Abstract
Members of the genus Aeromonas are ubiquitous in nature and have increasingly been implicated in numerous diseases of humans and other animal taxa. Although some species of aeromonads are human pathogens, their presence, density, and relative abundance are rarely considered in assessing water quality. The objectives of this study were to identify Aeromonas species within Lake Erie, determine their antibiotic resistance patterns, and assess their potential pathogenicity. Aeromonas strains were isolated from Lake Erie water by use of Aeromonas selective agar with and without tetracycline and ciprofloxacin. All isolates were analyzed for hemolytic ability and cytotoxicity against human epithelial cells and were identified to the species level by using 16S rRNA gene restriction fragment length polymorphisms and phylogenetic analysis based on gyrB gene sequences. A molecular virulence profile was identified for each isolate, using multiplex PCR analysis of six virulence genes. We demonstrated that Aeromonas comprised 16% of all culturable bacteria from Lake Erie. Among 119 Aeromonas isolates, six species were identified, though only two species (Aeromonas hydrophila and A. veronii) predominated among tetracycline- and ciprofloxacin-resistant isolates. Additionally, both of these species demonstrated pathogenic phenotypes in vitro. Virulence gene profiles demonstrated a high prevalence of aerolysin and serine protease genes among A. hydrophila and A. veronii isolates, a genetic profile which corresponded with pathogenic phenotypes. Together, our findings demonstrate increased antibiotic resistance among potentially pathogenic strains of aeromonads, illustrating an emerging potential health concern.
INTRODUCTION
Members of the genus Aeromonas are Gram-negative bacilli that are typically associated with freshwater environments, though they are also present in soil and clinical sources (1) and are symbionts of zebrafish (2), leeches (3), and dreissenid mussels (4). Within the past few decades, there has been a marked increase in Aeromonas research due to the clinical relevance of this genus in fish, humans, and other animal taxa (5). For example, Aeromonas salmonicida has received considerable attention from both fishery biologists and hatchery operators due to its ability to cause furunculosis, especially in commercially valuable salmonids (6, 7). A. salmonicida infections range from chronic to acute infections that, in some cases, lead to death within hours (7). Additionally, A. hydrophila and A. veronii have been identified as the etiologic agents of major fish kills worldwide (5) as well as epizootic ulcerative syndrome in several species of farmed fish (9).
Aeromonas species (most often A. caviae, A. hydrophila, and A. veronii) have received increased attention due to their association with a multitude of human diseases, ranging from subclinical conditions such as gastroenteritis to more severe extraenteric conditions, including wound infections and septicemia (1, 5, 10, 11). For example, A. hydrophila and A. veronii were the leading causes of skin and soft tissue infections found in survivors of the 2004 tsunami in Thailand (12). Aeromonads were also associated with pulmonary infections resulting from the tsunami (13) but are more typically identified as a cause of pneumonia in immunocompromised patients (14).
Due to the overuse of broad-spectrum antibiotics in clinical settings, agriculture, and fish hatcheries, there has been an increase in antibiotic resistance among disease-causing Aeromonas species (15). Members of the genus have developed a high level of resistance to commonly used antibiotics such as ampicillin (16), which has become so widespread that low levels of the antibiotic are used as a selective agent for the bacterium (17). There has also been an emerging resistance to other antibiotics, including tetracycline and ciprofloxacin, among Aeromonas species. The overuse of tetracycline as a broad-spectrum antibiotic within fish hatcheries and feedlots, as well as clinics, appears to have resulted in an increase in resistance (18). Although ciprofloxacin resistance is less common among environmental isolates, recent studies have indicated low levels of resistance among A. caviae, A. hydrophila, and A. sobria isolates, thus indicating that it is an emerging concern (15).
Identification and characterization of pathogenic and antibiotic-resistant strains of Aeromonas in aquatic environments are important steps in curtailing their spread and preventing further disease. The present study reports the prevalence of tetracycline and ciprofloxacin resistance among potentially pathogenic Aeromonas isolates from the Eastern Basin of Lake Erie. Additionally, virulence gene profiles were identified, along with hemolytic and cytotoxic phenotypes.
MATERIALS AND METHODS
Relative abundance and antibiotic resistance of Aeromonas in the environment.
Water samples were collected in Erie, PA, between 12 July and 10 November 2010, from the south pier of the shipping channel that connects Presque Isle Bay to Lake Erie (42°09.079′N, 80°04.738′W). Samples were collected 1 meter above the bottom substrate by use of a sewage sampler (model 990 A10; Wildlife Supply Co., Yulee, FL) and sterile 300-ml bottles. All samples were held on ice until processed. Samples of water (8 μl to 25 ml) were vacuum filtered through standard 0.45-μm nitrocellulose filters (Millipore, Billerica, MA) within 4 h of collection.
Filters were placed onto 60-mm by 15-mm tryptic soy agar (TSA; Difco, Lawrence, KS) plates to determine total CFU. Filters were also placed on ampicillin-dextrin agar (Hardy Diagnostics, Santa Maria, CA) with vancomycin (2 μg/ml) according to U.S. Environmental Protection Agency (EPA) methodology (19). Previous comparisons of 10 selective and differential media indicated that ampicillin-dextrin agar was most effective in differentiating Aeromonas from other waterborne bacteria (20). To further increase the selectivity of the ampicillin-dextrin agar, irgasan was added at a concentration of 5 μg/ml, i.e., the concentration contained in Aeromonas agar (Lab M, Bury, Lancashire, United Kingdom). Replica plating experiments on water collected at the site indicated that this modified agar (referred to here as ampicillin-dextrin agar with vancomycin and irgasan [ADA-VI]) reduced growth of non-Aeromonas organisms (colonies with coloration other than yellow) but did not affect the growth of presumptive Aeromonas organisms (yellow colonies) (data not shown). Volumes were plated onto TSA and ADA-VI plates in duplicate and incubated at 30°C for 24 h. Total numbers of cultivatable bacteria (total CFU on TSA) and presumptive Aeromonas isolates (yellow colonies on ADA-VI) (CFU/100 ml) were determined for each sample date. The relative abundance of presumptive Aeromonas was calculated by dividing the number of presumptive Aeromonas organisms by the total CFU and multiplying by 100.
Resistance of Aeromonas in the environment against tetracycline and ciprofloxacin was assessed by filter plating water samples onto ADA-VI supplemented with tetracycline (4 μg/ml) (21) or ciprofloxacin (0.16 μg/ml). The ciprofloxacin concentration was approximately midway between the MIC50 and MIC90 values reported by Ko et al. (15) for Aeromonas. Percent resistance of presumptive Aeromonas to each antibiotic was calculated by dividing the concentration of resistant Aeromonas organisms (CFU/100 ml) by the total concentration of Aeromonas organisms (CFU/100 ml) and multiplying by 100.
Isolation and confirmation of antibiotic resistance.
