Abstract
Lactobacillus plantarum has been used in human clinical trials to promote beneficial effects in the immune system, to alleviate intestinal disorders, and to reduce the risk of cardiovascular disease. It is also involved in many fermentation processes in the food industry. However, information on the fate of ingested L. plantarum is limited. In this study, 61 subjects received daily doses of fermented milk containing 2 × 1011 cells of L. plantarum Lp115 for different periods of time. The target microorganism was monitored in the fecal microbiota via quantitative PCR (qPCR). L. plantarum was detected and quantified in all of the subjects during the ingestion periods. The differences between the L. plantarum levels at time zero and during all the different ingestion periods were statistically significant (P = 0.001). However, at 15 and 45 days after discontinuing supplementation, the number of lactobacilli was reduced to the baseline level (those at time zero). A longer period with L. plantarum in the diet did not result in increased levels of this bacterium in the stool, based on postconsumption evaluations (P = 0.001). The qPCR method was specific and sensitive for L. plantarum quantification in such a complex microbial environment as the gastrointestinal tract.
INTRODUCTION
The human gastrointestinal tract (GIT) has a complex and dynamic microbiota. The GIT of each individual has a unique microbiota that varies according to age, health, and lifestyle (1–4). The microbial balance provides a barrier against pathogens and harmful food substances and has important protective functions promoting beneficial effects in the host (5).
Probiotic microorganisms are particularly associated with beneficial health effects in the hosts. These effects are commonly associated with adhesion of the probiotic microorganisms in the GIT. However, discussions in the literature claim that colonization by probiotic microorganisms is not necessary for beneficial effects in the host since the temporary persistence of allochthonous microbiota can promote such effects (3, 6).
For a long time, understanding of the role of specific microorganisms on GIT dysfunction or their beneficial effects on the balance of the intestinal microbiota has been limited due to a lack of detailed information regarding the development of the microbiota, its normal composition, and its changes in response to a specific diet. The major restricting factor of these studies may be attributed to the use of traditional plate culture methods involving only a very limited proportion of the microbial population (7–9).
However, currently, the use of PCR with other molecular tools and the increased number of sequenced genomes available in the GenBank/National Center for Biotechnology Information (NCBI) database have become important, and new tools have been developed to assess the intestinal microbiota and its effects on the host (3, 10–12).
Quantitative PCR (qPCR) is considered the most sensitive technique for analyzing rare targets because it enables the detection and quantification of a single copy of target DNA in some cases (10, 11). The technique has been successfully applied to quantify probiotic microorganisms in food samples (12, 13) and in studies involving the intestinal microbiota (8, 14, 15).
The consumption of Lactobacillus plantarum has been related to significant improvements in the health of humans and animals (16–19). This species was the first of the genus to have its genome sequenced (20). The metabolic pathways, regulatory mechanisms, and functions of many genes have been explored, and the findings have facilitated the exchange of information to obtain an understanding of the inherent effects of L. plantarum. Furthermore, the beneficial health effects of probiotics are considered strain specific. L. plantarum Lp115 was used in in vitro and in vivo studies to evaluate safety and functionality. The authors observed that the strain exhibited an adequate safety profile and strong survival in the GIT and had positive effects on the hosts (21, 22).
In this investigation, we quantified L. plantarum and evaluated its persistence in the GIT by qPCR of fecal samples from healthy subjects consuming fermented milk over different time periods of consumption and postconsumption.
MATERIALS AND METHODS
Subjects and experimental design.
A total of 61 healthy adult subjects consisting of 22 men and 39 women between the ages of 19 and 65 years were selected to participate in this study. The subjects did not have a history of gastrointestinal disease, and antibiotics were not administered during the investigation period or in the 2 months immediately preceding the study. During the study, there was no change in the habits or diet routines of the volunteers, except for the introduction of fermented milk. The study was approved by the Ethics Committee on Human Research at the State University of Londrina, Londrina, Paraná State, Brazil (process number 14995/2009).
The subjects were randomized into six independent groups with different time periods of consumption (0, 15, 30, 45, 60, and 90 days), and evaluations were performed at two times postconsumption (+15 and +45 days). The time preceding initial consumption (time zero) was used as the baseline for all of the groups. The five treatment groups received 80 ml of fermented milk containing L. plantarum Lp115 at 2 × 1011 cells/daily dose.
Fecal samples were collected at predetermined times (Fig. 1) and used for DNA extraction and qPCR quantification.
FIG 1.
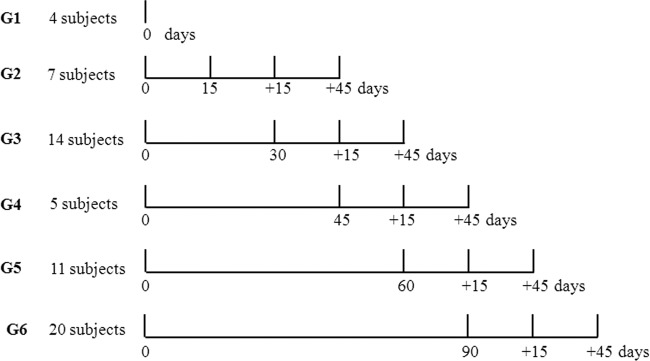
Experimental design for the different treatment groups (G2 to G6) and the control group (G1).
