Abstract
A new medium (lecithin and levofloxacin [LL] medium) is described for the isolation of Listeria monocytogenes from food samples. LL medium includes lecithin from soybeans for the detection of phosphatidylinositol-specific phospholipase C (PI-PLC) and phosphatidylcholine-specific phospholipase C (PC-PLC) produced by L. monocytogenes. Levofloxacin is incorporated to inhibit the growth of microorganisms other than L. monocytogenes, especially Bacillus cereus, shown to possess PI-PLC and PC-PLC activities. L. monocyogenes produced white colonies with a halo on LL medium, whereas Listeria innocua appeared as white colonies without a halo. Levofloxacin at 0.20 mg/liter completely inhibited the growth of B. cereus, while the growth of L. monocytogenes was unaffected. In the second phase of the study, the sensitivity and the specificity of LL medium were compared to those of modified Oxford agar (MOX) and two chromogenic media (Brilliance Listeria agar and CHROMagar Listeria), using a total of 250 food samples. From 200 unspiked food samples, the specificity of LL medium (96.0%) was superior to that of MOX (72.0%) and similar to the specificities of Brilliance Listeria agar (96.5%) and CHROMagar Listeria (94.5%). From 50 spiked food samples, LL medium and CHROMagar Listeria represented the highest sensitivities (96.0%), followed by Brilliance Listeria agar (92.0%) and MOX (54.0%). Also, LL medium showed the highest confirmation rate (98.8%), followed by Brilliance Listeria agar (98.7%), CHROMagar Listeria (98.3%), and MOX (52.0%). On the basis of its good specificity and cost effectiveness, LL medium is useful for the isolation of L. monocytogenes from food samples.
INTRODUCTION
The Gram-positive food-borne pathogen Listeria monocytogenes is a significant public health and food safety concern worldwide (1, 2). Infection of pregnant women, infants, the elderly, and immunosuppressed individuals with this pathogen can lead to listeriosis, a disease condition that can induce severe illnesses and relatively high mortality rates (3, 4). Infection has been associated with foods such as cheese, meat, milk, vegetables, and fish (5–9). Thus, effective methods for the isolation of L. monocytogenes from various foods are important to ensure food quality and safety.
A wide variety of selective and differential media have been developed for this purpose, including PALCAM, Oxford agar, and modified Oxford agar (MOX) (10–12). L. monocytogenes growing on these media is detected by the action of esculinase cleaving esculin (13). However, this metabolic enzyme is common to all Listeria species, so it does not distinguish L. monocytogenes from other nonpathogenic species of Listeria (14, 15). Therefore, subculture of multiple colonies is required to confirm the species, which takes at least a further 2 days (16). Listeria innocua in particular is often detected in foodstuffs and food environments (17). Also, L. innocua has been reported to grow faster than L. monocytogenes in enrichment broth, which makes detection of L. monocytogenes more difficult (18).
Several chromogenic media, including BBL CHROMagar Listeria, ALOA, Rapid'L. mono agar, CHROMagar Listeria, and Oxoid chromogenic Listeria agar (OCLA) have been introduced for the differentiation of L. monocytogenes from other Listeria spp. (14, 19–22). The detection of L. monocytogenes by chromogenic media usually involves cleavage of the substrate 5-bromo-4-chloro-3-indoxyl-β-d-glucopyranoside by β-d-glucosidase produced by Listeria spp., combined with l-α-phosphatidylinositol for the detection of phosphatidylinositol-specific phospholipase C (PI-PLC) and phosphatidylcholine-specific phospholipase C (PC-PLC) (14, 15, 23, 24). PI-PLC and PC-PLC, the major virulence factors, are only produced by pathogenic L. monocytogenes and Listeria ivanovii (15, 25). Chromogenic substrates have proven to be an efficient tool, utilizing specific enzymatic reactions of certain bacteria (2). However, chromogenic media can be expensive and may not be suitable for routine laboratory use (22). Also some Bacillus spp., especially Bacillus cereus, and Staphylococcus aureus are β-d-glucosidase, PI-PLC, and PC-PLC positive, and thus, they could grow on chromogenic media and produce colonies similar to those of L. monocytogenes (22, 26–30).
Recognizing the limits of currently used selective and differential media, it is desirable to develop an alternative medium with improved specificity while maintaining cost effectiveness.
The aim of this study was to develop a new selective and differential medium for the detection of L. monocytogenes (lecithin and levofloxacin [LL] medium) and to compare the specificity and the sensitivity of LL medium with those of conventional MOX and two chromogenic media by using stock cultures and food samples.
MATERIALS AND METHODS
Stock cultures.
The test bacteria used in this study (Tables 1 and 2) were obtained from the American Type Culture Collection (ATCC), the National Culture Collection for Pathogens (NCCP) (Osong, South Korea), the Korean Collection for Type Culture (KCTC), or the bacterial culture collection of the Food Hygiene Laboratory at Seoul National University (SNCC) (Seoul, South Korea). They were stored frozen at −80°C.
TABLE 1.