Presumptive Aeromonas organisms (i.e., yellow colonies) were picked randomly from filter plates containing ADA-VI (n = 54), ADA-VI with tetracycline (n = 50), and ADA-VI with ciprofloxacin (n = 20), streaked for isolation onto TSA plates, and then grown overnight in tryptic soy broth (TSB; Difco, Lawrence, KS). Glycerol (15%) frozen stocks were made from each overnight culture and stored at −80°C for later use. Each isolate was tested for cytochrome oxidase activity, trehalose fermentation, and indole production (19). Physiological tests with negative results were repeated to help ensure accuracy. Antibiotic resistance of isolates obtained from ADA-VI plates lacking or supplemented with tetracycline or ciprofloxacin was further evaluated by their ability to grow in TSB with tetracycline (4 μg/ml) or ciprofloxacin (0.16 μg/ml).
16S rRNA gene RFLP and gyrB sequence analysis.
Aeromonas species identification was performed following previous methodology (22). Briefly, bacterial DNA was obtained through lysis using BR-A buffer (GenScript, Piscataway, NJ). Universal primers (forward, 5′-AGAGTTTGATCATGGCTCAG-3′; and reverse, 5′-GGTTACCTTGTTACGACTT-3′) were used to PCR amplify a 1,465-bp 16S rRNA gene segment, using GoTaq Green master mix (Promega, Madison, WI). The amplicon was subsequently double digested with AluI and MboI (New England BioLabs, Ipswich, MA) at 37°C overnight and electrophoresed in a 4% MetaPhor agarose gel (Lonza, Rockland, ME) as previously described (22). Strains of A. hydrophila (ATCC 7966), A. caviae (ATCC 15468), and A. veronii (ATCC 9071) were used as positive controls. All restriction fragment length polymorphism (RFLP) patterns were further confirmed by gyrB sequence analysis (23). The neighbor-joining method (24) was used to compare 11 A. veronii, 6 A. hydrophila, and all other Aeromonas isolates to other Aeromonas reference strains. MEGA5 (25) was used to predict the evolutionary distances, using the maximum composite likelihood method (26). Figure S1 in the supplemental material demonstrates the phylogenetic analysis. For Aeromonas isolates for which gyrB and RFLP analyses revealed different species, gyrB results were used. All A. veronii and A. hydrophila RFLP patterns were in agreement with phylogenetic analysis based on gyrB sequence homology.
Pathogenicity in vitro.
Isolates were streaked onto TSA and 5% sheep blood agar plates to determine their hemolytic ability and were tested for cytotoxicity following a previously described protocol (27). Briefly, frozen stocks of isolates were plated onto ADA-VI and incubated at 30°C for 24 h. Isolated colonies from ADA-VI plates were used to make TSB cultures. After incubation at 30°C for 48 h, 1-ml aliquots were centrifuged at 12,000 × g for 30 min. Supernatants were then passed through a 0.2-μm syringe filter, diluted 1:5 in minimal essential medium (MEM) with 5% fetal bovine serum (FBS), and added to HeLa cells in a 96-well plate at 2 × 105 cells/ml. The negative control was TSB medium alone. Cytotoxicity was measured at 18 to 24 h postinoculation via an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (Sigma, St. Louis, MO) following the manufacturer's protocol. Briefly, MTT solution (5 mg/ml in phosphate-buffered saline [PBS]) was added to RPMI 1640 with 10% FBS and without phenol red for 2 h at 37°C to identify viable cells. MTT crystals were solubilized in 10% Triton X-100, 0.1 N HCl diluted in isopropanol. Cytotoxicity was determined by measuring the absorbance at 590 nm minus that at 650 nm, using an enzyme-linked immunosorbent assay (ELISA) plate reader and comparing the values to those of negative-control wells containing only HeLa cells and medium alone. Isolates were considered cytotoxic if their absorbance values were less than 50% the absorbance values of the negative controls.
Scanning electron microscopy.
Cytotoxic effects of Aeromonas isolates on HeLa cells were further investigated using scanning electron microscopy. Cell cultures were fixed in 100% ice-cold methanol for 10 min, with subsequent incubation with 1% OsO4 (Electron Microscopy Sciences, Hatfield, PA) overnight at room temperature. Samples were dehydrated with increasing concentrations of ethanol (10%, 25%, 50%, 75%, 90%, 100%, and 100% [vol/vol]) and stored in 100% ethanol at 4°C for future use. Samples were critical point dried under CO2, using a Ladd 28000 (Ladd Research Industries, Burlington, VT) critical point dryer, and then coated with gold with a Polaron Instruments E5100 (Polaron Instruments, Inc., Doylestown, PA) sputter coater. Samples were imaged using a Hitachi S-570 scanning electron microscope (Hitachi High-Technologies Corp., Tokyo, Japan) with a 25-kV acceleration voltage. Images were generated using an Orion digitizer (E.L.I. Microscopy, Charleroi, Belgium) connected to the microscope. The figure was composed using the Inkscape 0.47 vector graphics program. The final image was corrected for brightness and contrast after composition, and corrections were applied uniformly to all parts of the figure.
Multiplex analysis of virulence genes.
Multiplex analysis of virulence genes (lateral flagellum, nuclease, aerolysin, serine protease, glycerophospholipid cholesterol acyltransferase [GCAT], and lipase genes) was performed following a previously described protocol (28). Briefly, 1 μl of overnight culture was lysed in 20 μl of BR-A buffer (GenScript), and multiplex PCR was performed with 2× BacReady PCR master mix (GenScript) and six sets of primers (Table 1), specific for the virulence genes mentioned above. PCR amplification was performed under the following conditions: denaturation for 3 min at 94°C followed by 35 cycles of 94°C for 60 s, 64°C for 30 s, and 72°C for 45 s, with a final extension step at 72°C for 7 min. PCR products were analyzed using agarose gel electrophoresis.
TABLE 1.
Primer pairs used for PCR amplification of virulence genes and the 16S rRNA gene
| Target gene | Primer | Sequence (5′ → 3′) | Product size (bp) | GenBank accession no. (reference) | Reference |
|---|---|---|---|---|---|
| Lateral flagellum B | LatFB-F | GACCAGCAAGGATAGTGGGTTGGAG | 624 | AF348135 | 28 |
| LatFB-R | AAGCACCATCGCGTTGGTATAAGG | ||||
| Nuclease | Nucl-F | CAGGATCTGAACCGCCTCTATCAGG | 504 | AF004392 | 28 |
| Nucl-R | GTCCCAAGCTTCGAACAGTTTACGC | ||||
| Aerolysin | Aero-F | GAGCGAGAAGGTGACCACCAAGAAC | 417 | M16495 | 28 |
| Aero-R | TTCCAGTCCCACCACTTCACTTCAC | X65044 | |||
| Serine protease | SerP-F | ACGGAGTGCGTTCTTCCTACTCCAG | 211 | X67043 | 28 |
| SerP-R | CCGTTCATCACACCGTTGTAGTCG | AF159142 | |||
| GCAT | GCAT-F | CATGTCTCCGCCTATCACAACAAGC | 339 | X70686 | 28 |
| GCAT-R | CCAGAACATCTTGCCCTCACAGTTG | AF268080 | |||
| Lipase | Lip-F | GACCCCCTACCTGAACCTGAGCTAC | 155 | U63543 | 28 |
| Lip-R | AGTGACCCAGGAAGTGCACCTTGAG | ||||
| 16S rRNA | 16S-F | AGAGTTTGATCATGGCTCAG | 1,465 | 22 | |
| 16S-R | GGTTACCTTGTTACGACTT |
RESULTS
Relative abundance and antibiotic resistance of Aeromonas in the environment.
