Abstract
We present the results of a study using high-throughput whole-transcriptome sequencing (RNA-seq) and vibrational spectroscopy to characterize and fingerprint pathogenic-bacterium injury under conditions of unfavorable stress. Two garlic-derived organosulfur compounds were found to be highly effective antimicrobial compounds against Cronobacter sakazakii, a leading pathogen associated with invasive infection of infants and causing meningitis, necrotizing entercolitis, and bacteremia. RNA-seq shows changes in gene expression patterns and transcriptomic response, while confocal micro-Raman spectroscopy characterizes macromolecular changes in the bacterial cell resulting from this chemical stress. RNA-seq analyses showed that the bacterial response to ajoene differed from the response to diallyl sulfide. Specifically, ajoene caused downregulation of motility-related genes, while diallyl sulfide treatment caused an increased expression of cell wall synthesis genes. Confocal micro-Raman spectroscopy revealed that the two compounds appear to have the same phase I antimicrobial mechanism of binding to thiol-containing proteins/enzymes in bacterial cells generating a disulfide stretching band but different phase II antimicrobial mechanisms, showing alterations in the secondary structures of proteins in two different ways. Diallyl sulfide primarily altered the α-helix and β-sheet, as reflected in changes in amide I, while ajoene altered the structures containing phenylalanine and tyrosine. Bayesian probability analysis validated the ability of principal component analysis to differentiate treated and control C. sakazakii cells. Scanning electron microscopy confirmed cell injury, showing significant morphological variations in cells following treatments by these two compounds. Findings from this study aid in the development of effective intervention strategies to reduce the risk of C. sakazakii contamination in the food production environment and on food contact surfaces, reducing the risks to susceptible consumers.
INTRODUCTION
Cronobacter sakazakii is a motile, nonsporeforming Gram-negative facultative anaerobe (1). It was noted to be an opportunistic pathogen responsible for life-threatening forms of meningitis, sepsis, necrotizing colitis, bacteremia, and meningoencephalitis in preterm neonates and infants (2–6). The mortality rate for neonatal infections has been reported to be as high as 80% (7). C. sakazakii infections may also result in severe neurological sequelae such as hydrocephalus, quadriplegia, and retarded neural development in survivors (8). Epidemiological studies implicated powdered infant formula as a primary source of transmission (6); indeed, this bacterium was detected in 2.4% to 14.2% of powdered infant formula products (5). Antibiotic therapy with a combination of ampicillin and gentamicin is an efficient method to treat C. sakazakii infections (4). However, the emergence of strains resistant to antibiotics has led the design and use of novel antimicrobials. In recent years, plant-derived biologically active compounds, such as trans-cinnamaldehyde (2, 3), phenolic benzaldehyde (9), curcumin (10), and anthocyanin (11), have attracted more attention due to strong antimicrobial activity, particularly for effective inhibition of the survival and growth of multidrug-resistant bacteria (12).
Garlic (Allium sativum L.), besides its function as spice and food, has been used as medicinal plant for over 4,000 years. It contains more than 100 biologically active secondary metabolites (13). Previous research demonstrated that garlic may be effective against cardiovascular disease because of its antithrombotic, hypolipidemic, hypocholesterolemic, antihypertensive, antidiabetic, and antihyperhomocysteinemia effects (14, 15). Further, garlic possesses many other biological activities, including antimicrobial, antioxidant, anticancer, antimutagenic, antiasthmatic, and anti-inflammatory immune modulation activities and prebiotic activities (14–22). Evidence suggests that the biological and medical functions of garlic are mainly due to its high level of organosulfur compounds (15), while proteins derived from garlic do not have antimicrobial activity (23) and phenolic compounds make only a minor contribution (24).
The primary sulfur-containing constituents in garlic are S-alk(en)yl-l-cysteine sulfoxides, such as alliin [(+)-(S)-allyl-l-cysteine-sulfoxide] and γ-glutamyl cysteines (15, 16). When a raw garlic bulb is crushed, alliin and alliinase, which are stored in separate cellular compartments of the garlic clove, are mixed together. In the role of alliinase, alliin is further subjected to dehydration by pyridoxal phosphate and transformed to allyl sulfenic acid, pyruvic acid, and ammonia (16). Allyl sulfenic acid is unstable and very reactive at room temperature. With the elimination of water, two molecules of allyl sulfenic acid condense spontaneously to allicin (see Fig. S1A in the supplemental material), validated as an important substance for the medicinal properties of garlic (13, 15–17). However, allicin is an unstable compound which is rapidly decomposed into other stable sulfur-containing constituents. The manufacturing procedure and garlic varieties are two important factors that affect the categories and compositions of organosulfur compounds decomposed from allicin (15, 25). Specifically, ajoene [(E)- and (Z)-4,5,9-trithiadodeca-1,6,11-triene-9-oxides] is commonly found in oil-macerated/ether-extracted garlic oil, with E-ajoene usually present at levels twice as high as those of Z-ajoene, while steam-distilled garlic oil is mainly composed of lipid-soluble sulfur compounds, including diallyl sulfide, diallyl disulfide, and diallyl trisulfide, and of minor amounts of many other volatile compounds, such as allyl methyl disulfide and diallyl tetrasulfide (see Fig. S1B) (15, 16, 23).
Both ajoene and lipid-soluble sulfur compounds (e.g., diallyl sulfides) have been reported to display strong antimicrobial activity against a number of Gram-positive and Gram-negative bacteria, yeasts, and viruses (15, 16, 19, 22–30). We recently validated the hypothesis that diallyl sulfides could effectively inactivate planktonic cells of Listeria monocytogenes, Escherichia coli O157:H7 (31), and Campylobacter jejuni (24) and sessile C. jejuni cells in a biofilm (18). Velliyagounder et al. also confirmed the in vitro antimicrobial efficacy against Aggregatibacter actinomycetemcomitans (23). Further, Yoshida et al. confirmed that Z-ajoene has stronger antimicrobial activity than E-ajoene, indicating the impact of the conformation of the compound on its biological activity (22, 32). Previous to this study, researchers had not examined the effect of the antimicrobial activity of ajoene and diallyl sulfide on C. sakazakii.
In the current study, we examined the in vitro antimicrobial effect of ajoene and diallyl sulfide on C. sakazakii. Additionally, the mechanism of sublethal injury of C. sakazakii under conditions of stress from these two organosulfur compounds was characterized and fingerprinted on both the genotypic and phenotypic levels using whole-transcriptome sequencing (RNA-seq) and Raman spectroscopy, respectively. Raman spectroscopy is based on vibrational spectroscopy and can be applied to characterize the biochemical components of bacterial cell membranes (33) and bacterial metabolites (34); thus, it is a reasonable technological tool to determine bacterial stress and injury in response to unfavorable environmental conditions. In fact, we have recently conducted vibrational spectroscopy coupled with multivariate analyses to determine bacterial stress and injury induced by cold and freezing storage (35) and by the presence of metal oxide nanoparticles (36) and of bioactive compounds derived from plants (18, 24, 31).
Several studies have validated the idea that DNA microarray analysis, a method relying upon hybridization of targeted oligonucleotides to particular loci, could be applied to investigate the global gene expression and transcriptomic response of food-borne pathogens to specific stress, such as chlorine treatment (37, 38) and hydrogen peroxide treatment (39). However, the relatively low detection sensitivity of DNA-DNA hybridization has limited microarrays to detection of only the trace level of bacterial transcriptomic response to the unfavorable stress. For example, it is very challenging to apply the DNA microarray technique to detect the rare transcripts and noncoding regulatory RNAs (40). Recently, RNA-seq has been applied to study the bacterial transcriptome (41–43) and further characterize the changes in pathogenic bacterial gene expression during infection of host cells (44). Further, transcriptome sequencing of Salmonella enterica serovar Enteritidis under conditions of desiccation and starvation stress in oil was studied using RNA-seq (40).
To the best of our knowledge, this is the first report of a study that used both high-throughput genotypic (i.e., RNA-seq) and phenotypic (i.e., Raman spectroscopy) tools to systematically study bacterial injury under unfavorable conditions. We used C. sakazakii as an exemplary microorganism for the study. Our innovative analytical approaches were employed to develop a model for illustrating the mechanism for stress and injury of C. sakazakii under unfavorable conditions.
MATERIALS AND METHODS
Chemicals and reagents.
Diallyl sulfide (purity, 97%), crude diallyl disulfide (purity, 80%), and l-cysteine (purity, 97%) were purchased from Sigma-Aldrich (St. Louis, MO). Dimethyl sulfoxide (DMSO; purity, >99%) was obtained from Sangon Biotech Co., Ltd. (Shanghai, China).
Synthesis of ajoene.