Growth of Listeria spp. and B. cereus on medium with different levofloxacin concentrations
| Species | Strain | Growth on medium with levofloxacin concn (mg/liter) ofa: |
|||
|---|---|---|---|---|---|
| 0.25 | 0.20 | 0.15 | 0.10 | ||
| L. monocytogenes | ATCC 19114 | +++ | +++ | +++ | +++ |
| ATCC 19115 | +++ | +++ | +++ | +++ | |
| ATCC 15313 | +++ | +++ | +++ | +++ | |
| NCCP 10810 | + | +++ | +++ | +++ | |
| NCCP 10811 | +++ | +++ | +++ | +++ | |
| NCCP 10943 | +++ | +++ | +++ | +++ | |
| SNCC 1 | + | ++ | +++ | +++ | |
| SNCC 2 | +++ | +++ | +++ | +++ | |
| SNCC 3 | +++ | +++ | +++ | +++ | |
| SNCC 4 | +++ | +++ | +++ | +++ | |
| SNCC 5 | +++ | +++ | +++ | +++ | |
| SNCC 6 | +++ | +++ | +++ | +++ | |
| SNCC 7 | +++ | +++ | +++ | +++ | |
| SNCC 8 | +++ | +++ | +++ | +++ | |
| SNCC 9 | +++ | +++ | +++ | +++ | |
| SNCC 10 | +++ | +++ | +++ | +++ | |
| SNCC 11 | +++ | +++ | +++ | +++ | |
| SNCC 12 | +++ | +++ | +++ | +++ | |
| SNCC 13 | +++ | +++ | +++ | +++ | |
| SNCC 14 | +++ | +++ | +++ | +++ | |
| SNCC 15 | + | ++ | +++ | +++ | |
| SNCC 16 | +++ | +++ | +++ | +++ | |
| SNCC 18 | +++ | +++ | +++ | +++ | |
| SNCC 19 | +++ | +++ | +++ | +++ | |
| SNCC 21 | +++ | +++ | +++ | +++ | |
| SNCC 22 | ++ | +++ | +++ | +++ | |
| SNCC 23 | +++ | +++ | +++ | +++ | |
| SNCC 24 | ++ | +++ | +++ | +++ | |
| SNCC 25 | +++ | +++ | +++ | +++ | |
| L. ivanovii | NCCP 10953 | + | +++ | +++ | +++ |
| L. innocua | SNCC 1 | +++ | +++ | +++ | +++ |
| SNCC 2 | +++ | +++ | +++ | +++ | |
| L. welshimeri | NCCP 10965 | ++ | +++ | +++ | +++ |
| L. seeligeri | NCCP 10966 | − | − | − | − |
| L. grayi | NCCP 10879 | − | + | + | ++ |
| Bacillus cereus | ATCC 10876 | − | − | − | + |
| ATCC 13061 | − | − | + | ++ | |
| ATCC 14579 | − | − | − | + | |
| NCCP 11308 | − | − | + | + | |
| NCCP 10109 | − | − | + | + | |
| NCCP 11307 | − | − | + | + | |
| NCCP 11306 | − | − | + | + | |
| NCCP 11313 | − | − | − | − | |
| NCCP 11309 | − | − | − | − | |
| NCCP 10084 | − | − | − | − | |
| NCCP 11310 | − | − | − | − | |
| NCCP 11311 | − | − | − | − | |
| SNCC 1 | − | − | − | + | |
| SNCC 2 | − | − | − | + | |
| SNCC 3 | − | − | − | − | |
| SNCC 4 | − | − | − | − | |
| SNCC 5 | − | − | − | − | |
| SNCC 6 | − | − | − | − | |
| SNCC 7 | − | − | − | − | |
| SNCC 8 | − | − | − | − | |
| SNCC 9 | − | − | − | − | |
| SNCC 10 | − | − | − | − | |
| SNCC 11 | − | − | − | − | |
| SNCC 12 | − | − | − | − | |
−, no growth; +, growth in the original streaked area; ++, growth in the second streaked area; +++, growth in all streaked areas.
TABLE 2.
Evaluation of the performance of each medium with stock cultures
| Culture and parameter | Result for: |
|||
|---|---|---|---|---|
| MOX | Brilliance Listeria agar | CHROMagar Listeria | LL medium | |
| L. monocytogenes (n = 29) | ||||
| No. true positive | 29 | 29 | 29 | 29 |
| No. false negative | 0 | 0 | 0 | 0 |
| Non-L. monocytogenes (n = 76) | ||||
| No. false positive | 17 | 1 | 3 | 1 |
| No. true negative | 59 | 75 | 73 | 75 |
| Accuracy (%)a | 85.2 | 99.0 | 98.5 | 99.0 |
| Exclusivity (%)b | 77.6 | 98.7 | 96.1 | 98.7 |
| Inclusivity (%)c | 100 | 100 | 100 | 100 |
[(True positive + true negative)/total] × 100.
[True negative/(true negative + false positive)] × 100.
[True positive/(true positive + false negative)] × 100.
Growth of test bacteria on basal medium containing different concentrations of levofloxacin.