Water samples were collected from Presque Isle Bay on eight dates between 12 July and 10 November 2010 and used to enumerate total cultivatable bacteria and presumptive Aeromonas organisms (Fig. 1). Abundances of total cultivatable bacteria (growth on TSA) ranged from 4,275 to 68,750 CFU/100 ml of water (mean = 25,672 CFU/100 ml of water; standard deviation [SD] = 20,909 CFU/100 ml of water). The spike in bacterial abundance on 13 September coincided with a dense algal bloom in the bay. Abundances of presumptive Aeromonas organisms (yellow colonies on ADA-VI) ranged from 475 to 36,750 CFU/100 ml of water (mean = 3,382 CFU/100 ml of water; SD = 2,753 CFU/100 ml of water), resulting in a prevalence range of 1.1% to 29.0% (mean = 16.0%; SD = 10.2%).
FIG 1.
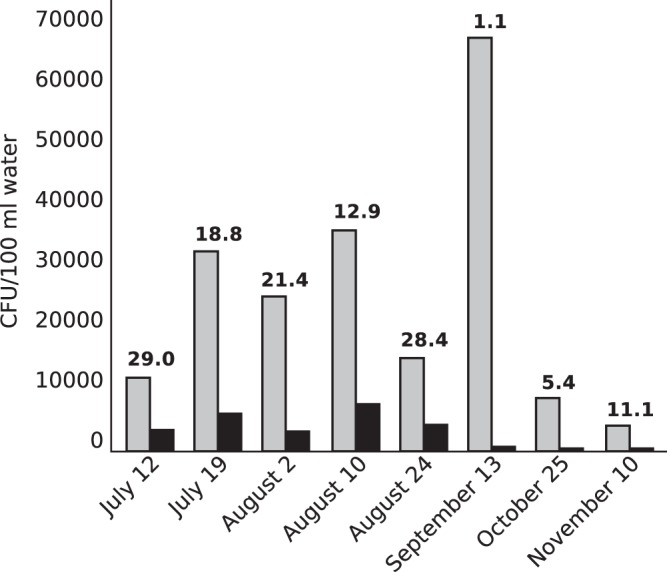
Prevalence of Aeromonas in Presque Isle Bay, Lake Erie. Abundances (CFU/100 ml water) of cultivatable bacteria (light bars) and presumptive Aeromonas organisms (dark bars) were determined by filter plating water samples onto tryptic soy agar and ADA-VI, respectively. The value above each pair of bars is the relative abundance (%) of presumptive Aeromonas. The mean relative abundance of Aeromonas at the site was 16.0% (SD = 10.2%) for the study period.
Resistance of presumptive Aeromonas organisms from Presque Isle Bay to tetracycline and ciprofloxacin (quantified by comparing growth on ADA-VI filter plates supplemented with tetracycline or ciprofloxacin to that on ADA-VI filter plates lacking both antibiotics) was relatively rare in the environment, comprising less than 1% of presumptive Aeromonas organisms across dates. On average, 0.53% (SD = 0.20%) of presumptive Aeromonas organisms were resistant to tetracycline, and 0.12% (SD = 0.02%) were resistant to ciprofloxacin.
Species identification and antibiotic resistance of Aeromonas isolates.
Species identification was performed on 124 isolates by using 16S rRNA gene RFLP analysis; 119 isolates demonstrated published Aeromonas RFLP patterns (22) and were further confirmed by gyrB sequence analysis. The presence and relative abundance of species varied among ADA-VI plates lacking additional antibiotics and those supplemented with tetracycline or ciprofloxacin (Fig. 2). Of the 54 isolates obtained from ADA-VI filter plates lacking additional antibiotics, 51 were confirmed as Aeromonas, and they belonged to six different species (A. allosaccharophila, A. bestiarum, A. jandaei, A. hydrophila, A. sobria, and A. veronii). The majority of Aeromonas organisms isolated from those plates were identified as A. veronii (80.3%). All 50 isolates from ADA-VI filter plates supplemented with tetracycline demonstrated previously described RFLP patterns (22). In contrast to the species distribution on ADA-VI filter plates lacking additional antibiotics, the majority of isolates from plates supplemented with tetracycline were identified as A. veronii (74.0%) or A. hydrophila (24.0%). Of the 20 isolates obtained from ADA-VI plates supplemented with ciprofloxacin, 18 exhibited previously published RFLP patterns (22) and were identified as either A. hydrophila (72.2%) or A. veronii (27.8%).
FIG 2.
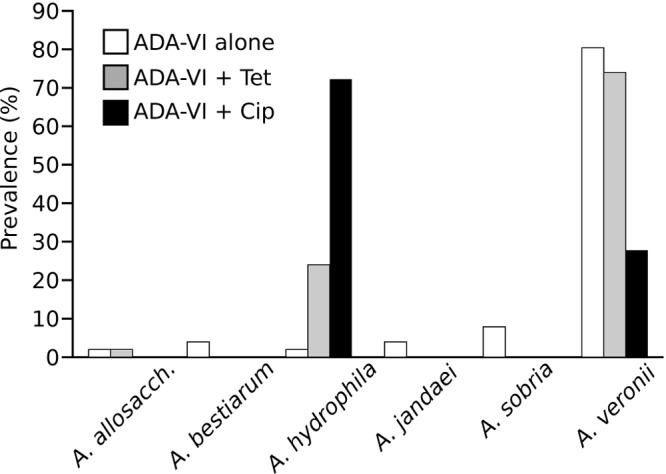
Prevalences of Aeromonas species among tetracycline- and ciprofloxacin-resistant populations. Isolates (n = 119 total) were randomly picked from filter plates containing ADA-VI (white bars; n = 51), ADA-VI supplemented with 4 μg/ml tetracycline (gray bars; n = 50), and ADA-VI supplemented with 0.16 μg/ml ciprofloxacin (black bars; n = 18). Species identity was determined using RFLP analysis of the 16S rRNA gene and was confirmed by gyrB sequencing analysis. A. allosacch., A. allosaccharophila.
Among the 26 A. hydrophila isolates, 96% demonstrated resistance to tetracycline, ciprofloxacin, or both antibiotics when grown in TSB supplemented with antibiotics (Table 2). Similarly, 50.6% of 83 A. veronii isolates demonstrated resistance to one or both of the antibiotics (Table 2). In contrast, of the 10 isolates that were identified as species other than A. hydrophila or A. veronii, only 1 (an isolate of A. allosaccharophila) demonstrated resistance (Table 2).
