Abstract
Lantibiotics are ribosomally synthesized, posttranslationally modified antimicrobial peptides. Their biosynthesis genes are usually organized in gene clusters, which are mainly found in Gram-positive bacteria, including pathogenic streptococci. Three highly virulent Streptococcus suis serotype 2 strains (98HAH33, 05ZYH33, and SC84) have been shown to contain an 89K pathogenicity island. Here, on these islands, we unveiled and reannotated a putative lantibiotic locus designated sui which contains a virulence-associated two-component regulator, suiK-suiR. In silico analysis revealed that the putative lantibiotic modification gene suiM was interrupted by a 7.9-kb integron and that other biosynthesis-related genes contained various frameshift mutations. By reconstituting the intact suiM in Escherichia coli together with a semi-in vitro biosynthesis system, a putative lantibiotic named suicin was produced with bactericidal activities against a variety of Gram-positive strains, including pathogenic streptococci and vancomycin-resistant enterococci. Ring topology dissection indicated that the 34-amino-acid lantibiotic contained two methyllanthionine residues and one disulfide bridge, which render suicin in an N-terminal linear and C-terminal globular shape. To confirm the function of suiK-suiR, SuiR was overexpressed and purified. In vitro analysis showed that SuiR could specifically bind to the suiA gene promoter. Its coexpression with suiK could activate suiA gene promoter in Lactococcus lactis NZ9000. Conclusively, we obtained a novel lantibiotic suicin by restoring its production from the remnant sui locus and demonstrated that virulence-associated SuiK-SuiR regulates its production.
INTRODUCTION
Lantibiotics, an abbreviated term for lanthionine-containing antibiotics, are gene-encoded antimicrobial peptides that are mainly produced by Gram-positive bacteria and are characterized for the presence of unusual amino acids such as lanthionine (Lan) and methyllanthionine (MeLan) in their sequences. Nisin, the prototype lantibiotic, has been widely used as a food preservative in the food industry without substantial occurrence of resistance (1). Lantibiotics are active at nanomolar concentrations against a wide range of Gram-positive bacteria, including some clinically important pathogens (2). Thus, they have been regarded as potential alternatives for conventional antibiotics, with promising applications in both the food and pharmaceutical industries (3).
The lantibiotic biosynthesis apparatus is encoded by gene clusters, which contain genes encoding precursor peptides, modification and processing enzymes, translocation proteins, regulatory components, and immunity proteins. Lantibiotics are initially synthesized as precursor peptides that consist of N-terminal leader peptides and C-terminal core peptides. Certain Ser/Thr residues in the core peptides are dehydrated and converted to dehydroalanine (Dha) and dehydrobutyrine (Dhb), which are subsequently conjugated with Cys via covalent bonds to form thioether bridges, namely, Lan and MeLan (4). Based on their structural features, lantibiotics are classified into type A and B groups, among which type A lantibiotics are elongated whereas type B lantibiotics are globular in structure (5). Type A lantibiotics are further divided into two subgroups. Type AI lantibiotics such as nisin, subtilin, and epidermin are elongated and amphipathic screw-shaped cationic peptides catalyzed by the cooperative actions of two distinct enzymes—dehydratase LanB and cyclase LanC (6). Type AII lantibiotics such as lacticin 481 and nukacin ISK-1 are N-terminal linear and C-terminal globular cationic peptides promoted by a bifunctional enzyme (LanM) that shows both dehydratase and cyclase activities (7). Mature lantibiotics are released following cleavage of the leader peptides by specific proteases. Leader peptides of type AII lantibiotics usually contain a double glycine (GG) motif that is processed by the LanT dedicated transporter protein (8). In typical cases, the production of lantibiotics is autoinduced via a two-component system (TCS) containing a LanK sensor histidine kinase and a LanR response regulator (4).
Lantibiotics are most frequently found to be produced by probiotic lactic acid bacteria for their traditional use in the food and diary industries (9). However, some of them have been lately discovered in various pathogenic micoorganisms (10). A typical instance is the two-component lantibiotic cytolysin that possesses both bactericidal and hemolytic activity, acting as an important virulence factor for Enterococcus faecalis (11). In addition, streptococci are prevalently known as producers of lantibiotics, many of which are pathogens. For example, Streptococcus pyogenes produces streptin and streptococcin A-FF22, S. mutans produces the three lantibiotics mutacin I, II, and III, and S. uberis produces the nisin-like lantibiotic nisin U (12). Although S. pneumoniae has not been found to produce any lantibiotics, a two-component lantibiotic from S. pneumoniae R6 has been synthesized by using nisin synthetic machinery (13). With increasing available genomic information, genome mining with LanM, LanBC, or BAGEL has unearthed a great deal of otherwise undefined lantbiotic loci in streptococci (14–16). The prevalence of lantibiotic gene clusters in pathogenic streptococci might improve their competiveness among related species and promote their colonization and infection of the human host, thus facilitating their pathogenesis (17).
As an important zoonotic pathogen, S. suis has 35 identified serotypes. S. suis serotype 2 (SS2), the most prevalent and virulent one, has been found in 20 countries and has caused several human infection outbreaks (18). Two recent large-scale outbreaks of human streptococcal toxic-shock-like syndrome (STSLS) caused by SS2 infections in China have already raised considerable international concern (19). The genomes of three STSLS-causing SS2 strains (98HAH33, 05ZYH33, and SC84) have been sequenced, and a unique 89-kb pathogenicity island (PAI), designated 89K, was located by comparative genomic analysis performed with other serotype 2 strains (20). On this 89K PAI, a two-component regulation system, salK-salR, was identified and shown to be crucial for the high virulence of SS2 because knockout of salK-salR eliminates its lethality, which could be restored by complementary expression of salK-salR (21). However, the mechanism underlying the function of SalK-SalR remains unknown.
Previous research indicated that SalK-SalR played a role in lantibiotic production, as it is operonically associated with the lantibiotic gene cluster (22). In the present study, we analyzed and characterized the genes neighboring salK and salR and reannotated a putative and novel lantibiotic gene cluster locating on 89K PAI in the aforementioned STSLS-causing SS2 strains. We recommended the gene cluster to be designated sui, in which a putative lantibiotic modification gene suiM was found to be interrupted by a 7.9-kb integron, suggesting its dysfunction in lantibiotic modification. Here, we recovered the function of intact suiM and biosynthesized a novel bioactive lantibiotic named suicin. Structure dissection elucidated that suicin was a type AII lantibiotic which showed an N-terminal linear and C-terminal globular structure. We also demonstrated that the virulence-associated SuiK-SuiR system was involved in regulating suicin biosynthesis, implying that role of SuiK-SuiR in SS2 virulence might be associated with its role in suicin production in SS2.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α and MC1061 were used as hosts for DNA cloning and BL21(DE3) for protein expression. The bacteria were incubated in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% sodium chloride) at 37°C, and 200 μg/ml erythromycin (Em) or 50 μg/ml kanamycin (Kan) was added if necessary. Lactococcus lactis NZ9000 was incubated in M17 medium (0.5% tryptone, 0.25% yeast extract, 0.5% soya peptone, 0.5% beef extract, 1.9% β-glycerophosphate disodium, 0.05% vitamin C, 0.025% magnesium sulfate) supplemented with 0.5% glucose (GM17), and 5 μg/ml Em was included if needed. S. suis HAbb was isolated from an STSS patient from the epidemic in Jiangsu (China), and its genomic DNA was provided by the Research Institute for Medicine of Nanjing Command in China. TCEP [tris(2-carboxyethyl) phosphine], dl-dithiothreitol (DTT), NEM (N-ethylmaleimide), and DiBAC4(3) [bis-(1,3-dibutylbarbituric acid) trimethine oxonol] were purchased from Sigma-Aldrich. Crude nisin Z was obtained from the Silver Elephant company (Zhejiang, China) and was applied to reverse-phase high-pressure liquid chromatography (RP-HPLC) using a C18 column to obtain pure nisin Z.