To determine optimum concentration of antibiotics, the growth of bacteria on basal medium containing four different concentrations of levofloxacin was tested. The ingredients of basal medium for this study were as follows: 8.9 g of pancreatic digest of casein (Neogen, Lansing, MI), 4.4 g of protease peptone (Difco, Sparks, MD), 4.4 g of yeast extract (Difco), 2.7 g of beef heart infusion (Difco), 4.4 g of sodium chloride (Samchun Chemical Co., Ltd., Pyeongtaeksi, South Korea), 15.0 g of lithium chloride (Samchun Chemical), and 15.0 g of Bacto agar (Difco) per liter. The ingredients were added to distilled water and sterilized at 121°C for 15 min. After sterilization, the medium was tempered to 48°C, and 10.0 mg of colistin sulfate (Difco) and 20.0 mg of moxalactam (Difco) were added to the basal medium. Prepared basal medium was supplemented with levofloxacin (Sigma-Aldrich, St. Louis, MO) at concentrations from 0.10 to 0.25 mg/liter at 0.05-mg/liter increments. Modified Oxford agar (MOX) was prepared with Oxford agar base (Difco) supplemented with modified Oxford antimicrobial supplement (Difco). The test bacteria (Table 1) were incubated in 10 ml of tryptic soy broth (TSB; Difco) at 37°C for 18 h. After incubation, one loopful of each strain was streaked onto MOX and each basal medium containing different concentrations of levofloxacin by the semiquantitative three-loop technique (24). This involves streaking three times on each test medium, using a sterile loop to make dilutions of that original streak. After incubation for 48 h at 37°C, the amount of growth on the agar is then reported semiquantitatively as “many” (+++), “moderate” (++), or “few” (+), respectively, depending on how far from the inoculum site colonies appear. The test bacteria that grow in all streaked areas would be reported as “many” (+++). The test bacteria that only grow in the original streak area would be reported as “few” (+).
Preparation of LL medium.
The ingredients of basal medium were added to 950 ml of distilled water and sterilized at 121°C for 15 min. After sterilization, the medium was tempered to 48°C, and 10.0 mg of colistin sulfate and 20.0 mg of moxalactam were added to the basal medium. Levofloxacin (0.2 mg) was also added to supplement the basal medium. Five grams of soya lecithin (Solae, St. Louis, MO) was dissolved in 50 ml of distilled water, sterilized at 121°C for 15 min, and added to the prepared medium. The medium was poured into 9-cm-diameter petri dishes.
Recovery of L. monocytogenes on MOX and LL medium.
Fifteen L. monocytogenes strains were grown in 10 ml of TSB at 37°C for 18 h. The cultures were 10-fold serially diluted with 9 ml of buffered peptone water (BPW; Difco), and 0.1 ml of diluents (10−4 to 10−7) was plated onto MOX and LL medium. Colony counts were recorded after incubation at 37°C for 48 h. This experiment was repeated independently three times.
Evaluation of the performance of LL medium with stock cultures.
The performance of LL medium was evaluated using stock cultures in comparison with those of MOX, Brilliance Listeria agar (Oxoid, Ltd., Basingstoke, United Kingdom), and CHROMagar Listeria (CHROMagar, Paris, France). Twenty-nine L. monocytogenes strains (Table 1) were selected for the inclusivity test, while 76 non-L. monocytogenes strains were used for the exclusivity test. The latter consisted of Listeria ivanovii (n = 1), L. innocua (n = 4), Listeria welshimeri (n = 1), Listeria seeligeri (n = 1), Listeria grayi (n = 1), B. cereus (n = 24), Bacillus circulans (n = 3), Staphylococcus aureus (n = 10), Staphylococcus epidermidis (n = 2), Staphylococcus xylosus (n = 1), unidentified isolates from MOX plates (n = 10), Escherichia coli (n = 8), Klebsiella pneumoniae (n = 4), Yersinia enterocolitica (n = 2), Hafnia alvei (n = 3), and Citrobacter freundii (n = 1). The test bacteria were grown in 10 ml of TSB at 37°C for 18 h. After incubation, one loopful of each strain was streaked onto each medium and incubated at 37°C for 24 to 48 h. The growth and colony morphology were observed after incubation.
Evaluation of the specificity and the sensitivity of LL medium using spiked and unspiked food samples.
