Abstract
The Bacillus subtilis cell wall is a dynamic structure, composed of peptidoglycan and teichoic acid, that is continually remodeled during growth. Remodeling is effected by the combined activities of penicillin binding proteins and autolysins that participate in the synthesis and turnover of peptidoglycan, respectively. It has been established that one or the other of the CwlO and LytE d,l-endopeptidase-type autolysins is essential for cell viability, a requirement that is fulfilled by coordinate control of their expression by WalRK and SigI RsgI. Here we report on the regulation of cwlO expression. The cwlO transcript is very unstable, with its degradation initiated by RNase Y cleavage within the 187-nucleotide leader sequence. An antisense cwlO transcript of heterogeneous length is expressed from a SigB promoter that has the potential to control cellular levels of cwlO RNA and protein under stress conditions. We discuss how a multiplicity of regulatory mechanisms makes CwlO expression and activity responsive to the prevailing growth conditions.
INTRODUCTION
Peptidoglycan, one of the principal components of the Gram-positive bacterial cell wall, is made of glycan strands cross-linked by short peptides. The glycan strands are composed of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues that vary in length among bacteria, averaging between 50 and 250 disaccharide units in bacilli (1). Peptide cross-links also vary in length and composition and often contain atypical amino acids. Peptidoglycan forms a flexible multilayered mesh-type structure outside the cytoplasmic membrane that contributes to cell shape, resists turgor pressure, and provides a platform for exposure of surface proteins. However, the cell wall is a dynamic structure that is continually remodeled during growth and division. Remodeling such a chemically complex structure while maintaining its integrity implies highly coordinated cell wall metabolic activity in the cytoplasmic and extracellular compartments.
Murein hydrolyases (autolysins) play a pivotal role in remodeling the cell wall during growth and division. Bacillus subtilis encodes approximately 35 autolysins with a range of enzymatic activities sufficient to cleave all peptidoglycan bonds (1, 2). Establishing the cellular role of each autolysin is hampered by significant redundancy at the enzyme activity level. For example, B. subtilis encodes seven d,l-endopeptidases, CwlO, LytE, CwlS, LytF, PgdS, YkfC, and YqgT (3). However, the motifs associated with the enzymatic domains differ, suggesting that each performs a distinct cellular role. The complexity of relationships between autolysins with similar enzymatic activity can be illustrated by the CwlO and LytE d,l-endopeptidases. The CwlO N-terminal domain is unique among autolysins and is required for interaction with the FtsEX ATP transporter that controls CwlO enzymatic activity (4, 5, 6). The LytE N-terminal domain is composed of three LysM motifs that are commonly found in variable number among autolysins (7, 8). CwlO and LytE can be individually deleted with only a slight phenotypic consequence: lytE null mutants are somewhat thinner than wild-type (WT) cells and form longer chains, whereas cwlO null mutants are wider than wild-type cells and can be somewhat bent (5). The cwlO and lytE genes are synthetically lethal, showing that the activity of one or the other enzyme is required for cell viability and that their cellular roles overlap (9, 10). However, CwlO and LytE can be distinguished by several attributes. CwlO is primarily involved in cell elongation and requires interaction with the membrane-located FtsEX ATP transporter-like complex and ATP hydrolysis for activity (4, 5, 6, 9). It is thought that this control mechanism restricts CwlO endopeptidase activity to the membrane-proximal region of the cell wall. Moreover, CwlO function requires the Mbl isoform of the actin-like cytoskeletal proteins for activity (5). LytE is involved in both elongation and cell division and appears to predominate under stress conditions (7, 11, 12). There are no reported additional protein requirements for LytE activity, but normal cellular function requires the MreBH isoform of the actin-like cytoskeletal proteins (13). Thus, while these two d,l-endopeptidases clearly perform overlapping central roles in cell elongation, there are nuanced differences in their cellular function.
Complex regulation of CwlO expression and activity might be expected in view of its pivotal role in cell wall elongation. Recent studies report control at the levels of transcription initiation and enzyme activity (5, 6, 9, 14). CwlO expression is activated by the WalRK two-component signal transduction system (TCS), its only known transcriptional regulator (Fig. 1A). Promoter activity of cwlO is high for a short period after inoculation of cells into fresh medium, is maintained at an intermediate level during exponential growth, and is reduced to <5% of maximal activity when cells enter stationary phase (15). Since WalRK is active only in exponentially growing cells, such control restricts CwlO expression to this cellular state, consistent with its proposed role in cell elongation. While all autolysins have the potential to disrupt cell wall integrity with lethal consequences, the multiplicity of controls on CwlO expression and enzymatic activity suggests that it poses an especial hazard to cell viability. The requirements for association with FtsEX and ATP hydrolysis plus its dependence on the actin-like Mbl isoform probably combine to restrict CwlO d,l-endopeptidase activity to the inner layer of the cell wall at specific positions along the cell cylinder (5, 6). Such control of CwlO enzyme activity also ensures that when released from the cell, the protein is inactive and unable to hydrolyze the outer layers of its own cell wall or that of adjacent bacteria.
FIG 1.
