Abstract
Saccharomyces cerevisiae sake yeast strain Kyokai no. 7 has one of the highest fermentation rates among brewery yeasts used worldwide; therefore, it is assumed that it is not possible to enhance its fermentation rate. However, in this study, we found that fermentation by sake yeast can be enhanced by inhibiting mitophagy. We observed mitophagy in wild-type sake yeast during the brewing of Ginjo sake, but not when the mitophagy gene (ATG32) was disrupted. During sake brewing, the maximum rate of CO2 production and final ethanol concentration generated by the atg32Δ laboratory yeast mutant were 7.50% and 2.12% higher than those of the parent strain, respectively. This mutant exhibited an improved fermentation profile when cultured under limiting nutrient concentrations such as those used during Ginjo sake brewing as well as in minimal synthetic medium. The mutant produced ethanol at a concentration that was 2.76% higher than the parent strain, which has significant implications for industrial bioethanol production. The ethanol yield of the atg32Δ mutant was increased, and its biomass yield was decreased relative to the parent sake yeast strain, indicating that the atg32Δ mutant has acquired a high fermentation capability at the cost of decreasing biomass. Because natural biomass resources often lack sufficient nutrient levels for optimal fermentation, mitophagy may serve as an important target for improving the fermentative capacity of brewery yeasts.
INTRODUCTION
Sake is a traditional Japanese alcoholic beverage produced from steamed rice and koji. During the manufacturing process, glucose is produced (saccharification) from the starch present in rice by the actions of enzymes produced by the koji fungus Aspergillus oryzae. Glucose is fermented to ethanol by Saccharomyces cerevisiae sake yeast strains (1). Sake contains the highest ethanol concentration of all the brewed alcoholic beverages worldwide. This high ethanol concentration is generated by technologies that include successive addition of enzymes and nutrients derived from koji during sake brewing (2, 3), a 3-step pitching process, brewing in winter, and the historical selection of high-ethanol-producing sake yeast strains (1). Sake yeast strains have been selected through a long history of cultivation, ranging from 100 to 400 years. The most frequently used sake yeast at present is Kyokai no. 7 (K7), which was isolated from sake mash in 1946 (4, 5). This strain produces a high concentration of ethanol, because it lacks functions of proteins encoded by MSN4, PPT1, and RIM15, which are required to mount a stress response (6–8). For this reason, researchers in this field believe that it is difficult to further augment the fermentation rate of this sake yeast.
Although rice is used as a raw material to brew sake, the surface of rice contains many constituents such as amino acids that impart a heavy and complex taste to sake. Because Japanese consumers tend to prefer a light and clear taste, rice with a polished surface is used for sake brewing. Sake is categorized into two representative types depending on the extent of rice polishing; these types have been specified by the official guidelines of the Japanese government (http://www.nta.go.jp/shiraberu/senmonjoho/sake/hyoji/seishu/gaiyo/02.htm). When the weight of the removed surface is less than 30% or more than 40% of the total weight of rice, sake is categorized as either normal sake or premium Ginjo sake, respectively. Because sake yeast strains are cultured in the presence of low nutrient concentrations during Ginjo sake brewing (30% lower amino acid concentration than normal sake brewing) (9), sake yeast produces flavors imparted by ethyl caproate and isoamyl acetate (10).
Autophagy is a bulk degradative and recycling process involving the transport of cytoplasmic components and organelles to the vacuole (plant and fungal cells) or lysosome (mammalian cells); it is required for homeostasis and is induced under conditions of nutrient starvation (11). Mitophagy is a selective form of autophagy that specifically degrades mitochondria (12, 13) and plays critical roles in the pathogenesis of Parkinson's disease. Mitophagy is mediated by the activity of the serine/threonine protein kinase PTEN-induced putative kinase 1 (PINK1) and the ubiquitin ligase Parkin (PARK2) (14, 15). PINK1 phosphorylates mitofusin 2 (MFN2), which functions as a Parkin receptor for culling damaged mitochondria in response to mitochondrial depolarization (16). Although yeasts undergo autophagy during wine fermentation (17–19), selective modes of autophagy, such as mitophagy, have not been reported. Moreover, there are no published studies on the relationship between mitophagy and the fermentation characteristics of yeast.
Our laboratory focuses on the effects of mitochondrial activities and the metabolic engineering of sake yeast strains that influence their ability to ferment substrates (1, 20–26). Mitochondria depolarize during anaerobiosis, which corresponds to the conditions used for industrial alcoholic fermentation (27–29), and mitophagy occurs when mitochondrial electron potential decreases (30). Moreover, mitophagy is induced in yeast cells when the availability of nutrients is limited (11). Indeed, sake yeasts are cultured under such conditions during sake brewing, because the nutrient-rich surface of rice substrate is polished and removed. Therefore, we hypothesized that mitophagy plays a role in the fermentation characteristics of sake yeast.
In the present study, we demonstrate that mitophagy occurs during sake brewing and that the fermentative capacity is improved by inhibiting mitophagy. This novel approach will be valuable for improving the fermentative capacity of other brewery yeasts.
MATERIALS AND METHODS
Yeast strains.
The S. cerevisiae strains used in this study are listed in Table 1. BY4743 (MATa/α his3ΔΔ1/his3Δ1 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0ura3Δ0/ura3Δ0) and the atg8Δ, atg11Δ, and atg32Δ mutants (BY4743 background) were purchased from Life Technologies. The sake yeast strain K7 was purchased from the Brewing Society of Japan. The K7 haploid strain K7H868 was obtained by sporulating the K7 parental diploid strain and was selected according to its brewing performance, which is similar to that of the K7 parental diploid strain (31).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype or description | Source |
|---|---|---|
| BY4743 | MATa/α his3Δ0/his3Δ0 leu2Δ/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0; ura3Δ0/ura3Δ0 | Open Biosystems |
| BY4743 atg8Δ | BY4743 atg8Δ::kanMX | Open Biosystems |
| BY4743 atg11Δ | BY4743 atg11Δ::kanMX | Open Biosystems |
| BY4743 atg32Δ | BY4743 atg32Δ::kanMX | Open Biosystems |
| BY4743mitGFP | BY4743+pYX142mitGFP | 22 |
| BY4743 atg32ΔmitGFP | BY4743 atg32Δ+pYX142mitGFP | This study |
| K7H868 | Sake yeast MATa | 31 |
| K7H868 atg32Δ | K7H868 atg32Δ::kanMX | This study |
| K7RAK | Sake yeast RAK1536 MATa/α his3/his3 | 34 |
| K7RAKmitGFP | RAK1536+pRS413GPDmitGFP | This study |
| K7RAK atg32Δ | RAK1536 atg32Δ::kanMX/atg32Δ::NAT1 | This study |
| K7RAK atg32ΔmitGFP | K7RAK atg32Δ+pRS413GPDmitGFP | This study |
Media.