The conventional culture method described by the U.S. Food and Drug Administration (FDA) (31) was used for the microbiological analysis of spiked and unspiked food samples. In this study, a total of 250 food samples (200 unspiked food samples, 25 food samples spiked with L. monocytogenes, and 25 food samples spiked with L. monocytogenes and L. innocua) were tested for evaluation of the specificity and the sensitivity of LL medium. These food samples consisting of ground beef, ground pork, deli meat, smoked ham, sausage, luncheon meat, mixed salad, mixed sprouts, potatoes, lettuce, carrots, spinach, sweet potatoes, celery, radish, milk, cheddar cheese, and shrimp were purchased from local retail markets (Seoul, South Korea). All foods that were used in the spiked food sample test were previously screened to ensure they were Listeria free. To prepare spiked food samples, each strain of L. monocytogenes (ATCC 19114, 19115, and 15313) and L. innocua (SNCC 1, 2, and 3) was grown in 10 ml of TSB at 37°C for 18 h. Twenty-five subsamples (25 g or 25 ml) of each food were inoculated with 3 different L. monocytogenes strains at an inoculum level of 50 to 100 CFU/25 g or ml. Also, 25 subsamples of each food were inoculated with 3 different L. monocytogenes strains and 3 different L. innocua strains at an inoculum level of 50 to 100 CFU/25 g or ml. Spiked and unspiked food samples were introduced into a sterile stomacher bags (Labplas, Inc., Sainte-Julie, Quebec, Canada) containing 225 ml of buffered Listeria enrichment broth (BLEB; Difco) and homogenized with a stomacher (Easy Mix; AES Chemunex, Rennes, France) for 1 min. After preenrichment at 30°C for 4 h, acriflavine (final concentration, 10 mg/liter) (Sigma-Aldrich), nalidixic acid (final concentration, 40 mg/liter) (Sigma-Aldrich), and cycloheximide (final concentration, 50 mg/liter) (Sigma-Aldrich) were added, and the mixture was incubated at 30°C for 44 h. After enrichment, one loopful of each sample was streaked onto LL medium, MOX, Brilliance Listeria agar, and CHROMagar Listeria and incubated at 37°C for 24 to 48 h. After incubation, up to 5 colonies suspected of being L. monocytogenes on these media were selected for identification. Bacterial colonies were identified using the API Listeria test (bioMérieux, Marcy l'Etoile, France) and the Vitek 2 system (bioMérieux). Colonies suspected of being L. monocytogenes were defined as white colonies with a halo on LL medium, black colonies with a black halo on MOX, blue/green colonies with a halo on Brilliance Listeria agar, and blue colonies with a white halo on CHROMagar Listeria.
The specificity was evaluated by calculating the proportion of L. monocytogenes-negative samples correctly found to be negative (i.e., those that did not appear as presumptive L. monocytogenes colonies). The sensitivity was calculated as the proportion of L. monocytogenes-positive samples correctly found to be positive (22). The confirmation rate was determined as the ratio of the number of confirmed L. monocytogenes colonies (C) to the number of total suspect colonies tested (S), expressed as (C/S) × 100% (14).
Statistical analyses.
The significance of differences in colony counts was calculated by the Student t test, with a significance level of P < 0.05, using Microsoft Excel. The significance of differences in the specificity and the sensitivity was calculated by the McNemar tests, with a significance level of P < 0.05, using the SPSS software package (SPSS, Chicago, IL).
RESULTS
Growth of test bacteria on basal medium containing different concentrations of levofloxacin.
The growth of L. monocytogenes, L. ivanovii, L. innocua, Listeria welshimeri, Listeria seeligeri, Listeria grayi, and B. cereus was observed on basal medium containing different concentrations of levofloxacin (Table 1). Three strains of L. monocytogenes (NCCP 10810, SNCC 1, and SNCC 15) and L. ivanovii only grew in the original streaking area of basal medium containing 0.25 mg/liter of levofloxacin. Basal medium containing 0.20 mg/liter of levofloxacin achieved satisfactory growth (++ or +++) for all strains of L. monocytogenes strains, L. ivanovii, L. innocua, and L. welschimeri tested. L. seeligeri did not grow on all tested basal media, and basal medium containing 0.10 mg/liter of levofloxacin was the only medium that supported growth of L. grayi. Basal medium containing 0.20 mg/liter of levofloxacin completely inhibited the growth of all B. cereus strains. Levofloxacin (0.15 and 0.10 mg/liter) did not completely inhibit the growth of B. cereus.
Recovery of L. monocytogenes on MOX and LL medium.
Bacterial counts on LL medium were not significantly (P > 0.05) different from those on MOX (data not shown). On MOX and LL medium, colonies did not always have the typical appearance after 24 h of incubation (data not shown). At 48 h, all L. monocytogenes strains developed the correct morphology on these media (black with a black halo and white with a halo, respectively).
Evaluation of the performance of LL medium with stock cultures.
All tested L. monocytogenes strains produced typical L. monocytogenes colonies on each tested medium (inclusivity of 100%) (Table 2). The best exclusivity was accomplished by Brilliance Listeria agar and LL medium. Only L. ivanovii (n = 1) produced presumptive L. monocytogenes colonies on these media, and these two media achieved an exclusivity of 98.7%. CHROMagar Listeria achieved an exclusivity of 96.1%. One strain of L. ivanovii and two strains of B. cereus produced colonies similar to L. monocytogenes on CHROMagar Listeria. MOX showed an exclusivity of 77.6%. L. innocua (n = 4), L. welshimeri (n = 1), L. grayi (n = 1), and unidentified isolates (n = 10) produced black colonies with a black halo on MOX indistinguishable from those of L. monocytogenes. All tested media did not support the growth of L. seeligeri. Figure 1 shows colonies of L. monocytogenes, L. ivanovii, and L. innocua formed on MOX and LL medium. L. monocytogenes (Fig. 1A) and L. ivanovii (Fig. 1B) produced typical black colonies with a black halo on MOX. However, L. innocua (Fig. 1C) produced colonies similar to those of L. monocytogenes and L. ivanovii. L. monocytogenes (Fig. 1D) and L. ivanovii (Fig. 1E) appeared as white colonies with a halo on LL medium, whereas L. innocua produced white colonies without a halo (Fig. 1F).