Ajoene was synthesized from diallyl disulfide (compound 1) via 2 steps (20) as follows:
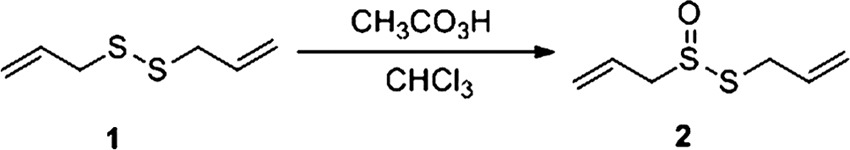 |
Peracetic acid (35%; 2.58 g, 11.87 mmol) was added dropwise via an addition funnel to a solution of commercially available diallyl disulfide (Sigma-Aldrich; 80%) (2.1 ml, 11.3 mmol) in chloroform (18.0 ml) at 0°C. The reaction mixture was stirred at 0°C for 30 min, and anhydrous sodium carbonate (4.0 g) was added in small portions during 30 min with vigorous stirring. The mixture was stirred for an additional 30 min at 0°C and then filtered through a pad of Celite and anhydrous magnesium sulfate. The filtrate was concentrated under a vacuum and purified by the use of a chromatography column to get allicin (compound 2) (1.2 g, 71% yield) as a light yellow liquid. 1H nuclear magnetic resonance (NMR) (CDCl3) levels of 6.30 to 5.04 (m, 6 H) and 3.94 to 3.65 (m, 4 H) were determined as follows; the NMR data are in accordance with previous publication (20):
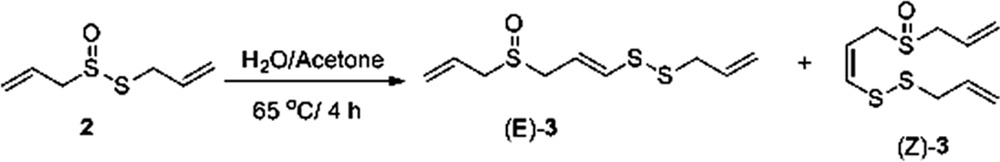 |
Allicin (compound 2) (1.0 g, 6.2 mmol) was dissolved in 40% aqueous acetone (10.0 ml), and the homogeneous solution was heated at 65°C for 4 h. The reaction mixture was diluted with 50% aqueous methanol (40.0 ml) and washed with hexane (5×; 20.0 ml). The aqueous methanolic layer was then saturated with ammonium sulfate and extracted with methylene chloride (20.0×; 2 ml). The methylene chloride extract was dried using magnesium sulfate, concentrated in vacuo, and purified by flash chromatography (silica gel/ethyl acetate), affording 0.10 g (21% yield) of Z-4,5,9-trithiadodeca-1,6,1l-triene (compound 3) (Z/E > 9/1). 1H NMR (CDCl3) levels of 6.55 (d, J = 9.0 Hz, 1 H), 6.14 to 5.36 (m, 7 H), and 3.62 to 3.23 (m, 6 H), which are in accordance with a previous publication (20), were determined.
Bacterial strains and culture methods.
Five strains of C. sakazakii (ATCC BAA-894, ATCC 12868, ATCC 29004, IQCC 10423, and IQCC 10449) were used in this study. All strains were separately cultivated overnight in 5 ml Luria-Bertani (LB) broth (BD Difco) at 37°C without shaking to achieve a concentration of ca. 9 log CFU/ml, and 1 ml of each bacterial culture was centrifuged at 8,000 × g for 10 min at 4°C. The supernatant was discarded, and the bacterial pellets were washed twice with sterile phosphate-buffered saline (PBS) (pH ∼7.0 to ∼7.2), containing NaCl (9 g), Na2HPO4 (13.76 g), and NaH2PO4 (1.794 g) (per liter of distilled water). Equal volumes of the resultant culture suspensions, each containing one of the five strains, were then combined to form a cocktail, and the cocktail was subsequently added into sterile PBS and tryptic soy broth (TSB; BD Bacto), respectively. An initial concentration of approximately 5 log CFU/ml was achieved by serial dilution in PBS or TSB.
Antibacterial effects of garlic-derived organosulfur compounds on C. sakazakii.
The synthesized Z-ajoene was dissolved in (filter-sterilized) DMSO at the initial molar concentration of 0.2 M and stored at 4°C without light exposure. Different volumes of Z-ajoene and diallyl sulfide were added into the bacterial culture systems, resulting in concentrations of Z-ajoene of 0.0777, 0.777, 3.88, and 7.77 mM and of diallyl sulfide of 0.777, 3.88, 7.77, and 77.7 mM. Control groups included bacterial cultures without addition of organosulfur compounds and bacterial cultures with addition of DMSO (0.55 M) only. Each inoculated sample was mixed well by a vortex procedure and incubated statically at 4, 22, and 37°C for 0, 1, 2, 4, 8, and 12 h (both PBS and broth samples). At each testing time, the bacterial samples were serially diluted in sterile PBS and spirally plated on tryptic soy agar (TSA; BD Difco). After incubation at 37°C for 24 h, viable cells were enumerated. All the tests were repeated in duplicate for one experiment, and the experiments were individually repeated at least three times.
Antibacterial effects of garlic-derived organosulfur compounds supplemented with l-cysteine on C. sakazakii.
The 3.88 mM concentration was selected as the initial concentration of both ajoene and diallyl sulfide in the bacterial culture systems, and equal amounts of l-cysteine were immediately supplemented into the two systems to counteract the effects of the organosulfur compounds. The control groups consisted of bacterial suspensions with equal molar concentrations of DMSO and without any additives. C. sakazakii cultures were incubated at 37°C for 0, 1, 2, 4, 8, and 12 h in PBS. At each sampling time, the cultures were serially diluted with sterile PBS and spiral plated on TSA, and the numbers of viable cells were determined after incubation at 37°C for 24 h. All experiments were repeated in triplicate.
RNA-seq and real-time qPCR.
Triplicate 7-ml samples of C. sakazakii at an optical density at 540 nm of 0.3 (OD540) were treated with 3.88 mM ajoene, 3.88 mM diallyl sulfide, or vehicle (i.e., DMSO) for 30 min at 22°C. The bacterial cultures were centrifuged at 8,000 × g for 5 min at 4°C, and the supernatants were discarded. Total RNA was extracted using an Ambion Bacterial RiboExpress RNA extraction kit (Life Technologies, Grand Island, NY). A portion of the extracted RNA was used to generate cDNA (Thermoscript III; Life Technologies), and quantitative PCR (qPCR) was performed on triplicate cDNA samples using Power SYBR green PCR Master Mix (Applied Biosystems, Warrington, United Kingdom) and an ABI Prism 7000 Fast instrument (Life Technologies). The rRNA in the sample was removed using a Ribo-Zero rRNA removal kit (Gram-negative bacteria) (Epicentre, Madison, WI). The purified mRNA was submitted to the Genomics Core of the Laboratory for Biotechnology and Bioanalysis, at Washington State University. The mRNA was sequenced using an Ion Torrent sequencing system (Life Technologies), and data were compiled and analyzed using CLC genomics workbench software (CLCBio, Cambridge, MA). The analyzed transcriptomes were sorted by false discovery rate (FDR) P values of less than 0.05 with a relative expression change of greater than 2-fold. The identified gene lists were submitted to DAVID for cluster analysis.
Raman instrumentation.
A confocal micro-Raman spectroscopic system was used in this study. This system includes a Raman spectrometer (Renishaw, Gloucestershire, United Kingdom), a Leica microscope (Leica Biosystems, Wetzlar, Germany) and a frequency-doubled, Nd:YAG (λ = 532 nm) laser (Coherent, Santa Clara, CA). The spectrometer has an entrance aperture of 50 μm and a focal length of 300 mm and is equipped with 1,200-line/mm grating. The Rayleigh scattering can be eliminated by the filters, and only the Raman scattered light photons were collected and dispersed by a diffraction grating and finally recorded as a spectrum by a 576-by-384-pixel charge-coupled-device (CCD) array detector. The size of each pixel is 16 by 16 μm. Gold-coated microarray chips covered with C. sakazakii samples left untreated or treated with 3.88 mM garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide) for 30 min at 22°C were mounted on a standard stage of the microscope, which was focused under the collection assembly, and spectra were recorded using a 20× Nikon objective (numerical aperture [NA] = 0.4, working distance [WD] = 3.9 mm) with a wave number range of 4,000 to 100 cm−1 in an extended mode. The measurement was conducted over 60 s of exposure time (3-s integration time, 20 accumulations) for 10 random spots on each sample with ∼2 mW of incident laser power. Prior to acquisition of each Raman spectrum, the bacterial sample was exposed to incident laser illumination for 30 s to reduce fluorescence background and achieve the highest signal-to-noise ratio (SNR). A Thorlabs PM100A optical power meter was used to measure and calibrate the incident laser power. WiRE 3.0 software (Renishaw, Gloucestershire, United Kingdom) was employed for instrumental control and spectral collection. The experiment was conducted in triplicate.
Spectral preprocessing.
The preprocessing of the raw Raman spectra can separate and subsequently eliminate the side effects which may influence the quality of spectroscope-based chemometric models and multivariate analyses. We first conducted a polynomial background fit (45) combined with baseline subtraction using identification and discrimination of minima via adaptive and least-squares thresholding (46) to remove fluorescence background derived from C. sakazakii cells on gold-coated microarray slides, Gaussian noise, white noise, CCD background noise, and cosmic spikes (47–49). Besides fluorescence background, most of the spectral interference is contributed by CCD background noise, which is generated due to the thermal fluctuations on the CCD detector. Spectral binning (2 cm−1) and smoothing (9-point Savitzky-Golay algorithm) were subsequently applied, followed by normalization of the spectra based upon the intensity of the C-H band in the wave numbers of 3,100 to 2,950 cm−1, the indication of the total biomass of C. sakazakii cells. Recent studies conducting Raman spectral normalization of C. jejuni (48) and E. coli and Staphylococcus spp. (50) validated that using this C-H vibrational band as the standard for normalization effectively removed the spectral fluctuation derived from the small focal volume of bacterial cells and yielded the best results for spectral baseline correction.
Spectral reproducibility, sensitivity, and specificity.