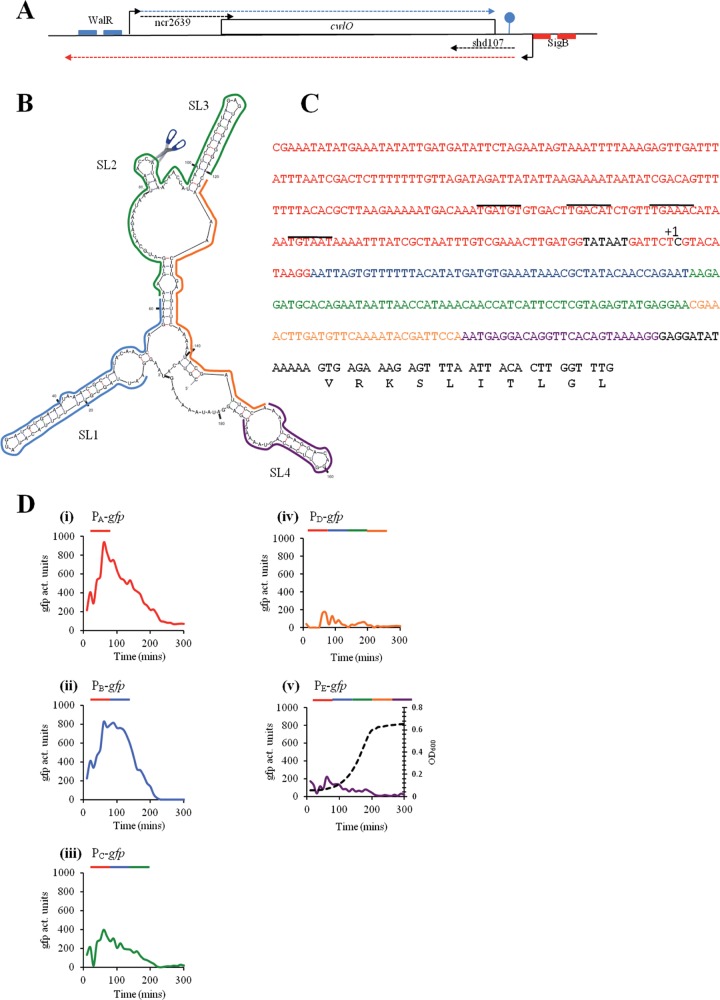
Organization of the cwlO locus and analysis of the leader sequence. (A) A schematic representation of the cwlO locus with its regulatory features. The cwlO open reading frame is represented by a rectangle. Initiation points of transcription are represented by bent arrows. The DNA recognition sequences for WalR and SigB are indicated by small blue and red boxes, respectively. Transcripts are indicated by broken lines, with the cwlO sense and antisense transcripts indicated by blue and red broken lines, respectively. The two small noncoding RNAs are designated by black broken lines. (B) A potential secondary structure for the cwlO leader sequence. The stem-loop structures are designated (SL1 to -4), the scissors represents the RNase Y cleavage site, and the colored lines delineate the leader regions present in the transcriptional fusions. (C) Sequence of the cwlO promoter, leader sequence, and beginning of the open reading frame. The Pribnow box and initiation point of transcription (+1) are shown in black, the promoter sequence is shown in red, the WalR recognitions sequences are overlined, and the regions of the leader sequence used in the transcriptional analysis are shown in blue, green, orange, and purple. (D) The regions of the leader sequence designated in panels B and C were fused to a promoterless gfpmut3 reporter gene and expression determined by measuring GFP activity throughout a phosphate limitation cycle. The sequence composition of the promoter-plus-leader region used in each transcriptional fusion is indicated by colored lines over each graph. A representative growth curve is shown in panel v. Time is in minutes after culture inoculation.
Several features of the cwlO locus suggest that expression is subject to additional regulation. The cwlO mRNA has a 187-nucleotide (nt) leader sequence with the potential to form secondary structure (Fig. 1B). Two noncoding RNAs (ncRNAs) have been reported at this locus (Fig. 1A): ncr2639 extends over the leader sequence and into the 5′ end of the structural gene on the sense strand, while shd107 extends over the 3′ end of the structural gene and terminator region on the antisense strand (16, 17). Furthermore, there is a putative SigB promoter located downstream of the cwlO terminator with the potential to transcribe an antisense cwlO transcript (Fig. 1A) (15, 18).
Here we report that CwlO expression is regulated at the level of RNA stability. The cwlO RNA transcript is highly unstable due to RNase Y cleavage within the 187-nucleotide leader sequence. Moreover, the level of cell-associated and secreted CwlO protein increases when CwlO expression is elevated by transcript stabilization. We discuss how a multiplicity of transcriptional and posttranscriptional controls can make CwlO expression responsive to the growth and environmental conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The oligonucleotides used in this study are listed in Table S1 in the supplemental material. Escherichia coli strain TG1 (19) was used for routine cloning. Strain EC101 (20) was used for propagating pGhost plasmids and strain BL21(DE3) for protein overproduction. E. coli strains were grown and maintained on Luria-Bertani (LB) medium with addition of ampicillin (100 μg/ml) as required. B. subtilis strains were grown in LB medium, high-phosphate defined medium (HPDM), or low-phosphate defined medium (LPDM) (21). Antibiotics were added when required at the following concentrations: chloramphenicol, 4 μg/ml; erythromycin, 2 μg/ml (to select for integrants) or 150 μg/ml (to verify insertion of erm into the chromosome); spectinomycin, 100 μg/ml; and kanamycin, 10 μg/ml. Growth was monitored using a UVmini-1240 UV-visible spectrophotometer (Shimadzu Instruments).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference, source, or construction |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIq lacZΔM15] | 19 |
| BL21(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | 41 |
| EC101 | TG1 containing a chromosomally encoded repA gene for pGhost replication | 20 |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| LS047 | Deletion of the SigB promoter that generates cwlO antisense RNA; no antisense detected | Insertion/excision of pLSI010 in strain 168 |
| LS095 | 168 ΔrsgI::kan | 12 |
| LS089 | ΔlytE::cat | 12 |
| LS090 | LS047 ΔlytE::cat | LFH ΔlytE::cat → LS047 |
| LS103 | ΔcwlO::cat | 12 |
| DN2021 | 168 cwlO::Δleader PcwlOgfp generated using pBaSysBioII | pDN2021 → 168 |
| DN2070 | 168WT cwlO::pDN2070 partial deletion of cwlO leader sequence PcwlOgfp generated using pBaSysBioII | pDN2070 → 168 |
| DN2071 | 168WT cwlO::pDN2071 partial deletion of cwlO leader sequence PcwlOgfp generated using pBaSysBioII | pDN2071 → 168 |