Indicator strains for antimicrobial assay were maintained in separate media as follows. Bacilli (Bacillus cereus, B. thuringiensis, B. subtilis, and B. lentus) were inoculated into LB medium at 37°C; Micrococcus flavus NCIB8166 was inoculated into S1 medium (0.8% tryptone, 0.5% yeast extract, 0.5% glucose, 0.2% disodium hydrogen phosphate, 0.5% sodium chloride, 0.1% Tween 20) at 30°C; streptococci (S. gordonii, S. mutans, and S. bovis) and enterococci (E. pernyi, E. faecalis, and E. durans) were cultured with BHI medium (1.75% brain heart infusion, 1% enzymatic digest of gelatin, 0.2% dextrose, 0.5% sodium chloride, 0.25% disodium phosphate) at 37°C; Lactococcus lactis were grown in GM17 medium at 30°C; lactobacilli (Lactobacillus acidophilus, Lb. casei, and Lb. curvatus) and Leuconostoc dextranicum were incubated into MRS medium (1% beef extract, 1% casein peptone, 0.5% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% ammonium citrate, 0.1% Tween 80, 0.01% magnesium sulfate, 0.005% manganese sulfate) at 30°C; and staphylococci (Staphylococcus aureus and S. epidermidis), Shigella dysenteriae, Shigella flexneri, and Pseudomonas aeruginosa were maintained with LB medium at 37°C. Solid media were prepared with 1.5% agar.
Plasmid construction and site-directed mutagenesis.
Molecular cloning techniques were carried out according to standard protocols (23). PCR was performed using TransStart FastPfu DNA polymerase (Transgene, China). Plasmids, PCR products, or DNA fragments were prepared or purified by the use of Axygen kits according to the respective instructions.
suiAM1 was amplified from genomic DNA of S. suis Habb with primers containing recognition sites of NdeI and BamHI and suiM2 with primers containing recognition sites of BamHI and XhoI, respectively. First, suiAM1 was ligated to pET28a to obtain pET-suiAM1. suiM2 was then introduced into pET-suiAM1 with a BamHI site between suiAM1 and suiM2. After the BamHI site was eliminated by site-specific mutagenesis as described later, pET-suiAM was obtained with intact suiM. The 477-bp fragment of suiT encoding the 159-amino-acid (aa) N-terminal protease domain was amplified with primers containing recognition sites of NcoI and XhoI and then introduced into pET-28a to obtain plasmid pET-suiT159. suiR was amplified with primers containing recognition sites of BamHI and XhoI and then introduced into pET28a to obtain pET-suiR. SuiT159 was N-terminally and SuiR was C-terminally 6×His tagged. A suiA promoter (PsuiA) was amplified from genomic DNA of S. suis Habb and gfp from pGFP (24). PsuiA and gfp were fused by overlapping PCR and introduced into pMG36e to obtain pMG36e-PsuiAgfp. suiKR and suiR were then amplified and introduced into pMG36e-PsuiAgfp in opposite orientations and were under the control of constitutive promoter P32, thus obtaining pMG36e-suiKRPsuiAgfp and pMG36e-suiRPsuiAgfp.
Site-specific ligase-independent mutagenesis (SLIM) was performed via a PCR method with primers containing corresponding mutations as referred to the protocols described by Chiu et al. (25).
Protein expression and purification.
E. coli BL21(DE3) transformed with expression vectors were incubated in 1 liter of LB broth at 37°C. At an optical density at 600 nm (OD600) of 0.6 to 0.7, IPTG (isopropyl β-d-1-thiogalactopyranoside) was added to the cell culture to a final concentration of 0.5 mM, and the cells were then induced for another 20 h at 16°C. Induced cells were harvested by centrifugation at 5,000 × g for 20 min and resuspended in 30 ml binding buffer (50 mM Na2HPO4, 500 mM NaCl, pH 7.4) plus 20 mM imidazole. After cells were lysed by sonication, centrifugation was performed at 10,000 × g for 30 min to separate supernatant and inclusion bodies. The supernatant was applied to immobilized metal affinity chromatography (IMAC) of an Ni2+ column in a stepwise wash with binding buffer plus 40 mM imidazole, and, finally, the target protein was eluted with binding buffer plus 500 mM imidazole. The inclusion bodies were resuspended in binding buffer containing 20 mM imidazole and 8 M urea and sonicated to dissolve. Following centrifugation to remove the remaining insoluble precipitates, the dissolved protein in 8 M urea was purified through IMAC by washing with gradient dilution of urea concentration and finally eluting with binding buffer containing 2 M urea and 500 mM imidazole.
His6-SuiA was mainly purified from inclusion bodies. The urea containing crude His6-SuiA was subjected to a reverse-phase HPLC system with a C4 semipreparative column (Dalian Elite, China) using a water-acetonitrile solvent system. The standard HPLC method consisted of a 15% to 30% acetonitrile gradient for 10 min followed by 30% to 65% acetonitrile gradient for 30 min at a flow rate of 1 ml/min. Peptides were detected by their absorbance at 220 and 280 nm. The purified peptides were lyophilized and dissolved in double-distilled water (ddH2O) for further experiments. SuiT159-His6 and His6-SuiR were purified from the supernatant with the methods described above. The protein concentration was quantified by the use of a standard bicinchoninic acid (BCA) assay kit (Thermo Scientific). Purified proteins were identified by the use of 16.5% Tricine–SDS-PAGE (26).
In vitro reconstitution of SuiT159-His6.
Purified His6-SuiA precursor peptides and their analogues were incubated with SuiT159-His6 under conditions of 50 mM Na2HPO4, 50 mM Na2SO4 (pH 7.4), and 1 mM DTT at 25°C for 3 h. SuiT159-His6 was then removed by heating at 60°C for 10 min and subsequent centrifugation at 12,000 × g for 10 min. Supernatant containing suicin was prepared for analysis by the use of a C18 ZipTip (Millipore, Germany) or purified by reverse-phase high-pressure liquid chromatography (RP-HPLC) using a C18 analysis column (Shimadzu, Japan). Purified suicin was lyophilized to dissolve in ddH2O and identified by 16.5% Tricine–SDS-PAGE.
Peptide modification and MS analysis.
Suicin and its mutants were subjected to treatment for chemical modification with NEM, which reacts with free thiols on Cys and results in a 125-Da increase per thiol. The NEM reaction was performed by incubation of purified sample with 1 mM TCEP and 10 mM NEM (pH 7.5) in an ice bath for 30 min. After the reaction, the samples were applied to a C18 ZipTip and detected by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using a model 4700 Proteomics Analyzer mass spectrometer (Applied Biosystems). Mass spectra were obtained in the positive reflectron mode for peptides smaller than 5 kDa and linear mode for peptides larger than 5 kDa. A CHCA (α-cyano-4-hydroxycinnamic acid) matrix was prepared by dissolving 5 mg CHCA in 1 ml of 50:50 acetonitrile/water containing 0.1% trifluoroacetic acid (TFA).
Antimicrobial assay.
Antimicrobial spectra of suicin were determined by an agar diffusion assay using the indicator strains shown (see Table 4) as described previously (27). Briefly, 25 μl of 10 μg/ml suicin was applied to 5-mm-diameter wells lawned with the indicator strain. Indicator strains were inoculated under appropriate conditions, and diameters of inhibition zone were determined by the use of a caliper. The MIC was determined for the sensitive strain M. flavus NCIB 8166 by a microdilution method using a 96-well plate as described by Levengood et al. (28).
TABLE 4.
Antimicrobial spectrum of suicin
| Indicator strain | No. of strains tested | Zone of inhibition (mm)a |
|---|---|---|
| Bacillus cereus | 3 | – |
| Bacillus brevis AS1.1165 | 1 | 7 |
| Bacillus subtilis 168 | 1 | 8 |
| Bacillus thuringiensis 1.1014 | 1 | – |
| Enterococcus faecalis V583 | 1 | 8 |
| Enterococcus durans 1.2023 | 1 | 6 |
| Enterococcus pernyi 1.1010 | 1 | 8 |
| Lactobacillus acidophilus 100-33 | 1 | 6 |
| Lactobacillus curvatus LTH1174 | 1 | 9 |
| Lactobacillus casei | 3 | – |
| Lactobacillus delbrueckii 8909 | 1 | – |
| Lactococcus lactis MG1363 | 1 | 7 |
| Leuconostoc dextranicum 181 | 1 | 11 |
| Micrococcus flavus NCIB8166 | 1 | 23 |
| Staphylococcus aureus 26122 | 1 | – |
| Staphylococcus epidermidis 1.1229 | 1 | – |
| Streptococcus bovis | 2 | – |
| Streptococcus mutans 1.2499 | 1 | – |
| Streptococcus gordonii 1.2496 | 1 | 6 |
| Escherichia coli DH5α | 1 | – |
| Pseudomonas aeruginosa | 1 | – |
| Shigella dysenteriae | 1 | – |
| Shigella flexneri | 1 | – |
Suicin (25 μl of 10 μg/ml) was applied to each indicator strain. The values stand for the diameter of the inhibition zone. –, no inhibition zone.