To propagate yeast cells, yeast extract-peptone-dextrose (YPD) medium containing 2% (wt/vol) Bacto peptone, 1% (wt/vol) Bacto yeast extract (Beckton Dickinson), and 2% (wt/vol) glucose was used. To propagate cells harboring mitochondrion-targeted green fluorescent protein (GFP), minimal synthetic medium containing a 0.67% (wt/vol) yeast nitrogen base without amino acids (Beckton Dickinson), 800 mg liter−1 complete supplement mixture Drop-out–HIS + 40 ADE (Formedium), and 2% (wt/vol) glucose was used. For fermentation tests, minimal synthetic medium containing a 0.67% (wt/vol) yeast nitrogen base without amino acids (Beckton Dickinson), 790 mg liter−1 complete supplement mixture Drop-out: Complete (Formedium), and 15% (wt/vol) glucose was used.
Construction of yeast mutants.
The two copies of ATG32 in the K7 strains were disrupted using a PCR-based method. To disrupt the first copy of ATG32, a DNA fragment containing kanMX as a selectable marker with the flanking regions of the ATG32 open reading frame was amplified using the forward primer atg32kanMXfw (5′-AGATCACCGTCTCTCTAGAGC-3′) and the reverse primer atg32kanMXrv (5′-TGCATTCACATTTACAGCGA-3′) and atg32Δ DNA (BY4743 background) as a template. The PCR product was used to transform the K7 strains, and cells transformed with the disrupted ATG32 gene were selected on plates containing 500 μg ml−1 G418. To disrupt the second copy of ATG32, a DNA fragment containing NAT1 as a selectable marker with the same flanking regions as the first gene was amplified using the primers atg32nat1fw (5′-TGAAGTCCTAATCACAAAAGCAAAAAAAATCTGCCAGGAACAGTAAACATCACATACGATTTAGGTGACAC-3′) and atg32nat1rv (5′-TAGTAAAAAAGTGAGTAGGAACGTGTATGTTTGTGTATATTGGAAAAAGGAATACGACTCACTATAGGGAG-3′) and pAG25 as a template. These amplicons were used to transform K7 strains as well. Transformants disrupted in the two copies of ATG32 were selected on plates containing 100 μg ml−1 nourseothricin. These strains were transformed with the plasmid pRS413GPDmitGFP. To construct the plasmid pRS413GPDmitGFP, a fragment containing the Su9 (1–69)-GFP fragment (GFP fused to a sequence encoding the first 69 amino acid residues of subunit 9 of the mitochondrial Fo ATPase of Neurospora crassa) (32) flanked by the oligonucleotide sequences 5′-ACTAGT-3′ on its 5′ end and 5′-GAATTC-3′ on its 3′ end was subcloned and was inserted into the SpeI-EcoRI-cleaved site of pRS413GPD. pRS413GPD is a plasmid (pRS413) with a glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (33). pRS413 is a low-copy-number autonomously replicating plasmid containing the HIS3 marker (35).
Sake brewing.
Sake was brewed according to a published method (22). Briefly, the yeast strains were cultured in YPD medium, centrifuged, and washed with distilled water. These cells (optical density at 600 nm [OD600], 200 units) were mixed with 60 g dried pregelatinized rice, 23 g dried pregelatinized koji (Tokushima Seiko Co.), 200 ml distilled water, and 45 μl of 90% lactic acid in a 500-ml glass beaker and incubated without shaking at 15°C for 14 days. The mash was stirred once 24 h after the start of incubation. For normal sake brewing, we used rice and koji with 30% of their surface removed. For Ginjo sake brewing, we used koji and rice with 50% and 60% of the surface removed, respectively. We monitored the progress of the fermentation by determining the loss of mass of the culture, which was calculated as the amount of CO2 evolved.
Fermentation test.
Yeast cells (3 × 107 cells) were inoculated with 100 ml of minimal synthetic medium containing 15% (wt/vol) glucose in a 300-ml Erlenmeyer flask equipped with an air lock on top of the flask. The medium was cultured statically at 30°C for 11 days, and mass was determined every day.
Ethanol concentration.
The ethanol concentrations of sake or fermented media were analyzed using a contact combustion system with an alcohol densitometer (Alcohol Checker YSA-200; Yazaki Meter Co. Ltd.) according to the manufacturer's instructions, as described previously (31).
CFU.
Samples were obtained from the sake mash at different time points and diluted by factors of 2,000, 10,000, and 50,000. One hundred microliters of the diluted samples was plated on YPD agar and incubated at 30°C for 2 days. We then counted the colonies formed.
Observation of mitochondria and vacuoles.
Sake mash was sampled at different time points, and yeast cells were recovered from a viscous white layer formed after centrifugation of the sake mash. Yeast cells were incubated with the liquid recovered from the sake mash containing 100 μM E-64 (Sigma-Aldrich) and 8 μM FM4-64 (Molecular Probes) in a 30°C water bath for 30 min, washed with the same liquid to remove free E-64 and FM4-64, incubated again in the liquid at 30°C for 90 min with mild shaking, washed with phosphate-buffered saline, and observed using a fluorescence microscope (Keyence BZ8000). To observe the three-dimensional structures of mitochondria and vacuoles, z-stack images were acquired.
Western blotting.
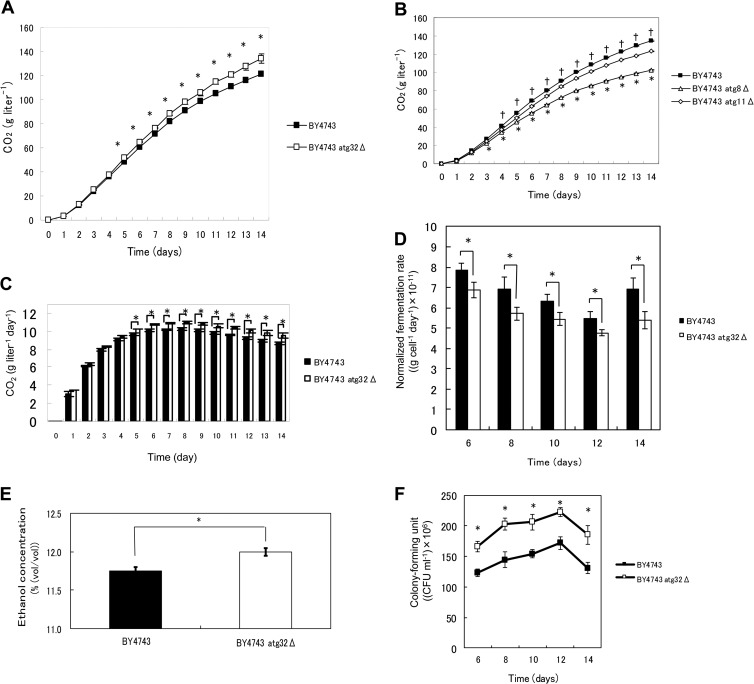
The stability of mitochondrial GFP was verified using Western blotting and an antibody against GFP. Cells of the atg32Δ sake yeast and its parent sake yeast strain were recovered from minimal synthetic medium incubated statically for 4 h or Ginjo sake mash brewed for 10 days. Cells were recovered from Ginjo sake mash as described above. Proteins were extracted using the Y-PER plus solution following the manufacturer's protocol (Thermo Scientific). The extracted protein solution was concentrated by extraction using chloroform-methanol-water (4:1:3), and the precipitated protein was solubilized in sterile water. The protein concentrations of the disrupted cells were determined using the Bradford method. Equal amounts of protein samples (20 μg) were applied to a 12.5% SDS-polyacrylamide gel and electrophoresed at 40 mA for 50 min. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (100 mA, 2 h). The membrane was blocked with skim milk (0.3% [wt/vol] in Tris-buffered saline-Tween 20 [TBST]). Incubation with the first antibody was performed overnight using an antibody against GFP (diluted 1:1,000) (mFX73; Wako Chemicals), and immune complexes were detected using an alkaline phosphatase-conjugated goat anti-mouse IgG (diluted 1:30,000) (Sigma-Aldrich). The membrane was visualized using the BCIP/NBT (5-bromo-4-chloro-3-indolyl-phosphate with nitro blue tetrazolium) liquid substrate system (Sigma-Aldrich).