FIG 1.
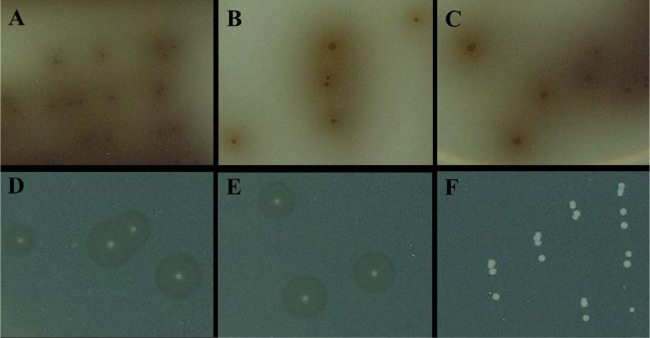
Colonies produced by L. monocytogenes, L. ivanovii, and L. innocua on MOX (A, B, and C) and LL medium (D, E, and F). L. monocytogenes (A), L. ivanovii (B), and L. innocua (C) appeared as black colonies with a black halo on MOX. On LL medium, L. monocytogenes (D) and L. ivanovii (E) produced white colonies with a halo, whereas L. innocua (F) produced white colonies without a halo.
Evaluation of the specificity and the sensitivity of LL medium using spiked and unspiked food samples.
Table 3 presents results of the bacteriological analysis of unspiked food samples. The number of negative samples was equal to the number of total food samples, because no L. monocytogenes strains were isolated on all tested media from 200 unspiked food samples. There were no significant differences among the specificities of Brilliance Listeria agar, CHROMagar Listeria, and LL medium. The specificities of Brilliance Listeria agar, CHROMagar Listeria, and LL medium were 96.5, 94.5, and 96.0%, respectively. The specificities of these three media were superior to the specificity of MOX (72.0%). Totals of 56, 7, 11, and 8 false-positive results were found on MOX, Brilliance Listeria agar, CHROMagar Listeria, and LL medium, respectively. From 50 spiked food samples, CHROMagar Listeria, and LL medium represented the highest sensitivities (96.0%), followed by Brilliance Listeria agar (92.0%) and MOX (54.0%) (Table 4). Also, LL medium showed the highest confirmation rate (98.8%), followed by Brilliance Listeria agar (98.7%), CHROMagar Listeria (98.3%), and MOX (52.0%).
TABLE 3.
Specificity of LL medium compared with those of MOX, Brilliance Listeria agar, and CHROMagar Listeria in the microbiological analysis of 200 unspiked food samples
| Medium | No. of results |
Specificity (%)a | |
|---|---|---|---|
| True negative | False positive | ||
| MOX | 144 | 56 | 72.0 |
| Brilliance Listeria agar | 193 | 7 | 96.5 |
| CHROMagar Listeria | 189 | 11 | 94.5 |
| LL medium | 192 | 8 | 96.0 |
(No. of true-negative results on the medium/no. of negative samples) × 100.
TABLE 4.
Evaluation of efficacy of each medium for the isolation of L. monocytogenes from 50 spiked food samples
| Medium | Sensitivity, %a | Confirmation rate, % (no. positive/total)b |
|---|---|---|
| MOX | 54.0 | 52.0 (130/250) |
| Brilliance Listeria agar | 92.0 | 98.7 (227/230) |
| CHROMagar Listeria | 96.0 | 98.3 (236/240) |
| LL medium | 96.0 | 98.8 (237/240) |
[No. of true positives/(no. of true positives + no. of false negatives)] × 100.
Percentage of suspect colonies tested and confirmed positive for L. monocytogenes.
DISCUSSION
Plating on selective and differential media, such as PALCAM, Oxford, MOX, and chromogenic agar, after preenrichment followed by selective enrichment in BLEB has been recommended for the isolation of L. monocytogenes from foods by the U.S. FDA (31). However, PALCAM, Oxford agar, and MOX, which use esculinase activity of Listeria spp., cannot differentiate L. monocytogenes from other Listeria spp. Also, several chromogenic media that use chromogenic compounds, such as 5-bromo-4-chloro-3-indoxyl-β-d-glucopyranoside and l-α-phosphatidylinositol, are expensive (22).
Oxoid chromogenic Listeria agar (OCLA) contains soya lecithin instead of phosphatidylinositol for the differentiation of L. monocytogenes from other Listeria spp. (22). The results suggest that soya lecithin is an appropriate alternative to phosphatidylinositol. Soya lecithin is a cheap and easily available product and could be used for the detection of PI-PLC and PC-PLC activity (22, 32). In this study, LL medium contains soya lecithin for the differentiation of L. monocytogenes from other Listeria spp. L. monocytogenes and L. ivanovii produced colonies (white colonies with a halo) distinct from those of other Listeria spp. on LL medium. Although L. ivanovii is less frequently encountered in foods, it is important to detect this organism, because some cases of human infection by L. ivanovii have been reported (33).
