Abstract
Salmonella spp. are among the major food-borne pathogens that cause mild diarrhea to severe bacteremia. The use of bacteriophages to control various food-borne pathogens, including Salmonella, has emerged as a promising alternative to traditional chemotherapy. We isolated the Siphoviridae family phage SSU5, which can infect only rough strains of Salmonella. The blocking of SSU5 adsorption by periodate treatment of host Salmonella cells and spotting and adsorption assays with mutants that contain various truncations in their lipopolysaccharide (LPS) cores revealed that the outer core region of the LPS is a receptor of SSU5. SSU5 could infect O-antigen (O-Ag)-deficient Salmonella mutants that developed by challenging of O-Ag-specific phages, and consequently, it delayed the emergence of the phage-resistant Salmonella population in broth culture when treated together with phages using O-Ag as a receptor. Therefore, these results suggested that phage SSU5 would be a promising auxiliary component of a phage cocktail to control rough strains of Salmonella enterica serovar Typhimurium, which might emerge as resistant mutants upon infection by phages using O-Ag as a receptor.
INTRODUCTION
Salmonella infection is considered one of the most important health problems worldwide. Salmonella causes mild diarrhea as well as severe diseases, including typhoid fever, enterocolitis, and bacteremia (1). Worldwide, it is estimated that there are more than 21 million and 5.4 million annual cases of typhoid fever and paratyphoid fever, respectively (2, 3). In addition, approximately 93.8 million cases of nontyphoidal Salmonella gastroenteritis with 155,000 deaths occur annually, and approximately 85% of these cases are food borne (4). Additionally, infants, the elderly, and immunocompromised people could be more at risk of Salmonella infection (http://www.cdc.gov/salmonella/general/technical.html). Recently, this risk has greatly increased with the emergence of a multidrug-resistant (MDR) Salmonella strain. MDR mutants that are frequently isolated from animal-derived foods and humans display resistance to cephamycins and extended-spectrum cephalosporins, as well as tetracycline, chloramphenicol, streptomycin, and sulfisoxazole (5). In addition, the rates and spectra of antimicrobial resistance are increasing among Salmonella isolates. Therefore, alternative treatments for Salmonella are urgently needed to control this food-borne pathogen effectively without concerns regarding the spreading of drug resistance.
Among these alternatives, the use of bacteriophages has emerged as a promising way to control several pathogens. Bacteriophages are viruses that specifically infect and lyse bacterial cells by taking over metabolic machineries of the host in lytic cycle such that phages are considered to possess the capability of serving as antimicrobial agents (6). Phages are ubiquitous in environments and have a long history of safe use (7). In contrast to antibiotics, phages are highly specific to their host; thus, it is believed that phages do not disturb the normal flora. In addition to the specific bacterial killing ability of phages, other special characteristics, such as the ability to self-dose and relatively easy handling, support the benefits of phage applications in various fields (7). Many studies have proven the ability of phages as biocontrol agents to control various food-borne pathogens, including Salmonella. For example, the numbers of Salmonella spp. in experimentally contaminated chicken frankfurters, raw packaged sliced roast beef, and cheddar cheeses were significantly reduced by phage Felix O1, P7, and SJ2 treatments, respectively (8–10).
However, similar to the case with antibiotics, the emergence of resistance in bacteria to bacteriophages was suggested as a drawback of phage applications (11). To overcome this problem, the use of cocktails that consist of various phages that utilize different bacterial receptors has been suggested because this method can delay or prevent the appearance of phage-resistant cells (11–15). Therefore, the isolation of varieties of phages that use diverse receptors is prerequisite for the development of such effective phage cocktails. The host receptors for phages in Gram-negative bacteria include many outer membrane proteins, flagella, pili, and lipopolysaccharides (LPSs) (16). In Salmonella phages, for example, phages bind to various structures on the bacterial surface, such as FhuA for ES18 (17), OmpC for Gifsy-1 and Gifsy-2 (18), BtuB for SPC35 (19), flagella for Φχ (20), and LPSs for several Salmonella phages, including P22, Felix O1, and SPC32H (21–23).
Although diverse receptors have been identified for Salmonella phages, as described above, all of the Salmonella phages that have previously been isolated by our group utilized LPS O antigens (O-Ag), outer membrane protein BtuB, or flagella as their receptors (24). To isolate phages that utilize other receptors, Salmonella enterica serovar Typhimurium mutants that were defective in btuB and/or rfbP were used in this study as host bacteria for phage screenings. As a result, the bacteriophage SSU5, which can infect only rough strains of Salmonella, was isolated and characterized. The receptor for SSU5 was revealed to be the core oligosaccharide (OS) region of the LPS, suggesting that SSU5 may be a valuable auxiliary component of effective phage cocktails for Salmonella biocontrol.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used for the host range test and the receptor determination are listed in Tables 1 and 2, respectively. The prophage-cured Salmonella enterica serovar Typhimurium LT2 strain [designated LT2(c)], which was obtained from the Cancer Research Center, Columbia, MO (25), and its derivatives were used to isolate and propagate bacteriophages. All bacterial strains were aerobically grown at 30°C or 37°C in Luria-Bertani (LB) broth or agar (supplemented with 1.5% agar).
TABLE 1.
Host range of phage SSU5
| Strain | SSU5 plaque formationa |
|---|---|
| Salmonella enterica subsp. enterica | |
| Serovar Typhimurium ATCC 14028s | (+) |
| Serovar Typhimurium NCTC 12023 | (+) |
| Serovar Typhimurium KCTC 1925 | + |
| Serovar Typhimurium ATCC 12023 | (+) |
| Serovar Typhimurium SL1344 | (+) |
| Serovar Typhimurium LT2(c) | (+) |
| Serovar Typhimurium UK1 | (+) |
| Serovar Typhi Ty 2-b | − |
| Serovar Paratyphi A IB 211 | − |
| Serovar Paratyphi B IB 231 | (+) |
| Serovar Paratyphi C IB 216 | (+) |
| Serovar Dublin IB 2973 | (+) |
| Salmonella enterica subsp. arizonae | |
| KCCM 41035 | + |
| KCCM 41036 | + |
| KCCM 41037 | − |
| Salmonella enterica subsp. indica KCCM 41759 | + |
| Salmonella enterica subsp. houtenae KCCM 41760 | + |
| Salmonella enterica subsp. diarizonae ATCC 41761 | (+) |
| Salmonella enterica subsp. salamae KCCM 41762 | (+) |
| Escherichia coli | |
| MG1655 | (+) |
| DH5α | − |
| DH10B | + |
| O157:H7 ATCC 43890 | − |
| Other Gram-negative bacteria | |
| Cronobacter sakazakii ATCC 29544 | + |
| Shigella flexneri 2a strain 2457T | + |
| Shigella boydii 1B 2474 | (+) |
+, clear plaque; (+), bacterial growth inhibition zone; −, no plaque.