Measurement of dry cell weight.
After fermentation, cells were collected by centrifugation, washed twice with sterile water, suspended in 1 ml sterile water, and added to 250-ml aluminum bottles. The bottles were weighed before adding the cells. The bottles were heated in an oven at 180°C overnight and then weighed. The differences between the weight of bottles before and after addition of cells have been presented as the dry cell weight. The dry cell weight was expressed as biomass.
Measurement of signal intensity of mitochondria.
Cells were photographed under a fluorescence microscope (Keyence BZ8000) at a magnification of ×1,000. Signal intensities of GFP images were determined using Dynamic Cell Count software (Keyence).
Measurement of cell area.
Cells were photographed under a microscope (Olympus BX53) at a magnification of ×1,000. The photos were processed using Image J software (National Institutes of Health). One hundred cells were outlined, and areas of the outlined cells were measured using the software.
Quantitative reverse transcriptase real-time PCR.
Yeast cells were collected from minimal synthetic medium or Ginjo sake mash by centrifugation. Total RNA was extracted from the cells using a hot phenol method (36). Total RNA was purified using the RNeasy minikit (Qiagen). Real-time PCR was performed using primers 5′-TGGTGTTCAATGCTTTTCAAG-3′ and 5′-CAAGGGTATCACCTTCAAACT-3′, the TaKaRa PrimeScript RT Master mix (Perfect Real Time) (TaKaRa Bio, Inc.), and the Light Cycler 480 system (Roche Diagnostics).
Statistical analysis.
The statistical significance of differences between the averages of two data groups with fewer than 30 samples was judged using an unpaired one-tailed Student t test without known deviations. The statistical significance of differences between the averages of two data groups with more than 30 samples was judged using the two-sample z-test (37).
RESULTS
Mitophagy occurs dependent on Atg32 in sake yeast during sake brewing.
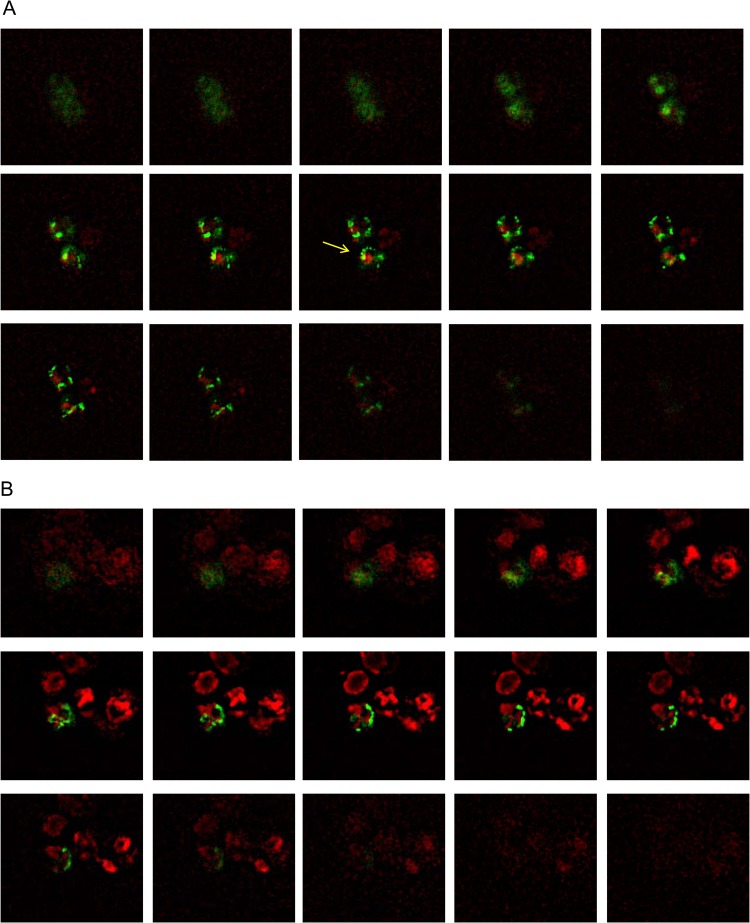
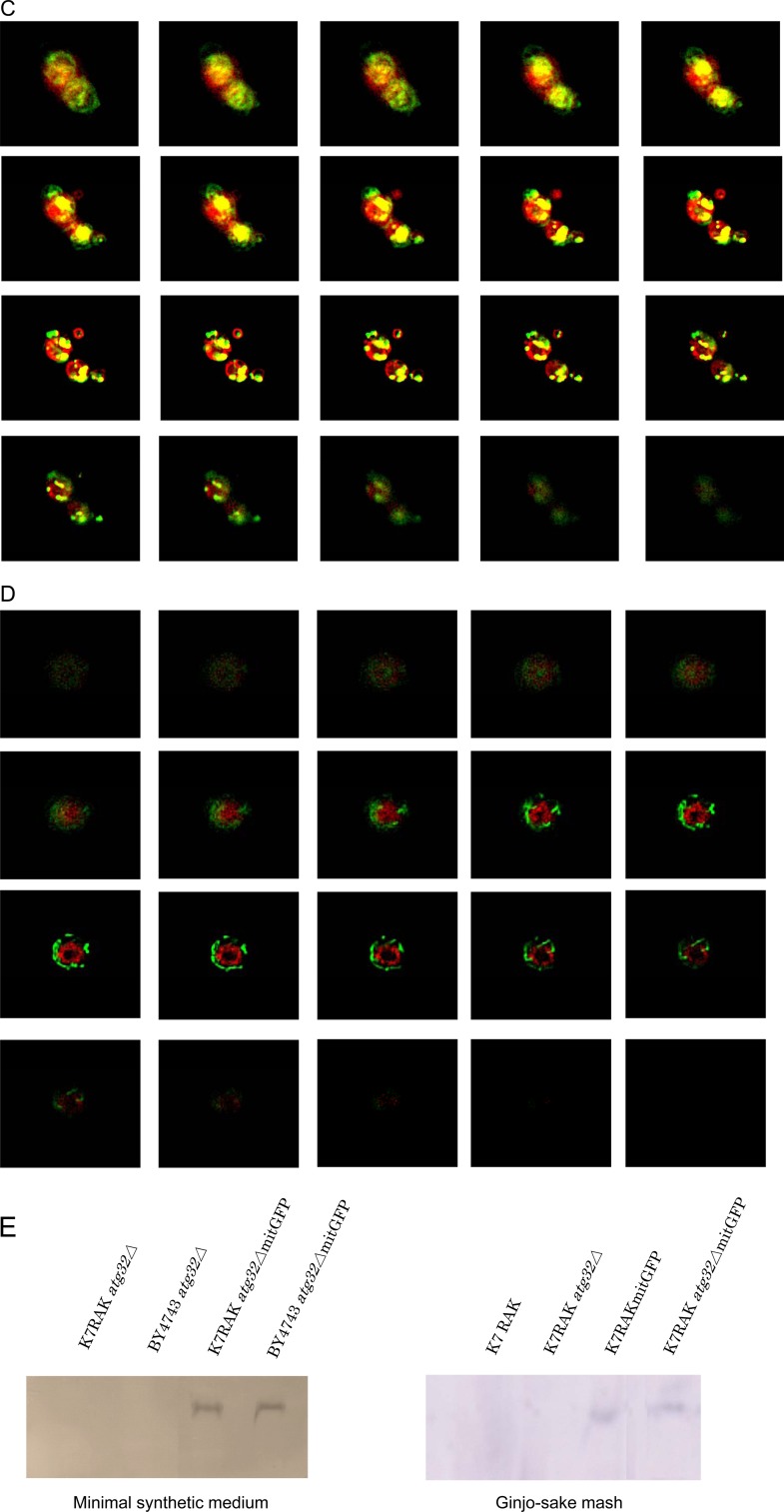
To determine whether mitophagy occurs in yeast cells during sake brewing, we generated a sake yeast strain disrupted in ATG32 and sake yeast strains that express GFP targeted to the mitochondria. ATG32 encodes a mitochondrial outer membrane protein, which recruits the autophagy adaptor protein Atg11p and the ubiquitin-like protein Atg8p to the mitochondrial surface to initiate mitophagy (11). A study reported that a mutant defective in Atg32 does not cause mitophagy (11). Sake was brewed using these strains, and mitochondrial and vacuolar structures were observed using a fluorescence microscope. Highly fragmented mitochondria and large swollen vacuoles were observed during the later phase of sake brewing, as reported previously (22, 38). In wild-type sake yeast cells analyzed during the brewing of normal sake, some portion of the