| DN2072 | 168WT cwlO::pDN2072 partial deletion of cwlO leader sequence PcwlOgfp generated using pBaSysBioII | pDN2072 → 168 |
| DN2073 | 168WT cwlO::pDN2073 partial deletion of cwlO leader sequence PcwlOgfp generated using pBaSysBioII | pDN2073 → 168 |
| DN3000 | Deletion of 141 nucleotides from the cwlO leader sequence | Insertion/excision of pDN3000 in strain168 |
| DN3005 | Deletion of 141 nucleotides from the cwlO leader sequence in strain LS047 | Insertion/excision of pDN3000 in LS047 |
| DN3015 | DN3000 ΔlytE::cat | LFHa ΔlytE::cat → DN3000 |
| DN3020 | Deletion of CwlO polyserine domain (ΔS295-S332 inclusive) using pGhost4 | Insertion/excision of pDN3020 in strain 168 |
| DN3021 | Deletion of CwlO N-terminal and polyserine domains (ΔE52-S332 inclusive) using PGhost4 | Insertion/excision of pDN3021 in strain168 |
| DN3022 | Deletion of CwlO polyserine domain (ΔS295-S340 inclusive) using pGhost4 ΔlytE::cat | LFH ΔlytE::cat → DN3020 |
| DN3025 | Deletion of amino acids 52–102 inclusive from the N terminus of CwlO (ΔN1) | Insertion/excision dif ΔN1 |
| DN3027 | Deletion of amino acids 231–288 inclusive from the N terminus of CwlO (ΔN3) | Insertion/excision dif ΔN3 |
| CCB034 | PspacrnjA(J1) pMAP65 erm kan | 42 |
| CCB294 | Δrny::spec amy::Pspacrny pMAP65 spec cm kan | 36 |
| CCB423 | 168 trpC2 Δrny::spec | 43 |
| CCB078 | ΔrnjB(J2)::spec | 42 |
| PB344 | ΔsigB::spec | 44 |
| Plasmids | ||
| pBaSysBioII | Plasmid to generate transcriptional GFP gene fusions at the gene locus | 22 |
| pLIS010 | pGhost4 construct to delete the sigB-dependent promoter of the cwlO antisense transcript | This study |
| pDN2021 | pBaSysBioII containing the WT cwlO promoter with the 5′ leader sequence deleted | This study |
| pDN2070 to -73 | pBaSysBioII constructs with incremental add back of the cwlO leader sequence (P1 to P4) | This study |
| pDN3000 | pGhost4 construct to delete 141 bp of the cwlO 5′ leader sequence | This study |
| pDN3020 | pGhost4 construct to delete polyserine domain of cwlO | This study |
| pDN3021 | pGhost4 construct to delete the N-terminal domain and the polyserine domain | This study |
| pLIS012 | pET21b clone expressing ′CwlO lacking the signal sequence | This study |
| pET21b | Plasmid for overexpression of His×6-tagged proteins | Novagen |
| pDG1661 | E. coli plasmid for ectopic integration at amyE containing cat | 45 |
| pGhost4+ | Temperature-sensitive replication derivative of pGK12 (erm) | 46 |
LFH, long-flanking-homology PCR.
Strains and plasmids.
To establish the contribution of the leader sequence to expression, we constructed a series of strains containing transcriptional fusions of the cwlO promoter, and of the cwlO promoter plus increasing amounts of leader sequence, with the gfpmut3 reporter gene. A DNA fragment carrying the cwlO promoter was amplified by PCR with primers yvcEF and yvcEdelLEADtscpR and cloned into plasmid pBaSysBioII following the ligation-independent cloning (LIC) procedure described by Botella et al. (22), generating plasmid pDN2021. Strain DN2021 was generated by integration of pDN2021 into the chromosome of wild-type strain 168 by a single-crossover event. To construct strains with the cwlO promoter plus increasing lengths of the leader sequence, plasmids pDN2070 to -2073 were constructed in a similar manner (i.e., with LIC-based cloning into pBaSysBioII) using primer pairs yvcEF/PyvcEdel1revLICtscp, yvcEF/PyvcEdel2revLICtscp, yvcEF/PyvcEdel3revLICtscp, and yvcEF/PyvcEdel4revLICtscp, respectively. Plasmids pDN2070 to -2073 were individually transformed into wild-type strain 168 to generate strains DN2070 through DN2073, in which the gfpmut3 reporter gene is expressed from the cwlO promoter only (DN2021, 238 bp of promoter DNA) and the cwlO promoter plus 53 bp (DN2070), 115 bp (DN2071), 141 bp (DN2072), and 167 bp (DN2073) of the leader sequence.
A markerless deletion of the cwlO leader sequence was generated by a procedure described previously (23). DNA fragments flanking the leader region (to be deleted) were amplified by PCR using primer pairs yvcEleadlacZF/PyvcEdelpGhupR (upstream fragment) and PyvcEdelpGhdwnF/PyvcEdelpGhdwnR (downstream fragment) and were then fused together by overlapping PCR using the flanking primer pairs (yvcEleadlacZF and PyvcEdelpGhdwnR). This fragment was then restricted with HindIII and EcoRI, ligated into pGHost4 that was similarly restricted, and transformed into E. coli strain EC101 to generate plasmid pDN3000. The pDN3000 plasmid was then transformed into B. subtilis wild-type strain 168, and the procedure of Biswas et al. (23) was followed to generate strain DN3000, containing a 141-bp markerless deletion within the cwlO leader sequence. This construct was verified by PCR. This strategy was also used to make strain DN3005, which contains a markerless deletion of the cwlO leader sequence in the LS047 strain background.
