Abstract
Buruli ulcer is an indolent, slowly progressing necrotizing disease of the skin caused by infection with Mycobacterium ulcerans. In the present study, we applied a redesigned technique to a vast panel of M. ulcerans disease isolates and clinical samples originating from multiple African disease foci in order to (i) gain fundamental insights into the population structure and evolutionary history of the pathogen and (ii) disentangle the phylogeographic relationships within the genetically conserved cluster of African M. ulcerans. Our analyses identified 23 different African insertion sequence element single nucleotide polymorphism (ISE-SNP) types that dominate in different areas where Buruli ulcer is endemic. These ISE-SNP types appear to be the initial stages of clonal diversification from a common, possibly ancestral ISE-SNP type. ISE-SNP types were found unevenly distributed over the greater West African hydrological drainage basins. Our findings suggest that geographical barriers bordering the basins to some extent prevented bacterial gene flow between basins and that this resulted in independent focal transmission clusters associated with the hydrological drainage areas. Different phylogenetic methods yielded two well-supported sister clades within the African ISE-SNP types. The ISE-SNP types from the “pan-African clade” were found to be widespread throughout Africa, while the ISE-SNP types of the “Gabonese/Cameroonian clade” were much rarer and found in a more restricted area, which suggested that the latter clade evolved more recently. Additionally, the Gabonese/Cameroonian clade was found to form a strongly supported monophyletic group with Papua New Guinean ISE-SNP type 8, which is unrelated to other Southeast Asian ISE-SNP types.
INTRODUCTION
Buruli ulcer (BU) is a slowly progressing necrotizing disease of the skin and subcutaneous tissue that is caused by infection with Mycobacterium ulcerans (1). BU is the third most common mycobacterial disease in humans, after tuberculosis and leprosy, and the least understood of the three (2). Even though the infection affects all age groups, at least half of all cases occur in children under age 15 years (3). More than 30 countries worldwide have reported (but not always confirmed) this emerging disease, with the highest incidence in West and Central Africa, where the disease occurs in foci among people living in rural marshes, wetlands, and riverine areas (1, 4). As proximity to these slow-flowing or stagnant water bodies is a known risk factor for M. ulcerans infection (5) and as M. ulcerans DNA has been detected in a variety of aquatic specimens (6, 7), it is generally believed that M. ulcerans is an environmental mycobacterium that can initiate infection after microtraumata of the skin (8). However, the exact mode of transmission and the environmental reservoir(s) of M. ulcerans remain largely unknown (9), as (i) culturing the slow-growing mycobacterium from an environmental source is particularly difficult (10) and (ii) the significance of the detection of M. ulcerans DNA by PCR in environmental samples remains unclear in the disease ecology of BU (6, 7, 11–16).
Multilocus sequence typing analyses (17) and subsequent whole-genome comparisons (18) have proved that M. ulcerans recently evolved from a Mycobacterium marinum progenitor by acquisition of the virulence plasmid pMUM001. This plasmid harbors genes required for the synthesis of the macrocyclic polyketide toxin mycolactone (19), which has cytotoxic and immunosuppressive properties that cause chronic ulcerative skin lesions with limited inflammation and thus plays a key role in the pathogenesis of BU (20). Both the acquisition of the plasmid and a reductive evolution (21, 22) led the generalist M. marinum to become a highly specialized mycobacterium that is more adapted to a restricted environment, such as that of a vertebrate host. Analysis of the genome sequence suggests that this new niche is likely to an obscure, aerated, osmotically stable, extracellular environment where slow growth, the loss of several immunogenic proteins, and production of mycolactone provided selective advantages (18, 22). Many of the changes in this evolutionary process were mediated by two insertion sequence elements (ISE), IS2404 and IS2606, that are present in the M. ulcerans genome in ≈200 and ≈90 copies, respectively (22). These short, mobile genetic DNA elements promote genetic rearrangements by modifying gene expression and sequestering genes, profoundly affecting mycobacterial genome plasticity (23). Increased ISE numbers are expected, as the aforementioned lifestyle shift causes many loci to become excessive, as they are no longer essential for survival in the new environment (24). Subsequent whole-genome comparisons (18) have shown furthermore that the resulting niche-adapted genomic signature was established in an M. ulcerans progenitor before its intercontinental dispersal.
Deciphering the structure of pathogenic bacterial populations is instrumental for the understanding of the epidemiology, global spread, and evolutionary history of bacterial infectious diseases. Moreover, understanding the population structure allows for studying meaningful bacterial differences that can affect disease control, including public health interventions, such as vaccination programs (25). Differences in the ratio of genetic variation caused by de novo mutations relative to recombination bring about a spectrum of different bacterial population structures, ranging from “clonal” (no recombination) to “nonclonal” (where a lot of recombination of alleles prevents the emergence of stable clones) (26). Because of the clonal population structure of M. ulcerans, conventional genetic fingerprinting methods have largely failed to genetically differentiate clinical disease isolates, complicating molecular analyses on the elucidation of the disease ecology and the population structure and evolutionary history of the pathogen (27). However, in 2009, Käser et al. (28) identified single nucleotide polymorphisms (SNPs) within M. ulcerans haplotype-specific IS2404 elements MUL_2990 and MUL_3871, which are located in region of difference 1 (RD1) and RD12, respectively (29). The identified SNPs differentiated multiple genotypes among isolates originating from one region in Ghana, resulting in the highest geographical resolution of genotyping achieved to date without the use of whole-genome sequencing. Given the apparent rarity of recombination in M. ulcerans, ISE-SNP types should contain sufficient phylogenetic signal to reconstruct recent evolutionary events on a continental scale. Hence, in the present study, we applied a redesigned form of the ISE-SNP typing technique as described by Käser et al. (28) to a vast panel of M. ulcerans isolates originating from multiple African disease foci to gain deeper insights into the population structure and evolutionary history of the pathogen and to continue to disentangle the phylogeographic relationships within the genetically conserved cluster of African M. ulcerans.
MATERIALS AND METHODS
A panel (n = 171) of 157 M. ulcerans clinical isolates and 14 clinical specimens with a quantification cycle Cq (IS2404) of ≤32 originating from disease foci in 11 different African countries was selected to assess the polymorphisms in the RD1- and RD12-associated haplotype-specific copies of IS2404 (Tables 1 and 2). Clinical specimens consisted of tissue fragments and swabs originating from ulcerated and nonulcerated BU lesions. These surplus samples had been collected for routine diagnostic purposes and for rechecking for quality control. All isolates and specimens were selected from the comprehensive mycobacterial collection of the Institute of Tropical Medicine (ITM) and were chosen to maximize temporal and spatial diversity within countries in which more than 20 isolates/specimens were available. Isolates and specimens were processed and analyzed for bacterial polymorphisms without use of any patient identifiers, except for country and village of origin if this information was available.
TABLE 1.