mitochondria (green) fused with the vacuolar membrane (red) to generate yellow signals in z-stack images (Fig. 1A, yellow arrow). In contrast, in the atg32Δ sake yeast, mitochondria (green) were located distant from the vacuolar membrane (red), and virtually no yellow signal was detected that would indicate fusion of mitochondria with the vacuolar membrane (Fig. 1B). We hypothesized that mitophagy would be more evident in cultures with limited nutrient concentration. Mitophagy was clearly observed in sake yeast during the brewing of Ginjo sake (a refined sake), likely because the medium contained low nutrient concentrations. In wild-type sake yeast, a significant portion of mitochondria (green) fused with the vacuolar membrane (red) to generate yellow signals (Fig. 1C), but this was not observed in atg32Δ sake yeast (Fig. 1D). The atg32Δ cells exhibited weaker mitochondrial signals than wild-type sake yeast during the brewing of Ginjo sake (for the wild type, average signal intensity = 75, standard deviation [SD] = 20, n = 172; for the atg32Δ strain, average signal intensity = 41, SD = 8, n = 201, two-sample z score = 67.9; P < 0.001). In contrast, there was no significant difference between the atg32Δ strain and its parent strain with respect to the amount of GFP (Fig. 1E) or in their levels of expression of GFP mRNA (data not shown) when cells were cultured under the conditions of Ginjo sake brewing. These results indicate that mitochondrial GFP, although expressed normally in the atg32Δ strain, cannot translocate to the mitochondria in the atg32Δ strain because of low mitochondrial electron potential, which is consistent with the report that this mutant accumulates dysfunctional mitochondria (39). Together, these results indicate that mitophagy occurs dependent on Atg32 and this mitophagy is required to maintain mitochondrial quantity in sake yeast during sake brewing.
FIG 1.
Mitochondrial and vacuolar morphological features of sake yeast during sake brewing. Sake yeast strains K7RAKmitGFP (A, C) and K7RAK atg32ΔmitGFP (B, D), which harbor mitochondrially targeted GFP, were used for brewing normal sake (A, B) and Ginjo sake (C, D). Sake mash was sampled in the later stage of brewing, i.e., at 10 days (A and B) or 6 days (C and D). Green signals indicate mitochondrially targeted GFP. Red signals indicate the vacuolar membrane stained with 8 μM FM4-64. Yellow signals indicate colocalization of mitochondria and vacuolar membranes. Z-stack images were obtained using a fluorescence microscope (Keyence BZ8000). (E) Stability of mitochondrial GFP in K7RAKmitGFP and K7RAK atg32ΔmitGFP. Cells were collected from culture in minimal synthetic medium incubated for 4 h or Ginjo sake mash brewed for 10 days. Proteins extracted from the cells (20 μg) were analyzed using Western blotting with an anti-GFP antibody.
Enhanced ethanol fermentation by the atg32Δ laboratory strain.