Fecal samples.
The subjects were instructed to collect approximately 5 g of feces in individual sterile containers. The samples were immediately cooled at 5°C and delivered to the laboratory within 12 h. All of the samples were homogenized, fractionated into three sterile tubes with 1.5 g of sample each, and frozen at −80°C until DNA extraction.
Reference strains and growth conditions.
In this study, L. plantarum strain Lp115 (Danisco, Cotia, SP, Brazil) was included in the diet of the volunteers and assessed as the target microorganism. L. plantarum Lp115 was cultured for 12 h in MRS broth (Oxoid, Cambridge, United Kingdom) with an Anaerobac system (Probac, São Paulo, Brazil) at 37°C under anaerobic conditions.
Bacillus thuringiensis 407 was added to each fecal sample and used as an internal reference in the qPCRs. The bacteria were cultured at 30°C in a shaker until the optical density at 600 nm reached 0.6 in LB broth, which consisted of 10 g of tryptone (Himedia, Mumbai, India), 5 g of yeast extract (Himedia, Mumbai, India), 5 g of NaCl (Vetec, Rio de Janeiro, Brazil), and distilled water to bring the volume to 1 liter. The B. thuringiensis 407 culture was centrifuged, and the biomass was suspended in a 0.85% (wt/vol) saline solution with 50% (vol/vol) glycerol (Synth, São Paulo, Brazil). The bacteria were aliquoted into samples with defined counts (4.3 × 103 cells/μl) and frozen at −80°C, and 10 μl of this culture was added to all the fecal samples before the DNA extraction for use as an internal reference in the qPCRs and to evaluate the efficiency of the DNA extraction method.
Extraction of DNA from bacteria.
Genomic DNA was extracted from serial dilutions of the L. plantarum and B. thuringiensis cultures according to the protocol previously described (8), with minor modifications. The cultures were centrifuged at 10,000 × g for 5 min. The cell pellet was washed two times with TE (10 mM Tris HCl, 1 mM EDTA, pH 8.0) buffer and suspended in 200 μl of TE buffer (reagents from Synth, São Paulo, Brazil).
The bacterial suspension was centrifuged at 10,000 × g for 10 min. The total genomic DNA in the supernatant was stored at −20°C until use.
Extraction of DNA from fecal samples.
Two fecal samples (200 mg) were extracted from each subject per consumption period (Fig. 1), for a total of 350 samples. The DNA from the fecal samples was extracted using a stool minikit (Qiagen, CA) according to the manufacturer's protocol.
The isolation of DNA from the stool sample for detection of microorganisms was conducted because conditions were optimized to increase the ratio of nonhuman DNA. The fecal suspension was heated to 95°C for 5 min because the target microorganisms were Gram positive. ASL stool lysis buffer (Qiagen) was added to each stool sample, and the mixture was added to 10 μl of the Bacillus thuringiensis 407 suspension (4.3 × 104 cells/10 μl). After this procedure, the DNA was eluted in 200 μl of AE elution buffer (Qiagen) and was stored at −20°C until use.
Oligonucleotides.
The primers and probes used for qPCR (Table 1) were designed using the Primer Express program (version 3.0; Applied Biosystems, São Paulo, Brazil). For detection of L. plantarum, primers LPrecAF and LPrecAR, which were 23 and 18 bp, respectively, were used (23). The same region of the recA gene was used to design a reporter probe labeled with the 6-carboxyfluorescein (FAM) fluorophore at the 5′ end and the minor groove binder (MGB) quencher at the 3′ end. For discrimination of B. thuringiensis, nucleotide sequences from the plcR gene of B. thuringiensis strain 407 available from the NCBI database were selected to design a specific primer pair. The same region of the gene was used to design a probe labeled with a 2′-chloro-7′-phenyl-4-dichloro-6-carboxyfluorescein (VIC) reporter at the 5′ end and MGB at the 3′ end according to the TaqMan technology (Applied Biosystems, CA).
TABLE 1.
Sequences and positions of the primers and probes used in this study
| Primer name | Sequence (5′–3′) | Positiona | Amplicon size (bp) |
|---|---|---|---|
| LPrecAF | GTGGTGCGGTCGATATTTTAGTT | 412–434 | 108 |
| LPrecAR | TCAGCCGCGCTTGTAACC | 502–519 | |
| Probe LP-FAMb | AATGGGTGACGCACACGTTG | 482–501 | |
| BTplcRF | TTTGTCCAATTTTTGAGCATGAA | 253–276 | 70 |
| BTplcRR | GTGCTTTCGTTACATTCGGTCTT | 301–324 | |
| Probe BT-VICb | AGCTTCTGTTGATAAAGGA | 277–296 |
The positions of the LP primers are relative to the partial consensus sequences of the recA gene from 20 Lactobacillus strains, and the positions of the BT primers are from the sequence of the plcR gene of B. thuringiensis 407.