Some Bacillus spp., especially Bacillus cereus, are β-d-glucosidase, PI-PLC, and PC-PLC positive (27, 29, 30); thus, they could grow on chromogenic media and produce colonies similar to those of L. monocytogenes (24, 30). Therefore, it is necessary to inhibit growth of B. cereus to ensure the specificity of the selective medium. In a previous study (34) of 42 isolates of B. cereus, the MIC50 and MIC90 of levofloxacin were 0.125 and 0.25 mg/liter, respectively. Forty-two strains of L. monocytogenes were more resistant to levofloxacin: the MIC50 and MIC90 of levofloxacin were 0.5 and 1 mg/liter, respectively (35). LL medium contains levofloxacin in addition to colistin sulfate and moxalactam to inhibit the growth of B. cereus. A levofloxacin concentration from 0.25 to 0.20 mg/liter completely inhibited growth of all B. cereus strains tested. LL medium includes 0.20 mg/liter of levofloxacin, because the growth of some L. monocytogenes strains and L. ivanovii was inhibited on basal medium containing 0.25 mg/liter of levofloxacin. The recovery of L. monocytogenes on LL medium was compared with that on MOX to evaluate the effect of additional antibiotics on the sensitivity of the medium (data not shown). Counts of L. monocytogenes on LL medium reached approximately the same level as those on MOX; thus, levofloxacin could be used as a selective agent in LL medium.
In this study, the specificity and the sensitivity of LL medium were compared with those of conventional MOX and two chromogenic media (Brilliance Listeria agar and CHROMagar Listeria) using stock cultures and food samples. Brilliance Listeria agar (formerly OCLA) contains soya lecithin instead of phosphatidylinositol. LL medium completely suppressed the growth of non-Listeria Gram-positive and Gram-negative bacteria (Table 2). LL medium also inhibited the growth of S. aureus, S. epidermidis, and S. xylosus. Staphylococcus spp. that have PI-PLC and PC-PLC activities can give the appearance of false-positive colonies on chromogenic medium that contains soya lecithin instead of phosphatidylinositol (22, 26, 36). The growth of some unidentified bacteria isolated from food samples and producing black colonies with a black halo on MOX was also completely inhibited on LL medium. Brilliance Listeria agar and CHROMagar Listeria also successfully inhibited the growth of non-Listeria Gram-positive and Gram-negative bacteria, but two B. cereus strains formed colonies similar to those of L. monocytogenes on CHROMagar Listeria.
When 200 unspiked food samples were examined, no L. monocytogenes was isolated on any of the media tested. The incidence of L. monocytogenes from domestic and imported foods in South Korea was lower than that reported in previous studies from other countries (37). Of 1,537 domestic and imported food products, L. monocytogenes was isolated from 122 food samples (7.9%). Also Jo et al. (38) reported L. monocytogenes was not isolated from 96 samples of fresh-cut produce or organic vegetables. L. innocua comprised the majority of presumptive Listeria colonies on MOX (data not shown). Especially, L. innocua was frequently detected in beef and pork samples. When 50 spiked food samples were tested, the sensitivity and the confirmation rate of MOX were much lower than those of LL medium, Brilliance Listeria agar, and CHROMagar Listeria. From 25 samples that were spiked with L. monocytogenes and L. innocua, colonies of L. monocytogenes were hardly differentiated from those of L. innocua on MOX. L. innocua has been known to frequently outgrow L. monocytogenes in enrichment broth (18). The ability to detect small numbers of L. monocytogenes colonies among colonies of L. innocua is an important advantage of LL medium.
In conclusion, the LL medium presented in this study has higher specificity and sensitivity than MOX. Especially, LL medium easily differentiates L. monocytogenes and L. ivanovii from L. innocua, which frequently produces false-positive results on conventional PALCAM, Oxford agar, and MOX. Also, LL medium showed similar specificity and sensitivity to Brilliance Listeria agar and CHROMagar Listeria with reduced cost. The cost of LL medium ($33.70/liter) is lower than those of chromogenic media (Brilliance Listeria agar, $63.98/liter; CHROMagar Listeria, $63.50/liter). LL medium could also inhibit the growth of PI-PLC- and PC-PLC-positive bacteria, such as B. cereus and Staphylococcus spp., which cause false-positive results on some chromogenic media. Due to the good specificity of LL medium, the labor and time necessary for further confirmation tests may be applied to fewer colonies. Also, the cost-effectiveness of LL medium would make it suitable for routine laboratory use. Although further study is required to evaluate the growth of more Listeria spp. on LL medium, LL medium may provide a valuable addition to the array of selective media available for the detection of L. monocytogenes from food.
ACKNOWLEDGMENTS
This research was supported by the Public Welfare & Safety Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2012M3A2A1051679). This research also was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (0635-20120004).