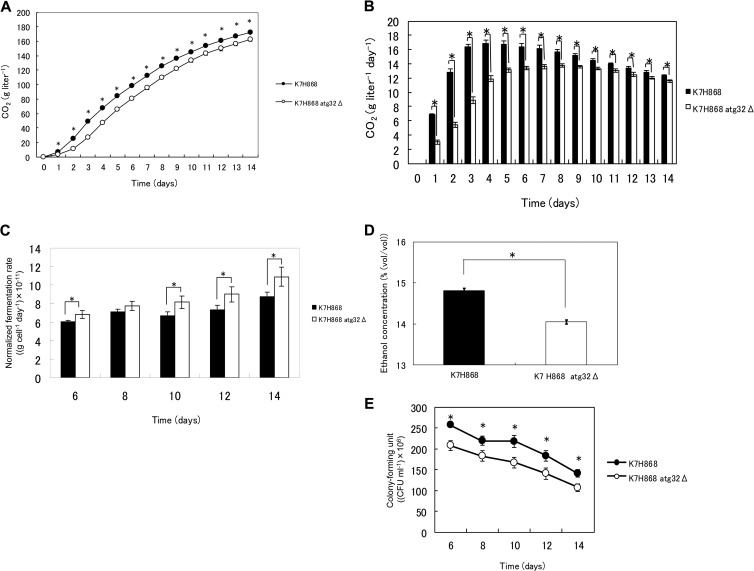
Because the above results indicated clearly that mitophagy occurred in sake yeast during sake brewing, we hypothesized that mitophagy may affect the characteristics of brewed sake. Therefore, sake was first brewed with the atg32Δ and its parent laboratory yeast strains. Analysis of the ability of the atg32Δ laboratory strain to produce CO2 indicated that it has an improved fermentation (P < 0.05; Fig. 2A). This result indicated that mitophagy plays a role in ethanol fermentation. We then asked whether inhibiting general autophagy would have the same effect. To address this question, we brewed sake with mutants defective in genes required for general autophagy (atg11Δ and atg8Δ laboratory yeast strains). However, in contrast to what was seen for the atg32Δ mutant, fermentation by these mutants was significantly reduced (P < 0.05; Fig. 2B), which is consistent with the results of previous studies of wine fermentation (40). These results indicate that disrupting mitophagy, but not general autophagy, enhances fermentation. To determine the basis for this effect, we assessed the fermentation profiles of atg32Δ mutants and found that the CO2 production rate (Fig. 2C), the final ethanol concentration (Fig. 2E), and CFU (Fig. 2F) were significantly higher than those of the parental strain (P < 0.05). The maximum CO2 production rate of the parent laboratory strain was 10.3 ± 0.167 g liter−1 day−1, and that of the atg32Δ mutant was 11.0 ± 0.146 g liter−1 day−1, which represents an increase of 7.50% relative to that of the parent strain (P < 0.05). These results indicate that the atg32Δ mutant has an enhanced ethanol fermentation rate. The normalized CO2 production rate of the atg32Δ strain per cell was significantly lower than that of the parent strain (P < 0.05) (Fig. 2D), suggesting that the metabolic competence of the atg32Δ mutant per cell did not mediate its high fermentation rate.
FIG 2.
Fermentation profiles of the atg32Δ strain and its parent laboratory yeast during the production of normal sake. (A) CO2 evolution by atg32Δ during sake brewing. Closed black and open symbols represent the results for the BY4743 and BY4743 atg32Δ strains, respectively. (B) CO2 evolution by atg8Δ and atg11Δ strains during sake brewing. Closed black symbols represent the results for BY4743, triangle symbols represent the results for BY4743 atg8Δ, and diamond symbols represent the results for BY4743 atg11Δ. The results are expressed as the weight loss of the mash, which represents the weight of CO2 (n = 3; †, statistical difference between BY4743 and BY4743 atg8Δ; *, statistical difference between BY4743 and BY4743 atg11Δ, P < 0.05, unpaired one-tailed Student's t test). (C) Fermentation rate of atg32Δ mutant (in g CO2 day−1). (D) Normalized fermentation rate of atg32Δ mutant (in g CO2 cell−1 day−1). (E) The final ethanol concentration in a culture of atg32Δ strain (%, vol/vol). (F) CFU of atg32Δ strain during sake brewing. Closed black and open symbols or boxes represent the results for BY4743 and BY4743 atg32Δ, respectively. The results are expressed as the means ± standard errors of the means (SEM) of three independent brewing experiments initiated with respective starter cultures (n = 3; *, P < 0.05, unpaired one-tailed Student's t test).
Enhanced ethanol fermentation by the atg32Δ sake yeast mutant when cultured under Ginjo sake (a refined sake) brewing conditions and also in minimal synthetic medium.
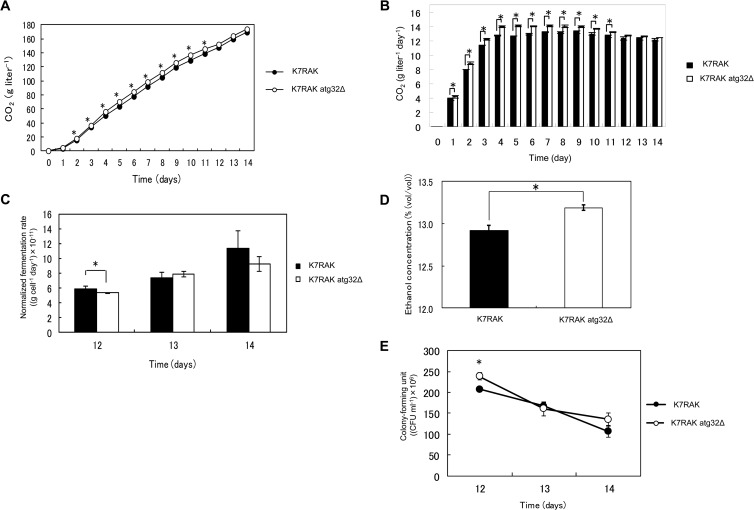
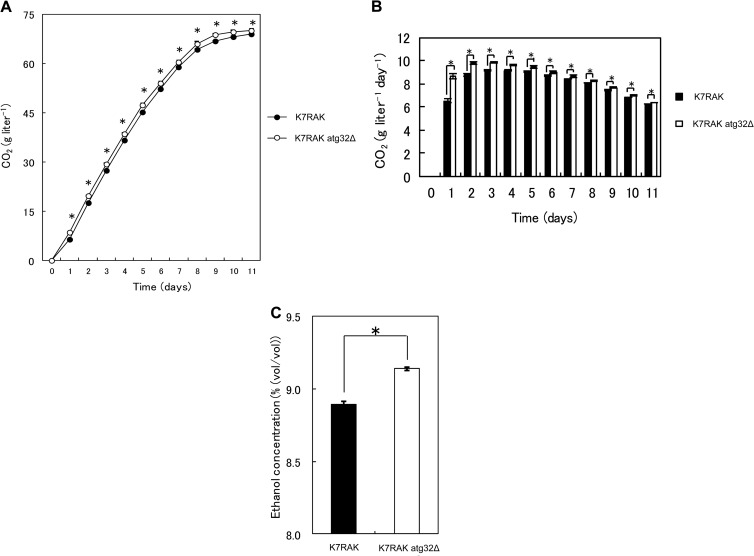
In order to elucidate if atg32Δ sake yeast also shows improved fermentation profile, the fermentation profile of atg32Δ sake yeast during the brewing of normal sake was investigated. In contrast to the atg32Δ laboratory strain, the atg32Δ sake yeast strain (K7H868, a haploid strain) showed a significantly (P < 0.05) lower ability to produce CO2 (Fig. 3A), CO2 production rate (Fig. 3B), final ethanol concentration (Fig. 3D), and CFU (Fig. 3E) than its parent sake yeast strain. The maximum CO2 production rates by the parent and atg32Δ sake yeast strains were 16.8 ± 0.470 g liter−1 day−1 and 13.6 ± 0.293 g liter−1 day−1, respectively, representing a 19.1% decrease relative to that of the parent strain (P < 0.05). Intriguingly, the normalized CO2 production rate per cell of atg32Δ sake yeast was higher than that of its parent strain under this condition (Fig. 3C), suggesting the metabolic impact of the atg32Δ mutation. Increase of fermentation and final ethanol concentration during the brewing of normal sake were not observed in atg32Δ diploid sake yeast strains either (see Fig. S1 in the supplemental material). These data led us to conclude that laboratory and sake yeasts may respond differently to nutrient concentrations, as described previously (41), because laboratory yeasts have long been cultured in media very rich in nutrients such as YPD, and sake yeasts have long been cultured in nutrient-poor media that contain polished rice. Therefore, sake yeast should exhibit the same response as laboratory yeast in the presence of low nutrient concentrations similar to those used for Ginjo sake brewing. Consistent with this hypothesis, under the conditions used to produce Ginjo sake, the atg32Δ sake yeast strain showed significantly (P < 0.05) increased CO2 production (Fig. 4A), fermentation rate (Fig. 4B), final ethanol concentration (Fig. 4D), and CFU (Fig. 4E) compared to the parent strain. The maximum CO2 production rate of the parent sake strain was 13.2 ± 0.179 g liter−1 day−1, and that of the atg32Δ mutant was 14.1 ± 0.126 g liter−1 day−1, representing an