Probes LP-FAM and BT-VIC were used to detect L. plantarum Lp115 and B. thuringiensis, respectively, and contained different reporter dyes, FAM and VIC, respectively, attached to the 5′ ends. The minor groove binder (MGB) was used as the quencher attached to the 3′ end of each probe.
Conventional multiplex PCR.
PCR was performed using 25 μl of a reaction mixture containing 50 ng of the genomic DNA each from the lactic acid bacteria (LAB) and B. thuringiensis. All of the strains were derived from culture collections or were trade strains: L. casei and L. acidophilus (C. Hansen, Brazil), L. pentosus ATCC 8041T, L. paraplantarum DSM 10841, L. plantarum Lp115, and B. thuringiensis 407. The LAB strains were grown anaerobically in MRS medium (Oxoid, Cambridge, United Kingdom) with an Anaerobac system (Probac, São Paulo, Brazil) and incubated at 37°C for 48 h.
The PCR conditions were those described by Costa et al. (23), with 2.0 μM BTplcR primers (Table 1) added for the multiplex PCR (all reagents were from Invitrogen, São Paulo, Brazil).
The PCR was conducted in a thermocycler (PT-100; MJ Research, Watertown, MA). After the amplification, 20 μl of the sample from each PCR mixture was added to 3.0 μl of loading buffer (0.25% [wt/vol] bromophenol blue, 0.15% [wt/vol] Ficoll [Merck, Darmstadt, Germany]), and the mixture was electrophoresed in an agarose gel at 2% (wt/vol) in TEB buffer (89 mM boric acid, 89 mM Tris, 2.5 mM EDTA, pH 8.3), stained with 0.8 μl of Sybr Safe (Invitrogen, OR), run at 3 V/cm, visualized, and registered as a photodocument (L-PIX HE; Locus, Campo Belo, S.P., Brazil).
qPCR analyses.
Samples were amplified with primers LPrecAF and LPrecAR, specific for the recA genes of L. plantarum (23), and primers specific for the plcR gene of B. thuringiensis strains (Table 1), used as an internal reference of the amplification (24).
The detection of internal reference and target genes as calibration standards and evaluation of the efficiency of the qPCR system were performed with a multiplex reaction mixture containing 12.5 μl of TaqMan universal PCR master mix (Applied Biosystems/Roche, NJ) with the following components: 0.4 μmol of the BTplcR primers; 0.6 μmol of the LPrecA primers; 0.25 μmol of the TaqMan hybridization probes labeled with a VIC or FAM reporter dye at the 5′ end for detection of B. thuringiensis and L. plantarum, respectively; 1.0 μl of bovine serum albumin at 2.5 μg/μl (Sigma-Aldrich, São Paulo, Brazil); 2.5 μl of each DNA sample; and ultrapure water (Teuto, Goias, Brazil) to complete the final volume to 25.0 μl. The detection of bacteria in the qPCR analysis was performed under the same conditions; however, 5 μl of DNA extracted from the fecal samples was used.
The reactions were performed on a model 7300 real-time PCR thermocycler (Applied Biosystems, CA), following the manufacturer's instructions, using a TaqMan system. After the initial steps at 50°C for 2 min (uracil N-glycosylase activity) and at 95°C for 10 min (activation of the AmpliTaq Gold polymerase), a two-step program of 95°C for 15 s and 60°C for 1 min was conducted for 40 cycles. The data were collected in the last phase of the PCR (extension phase), and the results were analyzed by use of a sequence detection program (PerkinElmer, MA).
Standard curves were obtained for cultures of Lp115 and B. thuringiensis 407 in broth diluted in peptone water. The dilutions were plated on solid medium, and the counts of the colonies ranged from 104 to 109 cells/ml for Lp115 and 102 to 107 cells/ml for B. thuringiensis 407. Each of the dilutions was used for DNA extraction, as described above. The calibration standards and the efficiency of the qPCR system were performed by plotting the cycle threshold (CT) values from serial dilutions of the L. plantarum Lp115 and B. thuringiensis 407 cultures as linear functions of the base 10 logarithm (Fig. 2). The initial number of bacteria was determined by plate counting, as described above.
FIG 2.
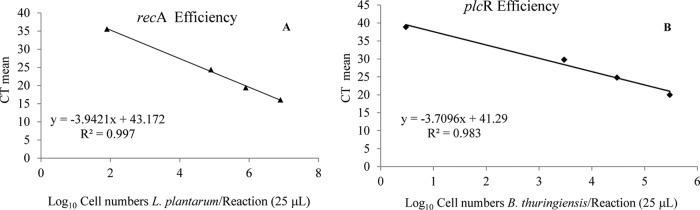
Efficiency of the multiplex PCR system for detection of the target gene (recA) (A) and the normalizer gene (plcR) (B). The calibration curves were obtained by plotting the CT values as a function of the log10 cell concentration by serial dilution (number of cells per 25-μl reaction mixture). The efficiency (E; in percent) was calculated by the equation 10−1/slope × 100. Data points represent those from three independent repetitions.