TABLE 2.
Investigation of the SSU5 receptor by spotting assay with several Salmonella mutants
| Host strain name or genotype | Defective structure(s) | SSU5 plaque formationa |
|---|---|---|
| LT2(c) | (+) | |
| ΔrfbP | O-Ag | + |
| ΔflgK | Flagella | (+) |
| ΔbtuB | Outer membrane protein BtuB | (+) |
| ΔfhuA | Outer membrane protein FhuA | (+) |
| ΔompC rfbP | Outer membrane protein OmpC, O-Ag | + |
| ΔlamB rfbP | Outer membrane protein LamB, O-Ag | + |
| ΔompF rfbP | Outer membrane protein OmpF, O-Ag | + |
| ΔflgK rfbP | Flagellar, O-Ag | + |
| ΔompA rfbP | Outer membrane protein OmpA, O-Ag | + |
| ΔompW rfbP | Outer membrane protein OmpW, O-Ag | + |
| ΔphoE rfbP | Outer membrane protein PhoE, O-Ag | + |
| ΔrfbP tolC | Outer membrane protein TolC, O-Ag | + |
| ΔbtuB rfbP | Outer membrane protein BtuB, O-Ag | + |
+, clear plaque; (+), bacterial growth inhibition zone.
Bacteriophage isolation, purification, and stock preparation.
Sewage, slurry, soil, and fecal samples for the screening of Salmonella-specific bacteriophages were collected from Seoul, Yangju, and Suwon in South Korea. Twenty-five grams of each solid sample was homogenized in 225 ml of sterile Butterfield's phosphate-buffered dilution water (312.5 μM KH2PO4 adjusted to pH 7.2) in sterile bags. Twenty-five milliliters of each suspension or liquid sample was added to equal volumes of 2× LB broth and incubated for 24 h, with uniform shaking, at 37°C. Chloroform (1%, final concentration) was added to the culture and incubated for an additional 5 min at 37°C, with shaking. After the removal of bacterial cell debris by centrifugation at 9,000 × g at 4°C for 10 min and by filtration using 0.22-μm-pore-size filters (Millipore, Billerica, MA), 10 ml of each filtrate was mixed with 50 ml of LB broth, which was inoculated with an overnight-grown culture of an appropriate host Salmonella strain (1%, final concentration), and then the mixture was incubated at 37°C for 12 h, with shaking. The culture was centrifuged and filter sterilized as described above.
The presence of phages in the filtrates was determined by the standard double-agar overlay method as described previously (19). Briefly, 100-μl filtrates and 5 ml of molten LB soft agar (LB broth supplemented with 0.4% agar) that had been inoculated with 100 μl of host Salmonella culture were mixed and plated on solidified 1.5% LB agar. Each single plaque that formed after an overnight incubation at 37°C was picked with a sterile pipette tip and eluted in SM buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4). The eluate was overlay plated again, and the formed single plaque was picked. This step was repeated at least three times to purify single bacteriophages.
To propagate the purified phages, a culture of an appropriate host Salmonella mutant (optical density at 600 nm [OD600] = ∼0.5) was infected with phages at a multiplicity of infection (MOI) of 1 and incubated at 37°C for 4 h. The cleared bacterial culture was centrifuged (9,000 × g for 10 min) and filtrated (0.22-μm-pore-size filters), and then phage particles in the filtrate were precipitated by mixing with 10% (wt/vol) polyethylene glycol (PEG) 6000 (Junsei) in 1 M sodium chloride (final concentration). Finally, CsCl density gradient ultracentrifugation (himac CP 100β; Hitachi, Japan) with different CsCl steps (step densities = 1.3, 1.45, 1.5, and 1.7 g/ml) at 78,500 × g for 2 h was performed at 4°C. A band of viral particles was recovered, dialyzed using standard dialysis buffer (10 mM NaCl, 10 mM MgCl2, and 50 mM Tris-HCl, adjusted to pH 8.0), and stored at 4°C until further use.
Determination of the bacteriophage host range.
In total, 100 μl of each test bacterial culture was added to 5 ml of molten LB soft agar (0.4% agar), and the mixture was overlay plated on a 1.5% LB agar plate to prepare the bacterial lawn. After solidification, 10-μl volumes of serially diluted (10-fold dilution) SSU5 phage suspensions were spotted, and then the plates were incubated at 37°C. The SSU5 susceptibility of tested bacteria was determined based on the single clear plaque or bacterial growth inhibition zone on the plates.
TEM.
The CsCl-purified, high-titer (more than 1010 PFU/ml) phages in SM buffers were examined by transmission electron microscopy (TEM). Each phage sample was placed on carbon-coated copper grids and negatively stained with 2% aqueous uranyl acetate (pH 4.0) for 1 min. Electroscope microscopy was performed using a transmission electron microscope (LIBRA 120; Carl Zeiss) at 80 kV at the National Academy of Agricultural Science (Suwon, South Korea). Bacteriophages were identified and classified into their relative families according to the guidelines of the International Committee on Taxonomy of Viruses (26) based on the virion morphology.
Construction of Salmonella mutants.
Salmonella strain LT2(c) mutants with deletions of specific genes were constructed by the lambda red recombination system (27). The kanamycin resistance (Kmr) cassette from plasmid pKD13 was amplified using primers that contained 40-nucleotide homologue sequences of each deletion-target gene and 20-nucleotide priming sequences of pKD13. The wild-type (WT) or rfbP gene deletion mutant (28) expressing exo, bet, and gam from plasmid pKD46 was transformed with the resulting PCR product. Finally, the Kmr cassette of the positive transformants was removed by introducing the pCP20 plasmid (27). Specific gene deletions were confirmed by PCR and subsequent DNA sequencing.