Spectral reproducibility from three independent experiments was investigated by calculating the differentiation index (Dy1y2) value using the following equations:
Lower Dy1y2 values indicate better spectral reproducibility, which is a critical parameter to determine intralaboratory reliability and to evaluate the possibility of establishing the chemometric models. In general, Dy1y2 values of less than 1,000 indicate satisfactory spectral reproducibility (24, 48, 49).
Spectral sensitivity and specificity from three independent experiments were determined using the Wards cluster algorithm at the cutoff value established at 99% similarity for the classification model (i.e., principal component analysis [PCA]) (49).
Second-derivative transformation analysis of spectra.
The second-derivative transformed Raman spectrum is the result of applying a derivative transform to the raw Raman spectrum, and this algorithm function may swing with greater amplitude than that of the raw spectrum to magnify the minor spectral variation and separate out overlapping bands (51). In addition, it could be an ideal noise filter because the changes in the spectral baseline have an insignificant impact on second derivatives (49). It was calculated with the following formula (52):
where f(i + g) stands for the sample at index i and gap offset g.
Classification chemometric models.
An unsupervised principal component analysis (PCA) model was conducted to investigate the variations in the samples without a priori knowledge about C. sakazakii with or without treatment of selective garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide). The Mahalanobis distance in the PCA model is defined as the distance between groups in units of within-group standard deviations and was calculated using the following equation (24, 31):
where S is the pooled estimate of the within-group covariance and x1 and x2 are the mean vectors for the two groups.
A naive Bayesian classifier was employed to classify spectrum based upon the features defined on selected principal components (PCs) from PCA. The posterior of a category given features X of an observation is obtained by the Baye's theorem:
Here, p(X|wi) is assumed as a multivariate normal distribution with means and covariance estimated from samples in each category. The mean is estimated as follows:
where Ni represents the sample numbers in wi category and n denotes the numbers of features. The covariance matrix is as follows:
The entries are estimated as follows:
where l indicates distinct samples in the category of wi and l = 0, 1, 2, …, Ni; xlj represents the jth feature of the lth sample in the category of wi; xlk represents the kth feature of the lth sample in the category of wi; and
and
indicate the jth feature and kth feature of the Nj sample in the category of wi, respectively.
The prior of each category is proportional to the number of samples in each category as follows:
where i = 0, 1, 2, … and P(wi) represents the prior probability of the ith category; Ni represents the sample number of the ith category; and N represents the total sample numbers. New observations would be classified as the category with biggest posterior value, which is equivalent to finding the category wi with the biggest hi(X), and the quantity can be calculated robustly as follows:
The application of Bayesian probability to establish and cross-validate the chemometric classification models for vibrational spectroscopy of bacterial planktonic cells (48, 49, 53), Bacillus spores (54), and viruses (55) has been recently introduced, while the currently used Bayesian probability model was realized on the basis of the minimum error rate. The Matlab codes for programming the PCA and Bayesian probability approach to establish chemometric models are available in the supplemental material.
SEM analyses.
Scanning electron microscopy (SEM) was performed to examine morphological changes of C. sakazakii cells before and after treatment with a 3.88 mM concentration of the garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide) in sterilized PBS for 1 h at 22°C. Untreated and treated C. sakazakii cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. C. sakazakii cells were fixed with 2.5% glutaraldehyde overnight at 4°C. The samples were then rinsed twice with 0.1 M phosphate buffer. Postfixation was carried out using 1% osmium tetroxide for 1 h prior to dehydration in an ethanol series (25%, 50%, 70%, 80%, 90% [10 min for each concentration], and 100% ethanol dehydrate [twice for 15 min each time]), and the reaction mixture was freeze-dried in a Christ alpha 1-4 lyophilizer (Christ, Osterode, Germany). The samples were mounted onto SEM stubs and sputter coated with a thin layer of gold. The coated samples were examined under a Leo 1530 Gemini field emission scanning electron microscope using an accelerating voltage of 5 kV (Zeiss, Jena, Germany).
Statistical analysis.
Experiments were performed in at least three replicate trials. The results are expressed as the means of the results of three independent replicates ± the standard deviations. The differences between samples were shown to be significant (P < 0.05) by one-way analysis of variance (ANOVA) using Matlab.
RESULTS
Antibacterial effects of garlic-derived organosulfur compounds on C. sakazakii.
Both ajoene and diallyl sulfide showed obvious antimicrobial effects on C. sakazakii. In general, ajoene has a higher antimicrobial effect than diallyl sulfide (an example is shown in Fig. S2A in the supplemental material). As shown in Table S1 in the supplemental material, the bactericidal effects were enhanced with an increase in molar concentrations of garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide), and the antibacterial effect was also time dependent. When 0.777 mM ajoene was applied to inactivate bacterial cocktails in PBS at 37°C for 8 h, viable cells were not recovered. Along with the increase in the molar concentration of ajoene, the time for bacterial inactivation was reduced significantly (i.e., viable cells were reduced to nondetectable levels at 2 h for 3.88 mM and 1 h for 7.77 mM in PBS at 37°C). There was still a significant bactericidal effect even though the concentration of ajoene dropped to 0.0777 mM (see Table S1A). The bactericidal effect of ajoene at 22°C and 4°C was lower than that at 37°C, indicating that the volatility may contribute to the antimicrobial effect of ajoene. Although lower temperatures (e.g., 4°C) could alter the bactericidal effect of diallyl sulfide, the variation of the effect was not as remarkable as that seen with ajoene (see Table S1B). C. sakazakii cells could be completely inactivated in several minutes by the treatment with 77.7 mM diallyl sulfide at 4, 22, and 37°C.
When nutritional ingredients were present, the bactericidal effects of the garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide) decreased significantly compared to incubation in PBS alone, indicating a protective effect from nutrients in broth medium. This protective effect was more obvious with the treatment using ajoene than with that using diallyl sulfide, resulting in a larger difference in the bacterial survival numbers in the two different matrices (i.e., PBS and TSB). The suppressive (bacteriostatic) effects of garlic-derived organosulfur compounds on a cocktail of C. sakazakii cells in TSB are also shown in Table S1 in the supplemental material, and a representative example is shown in Fig. S2B. C. sakazakii cells were inactivated within the first several hours of treatment with 7.77 mM diallyl sulfide in TSB at 37°C; however, incubation for 8 h allowed trace (i.e., nondetectable [ND; see Table S1 in the supplemental material]) levels of the injured and viable bacterial cells to recover and rebound to a certain high level (∼8 log CFU/ml at 12 h). Previous work has shown that only very high concentrations of allicin prevent bacterial cells from overcoming the inhibition and resuming growth, indicating that they were able to metabolize the allicin to a noninhibitory compound (56). This could be a plausible explanation for our current observation of the recovery of C. sakazakii cells treated by diallyl sulfide, a decomposed derivative of allicin.
Antibacterial effects of garlic-derived organosulfur compounds together with l-cysteine on C. sakazakii.
Equal amounts of l-cysteine was added into the bacterial suspensions treated by garlic-derived organosulfur compounds (i.e., 3.88 mM ajoene and diallyl sulfide) in PBS at 37°C. Due to the addition of l-cysteine, the amounts of ajoene were decreased significantly because the sulfhydryl (−SH) group in l-cysteine is highly reactive with disulfide bonds in ajoene (see Fig. S3 in the supplemental material). Consequently, the majority of the antimicrobial activity of ajoene on C. sakazakii was diminished; however, the antimicrobial effect of diallyl sulfide did not change significantly (P > 0.05) (see Table S2 in the supplemental material).
Transcriptomic responses of C. sakazakii to ajoene and diallyl sulfide.
To better understand the effect of ajoene and diallyl sulfide treatment, we examined changes in gene expression due to sublethal injury. Samples of C. sakazakii were added to TSB containing 3.88 mM ajoene or diallyl sulfide and incubated for 30 min at 22°C. Bacteria treated with vehicle (DMSO) served as a negative control. The RNA was extracted, purified, and depleted of rRNA. Patterns in gene expression were determined using RNA sequencing.
The RNA-seq results provided an adequate depth of genome coverage with an average of 10 reads per gene and a median read number of 437. The RNA isolated and sequenced from untreated bacteria and that from bacteria treated with ajoene and diallyl sulfide produced similar qualities of reads. Collectively, reads from all treatment conditions resulted in detection of all 4,392 chromosomal genes, as well as both plasmids, comprising an additional 165 genes. The accuracy of the sequencing results was validated with selected use of qPCR, targeting groEL, dnaK, oxyR, and ribH. The responses from the individual biological experiments tested by qPCR agreed with the pooled sequencing results (data not shown).
To identify patterns of differential gene regulation, we determined the genes that were differentially regulated at least 1.5-fold with an FDR-corrected P value of less than 0.05 (Table S3). Using DAVID analysis with the default settings, the differentially regulated genes were functionally categorized (57). Annotation clusters with enrichment scores of greater than 2.0 and GO terms with a Benjamini FDR of less than 0.05 are reported (Table 1 and Table 2). Under these conditions, we found that treatment with either ajoene or diallyl sulfide resulted in large changes in gene expression profiles of C. sakazakii (Fig. 1). Treatments using each of the two garlic-derived organosulfur compounds lead to similar directional changes in gene regulation. Ajoene-treated C. sakazakii was found to contain 637 genes with increased expression and 373 downregulated genes. Treatment of C. sakazakii with diallyl sulfide resulted in 1,434 genes with increased expression and 268 genes with decreased expression.