Isolates used in this studya
| ISE-SNP type | Culture no. | Country of origin | Administration division |
Source | YOI | Remark | ||
|---|---|---|---|---|---|---|---|---|
| First level | Second level | Third level | ||||||
| 1 | ITM_940511 | Ivory Coast | Moyen-Cavally | Duékoué | Niambli | ITM | 1994 | |
| 1 | ITM_000483 | Ivory Coast | Moyen-Cavally | Duékoué | Niambli | ITM | 2000 | |
| 1 | ITM_000870 | Ivory Coast | Dix-Huit Montagnes | Zouan-Hounien | Ouyatouo | ITM | 2000 | |
| 1 | ITM_063519 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Cité Songololo | IME | 2006 | |
| 1 | ITM_071924 | Congo | Kouilou | Madingo-Kayes | Loukouala | ITM | 2007 | Originated from same patient as 071925 |
| 1 | ITM_071925 | Congo | Kouilou | Madingo-Kayes | Loukouala | ITM | 2007 | Originated from same patient as 071924 |
| 1 | ITM_072398 | DRC | Bas-Congo | Cataractes/Songololo | Bamboma/Mbanza-Manteke | IME | 2007 | |
| 1 | ITM_072401 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2007 | |
| 1 | ITM_072732 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2007 | |
| 1 | ITM_072733 | DRC | Bas-Congo | Cataractes/Songololo | Luima-Mayanga/Ngombe | IME | 2007 | |
| 1 | ITM_072734 | DRC | Bas-Congo | Cataractes/Songololo | Bamboma/Mbanza-Manteke | IME | 2007 | |
| 1 | ITM_072735 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Luvuvamu | IME | 2007 | |
| 1 | ITM_072840 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2007 | Originated from same patient as 072841 |
| 1 | ITM_072841 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2007 | Originated from same patient as 072840 |
| 1 | ITM_073453 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2007 | |
| 1 | ITM_073459 | Benin | Kouffo | Lalo | Ahojinako | CDTUB Lalo | 2007 | |
| 1 | ITM_073463 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Kisonga | IME | 2007 | |
| 1 | ITM_073477 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Cité Songololo | IME | 2007 | |
| 1 | ITM_073478 | Angola | Malanje | Marimba | Kafufu/Luremo (Kwango River) | IME | 2007 | |
| 1 | ITM_073479 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Kisonga | IME | 2007 | |
| 1 | ITM_082600 | DRC | Bas-Congo | Cataractes/Songololo | Kilueka/Nzundu | IME | 2008 | |
| 1 | ITM_100140 | DRC | Bas-Congo | Cataractes/Songololo | Lovo/Tole | IME | 2010 | |
| 1 | ITM_100141 | DRC | Bas-Congo | Cataractes/Songololo | Mayanga/Mpelo | IME | 2010 | Originated from same patient as 100141 |
| 1 | ITM_100142 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Luvuvamu | IME | 2010 | Originated from same patient as 100142 |
| 1 | ITM_100832 | DRC | Bas-Congo | Cataractes/Songololo | Palabala/Nkamuna | IME | 2010 | |
| 1 | ITM_100833 | DRC | Bas-Congo | Cataractes/Songololo | Mayanga/Mpelo | IME | 2010 | |
| 1 | ITM_032481 | DRC | Bas-Congo | Cataractes/Songololo | Luima/Nkondo-Kiomba | IME | 2003 | |
| 1 | ITM_040149 | Ghana | Ashanti | Asante Akim North | Agogo Presbyterian Hospital | ITM | 2003 | |
| 1 | ITM_991591 | Togo | Maritime | Vo | Anagali | ITM | 1999 | |
| 1 | ITM_050303 | Congo | Kouilou | ITM | 1979 | Originated from same patient as 050304 | ||
| 1 | ITM_050304 | Congo | Kouilou | ITM | 1979 | Originated from same patient as 050303 | ||
| 1 | ITM_960658 | Angola | Bengo | Dande | Caxito | ITM | 1996 | Originated from same patient as 960657 |
| 1 | ITM_960657 | Angola | Bengo | Dande | Caxito | ITM | 1996 | Originated from same patient as 960658 |
| 1 | ITM_072662 | Ghana | Ashanti | Asante Akim North | Ananekrom | KCCR | 2007 | |
| 1 | ITM_072646 | Ghana | Ashanti | Atwima Mponua | Abofrom | KCCR | 2007 | |
| 1 | ITM_072651 | Ghana | Ashanti | KMA | Kaase | KCCR | 2007 | |
| 1 | ITM_120140 | Cameroon | Adamawa Region | Maya-Banyo | Bankim/Mbondji II | CPC | 2011 | |
| 1 | ITM_030950 | Benin | Kouffo | Lalo | Adoukandji | CDTUB Lalo | 2003 | |
| 1 | ITM_030716 | Benin | Kouffo | Lalo | Tchito/Village Aboeti | CDTUB Lalo | 2003 | |
| 1 | ITM_102686 | Nigeria | Oyo State | Ibadan | Ibadan | ITM | 2010 | |
| 1 | ITM_083232 | Angola | Lunda Norte | Xa-Muteba | Kwango River | ITM | 2008 | |
| 1 | ITM_000869 | Ivory Coast | Moyen-Cavally | Duékoué | Guezon | ITM | 2000 | |
| 1 | ITM_990007 | Ivory Coast | Haut-Sassandra | Issia | Guetuzon II | ITM | 1998 | |
| 1 | ITM_991633 | Ivory Coast | Moyen-Cavally | Duékoué | Guezon | ITM | 1999 | |
| 2 | ITM_030791 | Benin | Kouffo | Lalo | Tchito/Gare | CDTUB Zagnanado | 2003 | |
| 2 | ITM_970680 | Benin | Mono | Houéyogbé | Sahoué | CDTUB Lalo | 1997 | |
| 2 | ITM_022876 | Benin | Kouffo | Lalo | Tohou | CDTUB Lalo | 2002 | |
| 2 | ITM_021434 | Benin | Kouffo | Klouékanmè | Adjassagon | CDTUB Lalo | 2002 | |
| 2 | ITM_012596 | Benin | Mono | Bopa | Lobogo | CDTUB Lalo | 2001 | |
| 2 | ITM_071938 | Benin | Kouffo | Lalo | Tandji | CDTUB Lalo | 2007 | |
| 4 | ITM_5150 | DRC | Bandundu | Kwilu | ITM | 1962 | ||
| 5 | ITM_940512 | Benin | Zou | Ouinhi | Ouokon | CDTUB Zagnanado | 1994 | |
| 5 | ITM_010157 | Benin | Zou | Zogbodomè | Domè-Houandougon | CDTUB Zagnanado | 2001 | |
| 5 | ITM_000951 | Benin | Zou | Zogbodomè | Domè-Houandougon | CDTUB Zagnanado | 2000 | |
| 5 | ITM_970435 | Benin | Ouémé | Bonou | Bonou | CDTUB Zagnanado | 1997 | Originated from same patient as 970301 |
| 5 | ITM_970301 | Benin | Ouémé | Bonou | Bonou | CDTUB Zagnanado | 1997 | Originated from same patient as 970435 |
| 5 | ITM_000479 | Benin | Zou | Zagnanado | Zagnanado/Doga | CDTUB Zagnanado | 2000 | |
| 5 | ITM_092100 | Benin | Zou | Zagnanado | Doga-Domè | CDTUB Zagnanado | 2009 | |
| 5 | ITM_083865 | Benin | Zou | Ouinhi | Tohoue/Hounnoumè | CDTUB Zagnanado | 2008 | |
| 5 | ITM_093013 | Benin | Zou | Ouinhi | Ouinhi/Monzoungoudo | CDTUB Zagnanado | 2009 | |
| 5 | ITM_093695 | Benin | Zou | Ouinhi | Ouinhi/Monzoungoudo | CDTUB Zagnanado | 2009 | |
| 5 | ITM_101300 | Benin | Zou | Ouinhi | Sagon/Adamè | CDTUB Zagnanado | 2010 | |
| 5 | ITM_101302 | Benin | Zou | Ouinhi | Dasso/Bossa | CDTUB Zagnanado | 2010 | |
| 5 | ITM_102554 | Benin | Zou | Ouinhi | Dasso/Agonkon | CDTUB Zagnanado | 2010 | |
| 5 | ITM_081919 | Benin | Zou | Ouinhi | Dasso/Yaago and Akantomè | CDTUB Zagnanado | 2008 | |
| 5 | ITM_092997 | Benin | Zou | Djidja | Oungbègame | CDTUB Zagnanado | 2009 | |
| 5 | ITM_080066 | Benin | Zou | Ouinhi | Sagon/Ayizè | CDTUB Zagnanado | 2008 | |
| 5 | ITM_070381 | Benin | Zou | Ouinhi | Dasso/Yaago | CDTUB Zagnanado | 2007 | |
| 5 | ITM_073151 | Benin | Zou | Ouinhi | Ouinhi/Monzoungoudo | CDTUB Zagnanado | 2007 | |
| 5 | ITM_070131 | Benin | Zou | Zagnanado | Dovi-Dove/Tévedji | CDTUB Zagnanado | 2007 | |
| 5 | ITM_092473 | Benin | Zou | Ouinhi | Tohoue/Midjannangon | CDTUB Zagnanado | 2009 | |
| 5 | ITM_082549 | Benin | Zou | Ouinhi | Tohoue/Akassa | CDTUB Zagnanado | 2008 | |
| 5 | ITM_090149 | Benin | Zou | Zagnanado | Dovi-Dove/Tévedji | CDTUB Zagnanado | 2009 | |
| 5 | ITM_091800 | Benin | Zou | Ouinhi | Tohoue/Gangban | CDTUB Zagnanado | 2009 | |
| 5 | ITM_083584 | Benin | Zou | Ouinhi | Ouinhi/Ahicon | CDTUB Zagnanado | 2008 | |
| 5 | ITM_9146 | Benin | Zou | Zagnanado | Kpedekpo/Loko-Alankpe | CDTUB Zagnanado | 1992 | |
| 5 | ITM_991721 | Benin | Atlantique | Toffo | Séhoué | CDTUB Zagnanado | 1999 | |