TABLE 2.
Antibiotic resistance phenotypes of Aeromonas species isolated from Lake Erie
| Aeromonas species | No. of isolates | No. (%) of isolates with phenotypea |
|||
|---|---|---|---|---|---|
| Tets Cips | Tetr Cips | Tets Cipr | Tetr Cipr | ||
| A. allosaccharophila | 2 | 1 (50.0) | 1 (50.0) | 0 (0) | 0 (0) |
| A. bestiarum | 2 | 2 (100.0) | 0 (0) | 0 (0) | 0 (0) |
| A. hydrophila | 26 | 1 (3.8) | 3 (11.5) | 1 (3.8) | 21 (80.8) |
| A. jandaei | 2 | 2 (100.0) | 0 (0) | 0 (0) | 0 (0) |
| A. sobria | 4 | 4 (100.0) | 0 (0) | 0 (0) | 0 (0) |
| A. veronii | 83 | 41 (49.4) | 32 (38.6) | 2 (2.4) | 8 (9.6) |
Tet, tetracycline; Cip, ciprofloxacin; s, susceptible; r, resistant.
Biochemical properties of Aeromonas isolates.
A variety of biochemical properties were observed among the 119 isolates confirmed as Aeromonas (Table 3). The typical Aeromonas phenotype (19) of oxidase activity, trehalose fermentation, and indole production (O+ T+ I+) was observed in 81 (68.1%) isolates. Of the remaining 38 isolates, 26 were positive for oxidase activity and trehalose fermentation but did not produce indole (O+ T+ I−). Analysis of RFLP patterns and gyrB sequences identified the majority of isolates with the O+ T+ I− phenotype as A. hydrophila. The absence of trehalose fermentation and indole production was found in 10 (8.4%) isolates, all of which were identified as A. veronii. Oxidase activity was absent in two isolates, identified as A. veronii and A. hydrophila, with the latter isolate also being negative for indole production. Overall, A. hydrophila isolates demonstrated the highest prevalence of atypical physiological phenotypes, with only 23.1% testing positive for cytochrome oxidase activity, trehalose fermentation, and indole production.
TABLE 3.
Biochemical properties of Aeromonas species isolated from Lake Erie
| Species | No. of isolates | No. (%) of isolates with physiologic profilea |
||||
|---|---|---|---|---|---|---|
| O+ T+ I+ | O+ T+ I− | O+ T− I− | O− T+ I+ | O− T+ I− | ||
| A. allosaccharophila | 2 | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| A. bestiarum | 2 | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| A. hydrophila | 26 | 6 (23.1) | 19 (73.1) | 0 (0) | 0 (0) | 1 (3.8) |
| A. jandaei | 2 | 1 (50.0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) |
| A. sobria | 4 | 3 (75.0) | 1 (25.0.0) | 0 (0) | 0 (0) | 0 (0) |
| A. veronii | 83 | 67 (80.7) | 5 (6.0) | 10 (12.0) | 1 (1.2) | 0 (0) |
| Total | 119 | 81 (68.1) | 26 (21.8) | 10 (8.4) | 1 (0.8) | 1 (0.8) |
O, cytochrome oxidase activity; T, trehalose fermentation; I, indole production.
Characterization of virulence phenotypes and virulence gene profiles.
Beta-hemolysis and cytotoxic effects of cell-free supernatants on human epithelial cells were evaluated to determine the potential pathogenicity of Aeromonas isolates. Virulence phenotypes were associated predominantly with two species: A. veronii and A. hydrophila (Table 4). Beta-hemolysis was exhibited in 73% of A. veronii isolates, with 63% of isolates within this species demonstrating cytotoxic activity against human epithelial cells. Prevalences of beta-hemolytic and cytotoxic phenotypes of A. hydrophila isolates were even higher, i.e., 100% and 92%, respectively. Cytotoxic activity appeared to be mediated via different virulence mechanisms, including a loss of membrane integrity with subsequent membrane blebbing (Fig. 3B) and pore-forming activities (Fig. 3C). Other species of aeromonads had markedly lower prevalences of potential pathogenicity, with only one isolate demonstrating hemolysis. However, the small sample sizes of A. allosaccharophila, A. bestiarum, A. jandaei, and A. sobria preclude detailed comparisons across species.
TABLE 4.
Prevalences of virulence phenotypes and gene profiles among Aeromonas species
| Aeromonas species and gene profile | No. of isolates (n = 119) | Beta-hemolysis |
Cytotoxicity |
Presence of virulence gene |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % of each species (no. of isolates) | % of each genotype (no. of isolates) | % of each species (no. of isolates) | % of each genotype (no. of isolates) | Nuclease | Aerolysin | GCAT | Serine protease | Lipase | ||
| A. allosaccharophila | 2 | 0 (0) | 0 (0) | |||||||
| Gene profile 1 | 2 | 0 (0) | 0 (0) | − | − | − | + | − | ||
| A. bestiarum | 2 | 0 (0) | 0 (0) | |||||||
| Gene profile 1 | 2 | 0 (0) | 0 (0) | + | + | + | − | + | ||
| A. hydrophila | 26 | 100 (26) | 92 (24) | |||||||
| Gene profile 1 | 23 | 100 (23) | 88 (21) | + | + | − | + | + | ||
| Gene profile 2 | 2 | 100 (2) | 100 (2) | + | + | − | + | − | ||
| Gene profile 3 | 1 | 100 (1) | 100 (1) | − | + | − | + | − | ||
| A. jandaei | 2 | 50 (1) | 0 (0) | |||||||
| Gene profile 1 | 1 | 100 (1) | 0 (0) | − | + | − | + | − | ||
| Gene profile 2 | 1 | 0 (0) | 0 (0) | − | − | − | − | − | ||
| A. sobria | 4 | 0 (0) | 0 (0) | |||||||
| Gene profile 1 | 2 | 0 (0) | 0 (0) | + | + | − | − | − | ||
| Gene profile 2 | 1 | 0 (0) | 0 (0) | − | + | − | − | − | ||
| Gene profile 3 | 1 | 0 (0) | 0 (0) | − | − | − | − | − | ||
| A. veronii | 83 | 73 (61) | 63 (52) | |||||||
| Gene profile 1 | 1 | 0 (0) | 0 (0) | + | + | + | + | + | ||
| Gene profile 2 | 2 | 100 (2) | 100 (2) | − | + | − | + | + | ||
| Gene profile 3 | 61 | 84 (51) | 74 (45) | − | + | − | + | − | ||
| Gene profile 4 | 8 | 38 (3) | 25 (2) | − | + | − | − | − | ||
| Gene profile 5 | 3 | 100 (3) | 67 (2) | − | − | − | + | − | ||
| Gene profile 6 | 8 | 25 (2) | 0 (0) | − | − | − | − | − | ||
FIG 3.