TABLE 1.
Regulation of genes in C. sakazakii treated by ajoene
| Regulation category and annotation cluster | Term and function | Fold enrichment | Benjamini FDR |
|---|---|---|---|
| Upregulation | |||
| 1 (enrichment score, 1.24) | IPR012287; homeodomain related | 2.21 | 1.00 |
| IPR012287; transcription | 1.62 | 0.80 | |
| IPR001647; transcriptional regulator, TetR like, DNA binding, bacterial/archaeal | 2.35 | 1.00 | |
| 2 (enrichment score, 1.18) | IPR002059; cold shock protein, DNA binding | 5.95 | 1.00 |
| IPR019844; cold shock protein conserved site | 5.95 | 1.00 | |
| SM00357; cold shock protein | 3.73 | 0.82 | |
| IPR011129; cold shock protein | 3.72 | 1.00 | |
| PIRSF002599; cold_shock_A | 5.62 | 1.00 | |
| IPR012156; cold shock protein, CspA | 5.58 | 1.00 | |
| IPR012340; Nucleic acid binding, OB-folda | 1.65 | 1.00 | |
| 3 (enrichment score, 1.16) | GO:0042592; homeostatic process | 2.68 | 0.97 |
| GO:0045454; cell redox homeostasis | 3.02 | 0.91 | |
| GO:0019725; cellular homeostasis | 2.39 | 0.98 | |
| GO:0015035; protein disulfide oxidoreductase activity | 3.17 | 1.00 | |
| GO:0016667; oxidoreductase activity, acting on sulfur group of donors | 2.16 | 1.00 | |
| IPR017936; thioredoxin like | 2.97 | 1.00 | |
| GO:0015036; disulfide oxidoreductase activity | 2.64 | 0.99 | |
| IPR012335; thioredoxin fold | 1.73 | 1.00 | |
| Downregulation | |||
| 1 (enrichment score, 5.50) | GO:0050136; NADH dehydrogenase (quinone) activity | 9.99 | 0.00 |
| GO:0003954; NADH dehydrogenase activity | 9.99 | 0.00 | |
| GO:0016655; oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor | 9.99 | 0.00 | |
| GO:0008137; NADH dehydrogenase (ubiquinone) activity | 9.86 | 0.00 | |
| GO:0016651; oxidoreductase activity, acting on NADH or NADPH | 5.38 | 0.00 | |
| GO:0048038; quinone binding | 7.82 | 0.00 | |
| 2 (enrichment score, 5.40) | GO:0001539; ciliary or flagellar motility | 7.94 | 0.00 |
| GO:0051674; localization of cell | 7.94 | 0.00 | |
| GO:0048870; cell motility | 7.94 | 0.00 | |
| GO:0006928; cell motion | 7.58 | 0.00 | |
| GO:0003774; motor activity | 8.23 | 0.00 | |
| GO:0019861; flagellum | 4.94 | 0.00 | |
| GO:0009288; flagellin-based flagellum | 5.62 | 0.00 | |
| GO:0044463; cell projection part | 6.58 | 0.00 | |
| GO:0044460; flagellum part | 6.58 | 0.00 | |
| GO:0044461; flagellin-based flagellum part | 6.58 | 0.00 | |
| GO:0009425; flagellin-based flagellum basal body | 7.80 | 0.00 | |
| GO:0042995; cell projection | 2.68 | 0.00 | |
| 3 (enrichment score, 3.01) | GO:0009425; flagellin-based flagellum basal body | 7.80 | 0.00 |
| GO:0030694; flagellin-based flagellum basal body, rod | 8.78 | 0.05 | |
| 4 (enrichment score, 2.34) | GO:0016661; oxidoreductase activity, acting on other nitrogenous compounds as donors | 7.17 | 0.03 |
| GO:0009325; nitrate reductase complex | 8.78 | 0.05 |
OB-fold, oligonucleotide/oligosaccharide binding-fold.
TABLE 2.
Regulation of genes in C. sakazakii treated by diallyl sulfide
| Regulation category and annotation cluster | Term and function | Fold enrichment | Benjamini FDR |
|---|---|---|---|
| Upregulation | |||
| 1 (enrichment score, 3.67) | GO:0051188; cofactor biosynthetic process | 1.62 | 0.01 |
| GO:0009108; coenzyme biosynthetic process | 1.71 | 0.01 | |
| GO:0042375; quinone cofactor metabolic process | 2.21 | 0.03 | |
| GO:0051186; cofactor metabolic process | 1.39 | 0.05 | |
| 2 (enrichment score, 2.96) | GO:0016051; carbohydrate biosynthetic process | 1.68 | 0.01 |
| GO:0000271; polysaccharide biosynthetic process | 1.73 | 0.01 | |
| GO:0008610; lipid biosynthetic process | 1.69 | 0.01 | |
| GO:0009103; lipopolysaccharide biosynthetic process | 2.05 | 0.05 | |
| GO:0008653; lipopolysaccharide metabolic process | 2.05 | 0.05 | |
| GO:0005976; polysaccharide metabolic process | 1.47 | 0.09 | |
| 3 (enrichment score, 2.83) | GO:0008360; regulation of cell shape | 2.34 | 0.01 |
| GO:0022604; regulation of cell morphogenesis | 2.34 | 0.01 | |
| GO:0000271; polysaccharide biosynthetic process | 1.73 | 0.01 | |
| GO:0009273; peptidoglycan-based cell wall biogenesis | 2.21 | 0.03 | |
| GO:0042546; cell wall biogenesis | 2.21 | 0.03 | |
| GO:0009252; peptidoglycan biosynthetic process | 2.27 | 0.04 | |
| GO:0006024; glycosaminoglycan biosynthetic process | 2.27 | 0.04 | |
| GO:0006023; aminoglycan biosynthetic process | 2.27 | 0.04 | |
| 4 (enrichment score, 2.52) | GO:0042375; quinone cofactor metabolic process | 2.21 | 0.03 |
| GO:0045426; quinone cofactor biosynthetic process | 2.18 | 0.04 | |
| GO:0006743; ubiquinone metabolic process | 2.73 | 0.05 | |
| Downregulation | |||
| 1 (enrichment score, 2.97) | GO:0005198; structural molecule activity | 3.97 | 0.02 |
| GO:0019843; rRNA binding | 6.51 | 0.01 | |
| GO:0003735; structural constituent of ribosome | 4.32 | 0.02 | |
| GO:0043232; intracellular non-membrane-bound organelle | 2.51 | 0.03 | |
| GO:0043228; non-membrane-bound organelle | 2.51 | 0.03 | |
| GO:0003723; RNA binding | 2.89 | 0.10 | |
| GO:0005840; ribosome | 3.15 | 0.04 | |
| GO:0030529; ribonucleoprotein complex | 3.08 | 0.03 | |
| 2 (enrichment score, 2.47) | GO:0042398; cellular amino acid derivative biosynthetic process | 6.87 | 0.16 |
| GO:0042401; biogenic amine biosynthetic process | 9.37 | 0.12 | |
| GO:0009309; amine biosynthetic process | 2.65 | 0.17 | |
| GO:0006576; biogenic amine metabolic process | 7.93 | 0.14 | |
| GO:0006577; betaine metabolic process | 20.61 | 0.25 | |
| GO:0006578; betaine biosynthetic process | 20.61 | 0.25 | |
| GO:0006575; cellular amino acid-derivative metabolic process | 4.12 | 0.31 |
FIG 1.
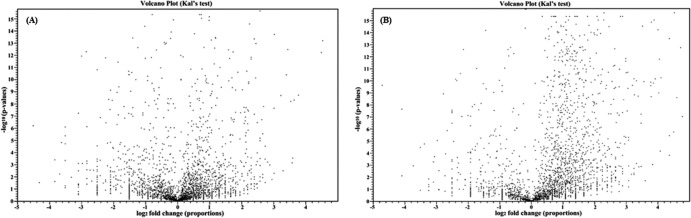
Treatment with either ajoene (A) or diallyl sulfide (B) resulted in a global change in gene expression in C. sakazakii.
Treatment with ajoene resulted in no significant pattern of increased gene expression as judged by Benjamini FDR (Table 1A). The downregulated genes were found to form four annotation clusters with an enrichment score of greater than 2.0. These clusters were NADH expression factors, flagellum and motility proteins, and nitrate reductases (Table 1B). The downregulation of both the nitrate reductases and the NADH expression factors seems to be related to a reaction to oxidative species. The downregulation of the flagellum is important as it contributes to the formation of the extracellular polymeric substances and serves as glue for the formation of biofilm (58).
Diallyl sulfide treatment results in the upregulation of coenzymes and quinones, cell shape and cell wall maintenance pathways, and lipopolysaccharide synthesis (Table 2A). These functional groups are all related to the bacterial membrane and may indicate an attempt to repair those structures. The genes that are downregulated in response to diallyl sulfide are RNA and amino acid biosynthesis biological clusters (Table 2B). This indicates a general decrease in the metabolic processes of this microorganism.