The fold changes in the levels of the probiotic after intervention were determined as ratios (after/before intervention) of the bacterial numbers in each DNA sample normalized to the total bacterial numbers (25); i.e., all of the samples were compared to the controls, which did not undergo any treatment (time zero), and the final amount of bacteria was estimated by relative quantification using calibration curves for the target and the internal reference by comparing the CT values to the standard curve. The amplification efficiency (E) and correlation coefficient (R2) were determined from the slope of the line derived from the standard curves. The value of E (in percent) was calculated using the equation 10−1/slope × 100 (24).
Duplicates of different samples were analyzed in each PCR run in three independent runs. DNA dilutions of L. plantarum and B. thuringiensis were used as positive controls in the reactions, and ultrapure water was used as a negative control.
Statistical analysis.
The groups were treated independently, and the results were correlated to those at time zero for statistical analysis. The relative differences between the measurements were determined according to the period of time of consumption (0, 15, 30, 45, 60, and 90 days) and postconsumption (+15 and + 45 days) on the basis of the numbers L. plantarum cells/g feces. The data were log10(x) transformed to fit the assumptions of normality and the homogeneity of variances. The Student-Newman-Keuls test was used to compare the means, and the F-test analysis of variance was significant at a level of 5%.
RESULTS
Subjects assessed.
In this study, we investigated 61 healthy male and female adults in groups with ages ranging from 17 to 25 (49.13%), 26 to 35 (33.33%), 36 to 50 (10.53%), and older than 50 (7.01%) years to obtain information regarding the quantity and maintenance of L. plantarum in the fecal microbiota after ingestion of fermented milk containing this probiotic bacterium.
Oligonucleotide specificity.
The specificities of both primer sets (target and control) as well as the probes (Table 1) were tested by evaluation for possible cross-reactivity or primer dimers by conventional PCR and qPCR. In both systems, amplification signals were observed for the target and for the internal control (normalizer) genes only when the DNA of L. plantarum and/or B. thuringiensis was present. This was clearly evidenced when the DNA from fecal samples of control subjects (group 1 [G1]; Fig. 1) and internal control DNA (from B. thuringiensis 407) was used in the reaction mixture and a positive signal was observed only for the plcR gene. Additionally, the conventional PCR also showed this specificity (Fig. 3).
FIG 3.
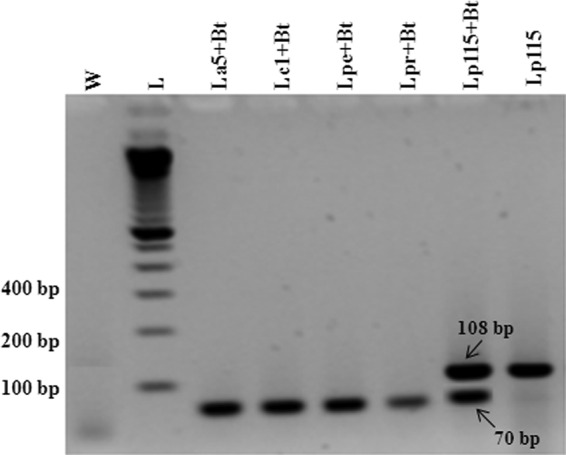
Electrophoresis of the 108- and 70-bp multiplex PCR products in a 2% agarose gel obtained from cultures of LAB strains and B. thuringiensis. Lanes: W, water (negative control); L, 100-bp DNA ladder; La5, L. acidophilus; Lc1, L. casei; Lpe, L. pentosus; Lpr, L. paraplantarum; Lp115, L. plantarum; Bt, B. thuringiensis.
System efficiency and quantitative detection of L. plantarum in fecal samples.
Data obtained from serial dilutions with cell concentrations ranging from 8 × 107 to 8 × 101 cells per reaction mixture for L. plantarum Lp115 and 3 × 105 to 3 × 101 cells per reaction mixture for B. thuringiensis 407 for both genes showed slope values ranging from −3.7 to −3.9. The efficiencies were 80% for the recA gene and 86% for the plcR gene (Fig. 2A and B).
The samples were analyzed before initial consumption for all 61 subjects, and detection of the target occurred only in two individual samples. During the intervention, L. plantarum was detected in the samples from all of the individuals in every probiotic consumption time period.
The absence of L. plantarum in most of the subjects at the baseline (time zero) and its detection during consumption demonstrated that a cross-reaction with related species present in the fecal microbiota did not occur. This was shown using conventional PCR (Fig. 3) and qPCR to be due to the presence of the target species (Lp115) or the reference species (B. thuringiensis 407) in the reaction mixtures.