| 5 | ITM_092472 | Benin | Atlantique | Toffo | Séhoué/Agaga | CDTUB Zagnanado | 2009 | |
| 5 | ITM_070383 | Benin | Ouémé | Dangbo | Dékin | CDTUB Zagnanado | 2007 | |
| 5 | ITM_070625 | Nigeria | Ogun State | Yewa North | Odja Odan | CDTUB Zagnanado | 2007 | |
| 5 | ITM_061509 | Benin | Zou | Zagnanado | Zagnanado | CDTUB Zagnanado | 2006 | |
| 5 | ITM_081676 | Benin | Plateau | Adja-Ouere | Tatonnoukon | CDTUB Zagnanado | 2008 | |
| 5 | ITM_081681 | Benin | Plateau | Issaba | Onigbolo | CDTUB Zagnanado | 2008 | |
| 5 | ITM_082696 | Benin | Ouémé | Adjohoun | Abato | CDTUB Zagnanado | 2008 | |
| 5 | ITM_091801 | Benin | Zou | Zogbodomè | Kpokissa/Hinzounmè | CDTUB Zagnanado | 2009 | |
| 5 | ITM_092101 | Benin | Ouémé | Dangbo | Gbéko | CDTUB Zagnanado | 2009 | |
| 5 | ITM_093694 | Benin | Ouémé | Dangbo | Gbéko | CDTUB Zagnanado | 2009 | |
| 5 | ITM_100126 | Benin | Zou | Zogbodomè | Kpokissa | CDTUB Zagnanado | 2010 | |
| 5 | ITM_951009 | Benin | Zou | Zagnanado | CDTUB Zagnanado | 1995 | ||
| 6 | ITM_5151 | DRC | Maniema | Kasongo | ITM | 1972 | ||
| 7 | ITM_970359 | Ghana | Ashanti | Amansie West | Manso-Afraso | ITM | 1997 | |
| 7 | ITM_970606 | Ghana | Ashanti | Amansie West | Yaw Kasakrom | ITM | 1997 | |
| 7 | ITM_970677 | Ghana | Ashanti | Amansie West | Manso Dominase | ITM | 1997 | |
| 7 | ITM_970678 | Ghana | Ashanti | Asante Akim North | Afrisre | ITM | 1997 | |
| 7 | ITM_970959 | Ghana | Ashanti | Amansie West | Manso-Afraso | ITM | 1997 | |
| 7 | ITM_970964 | Ghana | Ashanti | Amansie West | Offinho Asaman | ITM | 1997 | |
| 7 | ITM_971351 | Ghana | Ashanti | Atwima Mponua | Achiase | ITM | 1997 | |
| 7 | ITM_980063 | Ghana | Ashanti | Atwima Mponua | Achiase | ITM | 1998 | |
| 7 | ITM_940662 | Ivory Coast | Moyen-Cavally | Duékoué | Nanandi | ITM | 1994 | |
| 7 | ITM_990006 | Ivory Coast | Haut-Sassandra | Issia | Guetuzon I | ITM | 1998 | |
| 7 | ITM_990734 | Ivory Coast | Moyen-Cavally | Duékoué | Duékoué | ITM | 1999 | |
| 7 | ITM_991632 | Ivory Coast | Haut-Sassandra | Issia | Bediegbeu | ITM | 1999 | |
| 7 | ITM_072634 | Ghana | Ashanti | Asante Akim North | Adoniem | KCCR | 2007 | |
| 7 | ITM_072652 | Ghana | Ashanti | Atwima Mponua | Achiase | KCCR | 2007 | |
| 7 | ITM_072654 | Ghana | Ashanti | Atwima Mponua | Achiase | KCCR | 2007 | |
| 7 | ITM_072657 | Ghana | Ashanti | Atwima Mponua | Achiase | KCCR | 2007 | |
| 7 | ITM_072658 | Ghana | Western Region | Wassa West | Owusukrom | KCCR | 2007 | |
| 7 | ITM_072650 | Ghana | Ashanti | Atwima Nwabiagya | Kyereyase | KCCR | 2007 | |
| 7 | ITM_072630 | Ghana | Central | Upper Denkyira | Nkotumso | KCCR | 2007 | |
| 7 | ITM_072656 | Ghana | Ashanti | Atwima Mponua | Abompe | KCCR | 2007 | |
| 7 | ITM_072655 | Ghana | Ashanti | Atwima Mponua | Sireso | KCCR | 2007 | |
| 7 | ITM_072653 | Ghana | Ashanti | Atwima Mponua | Amadaa | KCCR | 2007 | |
| 7 | ITM_072645 | Ghana | Ashanti | Atwima Mponua | Achiase | KCCR | 2007 | |
| 12 | ITM_072814 | Benin | Ouémé | Dangbo | Gbéko | CDTUB Zagnanado | 2007 | |
| 13 | ITM_021433 | Benin | Kouffo | Lalo | Gnizoumè/Hangbanou | CDTUB Lalo | 2002 | |
| 13 | ITM_022045 | Benin | Kouffo | Lalo | Adoukandji/Yamontouhoué | CDTUB Lalo | 2002 | |
| 13 | ITM_022287 | Benin | Zou | Agbangnizoun | Kpota | CDTUB Lalo | 2002 | |
| 13 | ITM_022875 | Benin | Kouffo | Lalo | Gnizoumè | CDTUB Lalo | 2002 | |
| 13 | ITM_030717 | Benin | Kouffo | Lalo | Ahomadégbé | CDTUB Lalo | 2003 | |
| 13 | ITM_030718 | Benin | Kouffo | Lalo | Lalo | CDTUB Lalo | 2003 | |
| 13 | ITM_031892 | Benin | Kouffo | Lalo | Hlassamè | CDTUB Lalo | 2003 | |
| 13 | ITM_071804 | Benin | Kouffo | Lalo | Zalli | CDTUB Lalo | 2007 | |
| 14 | ITM_991590 | Togo | Maritime | Vo | Tchekpo Deve | ITM | 1999 | Originated from same patient as 000909 |
| 14 | ITM_000909 | Togo | Maritime | Vo | Tchekpo Deve | ITM | 2000 | Originated from same patient as 991590 |
| 14 | ITM_993354 | Togo | Maritime | Vo | Tchekpo Deve | ITM | 1999 | |
| 14 | ITM_042407 | Togo | Maritime | Vo | Kodji Kopé | ITM | 2004 | |
| 15 | ITM_070404 | DRC | Bas-Congo | Cataractes/Songololo | Kimpese/Cité-Kimpese | IME | 2007 | Originated from same patient as 070123 |
| 15 | ITM_070123 | DRC | Bas-Congo | Cataractes/Songololo | Kimpese/Cité-Kimpese | IME | 2007 | Originated from same patient as 070404 |
| 15 | ITM_092479 | DRC | Bas-Congo | Cataractes/Songololo | Kimpese/Cité-Kimpese | IME | 2009 | |
| 16 | ITM_990008 | Ivory Coast | Haut-Sassandra | Issia | Zakogbeu | ITM | 1998 | |
| 18 | ITM_070386 | Nigeria | Anambra State | Ayamelum | Ifite Ogwari | ITM | 2007 | |
| 19 | ITM_001211 | Ivory Coast | Dix-Huit Montagnes | Zouan-Hounien | Zouan-Hounien | ITM | 2000 | |
| 20 | ITM_020279 | Cameroon | Centre Region | Nyong-et-Mfoumou | Ayos | CPC | 2002 | |
| 20 | ITM_091067 | Gabon | Moyen-Ogooué | Ogooue et des Lacs | Junkville | ITM | 2009 | |
| 20 | ITM_110450 | Gabon | Moyen-Ogooué | Ogooue et des Lacs | Gravier | ITM | 2011 | |
| 22 | ITM_020280 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akolo | CPC | 2002 | |
| 22 | ITM_120542 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 23 | ITM_021081 | Cameroon | Centre Region | Nyong-et-Mfoumou | Obis | CPC | 2002 | |
| 23 | ITM_9102 | Cameroon | Centre Region | ITM | 1970 | |||
| 23 | ITM_9103 | Cameroon | Centre Region | ITM | 1970 | |||
| 23 | ITM_101500 | Gabon | Moyen-Ogooué | Ogooue et des Lacs | Lambaréné/Adaghe | ITM | 2010 | |
| 23 | ITM_110893 | Gabon | Moyen-Ogooué | Ogooue et des Lacs | Issac | ITM | 2011 | |
| 23 | ITM_120138 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2010 | |
| 23 | ITM_120139 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga/Wouma | CPC | 2011 | |
| 23 | ITM_120141 | Cameroon | Centre Region | Nyong-et-Mfoumou | Medjap | CPC | 2011 | |
| 23 | ITM_120142 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga/Ekolman | CPC | 2011 | |
| 23 | ITM_120534 | Cameroon | Centre Region | Nyong-Et-Soo | Bembé | CPC | 2011 | |
| 23 | ITM_120535 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 23 | ITM_120536 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga/Djo'o | CPC | 2011 | |
| 23 | ITM_120538 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 23 | ITM_120539 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 23 | ITM_120543 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 23 | ITM_120143 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
| 24 | ITM_051459 | Uganda | Northern Region | Adjumani | Adjumani | NCTC | 2005 | |
| 25 | ITM_120537 | Cameroon | Centre Region | Nyong-Et-Soo | Edjom | CPC | 2011 | |
| 26 | ITM_120540 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akam-Engali | CPC | 2011 | |
| 27 | ITM_120541 | Cameroon | Centre Region | Nyong-et-Mfoumou | Akonolinga | CPC | 2011 | |
Abbreviations: CDTUB, Centre de Dépistage et de Traitement de l'Ulcère de Buruli; CPC, Centre Pasteur du Cameroun; DRC, Democratic Republic of Congo; IME, Institut Médical Evangélique; KCCR, Kumasi Centre for Collaborative Research in Tropical Medicine; NCTC, National Collection of Type Cultures; PNLUB, Programme National de Lutte contre l'Ulcère de Buruli; YOI, year of isolation.
TABLE 2.