The difference between the two compounds in the patterns of gene expression is informative. Both treatments resulted in the downregulation of a set of 113 common genes, leaving 260 and 155 unique downregulated genes with ajoene and diallyl sulfide treatment, respectively. Further, 443 common genes were upregulated in both ajoene and diallyl sulfide treatments. Ajoene treatment resulted in the upregulation of 194 unique genes, while diallyl sulfide treatment resulted in the upregulation of 991 unique genes. The consistency of the regulation was seen when comparing the upregulated genes of one treatment condition with the downregulated genes of the other. However, 11 genes were upregulated following ajoene treatment and downregulated following diallyl sulfide treatment. Likewise, 55 genes were downregulated following ajoene treatment but upregulated by diallyl sulfide; among those genes are members of the glutamine synthesis pathway. Analysis of the 55 genes using DAVID showed a significant difference in the regulation results for two-component systems following the two treatments. Ajoene treatment resulted in the downregulation of five genes (ESA_01517, ESA_01523, glnG, glnA, and glnL), whereas diallyl sulfide treatment resulted in the upregulation of the same five genes. This indicates that ajoene reduces the ability of the bacteria to detect environmental changes whereas diallyl sulfide increases elements of environmental sensitivity, highlighting the effective differences between the two compounds.
Raman spectroscopy depicted C. sakazakii injury caused by ajoene and diallyl sulfide.
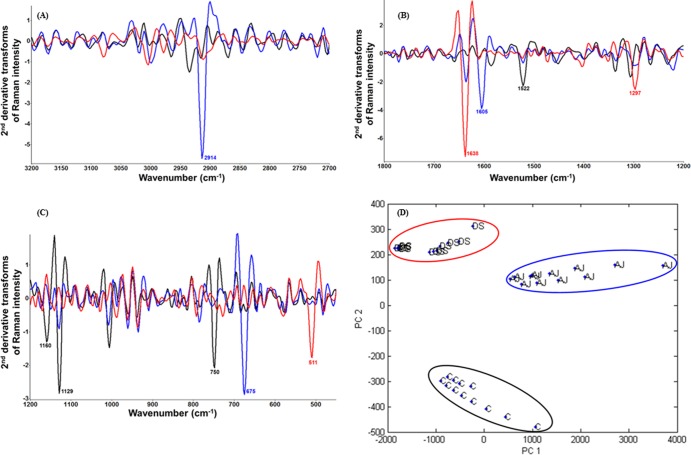
Raman spectra of untreated C. sakazakii and C. sakazakii treated by the same molar concentration (i.e., 3.88 mM) of ajoene and diallyl sulfide were recorded and then converted to the second-derivative transformed patterns. The second-derivative transformed Raman spectral features were compared to study the mechanism of C. sakazakii injury under conditions of unfavorable stress (Fig. 2A, B, and C). The area and height of each second-derivative transformed Raman band were calculated using Matlab, and only the significant (P < 0.05) band variations were characterized and analyzed (Table 3). For diallyl sulfide treatment, the band at 1,638 cm−1 is assigned to amide I (both α-helix and β-sheet) (33) and the band at 1,297 cm−1 is derived from CH2 deformation of fatty acids (59). The band at the wave number of 511 cm−1 originated from S-S disulfide stretching (18, 24, 31). For ajoene treatment, the band at 2,914 cm−1 is from CH of lipids and proteins (59) and the band at 1,605 cm−1 is assigned to phenylalanine, tyrosine, and C = C groups of proteins (60). The band at the wave number of 675 cm−1 is assigned to S-S disulfide stretching and ν(C-S) of amino acid methionine (60). Compared to those seen after the treatments by organosulfur compounds, the spectra derived from untreated C. sakazakii showed significant band variations at the wave numbers of 1,522 cm−1 and 1,160 cm−1 assigned to carotenoids on cell membrane (60), 1,129 cm−1 derived from acyl backbone in lipids (59), and 750 cm−1 assigned to tryptophan (60).
FIG 2.
C. sakazakii injury caused by diallyl sulfide and ajoene. C. sakazakii cells cultured overnight were statically treated with 3.88 mM ajoene (0.77 μl/ml) and 3.88 mM diallyl sulfide (0.5 μl/ml) in tryptic soy broth (TSB) at 22°C for 0.5 h. Panels A, B, and C show second-derivative transformations of Raman spectral features of untreated C. sakazakii (black) and of C. sakazakii treated with diallyl sulfide (red) and ajoene (blue), respectively. (D) Two-dimensional principal component analysis (PCA) for segregation of untreated C. sakazakii (C) and C. sakazakii treated with diallyl sulfide (DS) and ajoene (AJ). PCA was established using wave numbers of 1,800 to 400 cm−1 and 3,200 to 2,700 cm−1.
TABLE 3.
Band assignment of significant (P < 0.05) variations of second-derivative transforms of Raman spectral features of untreated C. sakazakii and C. sakazakii treated with diallyl sulfide and ajoene
| Treatment and wave no. (cm−1) | Assignment |
|---|---|
| Control | |
| 1,522 | Carotenoid peaks due to C-C & conjugated C = C band stretch |
| 1,160 | In-plane vibrations of the conjugated = C-C = β-carotene |
| 1,129 | ν(C-C) skeletal structure of acyl backbone in lipid |
| 750 | Symmetric breathing of tryptophan |
| Diallyl sulfide | |
| 1,638 | Amide I band (both α-helix and β-sheet) |
| 1,297 | CH2 deformation of fatty acids |
| 511 | S-S disulfide stretching band |
| Ajoene | |
| 2,914 | CH band of lipids and proteins |
| 1,605 | Phenylalanine, tyrosine, C = C (protein) |
| 675 | S-S disulfide stretching and ν(C-S) of amino acid methionine |
Therefore, ajoene and diallyl sulfide demonstrated some differences in the antimicrobial mechanisms against C. sakazakii planktonic cells on the basis of biochemical features. Initially, for the first phase, both diallyl sulfide and ajoene bind to thiol-containing proteins/enzymes to generate a disulfide stretching band, resulting in denaturation of proteins/enzymes in bacterial cells. This was in accordance with our previous study results showing that Raman spectroscopy could monitor the transmembrane transfer of sulfur-containing compounds into bacterial cells and that diallyl sulfides could freely penetrate through bacterial cell membrane and selectively bind to thiol-containing proteins, altering their structures (24, 31). Kyung also demonstrated that most sulfur-containing compounds derived from cysteine sulfoxides in Allium species are believed to inhibit microorganisms through an identical mechanism by reacting with the −SH groups of cellular proteins to generate mixed disulfides, as their antimicrobial activities are diminished and finally abolished by continuously adding cysteine, except that seen with allyl alcohol (28). That finding is supported by our current study, which showed that the significant “sulfur” band variations at wave numbers of 511 cm−1 and 675 cm−1 for C. sakazakii treated by diallyl sulfide and ajoene, respectively, and by l-cysteine could significantly reduce or eliminate the antibacterial effect of ajoene on C. sakazakii (see Table S2 in the supplemental material). However, different variations in protein bands of C. sakazakii treated by diallyl sulfide and ajoene indicated different second phases to alter the secondary structures of proteins. Specifically, diallyl sulfide mainly altered the α-helix and β-sheet of amide I (band at 1,638 cm−1) whereas ajoene mainly changed the structures of phenylalanine and tyrosine (band at 1,605 cm−1) (Table 3). Diallyl sulfide is derived from steam-distilled garlic oil, and ajoene originates from oil-macerated/ether-extracted garlic oil, and these two compounds have variations in chemical structures which may be the critical factor contributing to their different antimicrobial mechanisms (in phase II) and effects against bacteria (i.e., C. sakazakii).
The result of the PCA classification determined using the Raman spectral features in the wave numbers of 3,200 to 2,700 cm−1 and 1,800 to 400 cm−1 is shown in Fig. 2D. The spectra collected from the first two experiments were employed to construct the model, and the spectra recorded from the third experiment were used to validate the model. Clear segregation of PCA models indicated a significant (P < 0.05) difference between the groups of samples (i.e., untreated C. sakazakii and C. sakazakii treated with ajoene and diallyl sulfide), with the interclass distance ranging from 4.01 to 12.83 on the basis of Mahalanobis distance measurements calculated between the centroids of classes. Clusters with interclass distance values greater than 3 are considered to be significantly different from each other (24, 55). This PCA cluster model achieved a classification sensitivity of 93.2% and a specificity of 95.1% using Wards cluster algorithm at the cutoff value established at 99% similarity. Bayesian probability was then conducted to compare the first 10 significant features with PCs determined by using PCA. Good agreement in the results of these two methods was noted, indicating the robust nature of the PCA model for classification of different samples on the basis of Raman spectral features.
Electron microscope-examined morphological variations of C. sakazakii treated with ajoene and diallyl sulfide.
To correlate RNA-seq and Raman spectroscopic data with structural changes of C. sakazakii cells caused by treatment with garlic-derived organosulfur compounds, SEM data were collected for untreated and treated samples with 3.88 mM ajoene–PBS or diallyl sulfide–PBS for 1 h at 22°C. Figure S4 in the supplemental material shows that untreated bacterial cells had an intact cellular structure with well-defined membranes. Exposure to ajoene or diallyl sulfide resulted in morphological damage, such as the loss of cell structural integrity, cell deformation, and breakage of cell walls and membranes.