A similar approach was taken to delete the putative SigB-type promoter that drives expression of a cwlO antisense RNA. DNA fragments flanking the 32-base-pair SigB promoter-containing region (to be deleted) were generated by Phusion-mediated PCR using primer pairs yvcE_AS promoter up fwd (B)/yvcE_AS promoter up rev (upstream fragment) and yvcE_AS promoter do fwd/yvcE_AS promoter do rev (H) (downstream fragment). These fragments were fused by strand overlap PCR using the yvcE_AS promoter up fwd (B)/yvcE_AS promoter do rev (H) primer pair. This fragment was then restricted with BamHI-HindIII, cloned into BamHI-HindIII-restricted pGHost4, and transformed into E. coli strain EC101 to propagate plasmid pLIS010. Deletion of the SigB-type promoter from the chromosome of wild-type strain 168 was performed as described by Biswas et al. (23) to generate strain LS047, which was verified by PCR.
The pGhost4-based strategy of Biswas et al. (23) was employed to generate markerless in-frame deletions of regions within the N-terminal domain of the CwlO protein. The polyserine domain (extending from amino acid S295 to S340) was deleted by this procedure using primer pairs DPSyvcEUPF/DPSyvcEUPR (upstream flanking DNA fragment) and DPSyvcEdwnF/DNPSyvcEdwnR (downstream flanking DNA fragment), generating plasmid pDN3020 and strain DN3020, which has the desired deletion. This strategy was also used to generate strain DN3021 with an in-frame deletion of 288 amino acids in the amino-terminal domain of CwlO (the 45-amino-acid polyserine domain and 243 amino acids upstream of this domain) using primer pairs DNTyvcEUPF/DNTyvcEUPR (upstream flanking DNA fragment) and DNPSyvcEdwnF/DNPSyvcEdwnR (downstream flanking DNA fragment) to generate the plasmid pDN3021.
Strains LS090, DN3015, and DN3022 were generated by transforming strains LS047, DN3000, and DN3020, respectively, with a 2.5-kb DNA fragment (ΔlytE::cat) generated by PCR amplification of chromosomal DNA from strain LS089 with the primer pair lytEupfwd/lytEdorev.
The procedure described by Bloor and Cranenburgh (24) was employed to delete amino acids 52 to 102 (CwlOΔN1) and 231 to 288 (CwlOΔN3) within the N-terminal CwlO domain. DNA fragments flanking the N1 region (to be deleted) were generated by PCR using primer pairs cwlON1delUPFb/cwlON1delUPR (upstream fragment) and cwlON1delDWNF/cwlON1delDWNRb (downstream fragment). Likewise, DNA fragments flanking the N3 region (to be deleted) were generated by PCR using primer pairs cwlON3delUPFb/cwlON3delUPR and cwlON3delDWNF/cwlON3delDWNRb. These pairs of fragments were fused with a cassette containing the cat gene, encoding resistance to chloramphenicol, flanked by dif sites, amplified from plasmid template pDG1661 with the primer pair catdifffwd/catdiffrev, and transformed into B. subtilis wild-type strain 168, integrating into the chromosome by a double-crossover event, thereby replacing the region to be deleted with the antibiotic cassette. Selected integrants were grown in the absence of antibiotic for 24 h, streaked onto LB plates, and screened for antibiotic sensitivity, generated by excision of the antibiotic cassette. Deletion of CwlO regions N1 (strain DN3025) and N3 (strain 3027) was verified by diagnostic PCR and DNA sequencing.
Transcriptional analysis.
The transcriptional start point of the antisense cwlO transcript was mapped by 5′-end rapid amplification of cDNA ends (RACE) using total RNA (2 μg) isolated from strain 168 in post-exponential phase (T2) grown in LPDM. Primer yvcEASSP1 was used for first-strand cDNA synthesis, and primers yvcEASSP2 and yvcEASSP3B were used for subsequent nested amplification and sequencing according to the instructions in the 5′/3′ RACE second-generation kit (Roche). Direct sequencing of the final amplicon identified the transcriptional start point downstream of canonical SigB promoter elements.
Primer extension analysis was used to identify the site at which the cwlO leader sequence is processed. Primer extensions were performed with 10-μg total RNA samples isolated from strains 168, DN3000, CCB034, CCB078, and CCB294 grown with and without IPTG (isopropyl-β-d-thiogalactopyranoside) inducer as required. Primers yvcEPE4, yvcEPE5, and yvcEPE7 were labeled with 32P ATP and used to synthesize first-strand cDNA using SuperScript III reverse transcriptase as previously described (25).
Northern analysis was performed with total RNA isolated as previously described (25). To measure mRNA half-life, rifampin was added to cultures at a final concentration of 125 μg/ml and samples harvested at the designated times thereafter. Total mRNA was prepared, separated by electrophoresis, and transferred to nylon membranes, and cwlO mRNA and ascwlO mRNA were visualized using digoxigenin-labeled riboprobes. Riboprobes to selectively detect the sense and antisense transcripts were generated using template fragments amplified by PCR with T7 RNA polymerase using primers yvcEprobe fwd/yvcEprobe rev(T7) and ASprobefwd(T7)/ASproberev and T7 RNA polymerase according to the instructions in the Roche Northern kit.
Protein analysis.
CwlO protein without a signal sequence was purified from E. coli strain BL21(DE3) harboring plasmid pLIS012. A DNA fragment carrying the required region of cwlO was amplified by PCR using primer pair yvcE5′fwd(NdeI)/yvcEpET3′ and was inserted into the NdeI/XhoI sites of pET21b to generate plasmid pLIS012. Protein expression was performed as described by Howell et al. (26).
Western blot analysis was carried out essentially as previously described (26). Culture aliquots were harvested at the indicated time points and centrifuged to separate the cells from the medium. The protein content of cell lysates was quantitated using the bicinchoninic acid (BCA) protein assay kit (Novagen). Proteins in the medium samples were precipitated with ice-cold trichloroacetic acid (10% [vol/vol] final concentration), and the pellet was washed twice with ice-cold acetone and resuspended in King-Laemmli (27) sample buffer. The amount of precipitated protein loaded onto each lane of the gel was normalized to units of optical density at 600 nm (OD600) multiplied by volume.