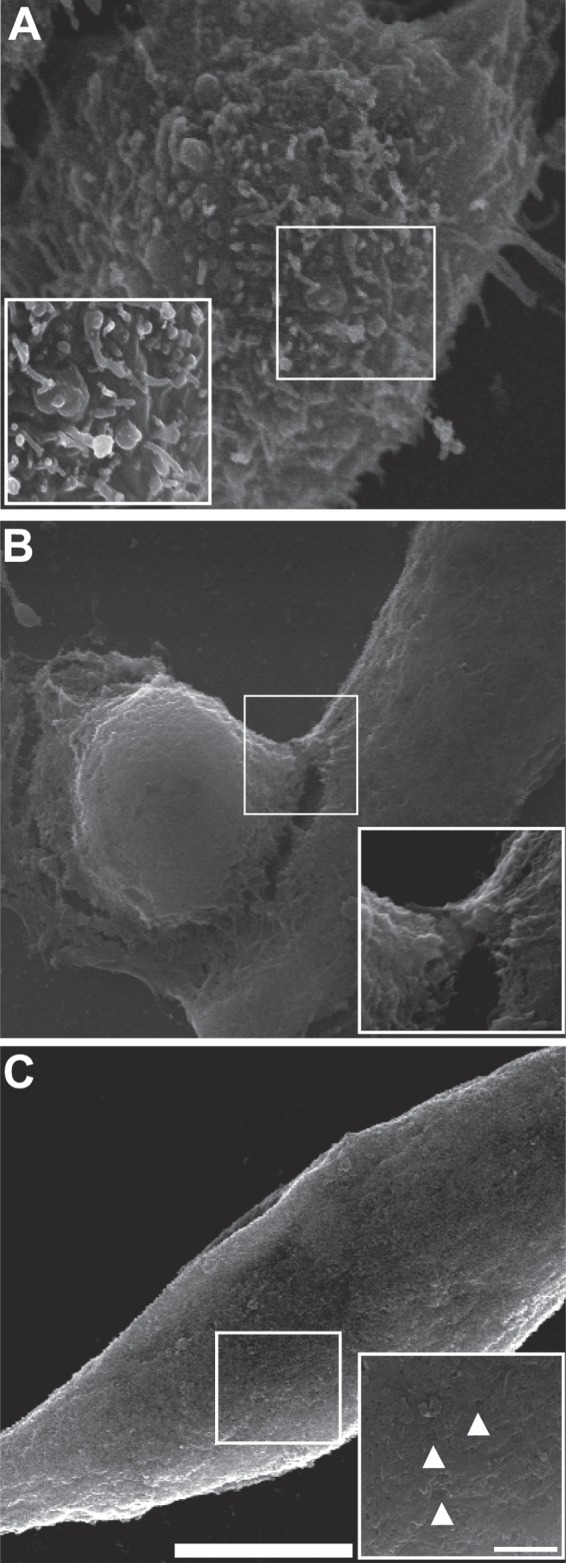
Cytotoxic effects of Aeromonas cell-free supernatants on human epithelial cells. HeLa cells were incubated with cell-free bacterial supernatants for 4 h and fixed to identify cytotoxic phenotypes. (A) Control HeLa cells. The inset shows an intact membrane. (B) HeLa cells following 4 h of incubation with Aeromonas supernatant, showing evident blebbing of the plasma membrane. The inset shows that the bleb is contiguous with the cell membrane. (C) HeLa cells following 4 h of incubation with another Aeromonas isolate supernatant, showing an irregular shape. The inset demonstrates pores in the plasma membrane (marked by arrowheads). Bars, 7.5 μm (main panels) and 1.5 μm (insets).
Identifying a virulence gene profile associated with hemolytic and cytotoxic phenotypes would aid in understanding Aeromonas pathogenesis. We tested for the presence of six different virulence genes (28) (lateral flagellum B, nuclease, aerolysin, serine protease, GCAT, and lipase genes) in each isolate. No isolates demonstrated the presence of the lateral flagellum gene (data not shown). Hemolytic and cytotoxic phenotypes were strongly associated with isolates harboring the aerolysin and serine protease genes (Fig. 4 and Table 4). However, determining which of those genes conferred virulence was difficult due to the presence of both serine protease and aerolysin genes in 100% of A. hydrophila isolates and 77% of A. veronii isolates (Fig. 4 and Table 4).
FIG 4.
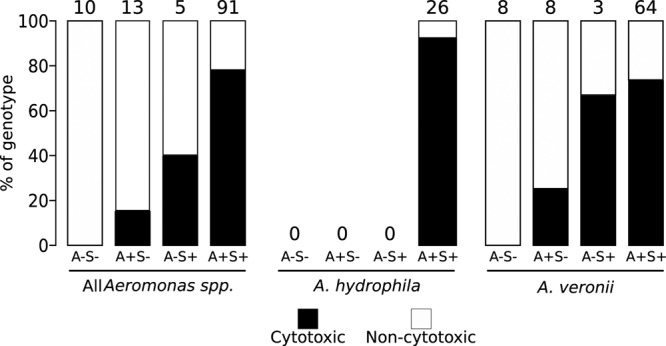
Aerolysin and serine protease association with cytotoxic phenotypes. Multiplex PCR was performed on all Aeromonas isolates, and the isolates were grouped into virulence gene profiles based on the presence or absence of aerolysin and serine protease. Cytotoxic activity is represented by dark bars, and noncytotoxicity is represented by white bars. The percent cytotoxicity against HeLa cells for each genotypic pattern was determined by dividing the number of cytotoxic isolates by the total number of isolates. The total numbers of isolates exhibiting the described genotypes are given above the bars. A, aerolysin; S, serine protease.
DISCUSSION
We evaluated antibiotic resistance among environmental isolates of Aeromonas, a genus of bacteria gaining attention due to its increasing prevalence as an etiologic agent in an array of diseases associated with both immunocompetent and immunocompromised individuals. This study is unique in that we were interested in comparing the prevalences of potentially pathogenic phenotypes and virulence gene profiles between antibiotic-resistant and -susceptible Aeromonas isolates. Few studies of Aeromonas have been performed in North America's Great Lakes, bodies of water that contain 20% of the earth's fresh surface water. Our findings identified Aeromonas species similar to those previously reported within zebra mussels obtained from Lake Erie (4), though a shortage of studies within the Great Lakes prevents detailed comparisons among studies.