Clinical specimens used in this studya
| ISE-SNP type | Sample no. | Country of origin | Administration division |
Source | YOI | ||
|---|---|---|---|---|---|---|---|
| First level | Second level | Third level | |||||
| 1 | BK121032 | Nigeria | Cross River State | Ogoja | TBL Hospital Monaiya | ITM | 2012 |
| 1 | BK120888 | DRC | Maniema | Kibombo | Likeri | PNLUB | 2012 |
| 1 | BK120890 | DRC | Maniema | Kasongo | Samba/Malela | PNLUB | 2012 |
| 1 | BK120891 | DRC | Maniema | Kasongo | Kankumba | PNLUB | 2012 |
| 1 | BK065361 | Nigeria | Enugu State | Igbo Eze North | Nkpo Hamida | ITM | 2006 |
| 5 | BK121025 | Nigeria | Ogun State | Abeokuta North | Abeokuta/Ijaye State Hospital | ITM | 2012 |
| 5 | BK121026 | Nigeria | Ogun State | Abeokuta North | Abeokuta/Ijaye State Hospital | ITM | 2012 |
| 5 | BK121031 | Nigeria | Ogun State | Yewa South | Oke-Odan/PHC Oke-Odan | ITM | 2012 |
| 17 | BK065369 | Nigeria | Ebonyi State | Ohaozora | Iburu | ITM | 2006 |
| 20 | BK105250 | Gabon | Nyanga | Douigni | Moussamou Kougou | ITM | 2010 |
| 21 | BK101660 | Gabon | Moyen-Ogooué | Ogooue et des Lacs Department | Lambaréné/Point V | ITM | 2010 |
| 23 | BK100901 | Gabon | Moyen-Ogooué | Ogooue et des Lacs Department | Lambaréné/Bellevue | ITM | 2010 |
| 23 | BK100900 | Gabon | Moyen-Ogooué | Ogooue et des Lacs Department | Lambaréné/Isaac | ITM | 2010 |
Abbreviations: DRC, Democratic Republic of Congo; PNLUB, Programme National de Lutte contre l'Ulcère de Buruli; YOI, year of isolation.
Based on conventional phenotypic and genotypic methods, bacterial isolates had previously been assigned to the species M. ulcerans. They had all tested positive for IS2404 via primers that amplify all copies of IS2404 routinely used for diagnostic PCR (30). Mycobacterial isolates were maintained for prolonged storage at ≤−70°C in Dubos broth enriched with growth supplement and glycerol. They were recultured on solid Löwenstein-Jensen medium. DNA was obtained by scraping 1 to 2 loopfuls of colonies into 400 μl of Tris-EDTA followed by heat inactivation at 100°C for 5 min and subsequent centrifugation to remove cellular debris. Clinical specimens were maintained (after decontamination) at ≤−18°C. The modified Boom DNA extraction procedure was carried out on all clinical specimens as previously described (31).
As the original ISE-SNP typing method described by Käser et al. (28) resulted in aspecific bands, short sequence reads, and high background signals, we redesigned and optimized primers and conditions for PCR and sequencing for application directly on clinical specimens. Primer pair RD1_SENSE (GGTGCTTAACGAAACGTGCTG) and RD1_ANTI_SENSE (ACGGGCTATCTGGAGAACGA) was designed to amplify a fragment of 1,431 bp in RD1 that comprises IS2404 (MUL_2990), while the primer pair RD12_SENSE (CGTTGGCGCGGTACAAGCTTCCCAA) and RD12_ANTI_SENSE (GATGGTCGCGGTGCTGCTTGCCCT) was used to amplify a 1,871-bp PCR product in RD12 that comprises IS2404 (MUL_3871). Primers were designed with Primer Premier 6 (Premier Biosoft, CA) and evaluated in silico with Amplify 3.1.4 (Bill Engels, University of Wisconsin). The PCR design was challenging, as only haplotype-specific copies of IS2404 (MUL_2990 and MUL_3871) were to be amplified and because the relevant regions were of considerable size (1,730 bp for RD1 and 1,905 bp for RD12). Although we reduced the size of the amplicons in both assays (by 299 bp for RD1 and by 48 bp for RD12), they still contained all the variable nucleotide positions described by Käser et al. (28). PCR mixtures contained 1.0 U of HotStarTaq polymerase (Qiagen, Hilden, Germany), 3.0 μl 10× PCR buffer, 6.0 μl Qsolution, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, and 0.5 μM each primer in a total volume of 30 μl. PCRs were carried out on a Biometra TProfessional thermal cycler under the following conditions: an initial denaturation step of 15 min at 95°C, followed by 40 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 65°C (RD1) or 70°C (RD12), and elongation for 2 min at 72°C, and ending with a final elongation step of 10 min at 72°C. PCR products were visualized with ethidium bromide on 1% agarose gels by electrophoresis (30 min, 100 V). PCR products were purified by automated gel excision. Bidirectional sequencing was performed at the Genetic Service Facility of the Flanders Institute for Biotechnology (GSF-VIB) on an Applied Biosystems 3730 DNA analyzer capillary sequencer with the ABI Prism BigDye Terminator cycle sequencing v3.1 kit and the PCR primers.
We estimated the bacterial load by a quantitative PCR (qPCR) for IS2404 as described by Fyfe et al. (32) on a set of 122 clinical specimens to determine the Cq (for IS2404) below which the optimized genotyping PCRs were always successful.
The sequences of RD1 and RD12 were concatenated to yield a 3,278-bp fragment and aligned using Clustal X v2.1 (33). Sequences were trimmed to an equal length, and all currently known ISE-SNP types (including 5 ISE-SNP types from Papua New Guinea, Australia, and Malaysia) (28) were added to this data set. We mapped SNPs according to the Agy99 bacterial reference chromosome (GenBank accession no. NC_008611). We constructed a neighbor-joining (NJ) tree based on p distances between ISE-SNP types (34) in MEGA v5 (35). Maximum parsimony (MP) and maximum likelihood (ML) trees were estimated in the same program by using a heuristic search with the tree bisection-reconnection branch-swapping algorithm and random addition of taxa. Relative branch support was evaluated with 1,000 bootstrap replicates (36) for the NJ and MP tree and 200 for the ML tree. Phylogenetic trees for ML analysis were inferred with the nucleotide substitution model selected within jModelTest v0.1.1 (37). Phylogenetic relationships were inferred with ISE-SNP 28 (strain ITM_030524) from Papua New Guinea as the outgroup, isolated from a patient who had never traveled outside of the region around Yarapos in the East Sepik Province (J. Taylor, personal communication). Trees were drawn using FigTree software (38).
A haplotype network was derived using the median joining algorithm after processing the data with the reduced median method as implemented within Network v4.6.1.0 with the default settings (39).
The open source geographic information system Quantum GIS (QGIS) (40) was used to generate the illustration of the geographical distribution of African M. ulcerans. The geographical locations of the residences of BU patients at the time of clinical visit were rendered as points. In the case where residence information was missing, we used the location of the hospital supplying the sample. A modification of the QGIS Python plugin Shift Points was used to modify this point shape file, in which point features with the same position overlapped. Point displacement rendered such features in a circle around the original “real” position. The river layer was translated from the River-Surface Water Body Network data set of the African Water Resource database of the Food and Agriculture Organization (FAO) of the United Nations (41). The administrative borders of countries were rendered from the Global Administrative Unit Layers data set of FAO (http://www.fao.org/geonetwork/srv/en/metadata.show?id=12691).
All statistical testing was performed in R v2.15.2 (42). The correlation between the number of isolates per country and the number of ISE-SNP types per country was checked by using Spearman's rank order correlation coefficient. To examine the relation between ISE-SNP types and the greater West African hydrological drainage basins, the Fisher exact test was used.
RESULTS
Primers and conditions for PCR and sequencing were redesigned and optimized from those described by Käser et al. (28) for application directly on clinical specimens (see Fig. S1 in the supplemental material). Isolates ITM_5150, ITM_5151 ITM_940511, ITM_940512, ITM_960658, ITM_940662, ITM_970359, and ITM_970680 were included in the panel to validate the redesigned assays. These isolates were also included in the panel of Käser et al. (28) and gave identical genotypes as with the redesigned assays.
As our collection of African M. ulcerans isolates did not represent all countries and their different regions where BU is endemic to the same extent, owing to the low sensitivity of culture, we fine-tuned the technique for application directly on clinical specimens by adjusting individual PCR component concentrations and optimizing the thermal PCR profile. We were thus able to deduce sequence information of clinical samples with a modest bacterial load corresponding to a Cq (IS2404) of ≤32. Failure of PCR amplification for specimens with Cq (IS2404) values of >32 was caused by a low mycobacterial DNA concentration.
Amplification and sequencing of IS2404 in MUL_2990 (RD1) and MUL_3871 (RD12) was successful for the entire collection of 157 (100%) clinical isolates (Tables 1 and 2). The optimized method also proved successful for all 14 clinical specimens analyzed, with a Cq (IS2404) of ≤32. A total of 75 (31 in MUL_2990 [RD1] and 44 in MUL_3871 [RD12]) variable nucleotide positions were identified, including four insertions/deletions (indels) (Fig. 1). This resulted in 28 ISE-SNP types, of which 23 were found on the African continent. Sixteen of these were newly identified types, while the other seven corresponded to the ISE-SNP types described by Käser et al. (28). The Papua New Guinean ISE-SNP type 28, used in the phylogenetic analyses as an outgroup, was also a novel type.