Polyclonal antiserum was raised against purified CwlO protein in New Zealand White rabbits according to the standard procedure used by Dundee Laboratories (Dundee, Scotland). The CwlO antibody was affinity purified using CwlO antigen coupled to CnBr-activated Sepharose as previously described (26).
LTQ-Orbitrap Velos MS for identification of CwlO peptides.
The extracellular proteome analysis was performed for wild-type B. subtilis grown in complete medium and harvested during the exponential growth phase at an OD540 of 0.8 and 1 h after transition into the stationary phase as described previously (28). The 25-kDa CwlO processing product and the 50-kDa CwlO protein were cut from the extracellular proteomes of exponential- and stationary-phase cells, respectively, and trypsin digested as described by Chi et al. (29). Tryptic peptides were subjected to reversed-phase column chromatography (self-packed C18 column; 100-μm inner diameter by 200 mm) operated on a Easy-nLC II instrument (Thermo Fisher Scientific, Waltham, MA). Elution was performed with a binary gradient of buffer A (0.1% [vol/vol] acetic acid) and buffer B (99.9% [vol/vol] acetonitrile [ACN], 0.1% [vol/vol] acetic acid) over a period of 100 min with a flow rate of 300 nl/min. Mass spectrometry (MS) and tandem MS (MS/MS) data were acquired with the LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source as described previously (28). CwlO was identified by searching all MS/MS spectra in “dta” format against a B. subtilis target-decoy protein sequence database (30) that was extracted from UniprotKB release 12.7 using Sorcerer-SEQUEST (SEQUEST v. 2.7 rev. 11 Thermo Electron including Scaffold_3_00_02; Proteome Software Inc., Portland, OR). The SEQUEST search was carried out with following parameters: a parent ion mass tolerance of 10 ppm and a fragment ion mass tolerance of 1.00 Da. Up to two tryptic miscleavages were allowed. Methionine oxidation (+15.994915 Da) and cysteine carbamidomethylation (+57.021464 Da) were set as variable modifications. Proteins were identified by at least two peptides, applying a stringent SEQUEST filter. SEQUEST identifications required at least ΔCn scores of greater than 0.10 and XCorr scores of greater than 2.2, 3.3, and 3.75 for doubly, triply, and quadruply charged peptides.
RESULTS
The RNA leader sequence acts negatively to regulate cwlO expression.
There are several features of the cwlO locus that suggest its expression is regulated by mechanisms in addition to WalRK-mediated control (Fig. 1A). Here we have focused our investigation on the 187-nucleotide cwlO leader sequence that is predicted by the mFold program to form a secondary structure as shown in Fig. 1B (31). To determine its contribution to cwlO expression, five transcriptional green fluorescent protein (GFP) gene fusions were constructed (PAgfp to PEgfp), each with increasing lengths of leader sequence extending beyond the 3′ end of the promoter region. The PAgfp fusion (strain DN2021) has the complete cwlO promoter and no leader sequence (red sequence in Fig. 1C). The PBgfp (strain DN2070), PCgfp (strain DN2071), PDgfp (strain DN2072), and PEgfp (strain DN2073) fusions have the complete cwlO promoter (red sequence) plus 60 bp (blue sequence), 122 bp (blue and green sequences), 146 bp (blue, green, and orange sequences), and 172 bp (blue, green, orange, and purple sequences), respectively, of the leader sequence fused to the gfpmut3 reporter gene (Fig. 1B and C). The leader sequence in PEgfp excludes the cwlO ribosome binding site (i.e., is shortened by 13 bp) to ensure that it is translated using the GFP gene ribosome binding site like the other fusions (Fig. 1C). Similar colors indicate the position of each sequence within the structure of the leader region (Fig. 1B). In exponentially growing cells, the PEgfp fusion (which contains the entire leader sequence) is expressed at a maximum level of ∼100 to 200 activity units, whereas the PAgfp fusion (with no leader sequence) is expressed at a level of ∼800 to 1,000 activity units (Fig. 1D, panels i and v). Importantly, expression of both fusions is still confined to exponentially growing cells, and it decreases to a very low level at the onset of stationary phase. Thus, the leader sequence has a negative effect on the level of cwlO expression but does not affect its temporal regulation.
Expression of the PBgfp fusion (∼800 activity units) is similar to that of PAgfp, showing that the blue leader region alone has little effect on cwlO expression (Fig. 1D, panel ii). However, addition of the green leader region (PCgfp fusion) reduces maximal expression 2-fold (∼400 activity units), while addition of the orange leader region (PDgfp) further reduces maximal expression to the same level as for the PEgfp fusion that has the complete leader sequence (Fig. 1D, panels iii, iv, and v). We conclude that there is a regulatory element partly or completely located within the green/orange region of the leader sequence that acts to decrease cwlO expression in exponentially growing cells.
The leader sequence destabilizes the cwlO mRNA.
To determine the mechanism by which the leader sequence negatively regulates expression, the abundances of cwlO transcript in wild-type strain 168 and in strain DN3000 (leaderless cwlO) were established. Our results (Fig. 2A) show that the leaderless cwlO transcript accumulates to a significantly higher level in DN3000 cells than does the wild-type transcript in strain 168 cells, a feature that is particularly evident at the 220- and 250-min points of exponential growth. This suggests that the leader sequence destabilizes the cwlO transcript. To test this hypothesis, the half-lives of the wild-type and leaderless cwlO transcripts in exponentially growing cells were measured. Our results (Fig. 2B) show that the wild-type cwlO transcript is very unstable, with a half-life of ∼1 min (left panel). In contrast, the leaderless cwlO transcript (strain DN3000) is very stable, with only a small decrease in transcript level during the first 4 min of the experiment (right panel). However, the transcript level decreases more rapidly thereafter, perhaps due to prolonged exposure to rifampin. A transcript half-life of ∼6 min is estimated for the duration (8 min) of the experiment. These results show that the leader sequence destabilizes the cwlO transcript in exponentially growing cells.
FIG 2.