Among aquatic isolates from the Eastern Basin of Lake Erie, Aeromonas comprised, on average, 16% of total cultivatable bacteria, with A. veronii (80%) being the most prevalent species. A. hydrophila, one of the most pathogenic Aeromonas species, comprised around 2% of the sample population. Estimates of antibiotic resistance among Aeromonas organisms were 0.53% for tetracycline and 0.12% for ciprofloxacin. However, antibiotic resistance was not uniformly distributed among species. Instead, over 98% of tetracycline- and ciprofloxacin-resistant isolates were identified as A. hydrophila or A. veronii, and 72% of isolates that were resistant to both antibiotics were identified as A. hydrophila. The higher prevalence of tetracycline resistance than ciprofloxacin resistance among Aeromonas organisms is in agreement with other environmental and clinical studies (15, 21, 29, 30) and highlights the antibiotic resistance in the genus, specifically among Aeromonas species that are associated with human disease.
The sources of antibiotic-resistant Aeromonas at our study site are not clear. However, combined sewer overflows (CSOs) in 2010 on three dates, all of which had rainfall events of >9 cm, resulted in 805,604 gallons of untreated water entering Presque Isle Bay and its outer harbor (City of Erie, Bureau of Sewers, Erie Wastewater Treatment Plant). More recent data from 2011 and 2012 showed CSOs of 1,364,014 gallons and 409,488 gallons, respectively. Although the city of Erie's treatment plant captured and treated over 99.9% of the 12,126.57 million gallons of wastewater that flowed into it in 2010 (well above the compliance requirement of 85%), CSOs that result from short-duration, high-intensity rain events may be a source of Aeromonas and other bacteria of human origin. Quantitative PCR on water from several tributary streams located near the city of Erie revealed the presence of human-derived Bacteroides at all study sites (Coastal Zone Management [CZM] projects CZM 2006-PE.10 and CZM 2007 PE.05). These findings suggest that contamination of tributary streams from sanitary sewer overflows, malfunctioning on-lot septic systems, municipal sewerage conveyance systems, and storm water discharges makes its way into Lake Erie and may serve as a source of human-derived bacteria at our study site (CZM projects CZM 2006-PE.10 and CZM 2007 PE.05). It is also possible that effluent from a trout hatchery operated by the Pennsylvania Fish and Boat Commission and several trout raceways operated by local sportsmen's organizations, along with agricultural runoff from within the watershed, may be sources of bacterial contamination originating in other animals; however, no point sources appear to have been identified to date. Long-term monitoring of Escherichia coli along the beaches of Presque Isle State Park and swimming advisories that typically occur after large runoff events indicate periodically high fecal coliform populations near the study site (31). The potential sources of bacterial contamination mentioned above are not unique to Lake Erie and suggest that antibiotic-resistant Aeromonas may be present in other areas of the Great Lakes.
Aeromonas populations at our study site may also be influenced by animals known to harbor them. Aeromonads, including A. hydrophila, are known to occur as part of the normal surface and gut floras of leeches (3, 32). Although leeches are probably present at our site, gut content analyses of bluegills (Lepomis macrochirus), pumpkinseeds (Lepomis gibbosus), and round gobies (Neogobius melanostomus) suggest that the population density is relatively low or that these species of fish avoid eating leeches (33, 34). Dreissenid mussels (Dreissena polymorpha and Dreissena rostriformis bugensis) from the Eastern Basin of Lake Erie have also been shown to harbor several species of Aeromonas (4), including A. hydrophila, A. veronii, A. sobria, A. media, A. jandaei, and A. salmonicida (T. A. Skwor and G. M. Andraso, unpublished data). Dense populations of dreissenids occur at our study site (34) and in most regions in the Great Lakes and may therefore serve as reservoirs for potentially pathogenic aeromonads (4).
In addition to the potential clinical consequences of increasing antibiotic resistance of Aeromonas, their ubiquitous nature may make them a reservoir and ecological vehicle for spreading antibiotic resistance to other bacteria, including E. coli (35). Antibiotic resistance genes among Aeromonas organisms have been associated with integron-associated R plasmids, and tetracycline resistance, in particular, has been associated with an array of different plasmids within A. hydrophila isolates (36). Ciprofloxacin resistance among environmental and clinical Aeromonas isolates has been attributed to plasmids (37, 38) as well as chromosomal mutations in quinolone resistance-determining regions of gyrA (39, 40).
The use of biochemical characters (oxidase activity, trehalose fermentation, and indole production) to identify presumptive Aeromonas has been adopted by the U.S. Environmental Protection Agency (19). Although the 119 isolates described in this study grew as yellow colonies on ADA-VI agar and were confirmed by published 16S rRNA gene RFLP patterns (22) and gyrB sequence analysis, only 81 (68.1%) had typical Aeromonas physiological properties. In particular, the majority of A. hydrophila isolates (73.1%) were indole negative, and one isolate was negative for oxidase activity and indole production. Deviation from typical Aeromonas physiological traits has been noted previously in A. hydrophila and other species; however, in that study, only one A. hydrophila isolate tested negative for indole production (1). Considering that all negative tests were repeated to ensure accuracy, a potential reason for these atypical findings among A. hydrophila isolates may be specific to antibiotic-resistant strains. In our study, we identified only one A. hydrophila isolate that was susceptible to both antibiotics among the 54 isolates obtained from ADA-VI plates lacking tetracycline and ciprofloxacin, thus preventing us from determining the typical physiological phenotype or phenotypes of susceptible strains.
The majority (25/26 isolates) of A. hydrophila isolates were obtained from ADA-VI plates with tetracycline or ciprofloxacin. After identifying antibiotic resistance, we examined their potentials to be pathogenic by their cytotoxic and hemolytic activities. We found that 92% of antibiotic-resistant A. hydrophila isolates were cytotoxic and 100% were hemolytic. Our results are consistent with previous work on A. hydrophila that demonstrated 98% of water isolates as cytotoxic and 97% as hemolytic (41). Another Aeromonas species associated with human disease, A. veronii, also demonstrated high prevalences of cytotoxicity (72%) and hemolysis (88%) in our study. The apparently lower prevalence of potentially pathogenic A. veronii isolates than of A. hydrophila isolates is consistent with previous work showing that only 61% of A. veronii water isolates were cytotoxic and hemolytic (41).
Numerous genes have been proposed to mediate pathogenic phenotypes of Aeromonas, specifically those associated with type II (42), III (43), and VI (44) secretion systems. The regulation of these secretion systems is mediated via quorum sensing and may be associated with polar and lateral flagellar activity (45–48). Type II secretion systems regulated by quorum sensing are involved in the secretion of multiple virulence factors, including serine protease and aerolysin (49, 50). In our study, serine protease and aerolysin genes were the most prevalent virulence genes among all cytotoxic and hemolytic Aeromonas isolates. The prevalence of both genes was 100% in A. hydrophila and 77% in A. veronii, corresponding with their high prevalence of pathogenic phenotypes (≥63%). Of the remaining 10 Aeromonas isolates, only one (an isolate of A. jandaei) exhibited a hemolytic phenotype which corresponded with the presence of both aerolysin and serine protease. The remaining isolates lacked pathogenic phenotypes and the presence of both aerolysin and serine protease. Clinically, both aerolysin and serine protease have demonstrated cytotoxic effects (51, 52), and their cooccurrence among A. hydrophila strains is more common in diseased than in healthy fish (53).