Bacteriophage adsorption assay.
A bacteriophage adsorption assay with various S. Typhimurium strains was performed according to a previous study (28), with some modifications. Briefly, the bacterial culture was harvested at an OD600 of 1.0, resuspended, and 10-fold diluted with LB broth. Phage SSU5 was added at an MOI of 0.01, and the adsorption proceeded at 37°C for 15 min. Then, 1-ml samples were collected at 0, 1, 5, 10, and 15 min, centrifuged at 16,000 × g for 1 min, and filtrated using 0.22-μm-pore-size filters (Millipore). The serially diluted filtrates were used to count unabsorbed phages by standard overlay plating using the ΔrfbP mutant as an indicator strain. The SSU5 titer in the control group, which contained bacterial cell-free LB broth, was considered an initial phage titer. The rate of adsorption of SSU5 to each strain was compared by calculating an adsorption constant (k) as follows: k = −ln (Pt/P0)/Nt, where Pt is phage titer at the time t (PFU/ml), P0 is initial phage titer (PFU/ml), N is bacterial cell density (CFU/ml), and t is time (min). The density of bacterial cells used was determined by the direct plate count on LB agar.
Periodate or proteinase K treatments.
The ΔrfbP mutant was treated with periodate or proteinase K to examine the effect of the treatments on SSU5 adsorption, according to methods that were described by Kiljunen et al. (29), with some modifications. Briefly, when the OD600 of the bacterial culture reached 1.0, 1 ml of the culture was collected by centrifugation at 16,000 × g for 1 min and washed with 1 ml of fresh LB broth. Proteinase K (0.2 mg/ml, final concentration) was added to the prepared sample and then incubated at 37°C for 2 h. For periodate treatment, 2 ml of the culture was harvested by centrifugation and washed with 1 ml of LB broth. The pellet was then treated with 1 ml of sodium acetate (50 mM, adjusted to pH 5.2) or sodium acetate containing either 10 or 100 mM periodate for 2 h in the dark. After the treatments, cells were washed at least three times with 1 ml of LB broth, which was adjusted to an OD600 of 0.1, and then the phage adsorption assay was performed as described above.
Bacterial challenge assay.
LB broth (50 ml), which was inoculated with 1% bacterial overnight culture (final concentration), was incubated aerobically at 37°C with constant shaking. When the OD600 reached ∼0.4, phages were added at an MOI of 0.1. During further incubation under the same conditions, the culture samples were collected every hour to monitor the bacterial growth by measuring the OD600. Instead of phage, SM buffer was added to the culture for a negative control. Experiments were repeated three times.
Isolation of S. Typhimurium mutants resistant to O-Ag-specific phages.
To isolate the phage-resistant Salmonella mutants, cultures of LT2(c) in the early log phase (OD600 = ∼0.5) were infected with Salmonella O-Ag-specific phage P22H5 or SSU14 at an MOI of 1 and then further incubated at 37°C for 24 h, with constant shaking. A loopful (10 μl) of phage-challenged culture was streaked on a fresh LB plate and incubated overnight at 37°C. To purify the phage-resistant colonies, a single colony was sequentially streaked on fresh LB plates at least 3 times. The putative phage-resistant colonies were subjected to the spotting assay to confirm the resistance to the corresponding phages and the susceptibility to SSU5.
Extraction and analysis of LPSs from phage-resistant S. Typhimurium mutants.
LPSs of P22H5- or SSU14-resistant S. Typhimurium mutants were extracted by the modified phenol-water extraction method (28). The extracted LPSs were separated on a 15% acrylamide slab gel by deoxycholate-acrylamide gel electrophoresis (DOC-PAGE) as described by Kim and Ryu (28). LPSs that were extracted from WT S. Typhimurium, as well as a purchased S. Typhimurium LPS (Sigma; catalogue no. L6511), were also electrophoresed as positive controls. The gel was fluorescently stained using a Pro-Q Emerald 300 lipopolysaccharide gel stain kit (catalogue no. P20495; Molecular Probes, Eugene, OR) according to the manufacturer's instructions and then visualized under UV light using a Red imaging system (Alpha Innotech Corporation, San Leandro, CA).
RESULTS
Isolation of phage SSU5.
In this study, we attempted to broaden the receptor spectrum of Salmonella bacteriophages because we isolated phages that use only three different types of receptors, such as O-Ag, BtuB, and flagella (24). Using the prophage-cured S. Typhimurium LT2 strain with either btuB or btuB and rfbP deletion mutations as host bacteria, 13 bacteriophages were isolated. The spotting assay for the rfbP, btuB, or flgK deletion mutants that were defective in O-Ag, BtuB, or flagella, respectively, revealed that five of the isolated phages utilized O-Ag and that seven of these phages utilized flagella as a receptor. Interestingly, one of these phages, named SSU5, formed clear plaques on the rfbP deletion strain but not on the btuB or flgK deletion strain as well as on the WT strain. Therefore, SSU5 was further characterized.
SSU5 morphology and host range.
TEM images of SSU5 revealed that this phage has an icosahedral head and noncontractile flexible tail, which indicated that SSU5 belongs to the family Siphoviridae (Fig. 1). The mean diameter of the head and the length of the tail were approximately 70 nm and 220 nm, respectively.
FIG 1.
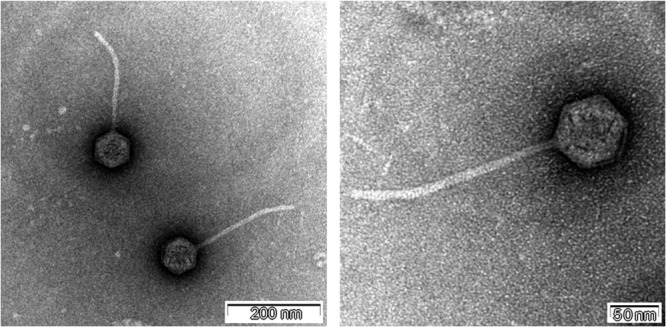
Transmission electron microscopy images of phage SSU5. SSU5 was negatively stained with 2% uranyl acetate and observed by TEM. The scale bar is at the bottom right corner of each image.