Determination of membrane potential.
Membrane potential disruption was monitored by DiBAC4(3), which is a sensitive slow-response membrane potential probe that measures potential-dependent changes by accompanied fluorescence changes (29). M. flavus NCIB8166 was grown in S1 medium at 30°C to an OD600 of ca. 0.6 to 0.8 followed by centrifugation (4°C and 5,000 × g for 5 min) to collect viable bacterial cells. Cells were then washed and resuspended in 5 mM HEPES buffer (5 mM HEPES, 0.4% glucose, 2.5 mM MgSO4, pH 7.8) to an OD600 of ca. 0.05 to 0.1. A 120-μl volume of the cell suspension was added to the wells of a 96-well microplate. DiBAC4(3) was added to achieve a final concentration of 1 μM and incubated at room temperature for 4 min before suicin or nisin Z was added. Then, 30 μl of suicin (25× or 50× MIC) was mixed in and the mixture was incubated for another 10 min. Nisin Z (50× MIC) was used as a positive control. Fluorescence was monitored by the use of a microplate reader (BioTek) at excitation and emission wavelengths of 493 and 516 nm, respectively.
EMSA.
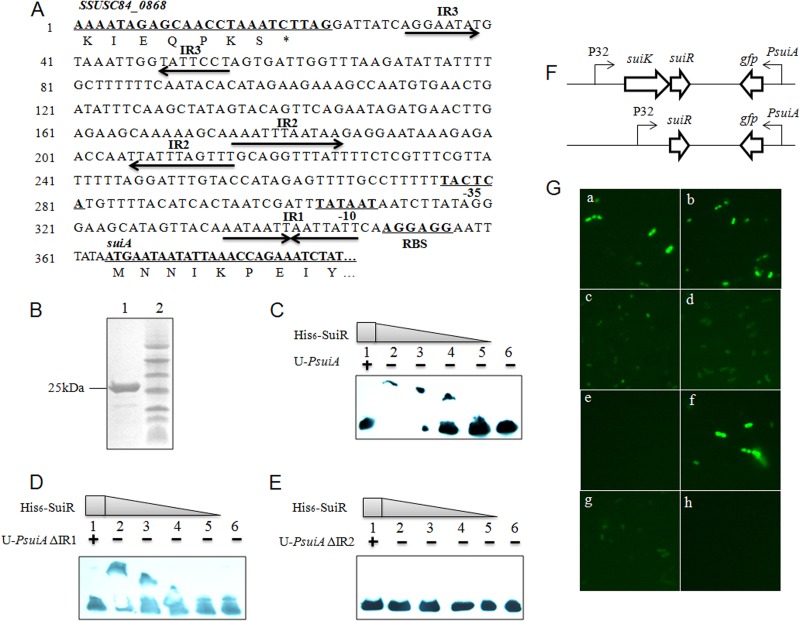
An electrophoretic mobility shift assay (EMSA) was performed to determine if SuiR could bind to the promoter of suiA. A 293-bp promoter region of suiA (PsuiA) was amplified with primers, and the 5′ primer was labeled with biotin. Biotin-PsuiA was purified from 1% agarose gel. Then, increasing concentrations of purified His6-SuiR ranging from 0 to 200 μg/ml were incubated with biotin-PsuiA at 20°C for 20 min. For a competition assay, a mixture that included a 100-fold excess of unlabeled PsuiA (U-PsuiA) was used. After the incubation, samples were applied to 4.5% nondenaturing polyacrylamide gel with 80 V for 1 to 1.5 h. Biotin-PsuiA was transferred to a nylon membrane, and streptavidin-horseradish peroxidase (HRP) conjugates were used to specifically bind biotin. Followed by X-ray film exposure, retarded biotin-labeled probes were visualized.
Activation of GFP by SuiK-SuiR.
The assay was modified from an assay described previously (24). Briefly, pMG36e-derived plasmids containing suiKR or their mutants were transformed into L. lactis NZ9000 by the use of an electroporation method as described by Holo and Nes (30). Transformants were incubated in GM17 at 30°C to an OD600 of ca. 0.4 to 0.5, and then suicin was added in concentrations ranging from 10 to 400 ng/ml for induction if needed. After induction was performed for 4 h, viable cells were collected to determine green fluorescent protein (GFP) fluorescence with a Zeiss Axio Imager A1 microscope.
RESULTS
Identification of genes that might be involved in biosynthesis of a putative lantibiotic suicin.
In three epidemic-causing and genome-sequenced S. suis serotype 2 (SS2) strains, 98HAH33 (accession number CP000408), 05ZYH33 (accession number CP000407), and SC84 (accession number FM252031), salK and salR, members of a two-component system which encode a histidine kinase and a response regulator, respectively, were identified on a unique 89K PAI. In S. suis SC84, SalK (YP_003024863) and SalR (YP_003024862) were named after SalK-SalR from S. salivarius because of their 26.0% and 41.0% identity with their counterparts from S. salivarius, respectively (31). Further analysis indicated that the SalK-SalR exhibited 34.3% and 55.3% identity with BovK-BovR from S. bovis, respectively. Like BovK-BovR, SalK in S. suis was presumed to be an eight-transmembrane kinase and SalR was a cytoplasmic regulator composed of an N-terminal signal receiver domain and a C-terminal HTH (helix-turn-helix) DNA binding domain. Both BovK-BovR of S. bovis and SalK-SalR of S. salivarius were demonstrated to be responsible for autoregulation of lantibiotic biosynthesis (24, 31). Thus, we speculated that SalK-SalR in S. suis serotype 2 might be involved in lantibiotic production.
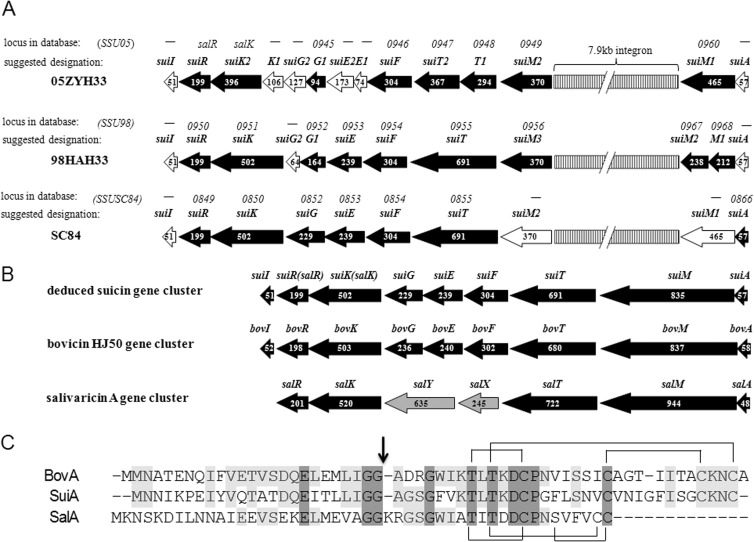
Based on the sequence analysis of the genes neighboring salK and salR, one putative lan-resembling locus containing 9 genes, which was designated sui, was revealed in the aforementioned SS2 strains (Fig. 1A). salK-salR was therefore renamed suiK-suiR here. The other 7 open reading frames (ORFs) were designated suiA, suiM, suiT, suiF, suiE, suiG, and suiI, respectively. Therefore, the potential lantibiotic was termed suicin. In these three sui loci, the putative suiA precursor gene and suiI immunity gene were omitted from annotation in 98HAH33 and 05ZYH33 whereas only suiI was omitted from SC84 in the NCBI database due to the small size or nonconserved nature of those three genes. Several interruptions or mutations existed in each gene cluster. The suiM gene was found to be disrupted by a 7.9-kb integron that most probably originated from E. faecalis (22), while another nucleotide deletion led to a frameshift of suiM in S. suis 98HAH33. In addition, there was one nucleotide deletion in suiG of S. suis 98HAH33 and one each in suiE, suiG, and suiK of 05ZYH33, leading to frameshifts of these genes. The disruption of suiK in strain 05ZYH33 had led to the absence of three N-terminal transmembrane domains. The deduced sui locus displayed organizational similarity to the bovicin HJ50 gene cluster (Fig. 1B), suggesting its primary function of suicin biosynthesis. The bovicin HJ50 gene cluster was resequenced and reannotated here on the basis of the finding that bovT was actually one gene instead of two separate genes (bovT and bovE) as first annotated (32). In addition, orf1 and orf2 were reannotated as bovE and bovG, and bovI, located downstream of bovR, was unveiled.