FIG 1.
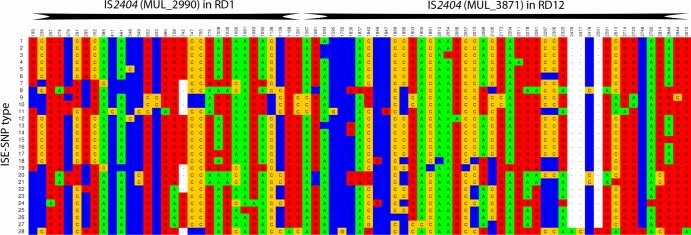
Sequence variation in two haplotype-specific concatenated IS2404 elements: MUL_2990 (RD1) and MUL_3871 (RD12). Only variable nucleotides in the aligned sequences are shown for all 28 ISE-SNP types. SNP position numbers are given according to the scheme described by Käser et al. (28), with position 1 corresponding to position 3313231 in RD1 and 1498 to position 4326896 in RD12, according to the Agy99 bacterial reference chromosome.
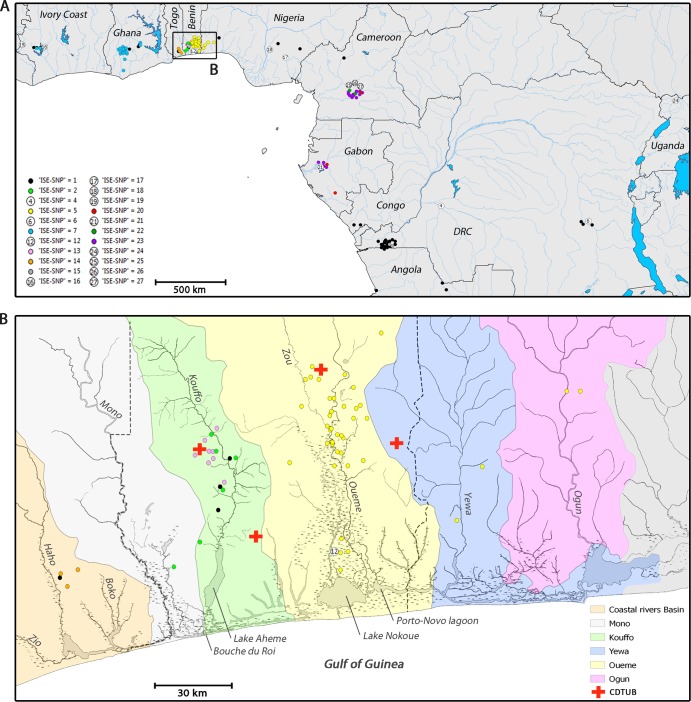
The Spearman's rank correlation coefficient showed a significant relationship between the number of isolates per country and number of ISE-SNP types identified per country (r[9] = 0.79; P < 0.01). We were able to identify all African ISE-SNP types described by Käser et al. (28) except for ISE-SNP type 3, which was found in the Greater Accra Region of Ghana, a region not covered by our panel. Some ISE-SNP types were common (types 1, 2, 5, 7, and 23), while others were represented by only one isolate/clinical specimen (types 4, 6, 12, 16, 17, 18, 19, 21, 24, 25, 26, and 27). Our panel included a number of linked isolates originating from the same patient. In all eight occurrences (Table 1), these linked isolates revealed the same ISE-SNP type. The geographical distribution of all African ISE-SNP types is shown in Fig. 2A. Most ISE-SNP types had a distinct restricted geographical localization. For example, all 41 isolates of ISE-SNP type 5 were recovered from an area in West Africa with a 60-km radius. Other African ISE-SNP types were more widely dispersed. ISE-SNP type 1, for instance, although also emerging in clusters in Central Africa, was identified throughout Central and West Africa. Furthermore, some regions harbored a multitude of different ISE-SNP types, while other regions yielded just one. In southern Benin, for example, the greatest variety of allelic patterns was found to have as many as five ISE-SNP types (viz. types 1, 2, 5, 12, and 13) circulating.
FIG 2.
(A) The geographical distribution of African M. ulcerans. The location of residence of the individual BU patients at the time of clinical visit was retrospectively correlated with the ISE-SNP typing results. ISE-SNP types represented by only one clinical isolate or specimen are depicted as numbers, while more common ISE-SNP types are color coded. (B) The uneven distribution of ISE-SNP types over the different greater hydrological drainage basins of West Africa.
We found a strong relationship (Fisher's exact test, P < 0.0001) between the distribution of ISE-SNP types and the greater West African hydrological drainage basins (Table 3; Fig. 2B). Hydrologically, this region can be divided into separate main drainage areas: the Mono, the Kouffo, the Oueme, the Yewa, the Ogun, and the Togolese Coastal Rivers Basin. The main rivers of these border-crossing basins all arise on the central west-African plateau and form broad fertile richly inundated plains when they reach the lowlands of the coastal regions, where areas where BU is endemic are concentrated. Here, the basins can be divided into an inland region drained by a network of freshwater rivers and streams that discharges into a region of extensive brackish-water swamps interconnected with lakes, narrow lagoons, and streams parallel to the coastline. Haplotype ISE-SNP 5 dominates in the areas of BU endemicity of the Oueme, the Yewa, and the Ogun basins. The haplotype is best represented along the Oueme and its last tributary, the Zou River, where most Beninese BU cases are reported (43). After the confluence, the Oueme traverses over 1,500 km2 of floodplains, after which the river discharges into Lake Nokoue, Porto-Novo Lagoon, and the coastal lagoons of Nigeria which are all interconnected by the numerous channels of the deltaic fan of the Oueme River. Two other drainage units, the Ogun and the Yewa, discharge in this same system of lagoons and streams. So, although the basins are separate drainage systems, they discharge into this collective interconnected system, which could potentially explain the observed shared distribution of haplotype ISE-SNP 5. Haplotypes ISE-SNP 2 and 13 dominate in the areas of BU endemicity of the Kouffo Basin (44, 45). After draining the regions of high BU endemicity of the commune of Lalo, the Kouffo discharges via Lake Aheme in the “western lagoonal complex,” which is in contact with the Gulf of Guinea at Bouche du Roi. Although this system is part of a semicontinuous line of narrow lagoons that runs behind the dunes along the entire coastal strip until the Ghanaian border, it is not in contact with the interconnecting drainage system of the Oueme Delta. The lower course of the Mono River forms the border between Togo and Benin and discharges in the same western lagoonal system as the Kouffo River. The Mono River basin, however, has no known areas of BU endemicity, despite similar riverine habitats. Even further west, in southern Togo, three small coastal rivers (the Boko, Haho, and Zio) form a third small basin. The basin encompasses a couple of regions of BU endemicity in which the Togolese haplotype ISE-SNP 14 is represented.
TABLE 3.
Distribution of ISE-SNP types over the hydrological drainage basins of southern Benin, southern Togo, and southwestern Nigeria
| ISE-SNP type | No. of times the type was found in basin |
|||||
|---|---|---|---|---|---|---|
| Coastal Rivers Basin | Mono | Kouffo | Oueme | Yewa | Ogun | |
| 1 | 1 | 0 | 3 | 0 | 0 | 0 |
| 2 | 0 | 1 | 5 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 37 | 2 | 2 |
| 12 | 0 | 0 | 0 | 1 | 0 | 0 |
| 13 | 0 | 0 | 8 | 0 | 0 | 0 |
| 14 | 3 | 0 | 0 | 0 | 0 | 0 |
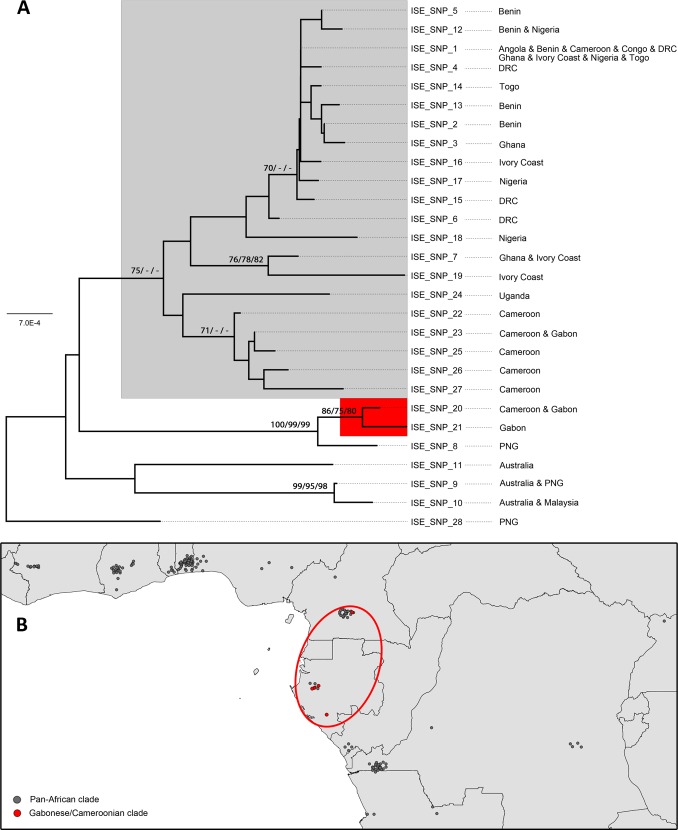
The NJ method yielded two well-supported sister clades within the African ISE-SNP types (Fig. 3A). The first clade comprised ISE-SNP types 20 and 21, which circulate in different regions of BU endemicity of Cameroon and Gabon; this clade also had high bootstrap support for the MP and ML analyses. A second pan-African clade comprised all other African ISE-SNP types (Fig. 3B). Support for other nodes within the pan-African clade was very low (bootstrap values of <70%), except in the NJ analysis for (i) a clade of ISE-SNP types 7 and 19 which circulate in Ghana and Ivory Coast, (ii) a clade of ISE-SNP types 22, 23, 25, 26, and 27, which all circulate in Cameroon and neighboring Gabon, and (iii) modest support in the NJ analysis for a clade of ISE-SNP types 1, 2, 3, 4, 5, 12, 13, 14, 15, 16, and 17, found throughout the continent. Mycobacterium ulcerans haplotypes from Australia and Southeast Asia were also included in the analysis, as these, together with African haplotypes, belong to the more virulent and distinct “classic” phylogenetic lineage (29, 46), relative to M. ulcerans isolates elsewhere. It is of particular interest that ISE-SNP type 8 from Papua New Guinea forms a strongly supported monophyletic group with ISE-SNP types 20 and 21 from Cameroon and Gabon and is distinctly unrelated to other Southeast Asian clinical isolates, which belong to ISE-SNP types 9, 10, and 28 (Fig. 3A). In contrast, other ISE-SNP types found in Papua New Guinea are related to Malaysian and Australian clinical isolates.