DISCUSSION
Garlic-derived organosulfur compounds, such as thiosulfinates and their derivatives, have been found to have a strong antimicrobial effect on a variety of pathogenic bacteria (28, 61). In the current study, two stable organosulfur compounds derived from decomposed thiosulfinates were tested for their antibacterial effect on C. sakazakii. The purity and stability of ajoene and diallyl sulfide were monitored using reverse-phase high-performance liquid chromatography (HPLC) (24, 25) during the course of the study. Both organosulfur compounds were stable for at least a month when kept in the dark at 4°C without light exposure. Both ajoene and diallyl sulfide have shown significant antimicrobial activity against C. sakazakii, and the antibacterial effect of ajoene was significantly higher than that of diallyl sulfide (see Table S1 and Fig. S2 in the supplemental material). A previous study confirmed that Z-ajoene has higher antimicrobial activity than E-ajoene (19). The MIC value of ajoene for C. sakazakii in the current study was slightly higher than 100 μg/ml, which was similar to the MICs for Gram-negative bacteria reported by Yoshida et al. (22) but higher than the ones for Helicobacter pylori reported by Ohta et al. (19). Naganawa et al. reported that ajoene was more effective in inactivating Gram-positive bacteria than Gram-negative bacteria (26). The antibacterial effects of ajoene on C. sakazakii cells in two different matrices (i.e., broth and PBS) and at different temperatures (i.e., 4, 22, and 37°C) were significantly (P < 0.05) different (see Table S1). This may have been due to the differences in solubility of ajoene in different solvents and at different temperatures affecting the antimicrobial activity.
Diallyl sulfide is present in garlic oil at a concentration ∼112 μg/g (23). In the current study, the MIC value of diallyl sulfide for C. sakazakii was about 500 μg/ml. Ross et al. (62) and O'Gara et al. (63) reported the significant antimicrobial effect of diallyl constituents in garlic oil on a variety of bacteria and also confirmed that the antimicrobial activity of the diallyl sulfides increased with the number of sulfur atoms, which was also recently validated by our group (24). A recent study has shown that the heat-stable components of garlic extract play a major role in the antimicrobial activity of garlic extract and that both diallyl sulfide and ajoene are thermally stable (23). Those authors have confirmed that diallyl sulfide could significantly inhibit the growth of Aggregatibacter actinomycetemcomitans even at a low concentration (i.e., 0.01 μg/ml). Allicin in the garlic extract lost all biological activity after heat treatment, but diallyl sulfide showed no significant difference of the antimicrobial effects before and after heat treatment.
When l-cysteine was added to C. sakazakii cultures with ajoene, the antimicrobial effect of ajoene was abolished rapidly. In contrast, l-cysteine did not affect the antibacterial effect of diallyl sulfide on C. sakazakii (see Table S2 in the supplemental material). This result indicated that the effective functional groups for the antimicrobial activity of ajoene and diallyl sulfide are different. Although diallyl sulfide and ajoene are similar in structure (e.g., consisting of allyl groups), the results suggest that the disulfide bond is important for the antimicrobial activity of ajoene. It used to be believed that thiosulfinates (i.e., ajoene) inhibit microorganisms because of their disulfide bond, which reacts generally with the −SH groups of cellular proteins. For example, Feldberg et al. validated that the in vitro antimicrobial mechanism of allicin was associated with immediate and total inhibition of RNA synthesis and general inhibition of −SH proteins/enzymes (56). When ajoene and l-cysteine exist simultaneously at neutral pH, they react quickly and produce a 1 molar equivalent of S-allyl mercaptocysteine (S-AMC) and E,Z-2-amino-9-oxo-4,5,9-trithaidodeca-6,11-dienoic acid (ajocysteine) (see Fig. S3) (64). This clearly highlighted that the vast majority of antimicrobial properties of ajoene were rapidly eliminated but that diallyl sulfide was not when l-cysteine was present. Cavallito et al. also reported that allicin could react with a −SH group of cysteine and forms S-allylmercaptocysteine (allyl-ss-cysteine) by cleaving the disulfide bond and replacing the allyl group with cysteine (65). l-Cysteine is a critical part of glutathione S-transferase (GST), which is one of the most common mechanisms that bacteria use to antagonize oxidative stresses (23). Jakobsen et al. (25) have found that synthesized ajoene loses activity in in vivo infectious models but that fresh garlic extract showed a much more pronounced effect, which may have been due to the presence of l-cysteine or GST.
Ajoene also contains a sulfinyl group (see Fig. S1B in the supplemental material), which was reported for antibacterial agents such as allicin. Therefore, the antimicrobial activity of ajoene is derived from the presence of both the disulfide bond and the sulfinyl group (26). There was no clear evidence to validate which structures contribute to the antimicrobial activity of diallyl sulfide. The antimicrobial effects of diallyl sulfide in two different matrices and at different temperatures, especially the antimicrobial effects of diallyl sulfide in TSB, were significantly different from those of ajoene, confirming that there were significant differences in the antimicrobial mechanisms of ajoene and diallyl sulfide.
Previous work has revealed that thiosulfinates and diallyl sulfide can easily penetrate the bacterial cell membrane (66) and bind to thiol-containing proteins in bacterial cells and form disulfide bonds (24, 31). An interesting observation in the current study was that both ajoene and diallyl sulfide could bind to −SH in proteins in bacterial cells (described in detail below). These results seemed to be contradictory with respect to the fact that cysteine could react only with ajoene rather than with diallyl sulfide, resulting in the variations in the effects of the antimicrobial activity of ajoene and diallyl sulfide on bacteria with the addition of cysteine. A plausible explanation is that the size of the molecules attached to −SH affects the binding of this functional group with organosulfur compounds to form a disulfide stretching bond.
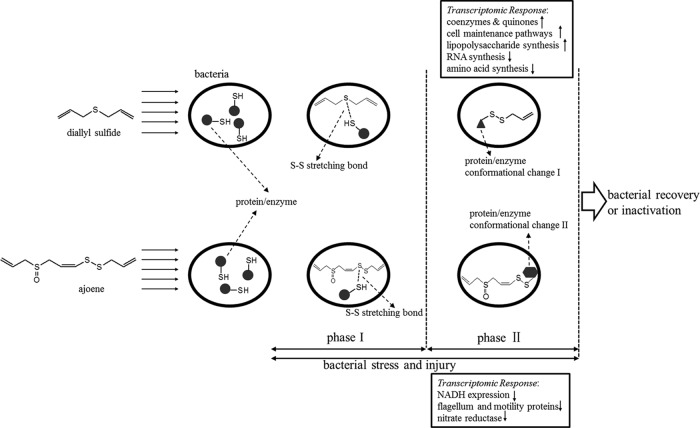
Here we have discussed in detail the mechanism of C. sakazakii stress and injury resulting from treatment with diallyl sulfide and ajoene (Fig. 3). Once the cells were treated with these garlic-derived organosulfur compounds, bacterial stress occurred.
FIG 3.
The mechanism of bacterial stress and injury by the treatment of diallyl sulfide and ajoene.
Although both diallyl sulfide treatment and ajoene treatment resulted in moderate upregulation of gene expression, the profiles were very different. There were fewer upregulated genes in ajoene-treated bacteria (637) than in diallyl sulfide-treated bacteria (1,434). Ajoene-treated bacteria were also found to have no significantly enriched annotation clusters that were upregulated. However, treatment with ajoene resulted in a significant decrease in expression of flagellum-related genes. This may indicate a specific bacterial response to ajoene. Diallyl sulfide is known to cause cell wall disruption (18). Therefore, it was not surprising to find that many of the genes that were upregulated belonged to biological clusters that are involved in cell wall synthesis. The downregulation of housekeeping genes may imply a state of decreased metabolic activity in response to the stress. Taken the results together, ajoene and diallyl sulfide result in very different bacterial responses, suggesting different mechanisms for the garlic-derived compounds.
A critical observation in this study was that two distinct phases of interaction between garlic-derived organosulfur compounds (i.e., ajoene and/or diallyl sulfide) and thiol-containing proteins were illustrated (Fig. 3). By using a confocal Raman microscope, we observed that ajoene and diallyl sulfide could penetrate the bacterial cell membrane and then combine with the −SH group of cellular proteins in bacteria to form an S-S stretching bond (bands at 511 cm−1 and 675 cm−1 in Fig. 2C), indicated as phase I. This subsequently altered the conformational structures of the relative enzymes/proteins (bands at 1,638 cm−1 and 1,605 cm−1 in Fig. 2B), resulting in malfunction, indicated as phase II. Ajoene and diallyl sulfide showed similar phase I results but different phase II results during the interaction with thiol-containing proteins in bacterial cells. Ajoene mainly altered the secondary structural features of phenylalanine and tyrosine (band at 1,605 cm−1), while diallyl sulfide changed the structural features of amide I for both α-helix and β-sheet (band at 1,638 cm−1).
It should be emphasized that the strain whose genome was sequenced (i.e., C. sakazakii ATCC 29004) and used in this study has undergone a series of phenotypic changes over time and may not be completely representative of the wild type. Further investigation using several different C. sakazakii strains is required to further test the generality of the current hypotheses.
In the current study, we systematically investigated bacterial injury using various analytical tools. Both RNA-seq and Raman spectroscopy are high-throughput techniques to independently characterize bacterial stress and injury under unfavorable conditions. Moen et al. used an explorative multifactor approach for investigating global survival mechanisms of C. jejuni under unfavorable environmental conditions (67). In that study, data were generated with DNA microarrays for information about gene expression patterns and with Fourier transform infrared (FT-IR) spectroscopy to study global macromolecular changes in cell. However, FT-IR spectroscopy was applied only to the wave numbers of 1,200 cm−1 to 900 cm−1 to specifically determine the variations in the amounts of polysaccharides and oligosaccharides on C. jejuni cell membranes under the nongrowth survival conditions. In contrast, RNA-seq has a lower detection limit and can be used to analyze the stresses more completely than DNA microarrays. Raman spectroscopy can investigate all the major macromolecular variations in the bacterial cell, such as polysaccharides, proteins, and lipids (Fig. 2 and Table 3). Further, the confocal technique of Raman microscope allows us to clearly demonstrate the disulfide bond formation during the interaction between garlic-derived organosulfur compound and thiol-containing proteins in cells, which could not be monitored using FT-IR spectroscopy. Therefore, complementary RNA-seq and confocal micro-Raman spectroscopy are ideal to study bacterial stress and injury under conditions of unfavorable treatments, such as those using plant-derived bioactive compounds as novel antimicrobials.