increase of 6.28% relative to that of the parent strain (P < 0.05). The atg32Δ sake yeast strain showed a normalized CO2 production rate that was not significantly different from that of its parent strain (Fig. 4C). Furthermore, during fermentation in minimal synthetic medium, the atg32Δ sake yeast strain exhibited significantly (P < 0.05) increased CO2 production (Fig. 5A), fermentation rate (Fig. 5B), and final ethanol concentration (Fig. 5C) relative to the parent strain. The maximum CO2 production rate of the parent sake strain was 9.14 ± 0.526 g liter−1 day−1, while that of atg32Δ strain was 9.83 ± 0.104 g liter−1 day−1, representing an increase of 7.44% (P < 0.05). The final ethanol concentration, CO2 production, cell/biomass, specific CO2 production rate, specific ethanol production rate, and ethanol yield of the atg32Δ strain were significantly higher than those of the parent sake yeast strain (P < 0.05), while the final biomass, OD600, and biomass yield of the atg32Δ mutant were significantly lower than those of the parent sake yeast strain (P < 0.05) (Table 2).
FIG 3.
Fermentation profiles of atg32Δ strain and its parent haploid sake yeast during the production of normal sake. (A) CO2 evolution during sake brewing. The results are expressed as the weight loss of the mash, which represents the weight of CO2. (B) Fermentation rate (in g CO2 liter−1 day−1). (C) Normalized fermentation rate (g cell−1 day−1). (D) Final ethanol concentration (%, vol/vol). (E) CFU during sake brewing. Closed black and open symbols or boxes represent the results for sake yeast K7H868 and K7H868 atg32Δ strains, respectively. The results are expressed as the means ± SEM of three independent brewing experiments initiated with respective starter cultures (n = 3; *, P < 0.05, unpaired one-tailed Student's t test).
FIG 4.
Fermentation profiles of atg32Δ strain and its parent sake yeast during the production of Ginjo sake. (A) CO2 evolution during sake brewing. The results are expressed as the weight loss of the mash, which represents the weight of CO2. (B) Fermentation rate (CO2 g liter−1 day−1). (C) Normalized fermentation rate (g CO2 cell−1 day−1). (D) Final ethanol concentration (%, vol/vol). (E) CFU during sake brewing. Closed black and open symbols or boxes represent the results for sake yeast strains K7RAK and K7RAK atg32Δ, respectively. The results are expressed as the means ± SEM of three independent brewing experiments initiated with respective starter cultures (n = 3; *, P < 0.05, unpaired one-tailed Student's t test).
FIG 5.
Fermentation profiles of atg32Δ sake yeast strain and its parent sake yeast during incubation in minimal synthetic medium. Sake yeast wild-type and atg32Δ strains were incubated in minimal synthetic medium containing 15% glucose, and their fermentation profiles were analyzed. (A) CO2 evolution during fermentation. The results are expressed as the weight loss of the culture, which represents the weight of CO2. (B) Fermentation rate (g CO2 liter−1 day−1). (C) Ethanol concentration on day 11 (%, vol/vol). Closed black and open symbols or boxes represent the results for sake yeast strains K7RAK and K7RAK atg32Δ, respectively. The results are expressed as the means ± SEM of eight independent fermentation experiments initiated with respective starter cultures (n = 8; *, P < 0.05; unpaired one-tailed Student's t test).
TABLE 2.
Fermentation characteristics of wild-type and atg32Δ sake yeast strains cultured in minimal synthetic mediuma
| Strain | Ethanol (g liter−1) | CO2 glucose (mol liter−1) | Residual/biomass (g liter−1) | CFU (liter−1) (×109) | Biomass (g liter−1) | OD600 | Cell (g−1) (×109) | qCO2 (mmol g−1 h−1) | qEthanol (mmol g−1 h1) | Yethanol/glucose (g g−1) | Ybiomass/glucose (mg g−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 70.2 ± 0.246 | 1.56 ± 0.00919 | ND | 11.6 ± 0.993 | 0.309 ± 0.0238 | 3.47 ± 0.0580 | 39.1 ± 4.02 | 20.0 ± 1.67 | 19.6 ± 1.62 | 0.468 ± 0.00174 | 2.06 ± 0.149 |
| atg32Δ mutant | 72.2 ± 0.132 | 1.58 ± 0.0116 | ND | 13.7 ± 1.85 | 0.228 ± 0.0212 | 3.12 ± 0.0612 | 62.2 ± 7.21 | 29.1 ± 4.22 | 28.8 ± 4.22 | 0.481 ± 0.000930 | 1.52 ± 0.132 |
| ** | * | * | ** | * | * | * | ** | * |
Values are means ± SEM of eight independent experiments initiated with respective starter cultures. Abbreviations and symbols: Yi/j, yield of production of constituent i on constituent j; qCO2, specific CO2 production rate (mmol CO2/g biomass/h); qEthanol, specific ethanol production rate (mmol ethanol/g biomass/h); ND, not detectable; *, P < 0.05; **, P < 0.01 as judged by unpaired one-sided Student's t test. Biomass indicates dry cell weight.
DISCUSSION
In the present study, we have shown that mitophagy occurs dependent on Atg32 in sake yeast during the brewing of Ginjo sake. The atg32Δ laboratory yeast strain showed improvement in fermentation characteristics such as the final ethanol concentration and fermentation rate. Fermentation was also enhanced in sake yeast under the conditions used for Ginjo sake as well as in minimal synthetic medium. Our present study demonstrates that the ethanol fermentation rate of sake yeast, the most active sake fermenter among brewery yeasts, can further be enhanced. The final ethanol concentration of minimal synthetic medium incubated with the atg32Δ strain was 2.76% increased relative to its parent sake yeast strain (Fig. 5C). These findings have significant implications for the bioethanol industry. Because natural biomass resources often lack sufficient nutrient levels required for optimal fermentation, this approach will be valuable for the production of bioethanol from natural biomass resources.