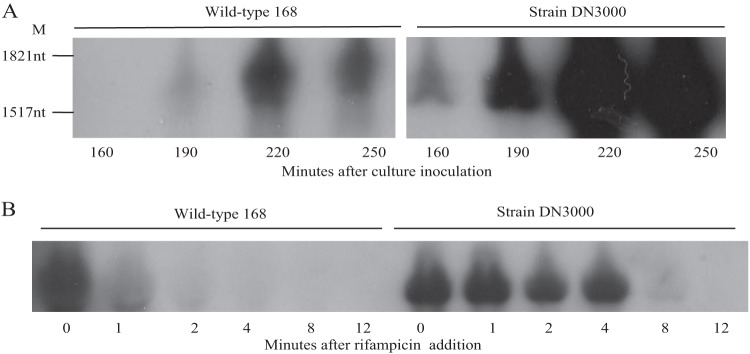
Effect of the cwlO leader sequence on cellular transcript levels. (A) RNA was harvested from cultures of wild-type strain 168 and strain DN3000 (in which the leader sequence is deleted) growing in HPDM at the times indicated and analyzed by Northern analysis. M, RNA size markers. (B) Determination of the half-lives of the wild-type and leaderless cwlO transcripts. Rifampin was added at T0 and RNA prepared from samples harvested subsequently at the indicated times. The cwlO transcript was detected by Northern analysis.
The cwlO transcript is destabilized by RNase Y cleavage at a stem-loop structure within the leader sequence.
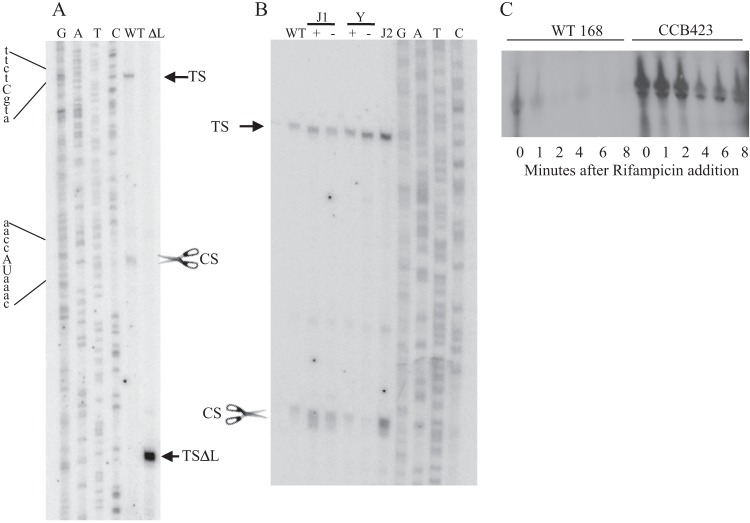
The potential of the leader sequence to form secondary structure suggests that it may play a role in processing of the cwlO transcript. This hypothesis was tested by primer extension analysis using primers located within the cwlO open reading frame (Fig. 3A). Two reverse transcripts were observed (Fig. 3A, WT): the larger transcript (TS) initiates at the base expected for transcription from the WalRK-activated σA-type promoter previously reported by Bisicchia et al. (9), while the smaller transcript (CS) initiates within the leader sequence, beginning with the sequence UAAAC located in SL2 (Fig. 1B). We have shown by transcriptional fusion analysis that the sequence between the −10 motif of the σA-type promoter and the cwlO initiation codon does not contain a promoter (Fig. 1C and data not shown). Therefore, we conclude that the CS band corresponds to cleavage of the cwlO leader at the ACCA/UAAA sequence (Fig. 1B). These reverse transcript endpoints (TS and CS) were verified with three separate primers (data not shown). A satisfying feature of this result is that the CS cleavage site is positioned within the region from nt 62 to 120 (green) of the leader, which was shown by transcriptional fusion analysis to be important for reduction of the cwlO transcript level during exponential growth (Fig. 1). Furthermore, both the location and nucleotide sequence of the B. subtilis cwlO CS cleavage site are conserved in the leader sequences of orthologues in Bacillus amyloliquefaciens and Bacillus atrophaeus (data not shown).
FIG 3.
Identification and analysis of RNase Y cleavage of the cwlO RNA leader sequence. (A) Primer extension analysis was performed on RNA harvested from exponentially growing cultures of wild-type strains 168 (WT) and DN3000 (ΔL) with the primers outlined in Materials and Methods. Samples were electrophoresed adjacent to a sequencing ladder (GATC). The transcription initiation sites (TS and TSΔL) and the cleavage site (CS with scissors) are shown. The sequence surrounding the initiation point of transcription (in lowercase letters) and the cleavage site (in uppercase letters) are show at the left. (B) The identity of the RNase that cleaves the cwlO leader sequence was determined using strains with a specific RNase deleted (RNase J2) or under conditional expression (RNase J1 and Y). Primer extension analysis was performed using primer PE4 on RNA extracted from exponentially growing cultures of the following strains: WT, wild-type strain 168; J1+, CCB034 (PspacrnjA) with IPTG; J1−, CCB034 (PspacrnjA) without IPTG; Y+, CCB294 (Pspacrny) with IPTG; Y−, CCB294 (Pspacrny) without IPTG; J2, CCB078 (ΔrnjB). Reverse transcripts were electrophoresed adjacent to a sequencing ladder (GATC). TS and CS are specified in panel A. (C) The contribution of RNase Y to cwlO transcript stability was determined by measuring the half-life of the transcript after addition of rifampin to exponentially growing wild-type (WT168) and Δrny (CCB423) cells. The level of RNA was determined by Northern analysis.