Footnotes
Published ahead of print 22 November 2013
REFERENCES
- 1.Todd ECD, Notermans S. 2011. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control 22:1484–1490. 10.1016/j.foodcont.2010.07.021 [DOI] [Google Scholar]
- 2.Pradhan AK, Ivanek R, Grohn YT, Bukowski R, Wiedmann M. 2011. Comparison of public health impact of Listeria monocytogenes product-to-product and environment-to-product contamination of deli meats at retail. J. Food. Prot. 74:1860–1868. 10.4315/0362-028X.JFP-10-351 [DOI] [PubMed] [Google Scholar]
- 3.Painter J, Slutsker L. 2007. Listeriosis in humans, p 85–109 In Ryser ET, Marth EH. (ed), Listeria, listeriosis, and food safety. Marcel Dekker, New York, NY [Google Scholar]
- 4.Swaminathan B, Gerner-Schmidt P. 2007. The epidemiology of human listeriosis. Microbes Infect. 9:1236–1243. 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- 5.Dalton CB, Austin CC, Sobel J, Hayes PS, Bibb WF, Graves LM, Swaminathan B, Proctor ME, Griffin PM. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100–105. 10.1056/NEJM199701093360204 [DOI] [PubMed] [Google Scholar]
- 6.Tham W, Ericsson H, Loncarevic S, Unnerstad H, Danielsson-Tham M. 2000. Lessons from an outbreak of listeriosis related to vacuum-packed gravad and cold-smoked fish. Int. J. Food Microbiol. 62:173–175. 10.1016/S0168-1605(00)00332-9 [DOI] [PubMed] [Google Scholar]
- 7.De Valk H, Vaillant V, Jacquet C, Rocourt J, Le Querrec F, Stainer F, Quelquejeu N, Pierre O, Pierre V, Desenclos JC, Goulet V. 2001. Two consecutive nationwide outbreaks of listeriosis in France, October 1999—February 2000. Am. J. Epidemiol. 154:944–950. 10.1093/aje/154.10.944 [DOI] [PubMed] [Google Scholar]
- 8.Lunden J, Tolvanen R, Korkeala H. 2004. Human listeriosis outbreaks linked to dairy products in Europe. J. Dairy Sci. 87:E6–E12. 10.3168/jds.S0022-0302(04)70056-9 [DOI] [Google Scholar]
- 9.Makino S, Kawamoto K, Takeshi K, Okada Y, Yamasaki M, Yamamoto S, Igimi S. 2005. An outbreak of food-borne listeriosis due to cheese in Japan, during 2001. Int. J. Food Microbiol. 104:189–196. 10.1016/j.ijfoodmicro.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 10.Curtis GDW, Mitchelaln RG, King AF, Griffin EJ. 1989. A selective differential medium for the isolation of Listeria monocytogenes. Lett. Appl. Microbiol. 8:95–98. 10.1111/j.1472-765X.1989.tb00231.x [DOI] [Google Scholar]
- 11.McClain D, Lee WL. 24 May 1989. revision FSIS method for the isolation and identification of Listeria monocytogenes from processed meat and poultry products. Laboratory communication no. 57. US Department of Agriculture, Food Safety and Inspection Service, Beltsville, MD [Google Scholar]
- 12.van Netten P, Perales I, van de Moosdijk A, Curtis GDW, Mossel DAA. 1989. Liquid and solid selective differential media for the detection and enumeration of L. monocytogenes and other Listeria spp. Int. J. Food Microbiol. 8:299–316 [DOI] [PubMed] [Google Scholar]
- 13.McLauchlin J. 1987. Listeria monocytogenes, recent advances in the taxonomy and epidemiology of listeriosis in humans. J. Appl. Bacteriol. 63:1–11. 10.1111/j.1365-2672.1987.tb02411.x [DOI] [PubMed] [Google Scholar]
- 14.Hegde V, Leon-Velarde CG, Stam CM, Jaykus LA, Odumeru JA. 2007. Evaluation of BBL CHROMagar Listeria agar for the isolation and identification of Listeria monocytogenes from food and environmental samples. J. Microbiol. Methods 68:82–87. 10.1016/j.mimet.2006.06.011 [DOI] [PubMed] [Google Scholar]
- 15.Leclercq A. 2004. Atypical colonial morphology and low recoveries of Listeria monocytogenes strains on Oxford, PALCAM, Rapid' L. mono and ALOA solid media. J. Microbiol. Methods 57:251–258 [DOI] [PubMed] [Google Scholar]
- 16.Greenwood M, Willis C, Doswell P, Allen G, Pathak K. 2005. Evaluation of chromogenic media for the detection of Listeria species in food. J. Appl. Microbiol. 99:1340–1345. 10.1111/j.1365-2672.2005.02734.x [DOI] [PubMed] [Google Scholar]
- 17.Johansson T. 1998. Enhanced detection and enumeration of Listeria monocytogenes from foodstuffs and food-processing environments. Int. J. Food Microbiol. 40:77–85. 10.1016/S0168-1605(98)00022-1 [DOI] [PubMed] [Google Scholar]
- 18.Zitz U, Zunabovic M, Domig KJ, Wilrich PT, Kneifel A. 2011. Reduced detectability of Listeria monocytogenes in the presence of Listeria innocua. J. Food. Prot. 74:1282–1287. 10.4315/0362-028X.JFP-11-045 [DOI] [PubMed] [Google Scholar]
- 19.Carles B, Jaquet C, Duthoit ML, Facon JP, Rocourt J. 1997. Evaluation d'un nouveau milieu de culture pour la detection rapide de Listeria monocytogenes dans les produits alimentaires: RAPID'L.MONO, p 52–53 Sanofi Information, Sanofi Diagnostics Pasteur Laboratory, Steenvoorde, France [Google Scholar]