One limitation of this study is that the virulence gene profiles of locally obtained clinical isolates are unknown. We were therefore unable to determine if the virulence genotypes seen in potentially pathogenic environmental isolates are the same genotypes found in local clinical settings. Deletion experiments with these virulence genes among Aeromonas isolates would also support their association with pathogenicity, especially considering a study that demonstrated that serine protease and GCAT were not associated with pathogenicity of A. salmonicida (54). However, the multitude of Aeromonas virulence genes and animal models utilized to study pathogenicity present difficulty in interpreting these findings.
Antibiotic resistance continues to increase among bacterial communities, but the association of resistance with pathogenic phenotypes has rarely been elucidated. It is imperative to determine if antibiotic-resistant strains are those capable of causing disease. In the studies described here, we identified the greatest prevalence of tetracycline and ciprofloxacin resistance in isolates of Aeromonas species previously associated with human disease: A. hydrophila and A. veronii. The high prevalence of cytotoxicity and hemolytic ability in those isolates suggests that they have the potential to cause human diseases such as wound infections and gastroenteritis if they are encountered by humans in local recreational waters. Due to their high prevalence in potentially pathogenic isolates, it appears that aerolysin and serine protease may play important roles in the pathogenicity of Aeromonas. Future studies are needed to identify the source of the antibiotic-resistant Aeromonas described herein to determine if human or agricultural waste is directly influencing bacterial resistance levels within the recreational waters of Lake Erie.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a minigrant through the Regional Science Consortium at the Tom Ridge Environmental Center, the Presque Isle Partnership, and Pennsylvania Sea Grant (T.S. and G.A.). Funding was also provided by the Great Lakes Innovative Stewardship through Education Network (GLISTEN), Gannon University faculty research grants (T.S.), a Sigma Xi undergraduate research grant (J.S.), and a Cooney-Jackman Endowed Professorship (G.A.).
We thank Sean Fouse, Tom Russo, John Little, Cassandra Wasson, and Jillian Rhoades for their laboratory assistance.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03645-13.
REFERENCES
- 1.Sinha S, Shimada T, Ramamurthy T, Bhattacharya SK, Yamasaki S, Takeda Y, Nair GB. 2004. Prevalence, serotype distribution, antibiotic susceptibility and genetic profiles of mesophilic Aeromonas species isolated from hospitalized diarrhoeal cases in Kolkata, India. J. Med. Microbiol. 53:527–534. 10.1099/jmm.0.05269-0 [DOI] [PubMed] [Google Scholar]
- 2.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. 2011. Evidence for a core gut microbiota in the zebrafish. ISME J. 5:1595–1608. 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nonomura H, Kato N, Ohno Y, Itokazu M, Matsunaga T, Watanabe K. 1996. Indigenous bacterial flora of medicinal leeches and their susceptibilities to 15 antimicrobial agents. J. Med. Microbiol. 45:490–493. 10.1099/00222615-45-6-490 [DOI] [PubMed] [Google Scholar]
- 4.Gu J-D, Mitchell R. 2002. Indigenous microflora and opportunistic pathogens of the freshwater zebra mussel, Dreissena polymorpha. Hydrobiologia 474:81–90. 10.1023/A:1016517107473 [DOI] [Google Scholar]
- 5.Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewart KV, Belanger JC, Williams J, Karakach T, Penny S, Tsoi SC, Richards RC, Douglas SE. 2005. Identification of genes differentially expressed in Atlantic salmon (Salmo salar) in response to infection by Aeromonas salmonicida using cDNA microarray technology. Dev. Comp. Immunol. 29:333–347. 10.1016/j.dci.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 7.Toranzo AE, Magarinos B, Romalde JL. 2005. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246:37–61. 10.1016/j.aquaculture.2005.01.002 [DOI] [Google Scholar]
- 8.Reference deleted.
- 9.Rahman M, Colque-Navarro P, Kuhn I, Huys G, Swings J, Mollby R. 2002. Identification and characterization of pathogenic Aeromonas veronii biovar sobria associated with epizootic ulcerative syndrome in fish in Bangladesh. Appl. Environ. Microbiol. 68:650–655. 10.1128/AEM.68.2.650-655.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deodhar LP, Saraswathi K, Varudkar A. 1991. Aeromonas spp. and their association with human diarrheal disease. J. Clin. Microbiol. 29:853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu XJ, Ferguson MR, Popov VL, Houston CW, Peterson JW, Chopra AK. 1998. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect. Immun. 66:3501–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiransuthikul N, Tantisiriwat W, Lertutsahakul K, Vibhagool A, Boonma P. 2005. Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin. Infect. Dis. 41:E93–E96. 10.1086/497372 [DOI] [PubMed] [Google Scholar]
- 13.Maegele M, Gregor S, Steinhausen E, Bouillon B, Heiss MM, Perbix W, Wappler F, Rixen D, Geisen J, Berger-Schreck B, Schwarz R. 2005. The long-distance tertiary air transfer and care of tsunami victims: injury pattern and microbiological and psychological aspects. Crit. Care Med. 33:1136–1140. 10.1097/01.CCM.0000163269.42524.50 [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay C, Bhargava A, Ayyagari A. 2003. Aeromonas hydrophila and aspiration pneumonia: a diverse presentation. Yonsei Med. J. 44:1087–1090 [DOI] [PubMed] [Google Scholar]
- 15.Ko WC, Yu KW, Liu CY, Huang CT, Leu HS, Chuang YC. 1996. Increasing antibiotic resistance in clinical isolates of Aeromonas strains in Taiwan. Antimicrob. Agents Chemother. 40:1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vila J, Ruiz J, Gallardo F, Vargas M, Soler L, Figueras MJ, Gascon J. 2003. Aeromonas spp. and traveler's diarrhea: clinical features and antimicrobial resistance. Emerg. Infect. Dis. 9:552–555. 10.3201/eid0905.020451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Want SV, Millership SE. 1990. Effects of incorporating ampicillin, bile salts and carbohydrates in media on the recognition and selection of Aeromonas spp. from faeces. J. Med. Microbiol. 32:49–54. 10.1099/00222615-32-1-49 [DOI] [PubMed] [Google Scholar]
- 18.Goni-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C. 2000. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl. Environ. Microbiol. 66:125–132. 10.1128/AEM.66.1.125-132.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. Environmental Protection Agency 2001. Method 1605: Aeromonas in finished water by membrane filtration using ampicillin-dextrin agar with vancomycin (ADA-V). Reference no. EPA-821-R-01-034. U.S. Environmental Protection Agency, Office of Water, Washington, DC [Google Scholar]