Primer extension analysis was also performed with RNA isolated from strain DN3000 (expressing the leaderless cwlO). A single reverse transcript (TSΔL) was observed, which initiates at the base expected for transcription from the previously identified σA-type promoter and is consistent with precise deletion of the expected 141 bases of the cwlO leader sequence. In agreement with the Northern analysis, the intensity of the reverse transcript generated with RNA harvested from DN3000 (leaderless cwlO) cells is higher than that generated with RNA from wild-type 168 (cwlO with leader) cells, despite transcription of both cwlO genes being directed by the same WalRK-controlled promoter (Fig. 3A). This primer extension analysis confirms that transcription of both the wild-type and leaderless cwlO transcripts initiates at the WalRK-controlled σA-type promoter and that control of cellular cwlO RNA levels occurs posttranscriptionally.
The putative RNA cleavage site (ACCA/UAAA) is located within a small stem-loop structure positioned between stem-loop structures SL1 and SL2 (Fig. 1B). To identify the RNase that cleaves the leader at this position, we performed primer extension analysis on RNA from strains depleted of RNase J1 (CCB034 PspacrnjA) or RNase Y (CCB294 Pspacrny) or with RNase J2 deleted (CCB078 ΔrnjB). The results shown were obtained with oligonucleotide yvcEPE7 (Fig. 3B) and confirmed with oligonucleotide yvcEPE4 (data not shown). Cleavage of the cwlO leader sequence occurs normally in cells depleted of RNase J1 (compare J1 lanes + and −) or deleted of RNase J2 (lane J2) but is significantly reduced in cells depleted of RNase Y (compare Y lanes + and −). This primer extension analysis also reveals that RNase Y cleavage results in a smear of bands initiating around the cleavage site (e.g., see CS in lane J1+ or J2 in Fig. 3B) that contrasts with the discrete band observed for the initiation point (TS) of transcription (Fig. 3A and B). It is likely that RNase Y either cleaves the leader sequence at several adjacent bases or cleaves at a single position which is then subject to exonucleolytic cleavage.
To establish if RNase Y cleavage within the leader sequence is the cause of cwlO transcript instability, we compared the half-lives of cwlO mRNA in wild-type strain 168 and in a strain (CCB423 Δrny) with the rny gene deleted. The full-length transcript has a half-life of ∼1 min in wild-type cells but of ∼4 min in the strain with rny deleted. It is clear that the cwlO message is stabilized in a strain lacking RNase Y (Fig. 3C). It is unlikely that the poor growth of strain CCB423 (Δrny) is the cause of cwlO transcript stabilization, since stabilization is also observed in strain DN3000, which grows at the same rate as wild-type strain 168.
We conclude that the cwlO mRNA is destabilized in vivo by RNase Y cleavage of the leader sequence at, or in the vicinity of, the ACCA/UAAA sequence.
Heterogeneous antisense cwlO transcripts are expressed from a σB-type promoter in stationary-phase cells.
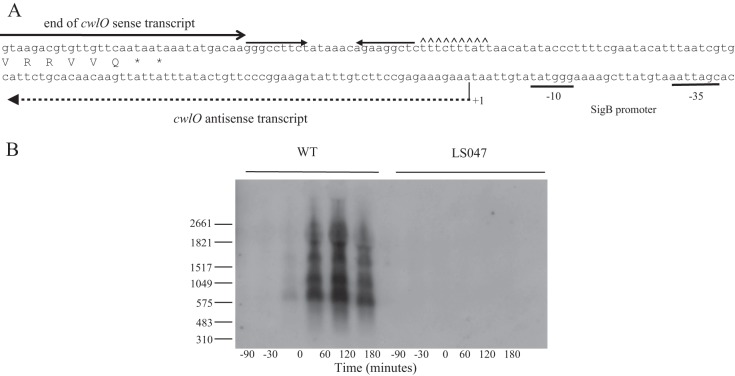
Transcriptome analysis of phosphate-limited cells suggested the presence of an antisense transcript that may be expressed from a putative σB-type promoter located downstream of the cwlO terminator (15, 18). This suggests an involvement of antisense RNA in regulating CwlO expression under stress conditions (Fig. 4A) (15, 18, 32). This was investigated by Northern analysis of RNA harvested from cells throughout a phosphate limitation cycle. The results show expression of antisense cwlO transcripts of heterogeneous length beginning at the onset (T0) of phosphate limitation (Fig. 4B). The two smaller transcripts (∼700 and ∼1,200 nucleotides) terminate within the cwlO open reading frame, while the two larger transcripts (∼1,700 and ∼2,200 nucleotides) extend beyond the cwlO control region into the upstream yvcD gene. The antisense transcript is not observed when the putative σB-type promoter is deleted (strain LS047) (Fig. 4B) or in a strain with the sigB gene deleted (PB344) (data not shown), confirming that it is transcribed by an RNA polymerase σB holoenzyme.
FIG 4.
Expression of cwlO sense and antisense transcripts. (A) Schematic diagram of the nucleotide sequence at the 3′ end of the cwlO operon. The cwlO terminator is shown by inverted arrows followed by carets located downstream of the open reading frame. The location of the cwlO sense transcript is indicated by a full arrow. The SigB promoter motifs are underlined. The initiation point of transcription of the antisense transcript is indicated by +1 and its length by a broken line. (B) Detection of the cwlO antisense RNA by Northern blotting. Transcription of the heterogeneous antisense transcript begins in wild-type cells (WT) growing in LPDM at the onset of phosphate limitation (T0), and the transcript persists for up to 3 h (T3). The transcript is not detected in phosphate-limited cells of strain LS047 (ΔPsigB), which has the SigB promoter deleted. RNA size markers are shown on the left.
In summary a heterogenous population of cwlO antisense RNA transcripts, the larger of which extends upstream from the cwlO promoter region, is expressed from a σB-type promoter in phosphate-limited cells.
CwlO protein levels are regulated by the cwlO mRNA leader.
We next sought to determine the contribution of the cwlO mRNA leader sequence to cellular CwlO protein levels. This was established by Western blot analysis using a CwlO-specific antibody. When growth is in HPDM or LPDM, the level of cell-associated CwlO protein is significantly higher in strain DN3000 (leaderless cwlO) than in wild-type strain 168 (Fig. 5A and B). Moreover, this high level in strain DN3000 results in CwlO being cell associated for longer after the onset of phosphate limitation, albeit at a low level. This shows that increased transcript stability extends the period of CwlO cell association (Fig. 5B).