In conclusion, we identified two garlic-derived organosulfur compounds (i.e., ajoene and diallyl sulfide) that have significant antimicrobial activity against C. sakazakii. RNA-seq was applied to determine the transcriptomic response of C. sakazakii treated by sublethal concentrations of ajoene and diallyl sulfide. The two compounds differed significantly in the patterns of induced gene expression. Confocal micro-Raman spectroscopy was used to fingerprint macromolecular variations in bacterial cells, and two distinct phases of interaction between organosulfur compounds and thiol-containing proteins in bacterial cells could be monitored. This research demonstrates the feasibility of combining different high-throughput analytical tools to study the mechanism of bacterial stress and injury under unfavorable conditions. Novel antimicrobial agents discovered from plants are potential resources to inactivate pathogens in the environment and on food contact surfaces to reduce food-borne diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds awarded to X.L. by the UBC new faculty start-up grants; funds awarded to S.W. by the Ministry of Science and Technology of China (2012CB720803); and funds awarded to M.E.K. by the National Institutes of Health (R56 AI088518-01A1) and the United States Department of Agriculture, National Institute of Food and Agriculture (2011-67015-30772). T.P.E. was supported by the National Institute of General Medical Sciencesy (T32GM008336).
The content is solely our responsibility and does not necessarily represent the official views of NIH or USDA.
We thank Chunrong Liu and Ming Xian of the Department of Chemistry, Washington State University, for the instruction of synthesis of ajoene and Mingyuan Zhong at Applied Mathematics, University of Washington, for critical discussion about Bayesian's modeling for spectroscopic analysis applied in the current study.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03460-13.
REFERENCES
- 1.Zhou X, Fu S, Gao J, Chen H. 2011Enterobacter sakazakii: an emerging foodborne pathogenic bacterium. Ann. Microbiol. 62:1–5 [Google Scholar]
- 2.Amalaradjou MA, Hoagland TA, Venkitanarayanan K. 2009. Inactivation of Enterobacter sakazakii in reconstituted infant formula by trans-cinnamaldehyde. Int. J. Food Microbiol. 129:146–149. 10.1016/j.ijfoodmicro.2008.11.016 [DOI] [PubMed] [Google Scholar]
- 3.Amalaradjou MA, Venkitanarayanan K. 2011. Effect of trans-cinnamaldehyde on inhibition and inactivation of Cronobacter sakazakii biofilm on abiotic surfaces. J. Food Prot. 74:200–208. 10.4315/0362-028X.JFP-10-296 [DOI] [PubMed] [Google Scholar]
- 4.Chenu JW, Cox JM. 2009. Cronobacter (‘Enterobacter sakazakii'): current status and future prospects. Lett. Appl. Microbiol. 49:153–159. 10.1111/j.1472-765X.2009.02651.x [DOI] [PubMed] [Google Scholar]
- 5.Kim H, Ryu JH, Beuchat LR. 2007. Effectiveness of disinfectants in killing Enterobacter sakazakii in suspension, dried on the surface of stainless steel, and in a biofilm. Appl. Environ. Microbiol. 73:1256–1265. 10.1128/AEM.01766-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedemann M. 2009. Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur. J. Clin. Microbiol. Infect. Dis. 28:1297–1304. 10.1007/s10096-009-0779-4 [DOI] [PubMed] [Google Scholar]
- 7.Bowen AB, Braden CR. 2006. Invasive Enterobacter sakazakii disease in infants. Emerging Infect. Dis. 12:1185–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsythe SJ. 2005. Enterobacter sakazakii and other bacteria in powdered infant milk formula. Matern. Child Nutr. 1:44–50. 10.1111/j.1740-8709.2004.00008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman M, Henika PR, Mandrell RE. 2003. Antibacterial activities of phenolic benzaldehydes and benzoic acids against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 66:1811–1821 [DOI] [PubMed] [Google Scholar]
- 10.De R, Kundu P, Swarnakar S, Ramamurthy T, Chowdhury A, Nair GB, Mukhopadhyay AK. 2009. Antimicrobial activity of curcumin against Helicobacter pylori isolates from India and during infections in mice. Antimicrob. Agents Chemother. 53:1592–1597. 10.1128/AAC.01242-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacombe A, Wu VC, Tyler S, Edwards K. 2010. Antimicrobial action of the American cranberry constituents; phenolics, anthocyanins, and organic acids, against Escherichia coli O157: H7. Int. J. Food Microbiol. 139:102–107. 10.1016/j.ijfoodmicro.2010.01.035 [DOI] [PubMed] [Google Scholar]
- 12.Wright GD, Sutherland AD. 2007. New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 13:260–267. 10.1016/j.molmed.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 13.Erkoç Ş, Sümer S, Erkoç F. 2003. Structural and electronic properties of ajoene molecule. J. Mol. Struct. (Theochem.) 631:271–276. 10.1016/S0166-1280(03)00259-8 [DOI] [Google Scholar]
- 14.Lee DY, Li H, Lim HJ, Lee HJ, Jeon R, Ryu JH. 2012. Anti-inflammatory activity of sulfur-containing compounds from garlic. J. Med. Food 15:992–999. 10.1089/jmf.2012.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corzomartinez M, Corzo N, Villamiel M. 2007. Biological properties of onions and garlic. Trends Food Sci. Technol. 18:609–625. 10.1016/j.tifs.2007.07.011 [DOI] [Google Scholar]
- 16.Ilic D, Nikolic V, Nikolic L, Stankovic M, Stanojevic L, Cakic M. 2011. Allicin and related compounds: Biosynthesis, synthesis and pharmacological activity. F.U. Phys. Chem. Technol. 9:9–20. 10.2298/FUPCT1101009I [DOI] [Google Scholar]
- 17.Kim YS, Kim KS, Han I, Kim MH, Jung MH, Park HK. 2012. Quantitative and qualitative analysis of the antifungal activity of allicin alone and in combination with antifungal drugs. PLoS One 7:e38242. 10.1371/journal.pone.0038242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Samuelson DR, Rasco BA, Konkel ME. 2012. Antimicrobial effect of diallyl sulphide on Campylobacter jejuni biofilms. J. Antimicrob. Chemother. 67:1915–1926. 10.1093/jac/dks138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohta R, Yamada N, Kaneko H, Ishikawa K, Fukuda H, Fujino T, Suzuki A. 1999. In vitro inhibition of the growth of Helicobacter pylori by oil-macerated garlic constituents. Antimicrob. Agents Chemother. 43:1811–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Block E, Ahmad S, Catalfamo JL, Jain MK, Apitz-Castro R. 1986. The chemistry of alkyl thiosulfinate esters. 9. Antithrombotic organosulfur compounds from garlic: structural, mechanistic, and synthetic studies. J. Am. Chem. Soc. 108:7045–7055. 10.1021/ja00282a033 [DOI] [Google Scholar]
- 21.Ishikawa K, Naganawa R, Yoshida H, Iwata N, Fukuda H, Fujino T, Suzuki A. 1996. Antimutagenic effects of ajoene, an organosulfur compound derived from garlic. Biosci. Biotechnol. Biochem. 60:2086–2088. 10.1271/bbb.60.2086 [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Iwata N, Katsuzaki H, Naganawa R, Ishikawa K, Fukuda H, Fujino T, Suzuki A. 1998. Antimicrobial activity of a compound isolated from an oil-macerated garlic extract. Biosci. Biotechnol. Biochem. 62:1014–1017. 10.1271/bbb.62.1014 [DOI] [PubMed] [Google Scholar]
- 23.Velliyagounder K, Ganeshnarayan K, Velusamy SK, Fine DH. 2012. In vitro efficacy of diallyl sulfides against the periodontopathogen Aggregatibacter actinomycetemcomitans. Antimicrob. Agents Chemother. 56:2397–2407. 10.1128/AAC.00020-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Rasco BA, Jabal JM, Aston DE, Lin M, Konkel ME. 2011. Investigating antibacterial effects of garlic (Allium sativum) concentrate and garlic-derived organosulfur compounds on Campylobacter jejuni by using Fourier transform infrared spectroscopy, Raman spectroscopy, and electron microscopy. Appl. Environ. Microbiol. 77:5257–5269. 10.1128/AEM.02845-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, Skindersoe ME, Rasmussen TB, Friedrich K, Uthe F, Jensen PO, Moser C, Nielsen KF, Eberl L, Larsen TO, Tanner D, Hoiby N, Bjarnsholt T, Givskov M. 2012. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 56:2314–2325. 10.1128/AAC.05919-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naganawa R, Iwata N, Ishikawa K, Fukuda H, Fujino T, Suzuki A. 1996. Inhibition of microbial growth by ajoene, a sulfur-containing compound derived from garlic. Appl. Environ. Microbiol. 62:4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cady NC, McKean KA, Behnke J, Kubec R, Mosier AP, Kasper SH, Burz DS, Musah RA. 2012. Inhibition of biofilm formation, quorum sensing and infection in Pseudomonas aeruginosa by natural products-inspired organosulfur compounds. PLoS One 7:e38492. 10.1371/journal.pone.0038492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyung KH. 2012. Antimicrobial properties of Allium species. Curr. Opin. Biotechnol. 23:142–147. 10.1016/j.copbio.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 29.San-Blas G, San-Blas F, Gil F, Marino L, Apitz-Castro R. 1989. Inhibition of growth of the dimorphic fungus Paracoccidioides brasiliensis by ajoene. Antimicrob. Agents Chemother. 33:1641–1644. 10.1128/AAC.33.9.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida S, Kasuga S, Hayashi N, Ushiroguchi T, Matsuura H, Nakagawa S. 1987. Antifungal activity of ajoene derived from garlic. Appl. Environ. Microbiol. 53:615–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X, Rasco BA, Kang DH, Jabal JM, Aston DE, Konkel ME. 2011. Infrared and Raman spectroscopic studies of the antimicrobial effects of garlic concentrates and diallyl constituents on foodborne pathogens. Anal. Chem. 83:4137–4146. 10.1021/ac2001498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida H, Katsuzaki H, Ohta R, Ishikawa K, Fukuda H, Fujino T, Suzuki A. 1999. An organosulfur compound isolated from oil-macerated garlic extract, and its antimicrobial effect. Biosci. Biotechnol. Biochem. 63:588–590. 10.1271/bbb.63.588 [DOI] [PubMed] [Google Scholar]