- 20.Handfield M, Simard P, Letarte R. 1996. Differential media for quantitative recovery of waterborne Aeromonas hydrophila. Appl. Environ. Microbiol. 62:3544–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravena-Roman M, Inglis TJJ, Henderson B, Riley TV, Chang BJ. 2012. Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob. Agents Chemother. 56:1110–1112. 10.1128/AAC.05387-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrell N, Acinas SG, Figueras MJ, Martinez-Murcia AJ. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35:1671–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martino ME, Fasolato L, Montemurro F, Rosteghin M, Manfrin A, Patarnello T, Novelli E, Cardazzo B. 2011. Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes. Appl. Environ. Microbiol. 77:4986–5000. 10.1128/AEM.00708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cumberbatch N, Gurwith MJ, Langston C, Sack RB, Brunton JL. 1979. Cytotoxic enterotoxin produced by Aeromonas hydrophila: relationship of toxigenic isolates to diarrheal disease. Infect. Immun. 23:829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nam IY, Joh K. 2007. Rapid detection of virulence factors of Aeromonas isolated from a trout farm by hexaplex-PCR. J. Microbiol. 45:297–304 [PubMed] [Google Scholar]
- 29.Igbinosa IH, Okoh AI. 2012. Antibiotic susceptibility profile of Aeromonas species isolated from wastewater treatment plant. ScientificWorldJournal 2012:764563. 10.1100/2012/764563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obi CL, Ramalivhana J, Samie A, Igumbor EO. 2007. Prevalence, pathogenesis, antibiotic susceptibility profiles, and in-vitro activity of selected medicinal plants against Aeromonas isolates from stool samples of patients in the Venda region of South Africa. J. Health Popul. Nutr. 25:428–435 [PMC free article] [PubMed] [Google Scholar]
- 31.Leslie H. 2007. Beach management by DCNR, p 16–28 In Beach closures at Presque Isle State Park: Past, Present, and Future Conference proceedings. US Department of Commerce, National Technical Information Service, Alexandria, VA [Google Scholar]
- 32.Worthen PL, Gode CJ, Graf J. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775–4781. 10.1128/AEM.00356-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andraso GM. 2005. Summer food habits of pumpkinseeds (Lepomis gibbosus) and bluegills (Lepomis macrochirus) in Presque Isle Bay, Lake Erie. J. Great Lakes Res. 31:397–404. 10.1016/S0380-1330(05)70271-9 [DOI] [Google Scholar]
- 34.Andraso GM, Ganger MT, Adamczyk J. 2011. Size-selective predation by round gobies (Neogobius melanostomus) on dreissenid mussels in the field. J. Great Lakes Res. 37:298–304. 10.1016/j.jglr.2011.02.006 [DOI] [Google Scholar]
- 35.Moura A, Oliveira C, Henriques I, Smalla K, Correia A. 2012. Broad diversity of conjugative plasmids in integron-carrying bacteria from wastewater environments. FEMS Microbiol. Lett. 330:157–164. 10.1111/j.1574-6968.2012.02544.x [DOI] [PubMed] [Google Scholar]
- 36.Borrego JJ, Morinigo MA, Martinez-Manzanares E, Bosca M, Castro D, Barja JL, Toranzo AE. 1991. Plasmid associated virulence properties of environmental isolates of Aeromonas hydrophila. J. Med. Microbiol. 35:264–269. 10.1099/00222615-35-5-264 [DOI] [PubMed] [Google Scholar]
- 37.Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14:231–237. 10.3201/eid1402.070677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han JE, Kim JH, Choresca CH, Jr, Shin SP, Jun JW, Chai JY, Park SC. 2012. A small IncQ-type plasmid carrying the quinolone resistance (qnrS2) gene from Aeromonas hydrophila. Lett. Appl. Microbiol. 54:374–376. 10.1111/j.1472-765X.2012.03208.x [DOI] [PubMed] [Google Scholar]
- 39.Figueira V, Vaz-Moreira I, Silva M, Manaia CM. 2011. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45:5599–5611. 10.1016/j.watres.2011.08.021 [DOI] [PubMed] [Google Scholar]
- 40.Han JE, Kim JH, Cheresca CH, Jr, Shin SP, Jun JW, Chai JY, Han SY, Park SC. 2012. First description of the qnrS-like (qnrS5) gene and analysis of quinolone resistance-determining regions in motile Aeromonas spp. from diseased fish and water. Res. Microbiol. 163:73–79. 10.1016/j.resmic.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 41.Khajanchi BK, Fadl AA, Borchardt MA, Berg RL, Horneman AJ, Stemper ME, Joseph SW, Moyer NP, Sha J, Chopra AK. 2010. Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76:2313–2325. 10.1128/AEM.02535-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bo JN, Howard SP. 1991. Mutagenesis and isolation of Aeromonas hydrophila genes which are required for extracellular secretion. J. Bacteriol. 173:1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burr SE, Stuber K, Wahli T, Frey J. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:5966–5970. 10.1128/JB.184.21.5966-5970.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. 2010. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192:155–168. 10.1128/JB.01260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlova EV, Popov VL, Sha J, Foltz SM, Erova TE, Agar SL, Horneman AJ, Chopra AK. 2008. Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 45:343–354. 10.1016/j.micpath.2008.08.007 [DOI] [PubMed] [Google Scholar]
- 46.Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr. Opin. Microbiol. 14:9–15. 10.1016/j.mib.2010.09.017 [DOI] [PubMed] [Google Scholar]
- 47.Maltz M, Graf J. 2011. The type II secretion system is essential for erythrocyte lysis and gut colonization by the leech digestive tract symbiont Aeromonas veronii. Appl. Environ. Microbiol. 77:597–603. 10.1128/AEM.01621-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sha J, Pillai L, Fadl AA, Galindo CL, Erova TE, Chopra AK. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446–6457. 10.1128/IAI.73.10.6446-6457.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khajanchi BK, Sha J, Kozlova EV, Erova TE, Suarez G, Sierra JC, Popov VL, Horneman AJ, Chopra AK. 2009. N-Acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155:3518–3531. 10.1099/mic.0.031575-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swift S, Lynch MJ, Fish L, Kirke DF, Tomas JM, Stewart GS, Williams P. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez LA, Ellis AE, Nieto TP. 1992. Purification and characterisation of an extracellular metalloprotease, serine protease and haemolysin of Aeromonas hydrophila strain B32: all are lethal for fish. Microb. Pathog. 13:17–24. 10.1016/0882-4010(92)90028-M [DOI] [PubMed] [Google Scholar]
- 52.Wong CY, Heuzenroeder MW, Flower RL. 1998. Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology 144:291–298. 10.1099/00221287-144-2-291 [DOI] [PubMed] [Google Scholar]
- 53.Hu M, Wang N, Pan ZH, Lu CP, Liu YJ. 2012. Identity and virulence properties of Aeromonas isolates from diseased fish, healthy controls and water environment in China. Lett. Appl. Microbiol. 55:224–233. 10.1111/j.1472-765X.2012.03281.x [DOI] [PubMed] [Google Scholar]
- 54.Vipond R, Bricknell IR, Durant E, Bowden TJ, Ellis AE, Smith M, MacIntyre S. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 66:1990–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