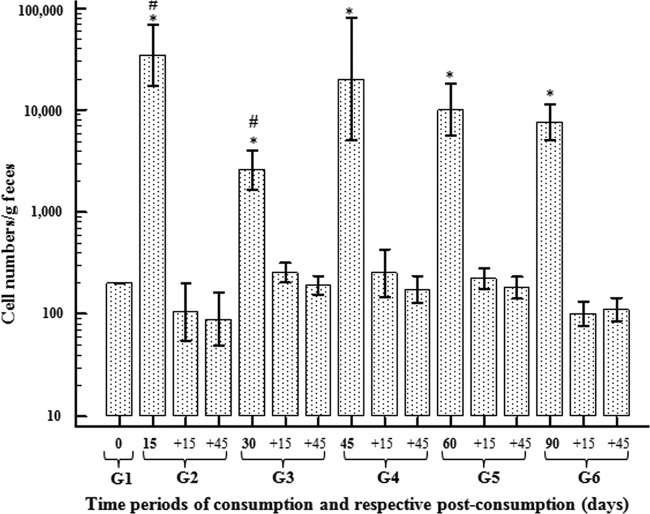
The means obtained for the subjects who consumed L. plantarum Lp115 for different periods of time were quantified (Fig. 4). A significant increase in the average numbers of L. plantarum cells in fecal samples over all of the consumption time periods compared to the number at the baseline was observed (P = 0.001). The maximum number of L. plantarum cells in the feces was observed in the subjects who consumed fermented milk for 15 days, with the number of L. plantarum cells/g feces reaching a level 17,500 times higher than that at time zero. However, it was decreased in the other time periods.
FIG 4.
Relative quantification of L. plantarum Lp115 cell numbers/g feces as a function of the consumption time periods (15, 30, 45, 60, and 90 days), followed by postconsumption time periods of +15 and +45 days. Quantification is represented as the mean value relative to that at time zero. Bars represent standard deviations. *, averages with a significant difference (P = 0.001) relative to the levels at time zero; #, averages with significant differences between time points.
There was a significant difference (P = 0.001) in cell numbers between groups that consumed fermented milk for 15 and 30 days (groups 2 and 3, respectively). Nevertheless, there were no significant differences among the other groups with different time periods of consumption.
During the posttreatment period (+15 and +45 days), the numbers of cells/g feces decreased to the level present at the baseline (Fig. 4). In order to compare the levels of L. plantarum between genders, we analyzed the data from qPCR for the different periods of consumption; however, no difference was observed between men and women.
DISCUSSION
Most studies on the beneficial effects of probiotics are based on individuals with several disease conditions. Understanding of the maintenance of these microorganisms in the intestinal microbiota or their benefits in healthy populations is limited (6, 26, 27). The study discussed herein considered a large number of healthy volunteers.
The qPCR methodology has been applied to quantify probiotics in foods and drugs in clinical trials. The sensitivity of the technique is a crucial advantage when small quantities of the target (28) or complex samples, such as intestinal microbiota, are analyzed.
In general, the quantification method in which quantities are expressed in numbers of CFU/ml using standard curves is the most widely used (8, 14, 15, 29). However, many of these studies do not use internal controls (normalizers) and/or do not demonstrate the efficiency of the qPCR system, which can lead to over- or underestimation of the data. Moreover, the use of an internal control is recommended to overcome the variability that can occur as a result of the method of DNA extraction and the occurrence of false-positive or false-negative results in PCRs (28, 30, 31). In this study, normalization was adopted, and suitable efficiencies were obtained for the systems involved, resulting in the detection of 8 × 101 cells of L. plantarum per reaction, which is equivalent to 2 × 104 cells/g.
The efficiency of PCR amplification systems is directly related to the slope of the curve. A value of −3.32 indicates a theoretical amplification with 100% efficiency (32). In the same qPCRs, the efficiencies of the different amplicons must be similar to avoid under- or overestimation of the results (33, 34). With the data obtained, the difference between the target and internal reference of 6% indicates that the systems are suitable. Additionally, the correlation coefficients (R2 values) are greater than 0.98 (Fig. 2), denoting a high correlation between the cycle threshold (CT) values and the log10 number of cells per reaction, confirming that the extraction method is efficient and that the amplification reactions had good repeatability.
We adopted a relative quantification method in which data are compared to those at the baseline (time zero). This method is suitable for demonstrating the proportion of the probiotic microorganism in the fecal microbiota after its introduction in the diet (Fig. 4). In this case, the quantification was based on the standard curves (Fig. 2A and B), wherein the input amount for unknown samples was calculated from the standard curve for a specific gene and normalized to the input amount for a reference gene, which was also calculated from the standard curve for that gene (35).
The absence of L. plantarum in the subjects' feces before probiotic consumption indicated that, in general, L. plantarum is not an endogenous constituent of the fecal microbiota in this population. Although L. plantarum is not commonly observed in the human fecal microbiota, its occurrence has been reported in some studies using phenotypic and genotypic approaches (36–39).
The quantification data showed that there was no significant difference among the groups during the consumption of the probiotic, except between groups 2 and 3 (Fig. 4).