- 20.Ottaviani F, Ottaviani M, Agosti M. 1997. Differential agar medium for Listeria monocytogenes. Ind. Aliment. 36:1–3 [Google Scholar]
- 21.Restaino L, Frampton EW, Irbe RM, Schabert G, Spitz H. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food. Prot. 62:244–251 [DOI] [PubMed] [Google Scholar]
- 22.Willis C, Baalham T, Greenwood M, Presland F. 2006. Evaluation of a new chromogenic agar for the detection of Listeria in food. J. Appl. Microbiol. 101:711–717. 10.1111/j.1365-2672.2006.02917.x [DOI] [PubMed] [Google Scholar]
- 23.Reissbrodt R. 2004. New chromogenic plating media for detection of pathogenic Listeria spp.—an overview. Int. J. Food Microbiol. 95:1–9. 10.1016/j.ijfoodmicro.2004.01.025 [DOI] [PubMed] [Google Scholar]
- 24.Stessl B, Luf W, Wagner M, Schoder D. 2009. Performance testing of six chromogenic ALOA-type media for the detection of Listeria monocytogenes. J. Appl. Microbiol. 106:651–659. 10.1111/j.1365-2672.2008.04039.x [DOI] [PubMed] [Google Scholar]
- 25.Mengaud J, Braun-Breton C, Cossart P. 1991. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor. Mol. Microbiol. 5:367–372. 10.1111/j.1365-2958.1991.tb02118.x [DOI] [PubMed] [Google Scholar]
- 26.Daugherty S, Low MG. 1993. Cloning, expression, and mutagenesis of phosphatidylinositol-specific phospholipase C from Staphylococcus aureus: a potential staphylococcal virulence factor. Infect. Immun. 61:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghelardi E, Celandroni F, Salvetti S, Barsotti C, Baggiani A, Senesi S. 2002. Identification and characterization of toxigenic Bacillus cereus isolates responsible for two food-poisoning outbreaks. FEMS Microbiol. Lett. 208:129–134. 10.1111/j.1574-6968.2002.tb11072.x [DOI] [PubMed] [Google Scholar]
- 28.Kloos WE, Wolfshohl JF. 1982. Identification of Staphylococcus species with the API STAPH-IDENT system. J. Clin. Microbiol. 16:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Netten P, Kramer JM. 1992. Media for the detection and enumeration of Bacillus cereus in foods: a review. Int. J. Food Microbiol. 17:85–99. 10.1016/0168-1605(92)90108-F [DOI] [PubMed] [Google Scholar]
- 30.Vlaemynck G, Lafarge V, Scotter S. 2000. Improvement of the detection of Listeria monocytogenes by the application of ALOA, a diagnostic, chromogenic isolation medium. J. Appl. Microbiol. 88:430–441. 10.1046/j.1365-2672.2000.00978.x [DOI] [PubMed] [Google Scholar]
- 31.Hitchins AD, Jinneman K. 2011. Detection and enumeration of Listeria monocytogenes in foods. Bacteriological analytical manual online. FDA, Washington, DC: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm071400.htm [Google Scholar]
- 32.da Silva Malheiros P, Micheletto YMS, da Silveira NP, Brandelli A. 2010. Development and characterization of phosphatidylcholine nanovesicles containing the antimicrobial peptide nisin. Food Res. Int. 43:1198–1203. 10.1016/j.foodres.2010.02.015 [DOI] [Google Scholar]
- 33.Guillet C, Join-Lambert O, Le Monnier A, Leclercq A, Mechaï F, Mamzer-Bruneel MF, Bielecka MK, Scortti M, Disson O, Berche P, Vazquez-Boland J, Lortholary O, Lecuit M. 2010. Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 16:136–138. 10.3201/eid1601.091155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luna VA, King DS, Gulledge J, Cannons AC, Amuso PT, Cattani J. 2007. Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 antimicrobials using Sensititre automated microbroth dilution and Etest agar gradient diffusion methods. J. Antimicrob. Chemother. 60:555–567. 10.1093/jac/dkm213 [DOI] [PubMed] [Google Scholar]
- 35.Salas C, Calvo J, Martínez-Martínez L. 2008. Activity of tigecycline against coryneform bacteria of clinical interest and Listeria monocytogenes. Antimicrob. Agents Chemother. 52:1503–1505. 10.1128/AAC.01129-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Low MG. 1981. Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus. Methods Enzymol. 71:741–746. 10.1016/0076-6879(81)71087-5 [DOI] [PubMed] [Google Scholar]
- 37.Baek SY, Lim SY, Lee DH, Min KH, Kim CM. 2000. Incidence and characterization of Listeria monocytogenes from domestic and imported foods in Korea. J. Food Prot. 63:186–189 [DOI] [PubMed] [Google Scholar]
- 38.Jo MJ, Jeong AR, Kim HJ, Lee N, Oh SW, Kim YJ, Chun HS, Koo M. 2011. Microbiological quality of fresh-cut produce and organic vegetables. Korean J. Food Sci. Technol. 43:91–97 http://www.koreascience.or.kr/journal/AboutJournal.jsp?kojic=SPGHB5 [Google Scholar]