FIG 1.
Schematic representation of sui loci and comparison of SuiA with BovA and SalA. (A) Interrupted sui loci in three virulent S. suis serotype 2 strains—S. suis 05ZYH33, 98HAH12, and SC84. Black horizontal arrows indicate genes annotated in the NCBI database, while white arrows represent omitted but reannotated genes. Numbers in the arrows indicate the numbers of amino acids contained in the proteins. Above each locus, the upper designations represent gene loci in each genomes and the lower designations represent recommended gene redesignations. (B) Comparison of deduced complete suicin gene cluster with bovicin HJ50 and salivaricin gene clusters. (C) Comparison of precursor peptide of SuiA with that of BovA and SalA. The vertical arrow indicates the GG cleavage site, and cross-linkages indicate the bridge patterns.
The suicin gene cluster in S. suis SC84 was comparatively complete and was assayed by alignment with bovicin HJ50 gene cluster (Table 1). suiA encoded a 57-aa precursor peptide, SuiA, consisting of a 24-aa leader peptide and 33-aa core peptide. SuiA exhibited characteristics of type AII lantibiotics, including a conserved Glu-8 and a double-glycine motif (GG) in the leader peptide and conserved dehydratable Thr residues and ring-forming Cys residues in the core peptide (Fig. 1C). SuiA and BovA shared 49.2% identity, and their core peptides shared 50.0% identity. Despite being disrupted, the putative complete SuiM shared 40.0% identity with the bovicin HJ50 modification enzyme BovM. SuiT was an ABC transporter protein with an N-terminal C39 cysteine protease domain that was postulated to be dedicated to lantibiotic leader peptide processing. It exhibited 41.4% identity with BovT. Downstream of suiT, there were three genes, suiFEG, encoding components of ABC transporters that shared 61.5%, 32.5%, and 38.9% identity with BovFEG, respectively, indicating their roles in immunity. Situated downstream of suiK-suiR, suiI encoded a 51-aa protein that showed 38.5% identity with BovI or TepI, which acted as a self-immunity protein from S. thermophilus SBT 1277 with respect to the lantibiotic thermophilin 1277, which is identical to bovicin HJ50 in terms of primary sequence as well as structure (33, 34). Thus, SuiI was speculated to exert an immunity function.
TABLE 1.
Reannotation and suggested designation of genes in sui locus of S. suis SC84
| Locus tag | Redesignation | Length(s) (aa) | Best hit (% identity/% similarity) | Functionality |
|---|---|---|---|---|
| SSUSC84-0866 | SuiA | 57 | BovA (49.2/66.1) | Precursor suicin |
| –b | SuiM1-SuiM2 | 465/370 | BovM (40.0/53.4)a | Modification |
| SSUSC84-0855 | SuiT | 691 | BovT(41.4/59.5) | Export |
| SSUSC84-0854 | SuiF | 304 | BovF(61.5/71.7) | Immunity |
| SSUSC84-0853 | SuiE | 239 | BovE (32.5/56.7) | Immunity |
| SSUSC84-0852 | SuiG | 229 | BovG (38.9/56.9) | Immunity |
| salK | SuiK | 502 | BovK (34.3/52.7) | Histidine kinase |
| salR | SuiR | 199 | BovR (55.3/70.4) | Response regulator |
| – | SuiI | 51 | BovI (38.5/50.0) | Immunity |
The putative intact SuiM was used for alignment with BovM.
–, no annotation in the genome of S. suis SC84.
We amplified the sui locus from the genomic DNA of SS2 strain S. suis Habb, which was isolated from an STSS patient in Jiangsu, China, and had virulence similar to that of sequenced SS2 strains (20). The sui locus was sequenced and turned out to be identical to that in S. suis SC84.
Biosynthesis of functional suicin in E. coli.
To obtain suicin, a semi-in vitro biosynthesis (SIVB) strategy was introduced (35), consisting of an in vivo modification via coexpression of suiA with suiM and an in vitro digestion via the peptidase domain of SuiT.
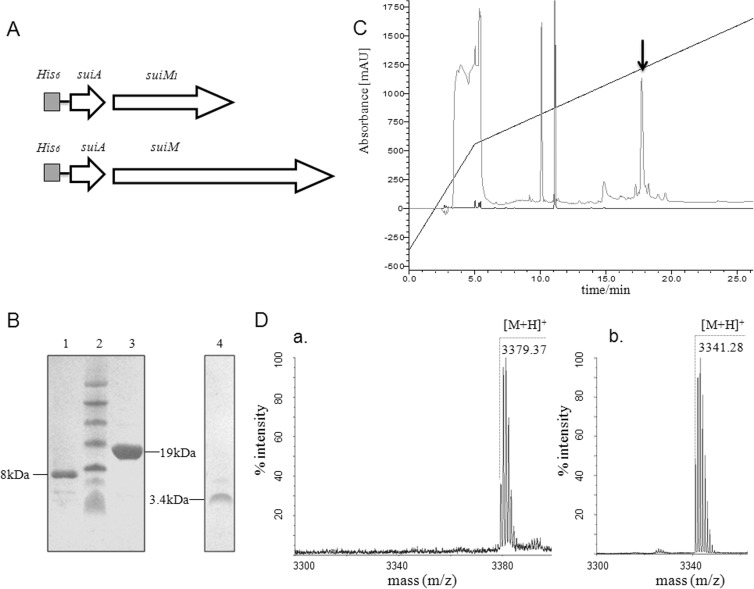
The two separate parts of suiM1 and suiM2 were linked to reconstitute the function of SuiM (Fig. 2A). Whether modified or not, His6-SuiA was mostly expressed in inclusion bodies (data not shown) and was subsequently purified by IMAC and C4 RP-HPLC (Fig. 2B). Mass spectrometry analysis revealed that the molecular mass of protonated His6-SuiA was 8,149.7 Da when SuiA was coexpressed with complete SuiM, resulting in a deviation of 39.5 Da from the theoretical mass of 8,189.2 Da. This indicated that SuiM modified SuiA and that two dehydrations had happened (Table 2). However, the mass of His6-SuiA coexpressed with only SuiM1 differed by 3.4 Da from the calculated mass, indicating that no dehydration had happened. The 3.4-Da difference might have been due to inaccurate detection of the linear mode in MS analysis of masses higher than 5 kDa or to spontaneous formation of disulfide bridges. The results presented above demonstrated that reconstituted complete SuiM, other than SuiM1, can fulfill its original function in vivo to modify the SuiA substrate.
FIG 2.
In vitro biosynthesis of suicin. (A) Schematic representation of coexpression combinations of precursor genes and modification genes. (B) Tricine-SDS-PAGE analysis: lane 1, His6-mSuiA; lane 2, protein marker; lane 3, SuiT159-His6; lane 4, suicin. (C) C18 RP-HPLC purification of suicin. The black arrow indicates the peak of suicin. mAU, milli-absorbance units. (D) MALDI-TOF MS analysis of unmodified SuiA core peptide (a) and suicin (b).
TABLE 2.