- 33.Lu X, Al-Qadiri HM, Lin M, Rasco BA. 2011. Application of mid-infrared and Raman spectroscopy to the study of bacteria. Food Bioprocess Technol. 4:919–935. 10.1007/s11947-011-0516-8 [DOI] [Google Scholar]
- 34.Palchaudhuri S, Rehse SJ, Hamasha K, Syed T, Kurtovic E, Kurtovic E, Stenger J. 2011. Raman spectroscopy of xylitol uptake and metabolism in Gram-positive and Gram-negative bacteria. Appl. Environ. Microbiol. 77:131–137. 10.1128/AEM.01458-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, Liu Q, Wu D, Al-Qadiri HM, Al-Alami NI, Kang DH, Shin JH, Tang J, Jabal JM, Aston ED, Rasco BA. 2011. Using of infrared spectroscopy to study the survival and injury of Escherichia coli O157:H7, Campylobacter jejuni and Pseudomonas aeruginosa under cold stress in low nutrient media. Food Microbiol. 28:537–546. 10.1016/j.fm.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 36.Lu X, Weakley AT, Aston DE, Rasco BA, Wang S, Konkel ME. 2012. Examination of nanoparticle inactivation of Campylobacter jejuni biofilms using infrared and Raman spectroscopies. J. Appl. Microbiol. 113:952–963. 10.1111/j.1365-2672.2012.05373.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75:6110–6123. 10.1128/AEM.00914-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Phillippy AM, Deng K, Rui X, Li Z, Tortorello ML, Zhang W. 2010. Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl. Environ. Microbiol. 76:5013–5024. 10.1128/AEM.00823-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570. 10.1128/JB.183.15.4562-4570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng X, Li Z, Zhang W. 2012. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 30:311–315. 10.1016/j.fm.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 41.Croucher NJ, Thomson NR. 2010. Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 13:619–624. 10.1016/j.mib.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, Yu L, Assefa SA, He M, Croucher NJ, Pickard DJ. 2009. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 5:e1000569. 10.1371/journal.pgen.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV. 2012. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 13:R23. 10.1186/gb-2012-13-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. 10.1016/j.chom.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieber CA, Mahadevan-Jansen A. 2003. Automated method for subtraction of fluorescence from biological Raman spectra. Appl. Spectrosc. 57:1363–1367. 10.1366/000370203322554518 [DOI] [PubMed] [Google Scholar]
- 46.Weakley AT, Griffiths PR, Aston DE. 2012. Automatic baseline subtraction of vibrational spectra using minima identification and discrimination via adaptive, least-squares thresholding. Appl. Spectrosc. 66:519–529. 10.1366/110-06526 [DOI] [PubMed] [Google Scholar]
- 47.Bocklitz T, Walter A, Hartmann K, Rosch P, Popp J. 2011. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 704:47–56. 10.1016/j.aca.2011.06.043 [DOI] [PubMed] [Google Scholar]
- 48.Lu X, Huang Q, Miller WG, Aston DE, Xu J, Xue F, Zhang H, Rasco BA, Wang S, Konkel ME. 2012. Comprehensive detection and discrimination of Campylobacter species by use of confocal micro-Raman spectroscopy and multilocus sequence typing. J. Clin. Microbiol. 50:2932–2946. 10.1128/JCM.01144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X, Samuelson DR, Xu Y, Zhang H, Wang S, Rasco BA, Xu J, Konkel ME. 2013. Detecting and tracking nosocomial methicillin-resistant Staphylococcus aureus using a microfluidic SERS biosensor. Anal. Chem. 85:2320–2327. 10.1021/ac303279u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rösch P, Harz M, Schmitt M, Peschke KD, Ronneberger O, Burkhardt H, Motzkus HW, Lankers M, Hofer S, Thiele H, Popp J. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71:1626–1637. 10.1128/AEM.71.3.1626-1637.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu X, Webb M, Talbott M, Van Eenennaam J, Palumbo A, Linares-Casenave J, Doroshov S, Struffenegger P, Rasco B. 2010. Distinguishing ovarian maturity of farmed white sturgeon (Acipenser transmontanus) by Fourier transform infrared spectroscopy: a potential tool for caviar production management. J. Agric. Food Chem. 58:4056–4064. 10.1021/jf9038502 [DOI] [PubMed] [Google Scholar]
- 52.Morrey JR. 1968. On determining spectral peak positions from composite spectra with a digital computer. Anal. Chem. 40:905–914. 10.1021/ac60262a006 [DOI] [Google Scholar]
- 53.Hamel L, Brown CW. 2012. Bayesian probability approach to feature significance for infrared spectra of bacteria. Appl. Spectrosc. 66:48–59 [Google Scholar]
- 54.Correa E, Goodacre R. 2011. A genetic algorithm-Bayesian network approach for the analysis of metabolomics and spectroscopic data: application to the rapid detection of Bacillus spores and identification of Bacillus species. BMC Bioinformatics 12:33. 10.1186/1471-2105-12-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X, Liu Q, Benavides-Montano JA, Nicola AV, Aston DE, Rasco BA, Aguilar HC. 2013. Detection of receptor-induced glycoprotein conformational changes on enveloped virions using confocal micro-Raman spectroscopy. J. Virol. 87:3130–3142. 10.1128/JVI.03220-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldberg R, Chang S, Kotik A, Nadler M, Neuwirth Z, Sundstrom D, Thompson N. 1988. In vitro mechanism of inhibition of bacterial cell growth by allicin. Antimicrob. Agents Chemother. 32:1763–1768. 10.1128/AAC.32.12.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 58.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 59.Naumann D. 2001. FT-infrared and FT-Raman spectroscopy in biomedical research. Appl. Spectrosc. Rev. 36:239–298. 10.1081/ASR-100106157 [DOI] [Google Scholar]
- 60.Movasaghi Z, Rehman S, Rehman IU. 2007. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42:493–541. 10.1080/05704920701551530 [DOI] [Google Scholar]
- 61.Kyung K, Lee Y. 2001. Antimicrobial activities of sulfur compounds derived from S-alk (en) yl-L-cysteine sulfoxides in Allium and Brassica. Food Rev. Int. 17:183–198. 10.1081/FRI-100000268 [DOI] [Google Scholar]
- 62.Ross Z, O'Gara EA, Hill DJ, Sleightholme H, Maslin DJ. 2001. Antimicrobial properties of garlic oil against human enteric bacteria: evaluation of methodologies and comparisons with garlic oil sulfides and garlic powder. Appl. Environ. Microbiol. 67:475–480. 10.1128/AEM.67.1.475-480.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Gara EA, Hill DJ, Maslin DJ. 2000. Activities of garlic oil, garlic powder, and their diallyl constituents against Helicobacter pylori. Appl. Environ. Microbiol. 66:2269–2273. 10.1128/AEM.66.5.2269-2273.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaschula CH, Hunter R, Parker MI. 2010. Garlic-derived anticancer agents: structure and biological activity of ajoene. Biofactors 36:78–85 [DOI] [PubMed] [Google Scholar]
- 65.Cavallito CJ, Buck JS, Suter C. 1944. Allicin, the antibacterial principle of Allium sativum. II. Determination of the chemical structure. J. Am. Chem. Soc. 66:1952–1954 [Google Scholar]
- 66.Miron T, Rabinkov A, Mirelman D, Wilchek M, Weiner L. 2000. The mode of action of allicin: its ready permeability through phospholipid membranes may contribute to its biological activity. Biochim. Biophys. Acta 1463:20–30. 10.1016/S0005-2736(99)00174-1 [DOI] [PubMed] [Google Scholar]
- 67.Moen B, Oust A, Langsrud Ø, Dorrell N, Marsden GL, Hinds J, Kohler A, Wren BW, Rudi K. 2005. Explorative multifactor approach for investigating global survival mechanisms of Campylobacter jejuni under environmental conditions. Appl. Environ. Microbiol. 71:2086–2094. 10.1128/AEM.71.4.2086-2094.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.