The lowest level of L. plantarum/g feces, which was found for the 30-day period of consumption (1,300 times higher than that at time zero), is not likely related to experimental error because DNA extraction from stool for testing by qPCR was conducted randomly, and the results were repeated with the biological duplicate. Several DNA samples, including the samples obtained on day 15 of consumption, were reextracted, and the target and normalizer genes were detected at the same levels observed in the first experiments.
The variation in the results for samples taken on days 15 and 30 (Fig. 4) could indicate that the exogenous species were not able to install in the GIT with only 15 days of probiotic consumption. Nevertheless, the bacteria were able to colonize and remain in the GIT after continuous intake (30 days), since they were detected at a lower level in the fecal samples at day 30. In contrast, the settlement and multiplication of Lp115 in the GIT during other time periods of continuous consumption could have caused an increase in the detection rate in the samples. However, only further studies can confirm the factors behind the occurrence of this phenomenon. This variability can also be explained by considering the specific characteristics of each individual, including the microbiota composition, genetic factors, diet, the secretion of mucus, digestive enzymes, and intestinal peristalsis (3). Although few data regarding this variance exist in the literature, similar research data show that some strains can adapt to drastic conditions, such as the GIT, and can compete and establish themselves in this environment (22, 40, 41).
Consumption of L. plantarum for the longest length of time did not increase the levels of detection in the feces. Moreover, the detection levels decreased after probiotic consumption was discontinued, and there were no significant differences relative to the levels at the baseline. Although the daily intake (2 × 1011 cells per dose) was much higher than the level detected in the feces (104 cells/g), our data do not indicate the destination of the bacteria after intake and in the postconsumption periods. Most of the ingested bacteria could possibly have adhered to the intestinal mucosa or been eliminated and the DNA degraded, preventing qPCR values from reaching greater than 104 cells. On the other hand, this strain may not be able to survive passage through the gastrointestinal tract and into feces.
Other studies using similar approaches with members of the genus Lactobacillus reported similar results for the species L. rhamnosus DR20, L. helveticus GCL1001, and L. casei Shirota (17, 42, 43). The authors argued that some probiotics do not persist permanently in the digestive tract after consumption ends (6, 44) or the microorganisms are not detected in fecal samples. Nevertheless, this could not be proved with the fecal microbiota evaluated in the present study because no biopsy of the intestinal mucosa was performed and the rate of mortality of the microorganisms along the GIT is high.
On the other hand, it has been stated in the literature that the establishment or colonization of the GIT is species or strain dependent and that effective colonization occurs more easily in neonates (2, 44–46) because the microbiota of the gut is not yet fully formed. Moreover, the intestinal microbiota in healthy adults appears to be stable and well defined at the genus level and even the species level. However, considerable variation in the composition of the fecal microbiota has been reported, and dietary components can influence this variability (2, 47).
Although fecal samples are readily available for study, it is questionable whether the microorganisms in feces are representative of the population of microbiota because they vary throughout the GIT (2). For more precise information about the gut microbiota, samples collected during endoscopic or surgical procedures are more accurate. However, these procedures are discouraged and rarely used in research, for obvious reasons (2, 3, 6).
The quantities of L. plantarum in samples from the same subjects (biological duplicates) were similar during the consumption and the postconsumption time periods, which resulted in a low coefficient of variation. However, the range among individuals was very large (data not shown). This may be related to the individual microbiota (3). These data show the reproducibility of the methodology used.
In conclusion, the quantification of L. plantarum Lp115 in the fecal microbiota of subjects during the period of consumption of fermented milk suggested that daily intake of probiotics is necessary to maintain levels in the GIT, since it was not possible to detect the bacteria in the feces after the cessation of consumption of fermented milk.
The relative quantification using qPCR in association with an internal reference control guarantees the precise quantification of the amount of bacteria in fecal samples. The method that uses these tools to quantify pathogens and probiotics in food samples and population microbial studies is convenient due to its simplicity, specificity, and high sensitivity and can be distinguished from conventional culture methods.
ACKNOWLEDGMENTS
We acknowledge the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process 140352/2008-2) for grants to G.N.C.
We gratefully acknowledge the Danisco, C. Hansen, and Duas Rodas companies for donations of bacterial cultures and flavoring. We also thank Fernando Macedo (UEL) for reading the manuscript and suggestions.