Modification of SuiA by SuiM1 and SuiM
| Precursor | LanM | Calculated molecular mass (Da) | Molecular mass by MS (Da) | Molecular mass Δ (Da) | No. of dehydrations |
|---|---|---|---|---|---|
| His6-SuiA | SuiM1 | 8,189.2 | 8,185.8 | 3.4 | 0 |
| His6-SuiA | SuiM | 8,189.2 | 8,149.7 | 39.5 | 2 |
Incubation of modified precursor peptide His6-mSuiA with C39 peptidase domain SuiT159-His6 released the mature lantibiotic suicin. Suicin was purified through C18 RP-HPLC with a retention time of 17.8 min (Fig. 2C). MS analysis of suicin showed an [M+H]+ of 3,341.28 Da (Fig. 2Db), which was a 38.35-Da decrease from the theoretical mass of core peptide of SuiA of 3,379.63 Da. This indicated that the cleavage site was right after a pair of glycine residues (Gly-2 and Gly-1), and the 38.35-Da decrease was thought to correspond to two dehydrations and one disulfide bridge formation. The His6-SuiA coexpressed with SuiM1 was also digested by SuiT159-His6, and the product exhibited an [M+H]+ of 3,379.37 Da (Fig. 2Da), which was in good agreement with theoretical mass of the core peptide of SuiA. This further indicated that His6-SuiA was unmodified by SuiM1.
Ring topology elucidation of suicin.
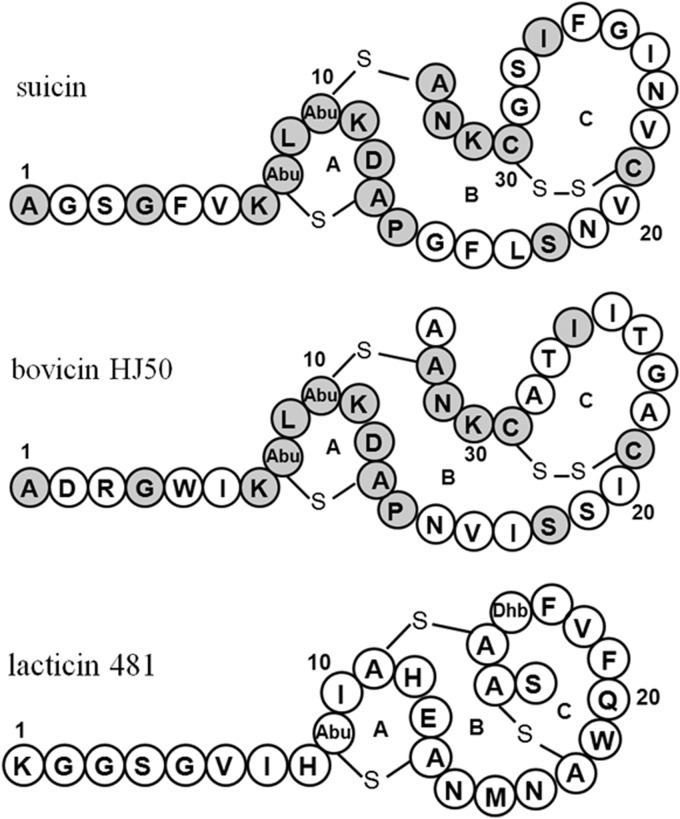
To dissect the ring topology of suicin, a series of suicin mutants were generated and subjected to NEM treatment and MS analysis (Table 3). The absence of dehydration in T8A/T10A demonstrated that the two dehydrations actually happened at Thr8 and Thr10. To dissect the bridge pattern, each Cys was mutated to Ala and the mutants were subjected to NEM treatment for conjugating free thiols. MS analysis showed that no Cys mutation had an influence on dehydration. However, C13A and C33A mutants had two NEM adducts, while C21A and C30A mutants had one. This indicated that Cys13 and Cys33 were involved in thioether ring formation, while Cys21 and Cys30 formed a disulfide bridge. This also indicated that mutant C13A has no influence on the thioether formation of Cys33 and vice versa. Furthermore, addition of two NEM molecules to mutant T10A/C33A was presumed to be attributable to Cys21 and Cys30. This indicated that the thioether bridges were formed by Thr10 and Cys33 as well as Thr8 and Cys13. Thus, the bridge pattern of suicin was dissected and found to contain two thioether bridges (rings A and B) and one disulfide bridge (ring C). Therefore, suicin is similar to bovicin HJ50 in structure (Fig. 3).
TABLE 3.
NEM modification of suicin and its mutants
| Suicin designation | Calculated molecular mass (Da) | Molecular mass by MS | PTMa | NEM addition (Da) | Molecular mass Δ (Da)b | No. of NEM |
|---|---|---|---|---|---|---|
| WT | 3,379.63 | 3,341.28 | 2H2O + S-S | 3,593.55 | 252.27 | 2 |
| T8A/T10A | 3,319.61 | 3,319.44 | –c | 3,819.87 | 500.43 | 4 |
| C13A | 3,347.66 | 3,309.47 | 2H2O | 3,561.55 | 250.08 | 2 |
| C21A | 3,347.66 | 3,311.52 | 2H2O | 3,436.58 | 125.06 | 1 |
| C30A | 3,347.66 | 3,311.53 | 2H2O | 3,436.60 | 125.07 | 1 |
| C33A | 3,347.66 | 3,309.52 | 2H2O | 3,561.64 | 250.12 | 2 |
| T10A/C33A | 3,317.65 | 3,299.64 | 1H2O | 3,549.69 | 250.05 | 2 |
PTM, posttranslational modification.
Data represent the mass difference after NEM addition.
–, no modification.
FIG 3.
Structural comparison of suicin, bovicin HJ50, and lacticin 481. Dark gray circles indicate identical residues in suicin and bovicin HJ50.
Suicin displayed antimicrobial activities against various Gram-positive pathogens.
Suicin was tested against the series of Gram-positive and -negative strains listed in Table 4. Suicin exhibited inhibitory activity against some Gram-positive strains from bacilli, micrococci, lactobacilli, lactococci, streptococci, leuconostoc, and enterococci. In particular, suicin showed inhibition of E. pernyi, which is the causative agent of empty-gut disease of tussah, and of vancomycin-resistant E. faecalis V583, which has been clinically associated with urinary tract infection, bacteremia, and infective endocarditis (36). As expected, suicin exhibited no activity against Gram-negative strains. M. flavus NCIB8166 was the most sensitive strain and was used as an indicator for MIC determinations. Results showed that suicin inhibited growth of M. flavus NCIB8166 at concentrations as low as 0.195 μg/ml, which is its MIC for M. flavus NCIB8166. As a positive control, nisin Z exhibited better potency, with a MIC of 0.0325 μg/ml.
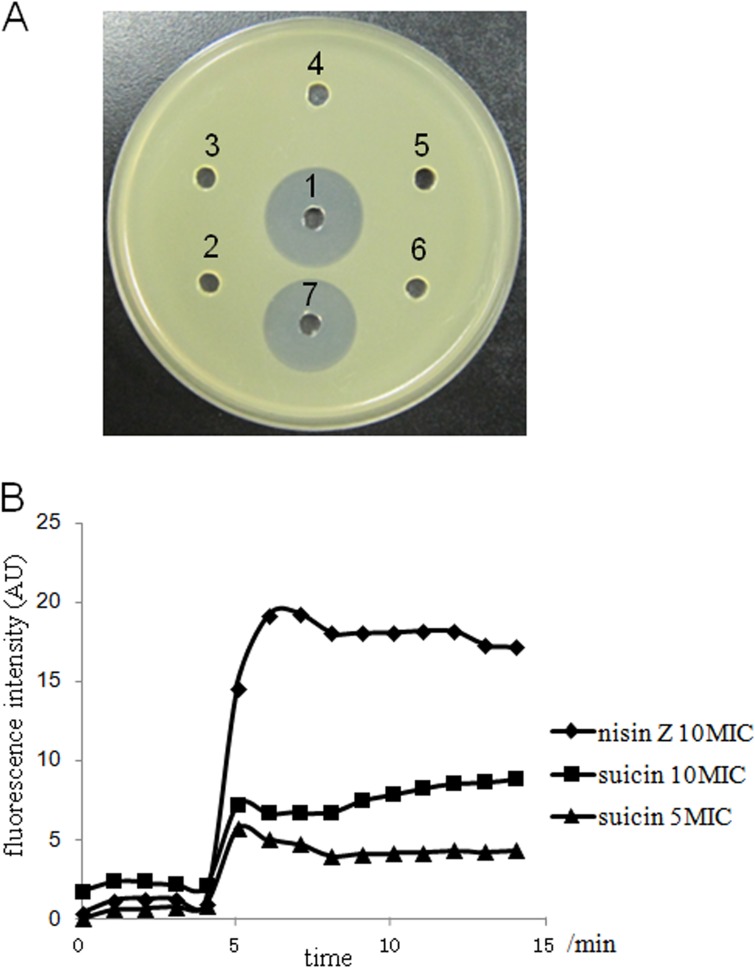
To elucidate whether thioether ring or disulfide bridge formation is required for the antimicrobial activity of suicin, a series of Cys mutants of suicin were purified and tested for inhibitory activity against M. flavus NCIB8166. As shown in Fig. 4A, while bovicin HJ50 and wild-type suicin markedly inhibited the growth of M. flavus NCIB8166, disruption of any thioether ring and/or disulfide bridge in suicin totally abolished its bactericidal activity against M. flavus NCIB8166, which was the most sensitive strain among all tested strains (Table 4). Therefore, both thioether rings and the disulfide bridge are essential for the function of suicin.