Although several mutations that augment the fermentation rate of sake yeast strains relative to those of other yeast strains have been reported, including MSN4, PPT1, and RLM1 (6–8), we are not aware of any report of a mutation that further augments ethanol fermentation. We have shown here that the atg32Δ mutation enhanced the ability of laboratory and sake yeast strains to produce ethanol during sake brewing and fermentation in minimal synthetic medium. A mutation that further enhances ethanol fermentation has not been reported for sake yeast, suggesting that deletion of ATG32 provides a novel approach for improving the fermentative capacity of sake yeast and potentially those of other strains. Moreover, ethanol tolerance did not differ between the atg32Δ mutant and its parent sake yeast strain (P > 0.05; see Fig. S2 in the supplemental material), suggesting that this strain would be a practical strain for industrial application. Enhancing fermentation output is the subject of intensive research on brewery yeasts (42, 43). An obvious benefit of these efforts includes reduced production costs, for example, of bioethanol and alcoholic beverages. Therefore, the present study should stimulate efforts to genetically manipulate brewery yeasts to increase ethanol productivity and final ethanol concentration.
The fermentation characteristics of the atg32Δ mutant suggest that the increased fermentative capability of this strain is caused by the increased carbon flux from glucose to ethanol per cell at the cost of decreasing the carbon flux from glucose to biomass. Consistent with this hypothesis, the average cell area of the atg32Δ mutant was 916 ± 20.4 arbitrary units of Image J software (n = 100), which was significantly smaller than that of its parent sake yeast (985 ± 30.8 arbitrary units of Image J software, n = 100; P < 0.01 as judged by the two-sample z-test). This finding may be explained by the difference in cell response of wild-type and atg32Δ cells exposed to stressful conditions induced by accumulation of fermented products and limited nutrient concentration. While wild-type cells use carbon flux to construct cell components by degrading mitochondria and recovering amino acids upon exposure to limited nutrient concentrations, atg32Δ cells lack this response, do not invest in carbon flux directed to biomass, and instead use the carbon flux toward ethanol.
Although autophagy has been observed under various wine-making conditions (17–19, 44), to our knowledge, no study has defined the role of mitophagy in alcoholic fermentation. The present study demonstrates that mitophagy occurs during alcoholic fermentation and its disruption enhances fermentation. Indeed, very recently, it has been shown that mitophagy occurs during fermentation, independent of respiration (13). Together, these studies indicate that mitophagy during alcoholic fermentation will be a new target in the development of fermentation technologies.
In summary, we have shown here that mitophagy occurs in sake yeast and that deleting the mitophagy gene augments this organism's capacity to produce ethanol. Considering that natural biomass resources often contain low concentrations of nutrients, this method will provide benefits in the production of bioethanol from biomass resources.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grant KAKENHI24580117 to H.K.
We thank Katsuhiko Kitamoto and Junichi Maruyama of the University of Tokyo for the valuable discussions and Rinji Akada of Yamaguchi University for providing yeast strain RAK1536.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03130-13.
REFERENCES
- 1.Kitagaki H, Kitamoto K. 2013. Breeding research on sake yeasts in Japan: history, recent technological advances, and future perspectives. Annu. Rev. Food Sci. Technol. 4:215–235. 10.1146/annurev-food-030212-182545 [DOI] [PubMed] [Google Scholar]
- 2.Hirata M, Tsuge K, Jayakody LN, Urano Y, Sawada K, Inaba S, Nagao K, Kitagaki H. 2012. Structural determination of glucosylceramides in the distillation remnants of shochu, the Japanese traditional liquor, and its production by Aspergillus kawachii. J. Agric. Food Chem. 60:11473–11482. 10.1021/jf303117e [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Izumi K, Nakahata E, Hirata M, Sawada K, Tsuge K, Nagao K, Kitagaki H. 2014. Quantitation and structural determination of glucosylceramides contained in sake lees. J. Oleo Sci. 63:15–23. 10.5650/jos.ess13086 [DOI] [PubMed] [Google Scholar]
- 4.Tsukahara T, Nakamura Y, Sato S, Tanaka Y. 1947. Brewing characteristics of Kyokai no. 6 and no. 7. J. Brew. Soc. Jpn. 42:26–27. 10.6013/jbrewsocjapan1915.42.10-12_26 [DOI] [Google Scholar]
- 5.Akao T, Yashiro I, Hosoyama A, Kitagaki H, Horikawa H, Watanabe D, Akada R, Ando Y, Harashima S, Inoue T, Inoue Y, Kajiwara S, Kitamoto K, Kitamoto N, Kobayashi O, Kuhara S, Masubuchi T, Mizoguchi H, Nakao Y, Nakazato A, Namise M, Oba T, Ogata T, Ohta A, Sato M, Shibasaki S, Takatsume Y, Tanimoto S, Tsuboi H, Nishimura A, Yoda K, Ishikawa T, Iwashita K, Fujita N, Shimoi H. 2011. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 18:423–434. 10.1093/dnares/dsr029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe M, Tamura K, Magbanua JP, Takano K, Kitamoto K, Kitagaki H, Akao T, Shimoi H. 2007. Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no. 11. J. Biosci. Bioeng. 104:163–170. 10.1263/jbb.104.163 [DOI] [PubMed] [Google Scholar]
- 7.Watanabe D, Araki Y, Zhou Y, Maeya N, Akao T, Shimoi H. 2012. A loss-of-function mutation in the PAS kinase Rim15p is related to defective quiescence entry and high fermentation rates of Saccharomyces cerevisiae sake yeast strains. Appl. Environ. Microbiol. 78:4008–4016. 10.1128/AEM.00165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe D, Wu H, Noguchi C, Zhou Y, Akao T, Shimoi H. 2011. Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress response components Msn2p and/or Msn4p. Appl. Environ. Microbiol. 77:934–941. 10.1128/AEM.01869-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iemura Y, Ohta Y, Hara S. 1995. The effect of type of rice and its polishing ratio on solubilization of rice during sake brewing. J. Brew. Soc. Jpn. 90:947–952 [Google Scholar]
- 10.Mason AB, Dufour JP. 2000. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast 16:1287–1298. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto K, Kondo-Okamoto N, Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17:87–97. 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 12.Kanki T, Klionsky DJ, Okamoto K. 2011. Mitochondria autophagy in yeast. Antioxid. Redox Signal. 14:1989–2001. 10.1089/ars.2010.3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiyama A, Kondo-Okamoto N, Okamoto K. 2013. Mitochondrial degradation during starvation is selective and temporally distinct from bulk autophagy in yeast. FEBS Lett. 587:1787–1792. 10.1016/j.febslet.2013.04.030 [DOI] [PubMed] [Google Scholar]