SSU5 exhibited specific growth inhibition against several Gram-negative bacteria (Table 1). This phage produced clear plaques in five Salmonella strains tested and a zone of growth inhibition in other Salmonella strains. Interestingly, Cronobacter sakazakii ATCC 29544, Shigella flexineri 2a strain 2457T, and Escherichia coli DH10B were also infected by SSU5, which suggested that an unknown common receptor for SSU5 might be shared among the bacteria that belong to the family Enterobacteriaceae.
Identification of the phage SSU5 receptor.
To clarify whether the receptor for SSU5 is a protein or carbohydrate, an SSU5 adsorption assay was performed after periodate or proteinase K treatment of the ΔrfbP mutant. Periodate degrades carbohydrate structures containing a 1,2-diol motif, such as oligosaccharides, whereas proteinase K can digest cell surface proteins, such as outer membrane proteins. The measurement of residual phage particles in the supernatant after phage adsorption indicated that the adsorption of SSU5 by Salmonella was significantly inhibited by the treatment of 10 or 100 mM periodate but not by proteinase K (Fig. 2). We also tested the SSU5 susceptibility of ΔrfbP mutants with various deletions in the genes associated with the synthesis of several outer membrane proteins that are known to be used as phage receptors; however, none of these mutants showed resistance (Table 2). These results indicate that the receptor for SSU5 may be the carbohydrates that are accessible by the phages only when O-Ag is not made. We speculate that the most probable receptor is a core oligosaccharide (OS); therefore, the effects of various mutations in genes involved in core OS biosynthesis on SSU5 infection were compared.
FIG 2.
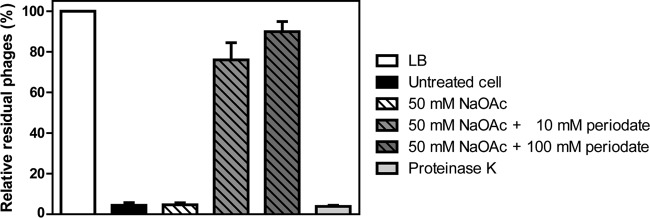
Effect of periodate and proteinase K treatments on SSU5 adsorption. An SSU5 adsorption assay was performed with the periodate- or proteinase K-treated ΔrfbP mutant strain. The untreated ΔrfbP cells in LB broth were used as the control (untreated cells). An SSU5 titer in cell-free LB broth was considered the 100% control (LB), and the residual phage titer in each sample after 15 min of adsorption at 37°C is represented by relative percentages. The means with SDs for three independent experiments are shown.
Effects of various core OS truncations on SSU5 adsorption.
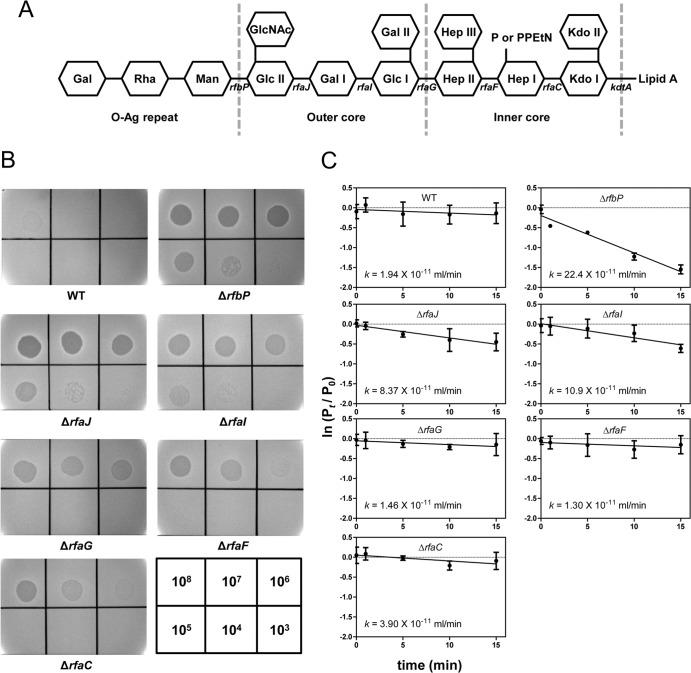
Mutants that had deletions in various genes involved in core OS biosynthesis, such as rfaJ, rfaI, rfaG, rfaF, or rfaC, were constructed and used to identify which moiety on the core OS is involved in the SSU5 adsorption. The core OS structures that resulted from each mutation are shown in Fig. 3A. Spotting assays with these mutant strains showed that SSU5 infectivity was decreased as the core OS was shortened (Fig. 3B). In particular, the efficiency of plating (EOP) of ΔrfaG, ΔrfaF, and ΔrfaC strains was significantly reduced compared with that of the ΔrfbP strain. Adsorption assays with the WT and with these mutants also revealed that ΔrfaG, ΔrfaF, and ΔrfaC mutants, as well as WT strain, exhibited significantly low SSU5 adsorption rates compared with that of the ΔrfbP mutant (P > 0.01; two-tailed t test [Fig. 3C]). Notably, the rough mutants that lost their entire outer core of LPSs showed the lowest adsorption constants, which were not significantly different from that of the WT. These results strongly suggest that the core OS, especially the outer core region, is important for SSU5 adsorption.
FIG 3.