FIG 3.
(A) Neighbor-joining tree showing the phylogenetic relationships between the 28 currently known ISE-SNP types of M. ulcerans, with haplotype ISE-SNP 28 from Papua New Guinea as an outgroup. Bootstrap values (if >70%) for the neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) analyses are indicated at the nodes as NJ/ML/MP. ISE-SNP types belonging to the pan-African clade and the Gabonese/Cameroonian clade are highlighted in gray and red, respectively. (B) Geographical distribution of the pan-African clade and the Gabonese/Cameroonian clade. The location of residence of the individual BU patients at the time of clinical visit was retrospectively correlated with the ISE-SNP typing results.
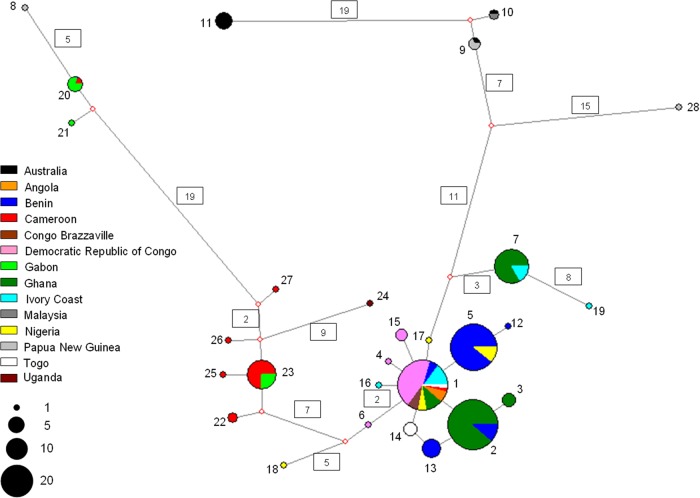
The phylogenetic network (Fig. 4) showed that a number of African ISE-SNP types are closely related and only differ in a single, or a few, mutational steps. However, other African ISE-SNP types are more distantly related and even differ in their number of mutational steps, which is similar to the number of mutational steps between African and non-African types. Analogous with the phylogenetic tree analysis, the pan-African clade is divided into two major clusters. The first major cluster comprises the common Central and West African ISE-SNP type 1 and several, closely related yet rarer ISE-SNP types. The second cluster comprises ISE-SNP types 22, 23, 25, 26, and 27, which are circulating in Cameroon and neighboring Gabon. The network also showed several more distantly related African haplotypes, of which most are relatively rare (types 18, 19, 20, 21, and 24) and one is common (type 7).
FIG 4.
Phylogenetic network showing patterns of descent among the 28 currently know ISE-SNP types of M. ulcerans in relation to their geographic origin. The network was derived by using the median joining algorithm after processing the data with the reduced median method as implemented within Network v.4.6.1.0. Each circle represents a unique ISE-SNP type, and the size of the circle is proportional to the number of individuals sharing that type. Numbers in boxes represent the number of mutational steps (if not given, then there was a single mutational step). Positions at which mutations occurred are given in Fig. 1. Color codes represent the country of origin, as shown in the key.
DISCUSSION
In this study, we applied an optimized ISE-SNP genotyping technique to a comprehensive panel of isolates from all African countries that ever yielded culture-confirmed BU cases. This analysis, unparalleled in size and scope, allowed us to assess the diversity and population structure across regions of BU endemicity on a continental scale and to explore the phylogenetic and phylogeographic relationships within the genetically conserved cluster of African M. ulcerans ISE-SNP types.
Analysis of polymorphisms in the RD1 and RD12 genomic regions, which have been determined to be among the most variable of the M. ulcerans bacterial chromosome (46), over our comprehensive sample panel spanning 11 African countries where M. ulcerans BU is endemic, identified 23 different African ISE-SNP types. The observed low level of polymorphisms (47), together with the characteristic geographical restriction of most ISE-SNP types, suggest a highly clonal population structure of African M. ulcerans. This is in agreement with the findings of Doig et al. (18), who found that clinical isolates from Ghana and Benin were only separated by an average pairwise distance of 160 SNPs over the entire 5.6-Mbp sequenced bacterial chromosome. This low sequence diversity is in strong contrast to that of other pathogens, like Helicobacter pylori, for which microevolution can be observed even within serial bacterial isolates from individual humans with prolonged infection (48). The genetic conservation among African M. ulcerans might reflect a short evolutionary history since its intracontinental dispersal, but it might also be explained by a low mutation rate. Reliable estimates of mutation rates are required to resolve these issues (47).
Closely related ISE-SNP types dominate in different areas of BU endemicity. The identified SNPs describe a phylogenetic path wherein these individual ISE-SNP types document the sequential accumulation of mutations from a common root. If we assume that (i) an ancestral ISE-SNP type will be more geographically dispersed than a more recently derived type and (ii) that the geographical distribution of the ISE-SNP types is not explained by selective effects, then this common root node is represented here by ISE-SNP 1, the most common type distributed over the entire continent. The different ISE-SNP types thereby represent the initial stages of clonal diversification through de novo mutations from this possibly ancestral type, after its intracontinental spread.
The unevenly distributed ISE-SNP types circulating within small regions of West Africa are furthermore suggestive of the existence of independent transmission clusters. We found a strong association between the distribution of ISE-SNP types and the greater West African hydrological drainage basins. Genetic differences between clinical isolates originating from two neighboring drainage areas in Benin have previously been reported (10, 18). It appears that geographic barriers (e.g., elevated regions and salt water) bordering these hydrological basins separated an ancestral genotype to a certain extent into discontinuous parts by the formation of a physical barrier to bacterial gene flow. Our data suggest that this resulted in differentiation by the slow accumulation of point mutational changes of the original founder clone (ISE-SNP 1) into different closely related types distributed over the various basins (Fig. 2B and Table 3). New ISE-SNP types derived from the founder type did not easily spread but formed focal transmission clusters associated with the hydrological drainage areas. Hence, BU infections in these areas probably resulted from locally confined transmission of a single circulating clone, with only occasional transfer of clones between basins. Our findings confirm a study of Röltgen et al. (49), in which a number of M. ulcerans haplotypes within the Densu hydrological basin of Ghana (with SNP typing based on whole-genome data) were differentiated, revealing similar focal transmission clusters within the basin itself. Hence, our findings provide additional evidence that both transmission and fine-grained evolutionary events play roles at the local level and we consequently hypothesize that potential reservoirs have limited mobility. Such a scenario would correspondingly account for the presence of villages where BU is endemic and those where it is not endemic that are in close proximity to each other (<10 km) within the same drainage basin (45).
Our phylogenetic analyses did not result in a fully resolved phylogenetic tree, since most nodes had low bootstrap support. Nevertheless, there was support for a “pan-African clade” and a “Gabonese/Cameroonian” sister clade. The ISE-SNP types from the pan-African clade are widespread throughout Africa, while the ISE-SNP types of the Gabonese/Cameroonian clade are much rarer and are found in a more restricted area (Fig. 3), which suggests that the latter clade evolved more recently. Alternatively, this may also be the result of a sampling artifact; indeed, the Spearman's rank correlation indicated that the higher the sampling effort per country, the more ISE-SNP types found. However, the entirety of the Gabonese/Cameroonian region in itself was well sampled, with five isolates/clinical samples belonging to the Gabonese/Cameroonian clade and 25 isolates/clinical samples to the pan-African clade (Tables 1 and 2; Fig. 3). Furthermore, the fact that we did not encounter ISE-SNP types of the Gabonese/Cameroonian clade in neighboring countries like the Democratic Republic of the Congo (DRC), where sampling was higher, also suggests that the ISE-SNP types belonging to the Gabonese/Cameroonian clade are not only rare but also have a limited distribution. Interestingly, the only ISE-SNP 1 isolate from Cameroon (ITM_120140) came from a patient from Bankim, a district located along the Mapé River (Sanaga Basin), while other studied isolates all came from around the Nyong River Basin. Bankim has been recently identified as an additional area of BU endemicity in Cameroon. However, whether BU was emerging in Bankim or constitutes a newly recognized preexisting disease focus remains unclear (50, 51).