FIG 4.
Growth inhibition and membrane potential disruption of Micrococcus flavus NCIB8166 by suicin. (A) Antimicrobial activity of suicin and its derivatives against M. flavus NCIB8166, with bovicin HJ50 as a positive control. To each hole, 25 μl of 5 μg/ml of each peptide was applied. 1, suicin; 2, unmodified core peptide of SuiA; 3, suicin C13A; 4, suicin C21A; 5, suicin C29A; 6, suicin C33A; 7, bovicin HJ50. (B) Effect of suicin and nisin on membrane potential of M. flavus NCIB8166. M. flavus NCIB8166 cells were treated with suicin or nisin Z with a time duration of 15 min, and fluorescence was recorded with excitation at 493 nm and emission at 516 nm. Diamond, nisin Z at 10-fold MIC (0.325 μg/ml); square, suicin at 10-fold MIC (1.95 μg/ml); triangle, suicin at 5-fold MIC (0.975 μg/ml).
Suicin could disrupt the membrane potential of sensitive cells.
It has been well established that lantibiotics, especially type A lantibiotics, primarily target and form pores in the cytoplasmic membrane of sensitive cells, thus resulting in disruption of the membrane potential and release of the intracellular components (4, 37). To determine whether suicin acts through disintegrating the membrane of sensitive cells, we monitored membrane potential changes of M. flavus NCIB8166 after application of suicin at 5- or 10-fold MIC. In the assay, we used a voltage-sensitive fluorescence dye, DiBAC4(3), to measure the membrane potentials. When the membrane potential is disrupted as a result of pore formation in the membrane, DiBAC4(3) enters the cell membrane and its fluorescence is enhanced (38). As shown in Fig. 4B, addition of suicin to the M. flavus NCIB8166 cells caused a robust increase of fluorescence in a concentration-dependent manner, indicating that the membrane potential of indicator cells was disrupted by suicin. This increase was also observed upon application of nisin (Fig. 4B), which is known to disrupt membrane potential with high potency (4). That the efficiency of suicin is lower than that of nisin in inducing fluorescence fluctuation might be due to the fact that the potency of suicin is lower than that of nisin in disrupting membrane potential. Therefore, forming pores in the membrane might be one important, if not the only, mode of action for suicin in inhibition of sensitive cell growth.
Virulence-associated SuiR could bind the suiA promoter.
As a TCS in the deduced sui locus, SuiK-SuiR was supposed to be responsible for autoregulation of suicin biosynthesis, though suicin production might be abolished by disruptions of the suicin gene cluster. To confirm that supposition, EMSA was conducted to identify the binding ability of His6-SuiR toward suiA promoter PsuiA. His6-SuiR was purified to homogeneity and identified by Tricine-SDS-PAGE analysis (Fig. 5B). His6-SuiR was then incubated with biotin-PsuiA and a 100-fold excess of unlabeled PsuiA (U-PsuiA). The transferability-retarded bands of biotin-PsuiA, which disappeared in the presence of excess unlabeled PsuiA, clearly indicated that His6-SuiR could specifically bind PsuiA (Fig. 5C).
FIG 5.
In vitro binding activity of His6-SuiR and in vivo activation of PsuiA by SuiK-SuiR. (A) Sequence analysis of suiA promoter PsuiA. (B) Purified His6-SuiR analyzed by Tricine-SDS-PAGE. Lane 1, His6-SuiR; lane 2, protein marker. (C) EMSA analysis of binding activity of His6-SuiR toward PsuiA. Lane 1, biotin-labeled PsuiA incubated with His6-SuiR and a 100-fold excess of unlabeled PsuiA (U-PsuiA); lanes 2 to 5, gradient dilution of His6-SuiR with biotin-labeled PsuiA; lane 6, biotin-labeled PsuiA only. (D and E) EMSA analysis of binding activity toward PsuiA ΔIR1 and ΔIR2, respectively. (F) Illustration of plasmids pMG36e-suiKRPsuiAgfp and pMG36e-suiRPsuiAgfp. (G) GFP visualization of L. lactis NZ9000 transformants with pMG36e-derived plasmids was examined with a fluorescence microscope. In the following, NZ-SuiK-SuiR and NZ-SuiR refer to L. lactis NZ9000 transformants of pMG36e-suiKRPsuiAgfp or pMG36e-suiRPsuiAgfp. a, NZ-SuiK-SuiR; b, NZ-SuiK-SuiR induced by 50 ng/ml suicin; c, NZ-SuiR; d, NZ-SuiK(H311A)-SuiR; e, NZ-SuiK-SuiR(D54A); f, NZ-SuiK-SuiR(D54E); g, NZ-SuiK-SuiR(PsuiAΔIR1); h, NZ-SuiK-SuiR(PsuiAΔIR2).
Sequence analysis of PsuiA indicated there were three inverted repeats (IR) located upstream of suiA (Fig. 5A). IR1 was located between a ribosome binding site (RBS) and a −10 box, and IR2 was located before a −35 box. IR3 was presumed to be the terminator of gene SSUSC84_0868. To determine whether IR1 or IR2 was the binding site of SuiR, EMSA analysis was conducted using the IR-deleted version of PsuiA (ΔIR). The mutations of ΔIR1 and ΔIR2 were performed via SLIM on the pMG36e-suiKRPsuiAgfp plasmid described later, and the corresponding probes were subsequently amplified with primers as used for biotin-PsuiA. The results showed that His6-SuiR could bind PsuiA ΔIR1 but not PsuiA ΔIR2 (Fig. 5D and E). This indicated that SuiR binds to the AT-rich inverted repeat IR2.
The SuiK-SuiR TCS was competent to activate PsuiA.
To determine if SuiK-SuiR could activate transcription of PsuiA, the SuiK-SuiR TCS was expressed in L. lactis NZ9000 and gfp was used as the reporter gene, whose expression is under the control of PsuiA (Fig. 5F). pMG36e-suiKRPsuiAgfp was transformed into L. lactis NZ9000, and green florescence was visualized, indicating GFP expression. However, GFP expression was independent of suicin induction (Fig. 5Ga and b), which might have been due to autophosphorylation of SuiK resulting from high-level expression of SuiK-SuiR driven by strong lactococcal promoter P32 (24). To see if SuiK was required for PsuiA activation, pMG36e-suiRPsuiAgfp was constructed and transformed into L. lactis NZ9000, which resulted in much lower GFP expression (Fig. 5Gc). This indicated that SuiK was necessary for SuiR phosphorylation to exert its full function, though other histidine kinases in L. lactis NZ9000 might have transferred a phosphoryl group to SuiR at a low level.
To further confirm the role of SuiK-SuiR signal transduction in activating PsuiA, analysis of alignment of SuiK-SuiR with LanK-LanR showed that there exists one conserved His residue (His311) in SuiK and an Asp (Asp54) in SuiR (data not shown). Then, site-directed mutagenesis targeting conserved residues was performed and the function of mutant SuiK-SuiR was assessed. We first found that PsuiA activation was inhibited by both H311A in SuiK and D54A in SuiR (Fig. 5Gd and e). However, a conservative mutation (D54E) of SuiR had no effect on the signaling potency of SuiK-SuiR (Fig. 5Gf). Furthermore, when the binding site of SuiR on PsuiA (IR2) was deleted, PsuiA ΔIR2 could not be activated by SuiK-SuiR any more (Fig. 5Gh). Paradoxically, though IR1 of PsuiA was not involved in SuiR binding to PsuiA in vitro (Fig. 5E), its deletion significantly attenuated PsuiA activation by SuiR in vivo (compare Fig. 5Gg with Fig. 5Ga). This indicated that IR1 and IR2 might play complementary roles in mediating binding of SuiR to PsuiA, though IR2 was more dominant.