- 14.Jin SM, Youle RJ. 2012. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 125:795–799. 10.1242/jcs.093849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Dorn GW., II 2013. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340:471–475. 10.1126/science.1231031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496:372–376. 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cebollero E, Rejas MT, González R. 2008. Autophagy in wine making. Methods Enzymol. 451:163–175. 10.1016/S0076-6879(08)03212-6 [DOI] [PubMed] [Google Scholar]
- 18.Cebollero E, Gonzalez R. 2006. Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines. Appl. Environ. Microbiol. 72:4121–4127. 10.1128/AEM.02920-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cebollero E, Carrascosa AV, Gonzalez R. 2005. Evidence for yeast autophagy during simulation of sparkling wine aging: a reappraisal of the mechanism of yeast autolysis in wine. Biotechnol. Prog. 21:614–616. 10.1021/bp049708y [DOI] [PubMed] [Google Scholar]
- 20.Kitagaki H. 2009. Mitochondrial-morphology-targeted breeding of industrial yeast strains for alcohol fermentation. Biotechnol. Appl. Biochem. 53:145–153. 10.1042/BA20090032 [DOI] [PubMed] [Google Scholar]
- 21.Kitagaki H, Kato T, Isogai A, Mikami S, Shimoi H. 2008. Inhibition of mitochondrial fragmentation during sake brewing causes high malate production in sake yeast. J. Biosci. Bioeng. 105:675–678. 10.1263/jbb.105.675 [DOI] [PubMed] [Google Scholar]
- 22.Kitagaki H, Shimoi H. 2007. Mitochondrial dynamics of yeast during sake brewing. J. Biosci. Bioeng. 104:227–230. 10.1263/jbb.104.227 [DOI] [PubMed] [Google Scholar]
- 23.Motomura S, Horie K, Kitagaki H. 2012. Mitochondrial activity of sake brewery yeast affects malic and succinic acid production during alcoholic fermentation. J. Inst. Brew. 118:22–26. 10.1002/jib.7 [DOI] [Google Scholar]
- 24.Horie K, Oba T, Motomura S, Isogai A, Yoshimura T, Tsuge K, Koganemaru K, Kobayashi G, Kitagaki H. 2010. Breeding of a low pyruvate-producing sake yeast by isolation of a mutant resistant to ethyl alpha-transcyanocinnamate, an inhibitor of mitochondrial pyruvate transport. Biosci. Biotechnol. Biochem. 74:843–847. 10.1271/bbb.90373 [DOI] [PubMed] [Google Scholar]
- 25.Kitagaki H, Takagi H. 2013. Mitochondrial metabolism and stress response of yeast: applications in fermentation technologies. J. Biosci. Bioeng. pii:S1389-1723(13)00352-6. 10.1016/j.jbiosc.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 26.Sasaki M, Oba T, Suenaga H, Sato M, Tsuruta H, Tsuge K, Yoshimura T, Koganemaru K, Kitagaki H. 2011. Breeding of a low pyruvate-producing ginjo sake yeast and its application to the fermentation industry. Seibutsu Kogaku Kaishi 89:222–227 [Google Scholar]
- 27.Larsen GA, Skjellegrind HK, Vinje ML, Berg-Johnsen J. 2008. Mitochondria are more resistant to hypoxic depolarization in the newborn than in the adult brain. Neurochem. Res. 33:1894–1900. 10.1007/s11064-008-9664-2 [DOI] [PubMed] [Google Scholar]
- 28.Lloyd D, Moran CA, Suller MTE, Dinsdale MG, Hayes AJ. 1996. Flow cytometric monitoring of rhodamine 123 and a cyanine dye uptake by yeast during cider fermentation. J. Inst. Brew. 102:251–259. 10.1002/j.2050-0416.1996.tb00910.x [DOI] [Google Scholar]
- 29.Larsen GA, Skjellegrind HK, Berg-Johnsen J, Moe MC, Vinje ML. 2006. Depolarization of mitochondria in isolated CA1 neurons during hypoxia, glucose deprivation and glutamate excitotoxicity. Brain Res. 1077:153–160. 10.1016/j.brainres.2005.10.095 [DOI] [PubMed] [Google Scholar]
- 30.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. 2005. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12:1613–1621. 10.1038/sj.cdd.4401697 [DOI] [PubMed] [Google Scholar]
- 31.Katou T, Kitagaki H, Akao T, Shimoi H. 2008. Brewing characteristics of haploid strains isolated from sake yeast Kyokai no. 7. Yeast 25:799–807. 10.1002/yea.1634 [DOI] [PubMed] [Google Scholar]
- 32.Westermann B, Neupert W. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- 33.Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto S, Ogura M, Aritomi K, Hoshida H, Nishizawa Y, Akada R. 2005. Isolation of auxotrophic mutants of diploid industrial yeast strains after UV mutagenesis. Appl. Environ. Microbiol. 71:312–319. 10.1128/AEM.71.1.312-319.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398–405 [DOI] [PubMed] [Google Scholar]
- 37.Chris Cheadle C, Vawter MP, Freed WJ, Becker KG. 2003. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5:73–81. 10.1016/S1525-1578(10)60455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izawa S, Ikeda K, Miki T, Wakai Y, Inoue Y. 2010. Vacuolar morphology of Saccharomyces cerevisiae during the process of wine making and Japanese sake brewing. Appl. Microbiol. Biotechnol. 88:277–282. 10.1007/s00253-010-2758-1 [DOI] [PubMed] [Google Scholar]
- 39.Kurihara Y, Kanki T, Aoki Y, Hirota Y, Saigusa T, Uchiumi T, Kang D. 2012. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287:3265–3272. 10.1074/jbc.M111.280156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piggott N, Cook MA, Tyers M, Measday V. 2011. Genome-wide fitness profiles reveal a requirement for autophagy during yeast fermentation. G3 (Bethesda) 1:353–367. 10.1534/g3.111.000836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homann OR, Cai H, Becker JM, Lindquist SL. 2005. Harnessing natural diversity to probe metabolic pathways. PLoS Genet. 1:e80. 10.1371/journal.pgen.0010080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marullo P, Mansour C, Dufour M, Albertin W, Sicard D, Bely M, Dubourdieu D. 2009. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 9:1148–1160. 10.1111/j.1567-1364.2009.00550.x [DOI] [PubMed] [Google Scholar]
- 43.Sasano Y, Takahashi S, Shima J, Takagi H. 2010. Antioxidant N-acetyltransferase Mpr1/2 of industrial baker's yeast enhances fermentation ability after air-drying stress in bread dough. Int. J. Food Microbiol. 138:181–185. 10.1016/j.ijfoodmicro.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 44.Orozco H, Matallana E, Aranda A. 2012. Oxidative stress tolerance, adenylate cyclase, and autophagy are key players in the chronological life span of Saccharomyces cerevisiae during winemaking. Appl. Environ. Microbiol. 78:2748–2757. 10.1128/AEM.07261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.