SSU5 adsorbed to the outer core OS of Salmonella LPS. (A) Schematic representation of the LPS structure of S. Typhimurium. Carbohydrates are shown as hexagons, and the relevant genes involved in the biosynthesis of various residues of LPS are indicated below the line connecting the hexagon. The mutation of each gene caused the elimination of the left residues marked with gene names on the structure (rfbP, undecaprenyl-phosphate galactosephosphotransferase; rfaJ, LPS 1,2-glycosyltransferase; rfaI, LPS 1,3-galactosyltransferase; rfaG, glucosyltransferase I; rfaF, ADP-heptose-LPS heptosyltransferase; rfaC, LPS heptosyltransferase I; kdtA, 3-deoxy-d-manno-octulosonic acid transferase). Abbreviations: Kdo, 3-deoxy-d-manno-octulosonic acid; Hep, heptose; Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; Rha, rhamnose; P, phosphate; PPEtN, pyrophosphorylethanolamine. (B) SSU5 spotting assay with mutants with truncations in the LPS core OS. Serially diluted (10-fold) SSU5 lysates were spotted on the lawns of indicated Salmonella mutants. The phage titer (PFU/ml) used for each spot is indicated in the grids at the bottom right corner. (C) Assay of adsorption of SSU5 to various core OS mutants. The adsorption constant (k) was calculated as described in the text. The means with SD of three independent assays are represented.
SSU5 infection of the mutants resistant to O-Ag-specific phages.
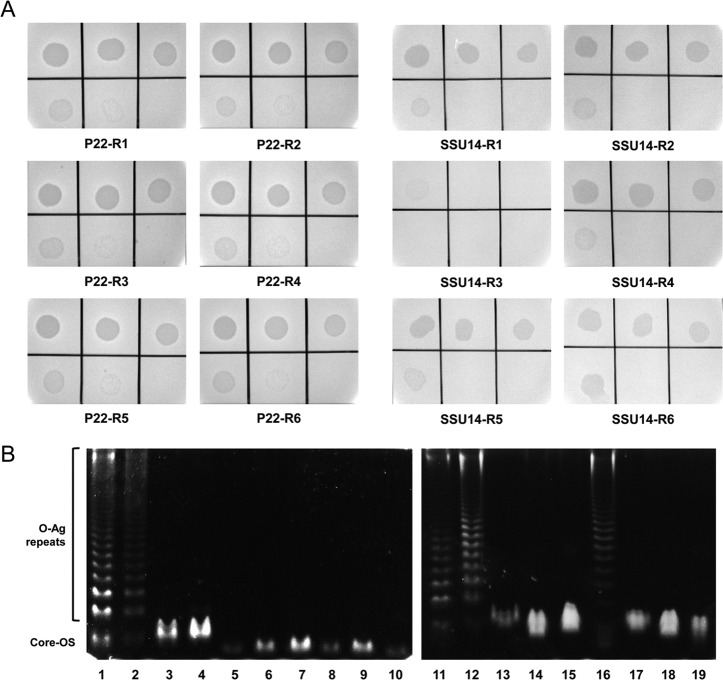
To avoid phage infections, bacteria have developed several types of phage defense mechanisms (30, 31). These include the loss or alteration of phage receptors on the bacterial cell surface (19, 28), superinfection exclusion by using homoimmune prophage repressors or specific superinfection exclusion proteins (e.g., SieA/B of P22) (32, 33), a restriction/modification system (34), abortive infection (35), and the CRISPR/Cas system (36). Because phage adsorption is the primary initial step for successful phage infection, many bacteria evade phage attachment by changing phage receptors on their cell surface. For example, infection with O-Ag-specific phages would result in the emergence of O-Ag-deficient resistant mutants. Because SSU5 recognized the core OS, we postulated that SSU5 could infect mutants that are resistant to O-Ag-specific phages, such as Salmonella phage P22H5 (a c2 mutant of P22) or phage SSU14, which was isolated along with SSU5 in the present study. To test this hypothesis, P22H5- or SSU14-resistant Salmonella mutants were isolated from independent challenges of WT LT2(c) with each phage (see Materials and Methods). Six mutants resistant to each phage were isolated, and their susceptibility to SSU5 was tested. As expected, all the isolated P22H5- or SSU14-resistant mutants, except one (SSU14-R3), were infected by SSU5, with significant EOPs compared with that of the WT strain (Fig. 4A).
FIG 4.
SSU5 infects Salmonella mutants that lost their O-Ag and are resistant to O-Ag-specific phages. (A) Serially diluted SSU5 lysate was spotted onto the lawns of isolated P22H5- or SSU14-resistant mutants (indicated as P22-R[number] or SSU14-R[number], respectively). Phage titers used are the same as for Fig. 3B. (B) DOC-PAGE of LPS extracted from P22H5- or SSU14-resistant mutants. LPSs that were extracted by the hot-phenol extraction method were electrophoresed on a 15% polyacrylamide slab gel. Gels were stained using the Pro-Q Emerald 300 lipopolysaccharide gel stain kit prior to visualization under UV light. Lanes 1 and 11, Salmonella LPS standard (purchased from Sigma); lanes 2 and 12, WT LT2(c); lane 3, S. Typhimurium KCTC 1925 (see Table 1); lanes 4 and 13, ΔrfbP; lanes 5 to 10, P22-R1 to -R6 in panel A; lanes 14 to 19, SSU14-R1 to -R6 in panel A.
Because the LPS structure is a main determinant in LPS-specific phage infection, LPSs were extracted from the mutants and analyzed by deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE). Remarkably, all the SSU5-susceptible mutants had lost their O-Ag. As shown in Fig. 4B, the WT LT2(c) strain had a complete LPS structure (ladder of various lengths of O-Ag with a core OS), which was similar to the commercial Salmonella LPS standard, whereas the ΔrfbP mutant and S. Typhimurium KCTC 1925, which are susceptible to SSU5 (Table 1), possessed only the core OS with no O-Ag repeats. These results indicate that SSU5 can recognize the exposed core OS of the resistant Salmonella mutants against O-Ag-specific phages. The only exception was mutant SSU14-R3, which possessed a complete LPS similar to WT (Fig. 4B), such that SSU5 might not be able to infect this mutant.
Bacterial challenge assay with phage cocktail.