Footnotes
Published ahead of print 22 November 2013
REFERENCES
- 1.Gill HS, Darragh AJ, Cross ML. 2001. Optimizing immunity and gut function in the elderly. J. Nutr. Health Aging 5:80–91 [PubMed] [Google Scholar]
- 2.Isolauri E, Salminen S, Ouwehand AC. 2004. Probiotics. Best Pract. Res. Clin. Gastroenterol. 18:299–313. 10.1016/j.bpg.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 4.Tiihonen K, Ouwehand AC, Rautonen N. 2010. Human intestinal microbiota and healthy ageing. Ageing Res. Rev. 9:107–116. 10.1016/j.arr.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 5.Jankovic I, Sybesma W, Phothirath P, Ananta E, Mercenier A. 2010. Application of probiotics in food products—challenges and new approaches. Curr. Opin. Biotechnol. 21:175–181. 10.1016/j.copbio.2010.03.009 [DOI] [PubMed] [Google Scholar]
- 6.Saarela M, Mogensen G, Fondén R, Mättö J, Mattila-Sandholm T. 2000. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84:197–215. 10.1016/S0168-1656(00)00375-8 [DOI] [PubMed] [Google Scholar]
- 7.Carey CM, Kirk JL, Ojha S, Kostrzynska M. 2007. Current and future uses of real time polymerase chain reaction and microarrays in the study of intestinal microbiota and probiotic use and effectiveness. Can. J. Microbiol. 53:537–550. 10.1139/W07-039 [DOI] [PubMed] [Google Scholar]
- 8.Gueimonde M, Tolkko S, Korpimki T, Salminen S. 2004. New real-time quantitative PCR procedure for quantification of Bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70:4165–4169. 10.1128/AEM.70.7.4165-4169.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoetendal EG, Rajilić-Stojanović M, de Vos WM. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605–1615. 10.1136/gut.2007.133603 [DOI] [PubMed] [Google Scholar]
- 10.Postollec F, Falentin H, Pavan S, Combrisson J, Sohi D. 2011. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 28:848–861. 10.1016/j.fm.2011.02.008 [DOI] [PubMed] [Google Scholar]
- 11.Palmer S, Wiegand A, Maldarelli F, Bazmi H, Mican J, Polis M, Dewar R, Planta A, Liu S, Metcalf J, Mellors J, Coffin J. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:4531–4536. 10.1128/JCM.41.10.4531-4536.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer M, Obermajer N, Matijašić BB, Rogelj I, Kmetec V. 2009. Quantification of live and dead probiotic bacteria in lyophilised product by real-time PCR and by flow cytometry. Appl. Microbiol. Biotechnol. 84:1137–1147. 10.1007/s00253-009-2068-7 [DOI] [PubMed] [Google Scholar]
- 13.Masco L, Vanhoutte L, Temmerman R, Swings J, Huys G. 2007. Evaluation of real-time PCR targeting the 16S rRNA and recA genes for the enumeration of bifidobacteria in probiotic products. Int. J. Food Microbiol. 113:351–357. 10.1016/j.ijfoodmicro.2006.07.021 [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto J, Matsuki T, Sasamoto M, Tomii Y, Watanabe K. 2008. Identification and quantification of Lactobacillus casei strain Shirota in human feces with strain-specific primers derived from randomly amplified polymorphic DNA. Int. J. Food Microbiol. 126:210–215. 10.1016/j.ijfoodmicro.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 15.Gopal PK, Prasad J, Gill HS. 2003. Effects of the consumption of Bifidobacterium lactis HN019 (DR10™) and galacto-oligosaccharides on the microflora of the gastrointestinal tract in the human subjects. Nutr. Res. 23:1313–1328. 10.1016/S0271-5317(03)00134-9 [DOI] [Google Scholar]
- 16.Hugenschmidt S, Schwenninger SM, Gnehm N, Lacroix C. 2010. Screening of a natural biodiversity of lactic and propionic acid bacteria for folate and vitamin B12 production in supplemented whey permeate. Int. Dairy J. 20:852–857. 10.1016/j.idairyj.2010.05.005 [DOI] [Google Scholar]
- 17.Naruszewicz M, Johansson ML, Zapolska-Downar D, Bukowska H. 2002. Effect of Lactobacillus plantarum 299v on cardiovascular disease risk factors in smokers. Am. J. Clin. Nutr. 76:1249–1255 [DOI] [PubMed] [Google Scholar]
- 18.Paolillo R, Carratelli CR, Sorrentino S, Mazzola N, Rizzo A. 2009. Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int. Immunopharmacol. 9:1265–1271. 10.1016/j.intimp.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 19.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, Mckenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. 2006. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 116:1310–1316. 10.1172/JCI21404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleerebezem M, Boekhorst J, Van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995. 10.1073/pnas.0337704100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paineau D, Carcano D, Leyer G, Darquy S, Alyanakian MA, Simoneau G, Bergmann JF, Brassart D, Bornet F, Ouwehand AC. 2008. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol. Med. Microbiol. 53:107–113. 10.1111/j.1574-695X.2008.00413.x [DOI] [PubMed] [Google Scholar]
- 22.Daniel C, Poiret S, Goudercourt D, Dennin V, Leyer G, Pot B. 2006. Selecting lactic acid bacteria for their safety and functionality by use of a mouse colitis model. Appl. Environ. Microbiol. 72:5799–5805. 10.1128/AEM.00109-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa GN, Villas-Bôas GT, Villas-Boas LA, Miglioranza LHS. 2011. In silico phylogenetic analysis of lactic acid bacteria and new primer set for identification of Lactobacillus plantarum in food samples. Eur. Food Res. Technol. 233:233–241. 10.1007/s00217-011-1508-7 [DOI] [Google Scholar]