DISCUSSION
The wide existence of lantibiotic gene clusters in bacteria, especially streptococci, has been extensively acknowledged in previous research, and the quantity of known gene clusters has recently been increased by genomic mining (12). However, disrupted or unannotated genes of small size or nonconserved nature might sometimes hamper context-based prediction by the use of such tools as BAGEL3 (15). As we identified here, the sui loci were probably overlooked because of the existence of interruptions or frameshift mutations or omissions of annotation. Following the clue of SuiK-SuiR sharing similarity with the TCS for lantibiotic regulation and comparison of its neighboring lan relics with bovicin HJ50 and salivaricin gene clusters, a primarily identical but disrupted sui locus was unveiled and reannotated in three SS2 genomes. Indeed, sui locus was potentially wide distributed in Chinese S. suis isolates because of its location on the 89K PAI (39, 40). Notably, similar situations were also observed in cytolysin determinants in E. faecalis and the sal locus in S. pyogenes such that intact or disrupted versions were unveiled in different strains (31, 41, 42). This implied that remnant lantibiotic loci like sui might actually or even widely exist in bacterial genomes, which might be overlooked with traditional mining approaches. Hence, by a combination of in silico prediction and in-depth comparative analysis, here the hidden remnant sui loci were rediscovered.
As suicin might not be produced by those sui loci containing SS2 due to disruption of the sui loci, especially that of the crucial modification gene suiM, suicin was therefore biosynthesized and characterized successfully by reconstituting the intact SuiM in E. coli and by subsequent processing via SuiT159 in vitro. As expected, SuiM1 could not produce any dehydrated SuiA, supporting the idea that disruption of lanM might abolish lantibiotic production (32, 43), whereas reconstituted intact SuiM produced SuiA with two dehydrations, indicating that its modification function had been restored. SuiT159, the peptidase domain of transporter SuiT, performed a digestion function at the double-Gly site to release mature suicin, which was confirmed by antimicrobial assay and MS analysis. Although authentic suicin was not produced, previous work has verified this strategy to obtain lantibiotics as genuine (35, 44). Suicin is, to date, the first lantibiotic originating from S. suis and showed remarkable inhibitory activity against various Gram-positive pathogens, implying a promising application as an antimicrobial agent. This work to obtain a new-to-nature lantibiotic, suicin, further exemplified the availability of applications to revitalize and thus broaden the range of lantibiotic resources from cryptic and, especially, remnant gene clusters.
Structural elucidation indicated that suicin contained two methyllanthionine residues and one disulfide bridge, exhibiting an N-terminal linear structure and a C-terminal globular structure characteristic of type AII lantibiotics. However, different from canonical type AII lantibiotics such as lacticin 481 and salivaricin A, suicin showed structural similarity with bovicin HJ50 in containing a disulfide bridge other than a thioether bridge in ring C (Fig. 3). The disulfide bridge is rare in lantibiotics but was demonstrated here to be critical for suicin activity and has been previously underlined in bovicin HJ50 (35). A previous study showed that bovicin HJ50 could disrupt the membrane integrity of sensitive strain M. flavus NCIB8166, resulting in potassium efflux (45). Here, we found that suicin also disintegrated the cell membrane of M. flavus NCIB8166, leading to the disruption of membrane potential (Fig. 4B). Suicin has a ring A identical to that of bovicin HJ50, which contains a TxS/TxD/EC motif (the first x is in most cases a hydrophobic residue, the latter x is undefined) conserved among type AII lantibiotics (37). This motif has been shown to mediate binding of type AII lantibiotics to lipid II, an essential intermediate during peptidoglycan synthesis and subsequent cell wall formation (37). Therefore, further study is needed to elucidate whether suicin could display bactericidal activity by inhibiting cell wall synthesis. Interestingly, by using SuiA as a drive sequence, at least 16 SuiA-like putative lantibiotic precursor peptides have been found in the NCBI database. Except for known bovicin HJ50-identical lantibiotics such as thermophilin 1277 (33) and macedovicin (46), the others are distributed in E. columbae, in Clostridium perfringens, and especially in bacillus strains. By using the above-mentioned semi-in vitro biosynthesis strategy, three new lantibiotics have been biosynthesized and characterized from pathogenic C. perfringens, B. cereus, and B. thuringiensis which were structurally elucidated to be similar to suicin and bovicin HJ50, including the conserved disulfide bridge (data not shown). Thus, along with suicin, they were grouped into bovicin HJ50-like lantibiotics, which represented a novel disulfide-containing subgroup of type AII lantibiotics.
SuiK-SuiR has been demonstrated to be essential for virulence of SS2 by regulating expression of virulence-associated factors (21, 47). Here, we found that SuiK-SuiR was also involved in transcriptional activation of promoter of structural gene suiA, displaying conventional function of LanK-LanR. SuiK was supposed to transfer the phosphoryl group to SuiR, initiating its binding to the promoter region of suiA at an AT-rich inverted repeat and consequentially activating GFP expression in our experiments. Based on sequence alignment (data not shown), His311 of SuiK and Asp54 of SuiR are predicted to be the primary sites of phosphorylation. It was verified that either mutation disabled the signal transduction from SuiK to SuiR. However, this signaling appears to be independent of suicin application, as PsuiA was efficiently activated in the absence of suicin (Fig. 5Ga and b). We speculate that it might be due to high-level expression of SuiK driven by strong lactococcal promoter P32, which leads to autophosphorylation of SuiK and signal leakage (24). Yet, SuiR alone was able to activate PsuiA transcription, though at low efficiency, as was also observed with BovR and SpaR (24, 48). Interestingly, the genome of an S. suis serotype 9 strain isolated in China was recently sequenced and shown to contain an intact sui locus (SSUD12_1302-SSUD12_1310) nearly identical to the one unveiled here (40). Although it has not been tested for lantibiotic production, the operonic association of suiK-suiR with the intact sui locus might underscore its connection to suicin production.
In conclusion, from a remnant gene cluster in highly virulent S. suis serotype 2, a bioactive type AII lantibiotic suicin was biosynthesized and characterized to contain a rare disulfide bridge. Suicin displayed inhibitory activities exclusively against Gram-positive bacteria, including pathogenic streptococci and vancomycin-resistant enterococci, making it an alternative candidate for combating against bacterial pathogens. Further, we found that the virulence-associated SuiK-SuiR is involved in activating promoter of suicin precursor gene. Thus, we propose that lan loci, especially disrupted ones, might evolve to extended their functions to control virulence-associated factors in S. suis serotype 2.
ACKNOWLEDGMENTS
We thank Zhizeng Sun at the University of Iowa Carver College of Medicine for critical reading of the manuscript. We are also very grateful to Tang Jiaqi in the Research Institute for Medicine of Nanjing Command in China for providing genomic DNA of S. suis Habb.
This research was supported by the National Natural Science Foundation of China (31070041) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-6, KSCX2-EW-Q-14, and KSCX2-EW-G-14).