We speculated that SSU5 could be a promising auxiliary biocontrol agent against Salmonella if this phage is used with other phages that utilize O-Ag as a receptor because of the ability of SSU5 to infect O-Ag-deficient Salmonella. SSU5 would retard or restrict the growth of Salmonella mutants that developed resistance to the O-Ag-specific phages. To verify this assumption, a phage cocktail, which consisted of SSU5 and SSU14, was tested in the bacterial challenge assay. Each phage or a cocktail of both phages was added to the S. Typhimurium LT2(c) culture at an MOI of 1, and the bacterial growth was periodically monitored by measuring the optical density at 600 nm. As expected, SSU5 did not affect the growth of WT LT2(c); however, SSU14 significantly inhibited the growth of Salmonella up to 7 h after phage infection (Fig. 5). The phage cocktail treatment further delayed the emergence and growth of a phage-resistant bacterial mutant compared with the SSU14 treatment, suggesting that SSU5 may have potential as an auxiliary phage cocktail component to more efficiently control Salmonella.
FIG 5.
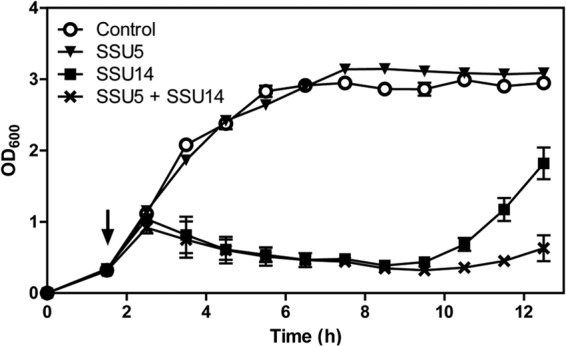
Bacterial challenge assay with SSU5, SSU14, or the phage cocktail, which consisted of both phages. WT LT2(c) cultures at the early exponential growth phase were infected by phages at an MOI of 1 (black arrow), and the OD600 was measured every hour to monitor the bacterial growth. Instead of phages lysate, SM buffer was added to the LT2(c) culture for use as a negative control. The means with SD of three independent experiments are shown.
DISCUSSION
In this study, phage SSU5, which does not utilize the bacterial O-Ag, BtuB, or flagella as a receptor, was isolated from a sewage sample using Salmonella mutants defective in BtuB and/or O-Ag as the host bacteria in the phage screening. Interestingly, SSU5 formed a bacterial growth inhibition zone on WT Salmonella strains but formed clear plaques on mutants that contained a ΔrfbP mutation (Table 2), suggesting that the host receptor for SSU5 is blocked by O-Ag. For some E. coli-specific phages, it was reported that long O-Ag chains of E. coli could be a barrier of accession to outer membrane protein (Omp) receptors (37). However, SSU5 did infect all of the potential Omp receptor-deficient mutants tested on the ΔrfbP background (Table 2), implying that a protein structure might not be utilized by SSU5 as the receptor. Indeed, the adsorption assay with periodate- or proteinase K-treated cells revealed that the SSU5 receptor is a carbohydrate moiety, potentially a part of the LPS, rather than a protein structure (Fig. 2). Considering the inability of SSU5 to infect WT Salmonella that possesses the intact S (smooth)-type LPS (Table 2 and Fig. 3), the inner part of the LPS that was masked by O-Ag was considered the receptor for SSU5. The spotting and adsorption assays demonstrated that the outer core region is the most important part for SSU5 adsorption because deep-rough mutants that possessed the inner core OS only exhibited a lower adsorption constant than that of the ΔrfbP mutant (Fig. 3C). This result was strikingly contrasted by that obtained with another core LPS-specific Yersinia phage, ΦA1122, which showed a higher adsorption constant when some distal residues of the core OS were removed (29), implying diversity in the recognition of bacterial OS moieties by phages.
Understanding the phage's host receptor is important for successful phage applications because the host receptor and the corresponding receptor binding protein (RBP) of phages are the main determinants for the phage's specificity. In addition, analysis of the host receptor is helpful for the preparation of an effective phage cocktail. One of the major concerns in phage applications is the emergence of phage-resistant mutants, which usually are developed by the prevention of phage adsorption through modifying or losing receptors, or through producing extracellular matrix or competitive inhibitors to mask or inhibit the receptor (30, 38). These changes, however, hardly occur simultaneously with all of the putative host receptors on the cell surface, such that a treatment of a phage cocktail consisting of various phages targeting different receptors was proposed as the effective biocontrol strategy (11–15, 39). Each phage in the cocktail could inhibit growth of mutants resistant to other phages, leading to more efficient control of target bacteria. Indeed, a phage cocktail that consisted of three different phages (GH-K1, GH-K2, and GH-K3) exhibited successful control of Klebsiella pneumoniae, with a significantly reduced mutation frequency in this pathogen (40), where each phage was isolated by a “step-by-step” approach that sequentially used a specific phage-resistant mutant as the host bacterium during the isolation of another phage(s).
Rough-specific phages, which could provide a broader host range than O-Ag-specific phages, are also feasible as a component of phage cocktails. In many Gram-negative bacteria, the O-Ag serotypes are highly diverse, whereas that of the core OS is relatively well conserved. E. coli and S. enterica show more than 170 O serotypes and 46 serogroups with modifications, respectively; however, only five core types are exhibited by E. coli, and two of these types resemble the core types of Salmonella (41). Therefore, one of the conserved core OS structures among genera in the family Enterobacteriaceae, including E. coli, Salmonella spp., Cronobacter spp., and Shigella spp., might allow SSU5 to infect C. sakazakii ATCC 29544 and S. flexneri 2a strain 2457T, as well as Salmonella (Table 2), suggesting the possibility of SSU5 controlling various pathogens other than Salmonella. The broad host spectrum of SSU5 was also supported by the fact that the SSU5 genome highly resembles the cryptic plasmids pHCM2 and pFra from S. Typhi CT18 and Yersinia pestis, respectively (42, 43).
In accordance with the spotting and adsorption assays, SSU5 could not inhibit the growth of WT LT2(c) in broth culture (Fig. 5), and thus, SSU5 cannot be solely used as a biocontrol agent. However, this phage can infect the Salmonella mutants resistant to O-Ag-specific phage P22H5 or SSU14 that were defective in O-Ag (Fig. 4A and B). The bacterial challenge assay with the phage cocktail consisting of SSU5 and SSU14 also revealed the ability of SSU5 to inhibit the growth of SSU14-resistant mutants (Fig. 5); SSU5 delayed the emergence of phage-resistant mutants and prolonged the period of growth inhibition.