- 24.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen N, Vogensen FK, Gøbel R, Michaelsen KF, Abu Al-Soud W, Sørensen SJ, Hansen LH, Jakobsen M. 2011. Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp. lactis Bi-07. FEMS Microbiol. Ecol. 75:482–496. 10.1111/j.1574-6941.2010.01024.x [DOI] [PubMed] [Google Scholar]
- 26.de Vrese M, Winkler P, Rautenberg P, Harder T, Noah C, Laue C, Ott S, Hampe J, Schreiber S, Heller K, Schrezenmeir J. 2006. Probiotic bacteria reduced duration and severity but not the incidence of common cold episodes in a double blind, randomized, controlled trial. Vaccine 24:6670–6674. 10.1016/j.vaccine.2006.05.048 [DOI] [PubMed] [Google Scholar]
- 27.Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. 2009. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics 124:e172–e179. 10.1542/peds.2008-2666 [DOI] [PubMed] [Google Scholar]
- 28.Böhm-Hofstätter H, Tschernutter M, Kunert R. 2010. Comparison of hybridization methods and real-time PCR: their value in animal cell line characterization. Appl. Microbiol. Biotechnol. 87:419–425. 10.1007/s00253-010-2580-9 [DOI] [PubMed] [Google Scholar]
- 29.Matijašić BB, Obermajer T, Rogelj I. 2010. Quantification of Lactobacillus gasseri, Enterococcus faecium and Bifidobacterium infantis in a probiotic OTC drug by real-time PCR. Food Control 21:419–425. 10.1016/j.foodcont.2009.07.001 [DOI] [Google Scholar]
- 30.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. 2006. The real-time polymerase chain reaction. Mol. Aspects Med. 27:95–125. 10.1016/j.mam.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 31.Dang W, Sun L. 2011. Determination of internal controls for quantitative real time RT-PCR analysis of the effect of Edwardsiella tarda infection on gene expression in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 30:720–728. 10.1016/j.fsi.2010.12.028 [DOI] [PubMed] [Google Scholar]
- 32.Ginzinger DG. 2002. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 30:503–512. 10.1016/S0301-472X(02)00806-8 [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Lu S, Smith AP, Moore D, Lee NM. 2010. Different real-time PCR systems yield different gene expression values. Mol. Cell. Probes 24:315–320. 10.1016/j.mcp.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Saint DA. 2002. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem. 302:52–59. 10.1006/abio.2001.5530 [DOI] [PubMed] [Google Scholar]
- 36.Adlerberth I, Ahrné S, Johansson ML, Molin G, Hanson LA, Wold AE. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 62:2244–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhanani AS, Gaudana SB, Bagchi T. 2011. The ability of Lactobacillus adhesin EF-Tu to interfere with pathogen adhesion. Eur. Food Res. Technol. 232:777–785. 10.1007/s00217-011-1443-7 [DOI] [Google Scholar]
- 38.Molin G, Jeppson B, Johansson ML, Ahrné S, Nobaek S, Ståhl M, Bengmark S. 1993. Numerical taxonomy of Lactobacillus spp. associated with healthy and diseased mucosa of the human intestines. J. Appl. Microbiol. 74:314–323. 10.1111/j.1365-2672.1993.tb03031.x [DOI] [PubMed] [Google Scholar]
- 39.Wang CY, Lin PR, Ng CC, Shyu YT. 2010. Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe 16:578–585. 10.1016/j.anaerobe.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 40.Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, Von Wright A. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Champs C, Maroncle N, Balestrino D, Rich C, Forestier C. 2003. Persistence of colonization of intestinal mucosa by a probiotic strain, Lactobacillus casei subsp. rhamnosus Lcr35 after oral consumption. J. Clin. Microbiol. 41:1270–1273. 10.1128/JCM.41.3.1270-1273.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito Y, Sakamoto M, Takizawa S, Benno S. 2004. Monitoring the cell number and viability of Lactobacillus helveticus GCL1001 in human feces by PCR methods. FEMS Microbiol. Lett. 231:125–130. 10.1016/S0378-1097(03)00951-0 [DOI] [PubMed] [Google Scholar]
- 43.Tannock GW, Munro K, Harmsen HJM, Welling GW, Smart J, Gopal PK. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578–2588. 10.1128/AEM.66.6.2578-2588.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyman M, Ménard S. 2002. Probiotic microorganisms: how they affect the intestinal pathophysiology. Cell. Mol. Life Sci. 59:1151–1165. 10.1007/s00018-002-8494-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langhendries JP, Detry J, Van Hees J, Lamboray JM, Darimont J, Mozin MJ, Secretin MC, Senterre J. 1995. Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full-term infants. J. Pediatr. Gastroenterol. Nutr. 21:177–181. 10.1097/00005176-199508000-00009 [DOI] [PubMed] [Google Scholar]
- 46.Haarman M, Knol J. 2006. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72:2359–2365. 10.1128/AEM.72.4.2359-2365.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santosa S, Farnworth E, Jones PJH. 2006. Probiotics and their potential health claims. Nutr. Rev. 64:265–274. 10.1111/j.1753-4887.2006.tb00209.x [DOI] [PubMed] [Google Scholar]