Footnotes
Published ahead of print 22 November 2013
REFERENCES
- 1.Hancock RE, Sahl H-G. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557. 10.1038/nbt1267 [DOI] [PubMed] [Google Scholar]
- 2.Piper C, Cotter PD, Ross RP, Hill C. 2009. Discovery of medically significant lantibiotics. Curr. Drug Discov. Technol. 6:1–18. 10.2174/157016309787581075 [DOI] [PubMed] [Google Scholar]
- 3.van Heel AJ, Montalban-Lopez M, Kuipers OP. 2011. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin. Drug Metab. Toxicol. 7:675–680. 10.1517/17425255.2011.573478 [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633–684. 10.1021/cr030105v [DOI] [PubMed] [Google Scholar]
- 5.Jung G. 1991. Lantibiotics—ribosomally synthesized biologically active polypeptides containing sulfide bridges and α,β-didehydroamino acids. Angew. Chem. Int. Ed. Engl. 30:1051–1068. 10.1002/anie.199110513 [DOI] [Google Scholar]
- 6.Koponen O, Tolonen M, Qiao M, Wahlström G, Helin J, Saris PE. 2002. NisB is required for the dehydration and NisC for the lanthionine formation in the post-translational modification of nisin. Microbiology 148(Pt 11):3561–3568 [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. 2005. Lacticin 481 synthetase phosphorylates its substrate during lantibiotic production. J. Am. Chem. Soc. 127:15332–15333. 10.1021/ja0543043 [DOI] [PubMed] [Google Scholar]
- 8.Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229–240. 10.1111/j.1365-2958.1995.tb02295.x [DOI] [PubMed] [Google Scholar]
- 9.Twomey D, Ross R, Ryan M, Meaney B, Hill C. 2002. Lantibiotics produced by lactic acid bacteria: structure, function and applications. Antonie Van Leeuwenhoek 82:165–185. 10.1023/A:1020660321724 [DOI] [PubMed] [Google Scholar]
- 10.Daly M, Cotter KPD, Hill C, Ross RP. 2012. Lantibiotic production by pathogenic microorganisms. Curr. Protein Pept. Sci. 13:509–523. 10.2174/138920312803582997 [DOI] [PubMed] [Google Scholar]
- 11.Coburn PS, Gilmore MS. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell. Microbiol. 5:661–669. 10.1046/j.1462-5822.2003.00310.x [DOI] [PubMed] [Google Scholar]
- 12.Nes IF, Diep DB, Holo H. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189–1198. 10.1128/JB.01254-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majchrzykiewicz JA, Lubelski J, Moll GN, Kuipers A, Bijlsma JJ, Kuipers OP, Rink R. 2010. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob. Agents Chemother. 54:1498–1505. 10.1128/AAC.00883-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Begley M, Cotter PD, Hill C, Ross RP. 2009. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 75:5451–5460. 10.1128/AEM.00730-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. 15 May 2013. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41:W448–W453. 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh A, O'Sullivan O, Ross RP, Cotter P, Hill C. 2010. In silico analysis highlights the frequency and diversity of type 1 lantibiotic gene clusters in genome sequenced bacteria. BMC Genomics 11:679. 10.1186/1471-2164-11-679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, O'Connor PM, Rossney A, Götz F, Hill C. 2010. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J. Bacteriol. 192:1131–1142. 10.1128/JB.01375-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang JQ, Wang CJ, Feng YJ, Yang WZ, Song HD, Chen ZH, Yu HJ, Pan XZ, Zhou XJ, Wang HR, Wu B, Wang HL, Zhao HM, Lin Y, Yue JH, Wu ZQ, He XW, Gao F, Khan AH, Wang J, Zhao GP, Wang Y, Wang X, Chen Z, Gao GF. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Normile D. 2005. Infectious diseases—WHO probes deadliness of China's pig-borne disease. Science 309:1308–1309. 10.1126/science.309.5739.1308a [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M. 2007. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, Ge J, Zheng F, Cao M, Dong Y, Liu D, Lin Y, Du H, Gao GF, Wang X, Hu F, Tang J. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. 10.1371/journal.pone.0002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Ni J, Teng K, Liu G, Qiao C, Huan L, Zhong J. 2011. Autoregulation of lantibiotic bovicin HJ50 biosynthesis by the BovK-BovR two-component signal transduction system in Streptococcus bovis HJ50. Appl. Environ. Microbiol. 77:407–415. 10.1128/AEM.01278-10 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Chiu J, March PE, Lee R, Tillett D. 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32:e174–e174. 10.1093/nar/gnh172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schägger H. 2006. Tricine–SDS-PAGE. Nat. Protoc. 1:16–22. 10.1038/nprot.2006.4 [DOI] [PubMed] [Google Scholar]
- 27.Sun Z, Zhong J, Liang X, Liu J, Chen X, Huan L. 2009. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob. Agents Chemother. 53:1964–1973. 10.1128/AAC.01382-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levengood MR, Knerr PJ, Oman TJ, van der Donk WA. 2009. In vitro mutasynthesis of lantibiotic analogues containing nonproteinogenic amino acids. J. Am. Chem. Soc. 131:12024–12025. 10.1021/ja903239s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168–1172. 10.1126/science.1185723 [DOI] [PubMed] [Google Scholar]
- 30.Holo H, Nes IF. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195–199 [DOI] [PubMed] [Google Scholar]
- 31.Upton M, Tagg J, Wescombe P, Jenkinson H. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931–3938. 10.1128/JB.183.13.3931-3938.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu G, Zhong J, Ni J, Chen M, Xiao H, Huan L. 2009. Characteristics of the bovicin HJ50 gene cluster in Streptococcus bovis HJ50. Microbiology 155:584–593. 10.1099/mic.0.022707-0 [DOI] [PubMed] [Google Scholar]
- 33.Kabuki T, Uenishi H, Seto Y, Yoshioka T, Nakajima H. 2009. A unique lantibiotic, thermophilin 1277, containing a disulfide bridge and two thioether bridges. J. Appl. Microbiol. 106:853–862. 10.1111/j.1365-2672.2008.04059.x [DOI] [PubMed] [Google Scholar]
- 34.Kabuki T, Kawai Y, Uenishi H, Seto Y, Kok J, Nakajima H, Saito T. 2011. Gene cluster for biosynthesis of thermophilin 1277—a lantibiotic produced by Streptococcus thermophilus SBT1277, and heterologous expression of TepI, a novel immunity peptide. J. Appl. Microbiol. 110:641–649. 10.1111/j.1365-2672.2010.04914.x [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Teng K, Huan L, Zhong J. 2011. Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol. Res. 166:146–154. 10.1016/j.micres.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 36.Paulsen I, Banerjei L, Myers G, Nelson K, Seshadri R, Read T, Fouts D, Eisen J, Gill S, Heidelberg J. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- 37.Islam MR, Nishie M, Nagao J, Zendo T, Keller S, Nakayama J, Kohda D, Sahl HG, Sonomoto K. 2012. Ring A of nukacin ISK-1: a lipid II-binding motif for type-A (II) lantibiotic. J. Am. Chem. Soc. 134:3687–3690. 10.1021/ja300007h [DOI] [PubMed] [Google Scholar]
- 38.Epps DE, Wolfe ML, Groppi V. 1994. Characterization of the steady-state and dynamic fluorescence properties of the potential-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol (Dibac4(3)) in model systems and cells. Chem. Phys. Lipids 69:137–150. 10.1016/0009-3084(94)90035-3 [DOI] [PubMed] [Google Scholar]
- 39.Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. 2011. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79:1670–1683. 10.1111/j.1365-2958.2011.07553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang A, Yang M, Hu P, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. 2011. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics 12:523. 10.1186/1471-2164-12-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankar N, Coburn P, Pillar C, Haas W, Gilmore M. 2004. Enterococcal cytolysin: activities and association with other virulence traits in a pathogenicity island. Int. J. Med. Microbiol. 293:609–618. 10.1078/1438-4221-00301 [DOI] [PubMed] [Google Scholar]
- 42.Shankar N, Baghdayan AS, Gilmore MS. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750. 10.1038/nature00802 [DOI] [PubMed] [Google Scholar]
- 43.Diep DB, Godager L, Brede D, Nes IF. 2006. Data mining and characterization of a novel pediocin-like bacteriocin system from the genome of Pediococcus pentosaceus ATCC 25745. Microbiology 152:1649–1659. 10.1099/mic.0.28794-0 [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Yang X, Garg N, van der Donk WA. 2011. Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 133:2338–2341. 10.1021/ja109044r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao H, Chen X, Chen M, Tang S, Zhao X, Huan L. 2004. Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 150:103–108. 10.1099/mic.0.26437-0 [DOI] [PubMed] [Google Scholar]
- 46.Georgalaki M, Papadimitriou K, Anastasiou R, Pot B, Van Driessche G, Devreese B, Tsakalidou E. 2013. Macedovicin, the second food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Food Microbiol. 33:124–130. 10.1016/j.fm.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 47.Shen X, Zhong Q, Zhao Y, Yin S, Chen T, Hu F, Li M. 2013. Proteome analysis of the two-component SalK/SalR system in epidemic Streptococcus suis serotype 2. Curr. Microbiol. 67:118–122. 10.1007/s00284-013-0343-4 [DOI] [PubMed] [Google Scholar]
- 48.Stein T, Borchert S, Kiesau P, Heinzmann S, Klöss S, Klein C, Helfrich M, Entian KD. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44:403–416. 10.1046/j.1365-2958.2002.02869.x [DOI] [PubMed] [Google Scholar]