LPS-defective Gram-negative bacteria can be brought to emergence by phage treatments or phase variations. Vibrio cholerae phage K139, which used the O1 antigen as a receptor, selected the phage-resistant mutants that were defective in the biosynthesis of the O1 antigen or core OS (44), and the treatment of three virulent S. Enteritidis phages generated the phage-resistant variants with rough phenotypes (45). As an adaptive method to maintain an in vivo population, yet not enough to cause diseases in the host, S. Typhimurium 798 exhibited two phenotypes by phase switching: a nonadhesive phenotype containing short O-Ag and an adhesive phenotype possessing long O-Ag (46). The recently revealed phase-variable expression of STM2209-STM2208 genes in S. enterica, which was responsible for the phase-variable alteration of O-Ag chain length, might be involved in this phenotype switching (47). Notably, clones with short O-Ag by the phase variation of STM2209-STM2208 genes were resistant to the O-Ag-specific phage P22. All of these R-type pathogens can be eliminated using R-type-specific phages, such as SSU5, and thus, these phages would be an auxiliary component of an effective phage cocktail.
One unexpected result in the present study was the isolation of an SSU14-resistant mutant that was simultaneously resistant to SSU5 (Fig. 4). This mutant, named SSU14-R3, contained a complete LPS structure similar to that of WT Salmonella. One possible explanation for SSU14 resistance in this mutant is some modifications in O-Ag, such as the phase-variable glucosylation of galactose residues in O-Ag by the LT2gtrABC1 glucosyltransferase gene cluster (28), such that SSU14, as well as SSU5, could not infect the mutant. However, SSU14 was capable of infecting Salmonella possessing the α-1,4-glucosylated O-Ag (data not shown), suggesting that an unknown genetic mutation(s) and/or physiological modification(s), that has to be revealed with further study, might have occurred in SSU14-R3. A regrown population in the phage cocktail-challenged culture at the late period (Fig. 5) might also have originated from this type of resistance mechanism(s). Therefore, the addition of phages that recognize host receptors other than O-Ag into phage cocktails might be necessary for more successful control of Salmonella.
It is generally considered that phages used for biocontrol or therapeutic agents should be virulent (48). Temperate phages, which are able to lysogenize the host bacteria, have the possibility of increasing virulence of the target bacteria through lysogenic conversion. In addition, the risk of horizontal gene transfer by phages (i.e., transduction) also has to be considered for practical phage applications: some phages have an ability to transfer the undesirable virulence or antibiotic resistance genes from target pathogens to other susceptible bacteria and subsequently influence pathogen evolution (49). However, the formation of clear plaques, indicative of virulent phages (50), suggested that SSU5 might be the virulent phage. A previous study (42) of the SSU5 genome also indicated that SSU5 might have virulent characteristics because it lacks the integrase gene. In addition, genes associated with potential virulence or antibiotic resistance were not found in the SSU5 genome. Furthermore, SSU5 failed to transduce the Kmr cassette from the Salmonella ΔrfbP btuB::Kmr strain to the recipient ΔrfbP strain under laboratory conditions (data not shown). Therefore, it appears that SSU5 hardly lysogenizes the target Salmonella and cannot transfer genes to other bacteria, suggesting that SSU5 would not cause undesirable bacterial evolution due to lysogenic conversion or transduction.
In conclusion, the newly isolated Salmonella phage SSU5 recognized the core LPS, specifically the outer core OS, for adsorption, providing SSU5 infectivity against rough strains. Consistent with this finding, SSU5 can eliminate most of the emerged mutants resistant to O-Ag-specific phages that exhibited rough phenotypes. Therefore, in spite of their individual limitations, SSU5 and other similar rough-specific phages would be useful as auxiliary components of phage cocktails with O-Ag-specific phages.
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (no. 20090078983) and “Cooperative Research Program for Agriculture Science & Technology Development (project no. PJ009842),” Rural Development Administration, Republic of Korea.
Footnotes
Published ahead of print 22 November 2013
REFERENCES
- 1.Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85:112–118. 10.1038/sj.icb.7100007 [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull. World Health Org. 82:346–353 [PMC free article] [PubMed] [Google Scholar]
- 3.Crump JA, Mintz ED. 2010. Global trends in typhoid and paratyphoid Fever. Clin. Infect. Dis. 50:241–246. 10.1086/649541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness' Studies 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 5.Winokur PL, Brueggemann A, DeSalvo DL, Hoffmann L, Apley MD, Uhlenhopp EK, Pfaller MA, Doern GV. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777–2783. 10.1128/AAC.44.10.2777-2783.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García P, Martinez B, Obeso JM, Rodriguez A. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479–485. 10.1111/j.1472-765X.2008.02458.x [DOI] [PubMed] [Google Scholar]
- 8.Bigwood T, Hudson JA, Billington C, Carey-Smith GV, Hememann JA. 2008. Phage inactivation of foodborne pathogens on cooked and raw meat. Food Microbiol. 25:400–406. 10.1016/j.fm.2007.11.003 [DOI] [PubMed] [Google Scholar]
- 9.Modi R, Hirvi Y, Hill A, Griffiths MW. 2001. Effect of phage on survival of Salmonella Enteritidis during manufacture and storage of cheddar cheese made from raw and pasteurized milk. J. Food Prot. 64:927–933 [DOI] [PubMed] [Google Scholar]
- 10.Whichard JM, Sriranganathan N, Pierson FW. 2003. Suppression of Salmonella growth by wild-type and large-plaque variants of bacteriophage Felix O1 in liquid culture and on chicken frankfurters. J. Food Prot. 66:220–225 [DOI] [PubMed] [Google Scholar]
- 11.Levin BR, Bull JJ. 2004. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2:166–173. 10.1038/nrmicro822 [DOI] [PubMed] [Google Scholar]
- 12.Gill JJ, Hyman P. 2010. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 11:2–14. 10.2174/138920110790725311 [DOI] [PubMed] [Google Scholar]
- 13.Goodridge LD. 2010. Designing phage therapeutics. Curr. Pharm. Biotechnol. 11:15–27. 10.2174/138920110790725348 [DOI] [PubMed] [Google Scholar]
