MG. 2012. Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J. Bacteriol. 194:2809–2818. 10.1128/JB.00088-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 26.Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. 2010. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49:8815–8817. 10.1021/bi1012854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giljanović J, Prkic A. 2010. Determination of coenzyme A (CoASH) in the presence of different thiols by using flow-injection with a UV/Vis spectrophotometric detector and potentiometric determination of CoASH using an iodide ISE. Molecules 15:100–113. 10.3390/molecules15010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riener CK, Kada G, Gruber HJ. 2002. Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 373:266–276. 10.1007/s00216-002-1347-2 [DOI] [PubMed] [Google Scholar]
- 29.Zheng Z, Gong Q, Liu T, Deng Y, Chen JC, Chen GQ. 2004. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl. Environ. Microbiol. 70:3807–3813. 10.1128/AEM.70.7.3807-3813.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabirova JS, Ferrer M, Lunsdorf H, Wray V, Kalscheuer R, Steinbuchel A, Timmis KN, Golyshin PN. 2006. Mutation in a “tesB-like” hydroxyacyl-coenzyme A-specific thioesterase gene causes hyperproduction of extracellular polyhydroxyalkanoates by Alcanivorax borkumensis SK2. J. Bacteriol. 188:8452–8459. 10.1128/JB.01321-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huijberts GN, Eggink G, de Waard P, Huisman GW, Witholt B. 1992. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl. Environ. Microbiol. 58:536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergman EN. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590 [DOI] [PubMed] [Google Scholar]
- 33.Clomburg JM, Vick JE, Blankschien MD, Rodriguez-Moya M, Gonzalez R. 2012. A synthetic biology approach to engineer a functional reversal of the beta-oxidation cycle. ACS Synth. Biol. 1:541–554. 10.1021/sb3000782 [DOI] [PubMed] [Google Scholar]
- 34.Chen M, Ma X, Chen X, Jiang M, Song H, Guo Z. 2013. Identification of a hotdog fold thioesterase involved in the biosynthesis of menaquinone in Escherichia coli. J. Bacteriol. 195:2768–2775. 10.1128/JB.00141-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. 2009. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 5:e1000605. 10.1371/journal.pcbi.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.