The Gabonese/Cameroonian clade was found to form a strongly supported monophyletic group with Papua New Guinean ISE-SNP type 8, which is distinctly unrelated to other ISE-SNP types found in Southeast Asia. With use of a different genotyping technique, the relatedness of a Papua New Guinean clinical isolate (not included in this study) to African rather than to Southeast Asian clinical isolates has been reported elsewhere (52). The process (historical events, restricted bacterial gene flow, etc.) that led to this intercontinental association of ISE-SNP haplotypes remains elusive.
In this report, we have analyzed a large collection of isolates representative of the African M. ulcerans population in order to characterize its population structure accurately and appropriately. The panel used in this study is, to our knowledge, the most comprehensive one studied so far. It covered disease foci from all 11 well-documented countries of BU endemicity, ranging from West, to Central, to East Africa. Six countries (Burkina Faso, Equatorial Guinea, Guinea, Kenya, Liberia, and South Sudan) that have reported a limited number of BU cases in the past (53) were not included in the study as we were unable to include specimens, or isolates, from them. Moreover, cases from the Central African Republic, Senegal, and Sierra Leone were never confirmed by laboratory tests (53). Although we tried to maximize spatial diversity within our panel, some countries are better represented than others, again due to the limited availability of clinical isolates. We might have missed some ISE-SNP types in these countries, because there was a significant relationship between the sampling effort per country and the amount of different ISE-SNP types identified per country. Because of all these limitations, we successfully optimized the genotyping PCR technique for application directly on clinical specimens, which allowed us to include clinical specimens from certain geographical regions of Gabon and Nigeria in which no M. ulcerans isolates were available (Table 2).
Nonetheless, the quality of these kinds of bacterial population studies largely depends on the quality of the patient information connected with the clinical isolates and specimens. Isolates ITM_070123 and ITM_070404 originated from the same patient, an Angolan refugee fleeing the civil war in his country (54), who was diagnosed before he was sheltered in humanitarian camps across the border in the DRC. We believe that ISE-SNP type 15, the haplotype to which his isolates belong, is North Angolan rather than Congolese, as another isolate, ITM_092479, also originated from a patient who likely is of Angolan origin. During the civil war, however, identity fraud was common in the camps in the DRC, causing misclassification of patient origins.
To our knowledge, ISE-SNP typing currently yields the greatest resolution within M. ulcerans, save for whole-genome sequencing. The method may be an easy, low-cost, powerful, reliable, and reproducible tool for reference laboratories to assist in the tracking of M. ulcerans ISE-SNP types for epidemiological studies on a continental scale (55).
Because African M. ulcerans shows such low genetic variation, further studies require a whole-genome approach to comprehensively evaluate the genetic diversity, the evolution, and the phylogenetic relatedness of African M. ulcerans and to delineate the exact origin and spread of the pathogen at the local and the continental levels. It is specifically the paucity of genetic diversity and the sequential order of the genetic changes that have occurred between individual isolates that render M. ulcerans such a promising model to reveal evolutionary bacterial mechanisms. Furthermore, given the comprehensive nature of full-genome data, sequences could also serve in large-scale microepidemiological studies that are focused on the elucidation of transmission pathways and relevant reservoirs of M. ulcerans. Indeed, different studies of mycobacterial genomics (18, 49, 56) have already shown that, at the whole-genome level, substantial genetic variation exists in African M. ulcerans, which can be exploited for phylogenetically robust strain classification. In order to capture as much diversity as possible and to minimize phylogenetic discovery bias (57) in such impending large sequencing endeavors, it will be desirable to select representative types from all the central and radial ISE-SNP types defined in this study.
Supplementary Material
ACKNOWLEDGMENTS
The present study pays tribute to the extensive collection of M. ulcerans isolates generated over decades by Françoise Portaels and her collaborators in countries where BU is endemic, who initiated and fueled research into the pathogenesis, diagnosis, and management of M. ulcerans disease.
Koen Vandelannoote was supported by a Ph.D. grant of the Flemish Interuniversity Council—University Development Cooperation (Belgium). Funding for this work was provided by the Stop Buruli Consortium, which is supported by the UBS Optimus Foundation, the European Community's Seventh Framework Programme under grant agreement 241500 (BURULIVAC), the European Commission (project INCO-CT-2005-051476-BURULICO), and the Fund for Scientific Research Flanders (Belgium; FWO grant G.0321.07N).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Tim Stinear for helpful discussions and critical comments on the manuscript. We thank Pim de Rijk, Krista Fissette, Elie Nduwamahoro, and Anita Van Aerde (ITM) for their excellent technical assistance. We thank three anonymous reviewers for their constructive and insightful comments, which helped us to improve the manuscript.
Footnotes
Published ahead of print 2 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02774-13.
REFERENCES
- 1.Portaels F, Silva MT, Meyers WM. 2009. Buruli ulcer. Clin. Dermatol. 27:291–305. 10.1016/j.clindermatol.2008.09.021 [DOI] [PubMed] [Google Scholar]
- 2.Walsh DS, Portaels F, Meyers WM. 2011. Buruli ulcer: advances in understanding Mycobacterium ulcerans infection. Dermatol. Clin. 29:1–8. 10.1016/j.det.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 3.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, Scott JT, Dramaix M, Portaels F. 2004. Mycobacterium ulcerans disease: role of age and gender in incidence and morbidity. Trop. Med. Int. Health 9:1297–1304. 10.1111/j.1365-3156.2004.01339.x [DOI] [PubMed] [Google Scholar]
- 4.Wagner T, Benbow ME, Burns M, Johnson RC, Merritt RW, Qi J, Small PL. 2008. A landscape-based model for predicting Mycobacterium ulcerans infection (Buruli ulcer disease) presence in Benin, West Africa. Ecohealth 5:69–79. 10.1007/s10393-007-0148-7 [DOI] [PubMed] [Google Scholar]
- 5.Jacobsen KH, Padgett JJ. 2010. Risk factors for Mycobacterium ulcerans infection. Int. J. Infect. Dis. 14:e677–e681. 10.1016/j.ijid.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 6.Portaels F, Elsen P, Guimaraes-Peres A, Fonteyne PA, Meyers WM. 1999. Insects in the transmission of Mycobacterium ulcerans infection. Lancet 353:986. 10.1016/S0140-6736(98)05177-0 [DOI] [PubMed] [Google Scholar]
- 7.Vandelannoote K, Durnez L, Amissah D, Gryseels S, Dodoo A, Yeboah S, Addo P, Eddyani M, Leirs H, Ablordey A, Portaels F. 2010. Application of real-time PCR in Ghana, a Buruli ulcer-endemic country, confirms the presence of Mycobacterium ulcerans in the environment. FEMS Microbiol. Lett. 304:191–194. 10.1111/j.1574-6968.2010.01902.x [DOI] [PubMed] [Google Scholar]
- 8.Meyers WM, Shelly WM, Connor DH, Meyers EK. 1974. Human Mycobacterium ulcerans infections developing at sites of trauma to skin. Am. J. Trop. Med. Hyg. 23:919–923 [DOI] [PubMed] [Google Scholar]
- 9.Merritt RW, Walker ED, Small PL, Wallace JR, Johnson PD, Benbow ME, Boakye DA. 2010. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl. Trop. Dis. 4(12):e911. 10.1371/journal.pntd.0000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K, de Rijk P, Elsen P, Fissette K, Fraga AG, Lee R, Mahrous E, Small PL, Stragier P, Torrado E, Van Aerde A, Silva MT, Pedrosa J. 2008. First cultivation and characterization of Mycobacterium ulcerans from the environment. PLoS Negl. Trop. Dis. 2(3):e178. 10.1371/journal.pntd.0000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gryseels S, Amissah D, Durnez L, Vandelannoote K, Leirs H, De Jonckheere J, Silva MT, Portaels F, Ablordey A, Eddyani M. 2012. Amoebae as potential environmental hosts for Mycobacterium ulcerans and other mycobacteria, but doubtful actors in Buruli ulcer epidemiology. PLoS Negl. Trop. Dis. 6(8):e1764. 10.1371/journal.pntd.00001764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williamson HR, Benbow ME, Nguyen KD, Beachboard DC, Kimbirauskas RK, McIntosh MD, Quaye C, Ampadu EO, Boakye D, Merritt RW, Small PL. 2008. Distribution of Mycobacterium ulcerans in buruli ulcer endemic and non-endemic aquatic sites in Ghana. PLoS Negl. Trop. Dis. 2(3):e205. 10.1371/journal.pntd.0000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt RW, Benbow ME, Small PLC. 2005. Unraveling an emerging disease associated with disturbed aquatic environments: the case of Buruli ulcer. Front. Ecol. Environ. 3:323–331 http://www.jstor.org/stable/3868566 [Google Scholar]