- 14.O'Flynn G, Ross RP, Fitzgerald GF, Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417–3424. 10.1128/AEM.70.6.3417-3424.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Kraft BL, Pan Y, Wall SK, Saez AC, Ebner PD. 2010. Development of an anti-Salmonella phage cocktail with increased host range. Foodborne Pathog. Dis. 7:1415–1419. 10.1089/fpd.2010.0621 [DOI] [PubMed] [Google Scholar]
- 16.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59:145–155 [PubMed] [Google Scholar]
- 17.Killmann H, Braun M, Herrmann C, Braun V. 2001. FhuA barrel-cork hybrids are active transporters and receptors. J. Bacteriol. 183:3476–3487. 10.1128/JB.183.11.3476-3487.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho TD, Slauch JM. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495–1498. 10.1128/JB.183.4.1495-1498.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050. 10.1128/AEM.02504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meynell EW. 1961. A phage, φχ, which attacks motile bacteria. J. Gen. Microbiol. 25:253–290. 10.1099/00221287-25-2-253 [DOI] [PubMed] [Google Scholar]
- 21.Hudson HP, Lindberg AA, Stocker BAD. 1978. Lipopolysaccharide core defects in Salmonella Typhimurium mutants which are resistant to Felix O-phage but retain smooth character. J. Gen. Microbiol. 109:97–112. 10.1099/00221287-109-1-97 [DOI] [PubMed] [Google Scholar]
- 22.Kim M, Ryu S. 2013. Antirepression system associated with the life cycle switch in the temperate Podoviridae phage SPC32H. J. Virol. 87:11775–11786. 10.1128/JVI.02173-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinbacher S, Miller S, Baxa U, Weintraub A, Seckler R. 1997. Interaction of Salmonella phage P22 with its O-antigen receptor studied by X-ray crystallography. Biol. Chem. 378:337–343 [DOI] [PubMed] [Google Scholar]
- 24.Shin H, Lee JH, Kim H, Choi Y, Heu S, Ryu S. 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS One 7:e43392. 10.1371/journal.pone.0043392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson M, Newman D, Helm RA, Dino A, Calcutt M, French W, Eisenstark A. 2009. Competition among isolates of Salmonella enterica ssp. enterica serovar Typhimurium: role of prophage/phage in archived cultures. FEMS Microbiol. Lett. 294:37–44. 10.1111/j.1574-6968.2009.01554.x [DOI] [PubMed] [Google Scholar]
- 26.Fauquet C, Fargette D. 2005. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol. J. 2:64. 10.1186/1743-422X-2-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M, Ryu S. 2012. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 86:411–425. 10.1111/j.1365-2958.2012.08202.x [DOI] [PubMed] [Google Scholar]
- 29.Kiljunen S, Datta N, Dentovskaya SV, Anisimov AP, Knirel YA, Bengoechea JA, Holst O, Skurnik M. 2011. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage φA1122. J. Bacteriol. 193:4963–4972. 10.1128/JB.00339-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 31.Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11:675–687. 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 32.Hofer B, Ruge M, Dreiseikelmann B. 1995. The superinfection exclusion gene (sieA) of bacteriophage P22: identification and overexpression of the gene and localization of the gene product. J. Bacteriol. 177:3080–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranade K, Poteete AR. 1993. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J. Bacteriol. 175:4712–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70:217–248 [DOI] [PubMed] [Google Scholar]
- 35.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899. 10.1073/pnas.0808832106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190. 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ley P, Degraaff P, Tommassen J. 1986. Shielding of Escherichia coli outer-membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J. Bacteriol. 168:449–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindberg AA. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241. 10.1146/annurev.mi.27.100173.001225 [DOI] [PubMed] [Google Scholar]
- 39.Filippov AA, Sergueev KV, He Y, Huang XZ, Gnade BT, Mueller AJ, Fernandez-Prada CM, Nikolich MP. 2011. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS One 6:e25486. 10.1371/journal.pone.0025486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, Zhao H, Gao Y, Song J, Lu R, Sun C, Feng X. 2012. A method for generation phage cocktail with great therapeutic potential. PLoS One 7:e31698. 10.1371/journal.pone.0031698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M, Kim S, Ryu S. 2012. Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J. Virol. 86:10894. 10.1128/JVI.01796-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kidgell C, Pickard D, Wain J, James K, Diem Nga LT, Diep TS, Levine MM, O'Gaora P, Prentice MB, Parkhill J, Day N, Farrar J, Dougan G. 2002. Characterisation and distribution of a cryptic Salmonella typhi plasmid pHCM2. Plasmid 47:159–171. 10.1016/S0147-619X(02)00013-6 [DOI] [PubMed] [Google Scholar]
- 44.Nesper J, Kapfhammer D, Klose KE, Merkert H, Reidl J. 2000. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182:5097–5104. 10.1128/JB.182.18.5097-5104.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santander J, Robeson J. 2007. Phage-resistance of Salmonella enterica serovar Enteritidis and pathogenesis in Caenorhabditis elegans is mediated by the lipopolysaccharide. Electron. J. Biotechnol. 10:627–632 http://www.redalyc.org/articulo.oa?id=173314726015 [Google Scholar]
- 46.Kwan LY, Isaacson RE. 1998. Identification and characterization of a phase-variable nonfimbrial Salmonella typhimurium gene that alters O-antigen production. Infect. Immun. 66:5725–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cota I, Blanc-Potard AB, Casadesus J. 2012. STM2209-STM2208 (opvAB): a phase variation locus of Salmonella enterica involved in control of O-antigen chain length. PLoS One 7:e36863. 10.1371/journal.pone.0036863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monk AB, Rees CD, Barrow P, Hagens S, Harper DR. 2010. Bacteriophage applications: where are we now? Lett. Appl. Microbiol. 51:363–369. 10.1111/j.1472-765X.2010.02916.x [DOI] [PubMed] [Google Scholar]
- 49.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602. 10.1128/MMBR.68.3.560-602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes W. 1970. The genetics of bacteria and their viruses: studies in basic genetics and molecules biology, 2nd ed, p 410–411 Blackwell Scientific, Oxford, United Kingdom [Google Scholar]