- 14.Marsollier L, Brodin P, Jackson M, Kordulakova J, Tafelmeyer P, Carbonnelle E, Aubry J, Milon G, Legras P, Andre JP, Leroy C, Cottin J, Guillou ML, Reysset G, Cole ST. 2007. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3(5):e62. 10.1371/journal.ppat.0030062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durnez L, Suykerbuyk P, Nicolas V, Barriere P, Verheyen E, Johnson CR, Leirs H, Portaels F. 2010. Terrestrial small mammals as reservoirs of Mycobacterium ulcerans in Benin. Appl. Environ. Microbiol. 76:4574–4577. 10.1128/AEM.00199-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddyani M, Ofori-Adjei D, Teugels G, De Weirdt D, Boakye D, Meyers WM, Portaels F. 2004. Potential role for fish in transmission of Mycobacterium ulcerans disease (Buruli ulcer): an environmental study. Appl. Environ. Microbiol. 70:5679–5681. 10.1128/AEM.70.9.5679-5681.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip MJ, Porter JL, Fyfe JA, Lavender CJ, Portaels F, Rhodes M, Kator H, Colorni A, Jenkin GA, Stinear T. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021–2029. 10.1128/JB.01442-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doig KD, Holt KE, Fyfe JA, Lavender CJ, Eddyani M, Portaels F, Yeboah-Manu D, Pluschke G, Seemann T, Stinear TP. 2012. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics 13:258. 10.1186/1471-2164-13-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. U. S. A. 101:1345–1349. 10.1073/pnas.0305877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small PL. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854–857. 10.1126/science.283.5403.854 [DOI] [PubMed] [Google Scholar]
- 21.Demangel C, Stinear TP, Cole ST. 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7:50–60. 10.1038/nrmicro2077 [DOI] [PubMed] [Google Scholar]
- 22.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PD, Davies JK, Jenkin GA, Small PL, Jones LM, Tekaia F, Laval F, Daffe M, Parkhill J, Cole ST. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192–200. 10.1101/gr.5942807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14:627–633 http://dx.doi.org/10.1016/j/gde.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 25.Musser JM. 1996. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg. Infect. Dis. 2:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spratt BG, Maiden MC. 1999. Bacterial population genetics, evolution and epidemiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roltgen K, Stinear TP, Pluschke G. 2012. The genome, evolution and diversity of Mycobacterium ulcerans. Infect. Genet. Evol. 12:522–529. 10.1016/j.meegid.2012.01.018 [DOI] [PubMed] [Google Scholar]
- 28.Käser M, Hauser J, Pluschke G. 2009. Single nucleotide polymorphisms on the road to strain differentiation in Mycobacterium ulcerans. J. Clin. Microbiol. 47:3647–3652. 10.1128/JCM.00761-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rondini S, Kaser M, Stinear T, Tessier M, Mangold C, Dernick G, Naegeli M, Portaels F, Certa U, Pluschke G. 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg. Infect. Dis. 13:1008–1015. 10.3201/eid1307.060205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guimaraes-Peres A, Portaels F, de Rijk P, Fissette K, Pattyn SR, van Vooren J, Fonteyne P. 1999. Comparison of two PCRs for detection of Mycobacterium ulcerans. J. Clin. Microbiol. 37:206–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durnez L, Stragier P, Roebben K, Ablordey A, Leirs H, Portaels F. 2009. A comparison of DNA extraction procedures for the detection of Mycobacterium ulcerans, the causative agent of Buruli ulcer, in clinical and environmental specimens. J. Microbiol. Methods 76:152–158. 10.1016/j.mimet.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 32.Fyfe JA, Lavender CJ, Johnson PD, Globan M, Sievers A, Azuolas J, Stinear TP. 2007. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl. Environ. Microbiol. 73:4733–4740. 10.1128/AEM.02971-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. 1985. Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 37.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 38.Rambaut A. 2013. FigTree, tree figure drawing tool, v. 1.3.1. Molecular evolution, phylogenetics and epidemiology. http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- 39.Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37–48 [DOI] [PubMed] [Google Scholar]
- 40.Quantum GIS. 2012. Quantum GIS Geographic Information System Open Source Geospatial Foundation Project. http://www.qgis.org/en/site/ [Google Scholar]
- 41.Jenness J, Dooley J, Aguilar-Manjarrez J, Riva C. 2007. African Water Resource Database. GIS-based tools for inland aquatic resource management. FAO, Rome, Italy: http://www.fao.org/docrep/010/a1170e/a1170e00.htm [Google Scholar]
- 42.Core Team R 2012. R: a language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 43.Sopoh GE, Johnson RC, Chauty A, Dossou AD, Aguiar J, Salmon O, Portaels F, Asiedu K. 2007. Buruli ulcer surveillance, Benin, 2003–2005. Emerg. Infect. Dis. 13:1374–1376. 10.3201/eid1309.061338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RC, Makoutode M, Sopoh GE, Elsen P, Gbovi J, Pouteau LH, Meyers WM, Boko M, Portaels F. 2005. Buruli ulcer distribution in Benin. Emerg. Infect. Dis. 11:500–501. 10.3201/eid1103.040597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson RC, Sopoh GE, Boko M, Zinsou C, Gbovi J, Makoutode M, Portaels F. 2005. Distribution of Mycobacterium ulcerans (Buruli ulcer) in the district of Lalo in Benin. Trop. Med. Int. Health 10:863–871 (In French.) 10.1111/j.1365-3156.2005.01465.x [DOI] [PubMed] [Google Scholar]
- 46.Käser M, Rondini S, Naegeli M, Stinear T, Portaels F, Certa U, Pluschke G. 2007. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol. Biol. 7:177. 10.1186/1471-2148-7-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53–70. 10.1146/annurev.micro.62.081307.162832 [DOI] [PubMed] [Google Scholar]
- 48.Morelli G, Didelot X, Kusecek B, Schwarz S, Bahlawane C, Falush D, Suerbaum S, Achtman M. 2010. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6(7):e1001036. 10.1371/journal.pgen.1001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roltgen K, Qi W, Ruf MT, Mensah-Quainoo E, Pidot SJ, Seemann T, Stinear TP, Kaser M, Yeboah-Manu D, Pluschke G. 2010. Single nucleotide polymorphism typing of Mycobacterium ulcerans reveals focal transmission of Buruli ulcer in a highly endemic region of Ghana. PLoS Negl. Trop. Dis. 4(7):e751. 10.1371/journal.pntd.0000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marion E, Landier J, Boisier P, Marsollier L, Fontanet A, Le Gall P, Aubry J, Djeunga N, Umboock A, Eyangoh S. 2011. Geographic expansion of Buruli ulcer disease, Cameroon. Emerg. Infect. Dis. 17:551–553. 10.3201/eid1703.091859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bratschi MW, Bolz M, Minyem JC, Grize L, Wantong FG, Kerber S, Njih Tabah E, Ruf MT, Mou F, Noumen D, Um Boock A, Pluschke G. 2013. Geographic distribution, age pattern and sites of lesions in a cohort of Buruli ulcer patients from the mape basin of cameroon. PLoS Negl. Trop. Dis. 7(6):e2252. 10.1371/journal.pntd.00002252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stinear T, Davies JK, Jenkin GA, Portaels F, Ross BC, Oppedisano F, Purcell M, Hayman JA, Johnson PD. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482–1487 http://jcm.asm.org/content/38/4/1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janssens P, Pattyn S, Meyers W, Portaels F. 2005. Buruli ulcer: an historical overview with updating. Bull. Seances Acad. R. Sci. Outre Mer 51:265–299 (In French) [Google Scholar]
- 54.Kibadi K, Tsakala M, Mputu-Yamba JB, Muyembe T, Kashongwe M, Imposso B, Nsiala A. 2003. Buruli ulcer in Angolese refugees in the Kimpese area, Lower Congo, D.R. Congo. Sante 13:39–41 (In French) [PubMed] [Google Scholar]
- 55.McGann H, Stragier P, Portaels F, Gascoyne Binzi D, Collyns T, Lucas S, Mawer D. 2009. Buruli ulcer in United Kingdom tourist returning from Latin America. Emerg. Infect. Dis. 15:1827–1829. 10.3201/eid1511.090460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi W, Kaser M, Roltgen K, Yeboah-Manu D, Pluschke G. 2009. Genomic diversity and evolution of Mycobacterium ulcerans revealed by next-generation sequencing. PLoS Pathog. 5(11):e1000580. 10.1371/journal.ppat.1000580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson T, Okinaka RT, Foster JT, Keim P. 2009. Phylogenetic understanding of clonal populations in an era of whole genome sequencing. Infect. Genet. Evol. 9:1010–1019. 10.1016/j.meegid.2009.05.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.