FIG 5.
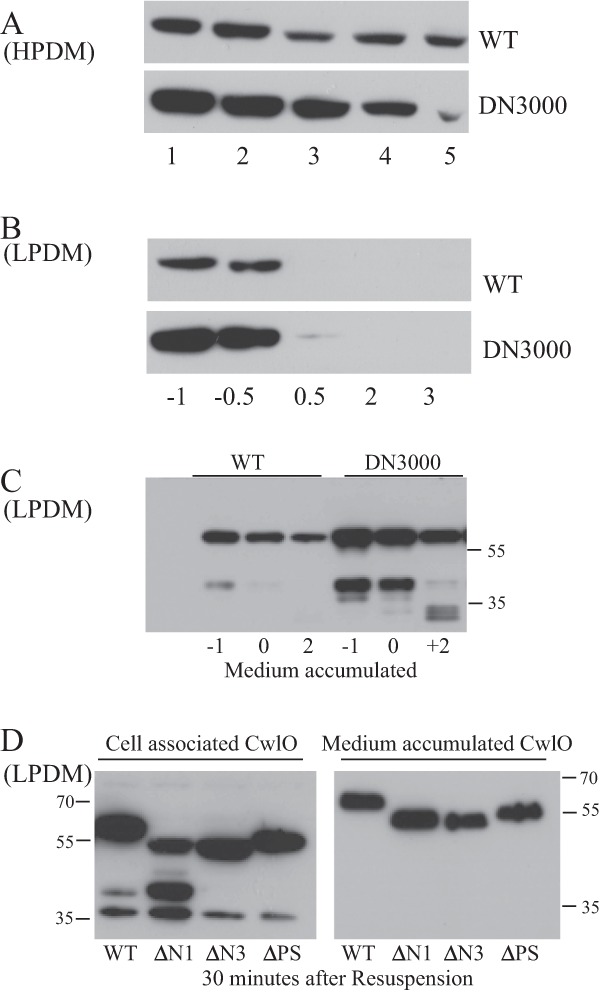
Determination of CwlO protein levels, both cell associated and accumulated in the medium. CwlO protein was detected by Western analysis using a specific antibody. (A) Cell extracts (15 μg total cell protein) were prepared from cells of wild-type (WT) and DN3000 (with the cwlO leader sequence deleted) strains grown in HPDM. Cells were growing exponentially (lanes 1 and 2), at the transition phase (lane 3), or in post-exponential growth phase (lanes 4 and 5). (B) Protein extracts (15 μg total cell protein) from cells grown in LPDM. Time is given in hours before (−) or after (+) the onset of phosphate limitation. (C) Accumulation of CwlO protein in the medium of cells grown in LPDM. The signal represents the amount of CwlO protein that had accumulated in 0.2 OD600 cell equivalent of medium. Time is measured in hours before (−) and after (+) the onset (0) of phosphate limitation. (D) Secretion of full-length (WT) and amino-terminally truncated (ΔN and ΔPS) CwlO variants from exponentially growing cells. Exponentially growing cultures (OD600, ∼0.4) of wild-type strain 168 (WT) and strains expressing CwlO variants (DN3025 [CwlOΔN1], DN3027 [CwlOΔN3], and DN3020 [CwlOΔPS]) were harvested, washed, and resuspended in the same volume of fresh medium. The amount of each cell-associated (left panel) and medium-accumulated (right panel) CwlO variant was determined at 30 min after resuspension and compared with that found in wild-type strain 168. Samples contained 20 μg of total cell protein and 0.2 OD600 cell equivalent of medium-accumulated protein. Molecular weight markers are shown.
Two forms of CwlO accumulate in the growth media of exponentially growing cells of wild-type strain 168 and strain DN3000 (Fig. 5C). The bulk of secreted CwlO protein is similar in size (∼50 kDa [note that CwlO migrates anomalously on SDS-polyacrylamide gels]) to the cell-associated protein. Mass spectrometry analysis shows that it lacks only the signal sequence, with the preprotein probably being cleaved at ASA (amino acids 28 to 30). The second CwlO species is ∼25 kDa in size and is detected only in the medium of exponentially growing cells, but it increases when CwlO is overexpressed (Fig. 5C). It is generated by cleavage of CwlO in the vicinity of amino acid 274, a processing event that separates the N-terminal (amino acids 1 to 290) and catalytic (amino acids 343 to 473) domains. Mass spectrometry analysis shows that the ∼25-kDa species comprises the carboxy-terminal NlpC/P60 domain that encodes the endopeptidase activity. Deletion of the cwlO leader sequence (DN3000) increases the level of the two species that accumulate in the medium, consistent with increased stability of the leaderless cwlO transcript (Fig. 5C).
An intact CwlO N-terminal domain is essential for function but not for cell association in vivo.
To establish how the N-terminal domain and polyserine domain (a domain also present in the LytF autolysin that may contribute to cell wall binding [33]) contribute to CwlO accumulation in the cell and the medium, three strains were constructed, each expressing a CwlO variant. Strains DN3025 (CwlOΔN1, with amino acids 52 to 102 deleted) and DN3027 (CwlOΔN3, with amino acids 231 to 288 deleted) express CwlO variants with deletions in the amino-terminal region, while strain DN3020 (CwlOΔPS, with amino acids 295 to 332 deleted) expresses a CwlO variant with the polyserine domain deleted. We repeatedly failed to construct a strain with amino acids 145 to 201 deleted. Taking advantage of the synthetic lethality of CwlO and LytE (9, 10), we tested the functionality of the CwlO variants by attempting to delete lytE in each strain. This was successfully achieved only in strain DN3020 (CwlOΔPS), showing that normal CwlO function in vivo requires an intact N-terminal domain but that the polyserine domain is dispensable.
We tested each CwlO variant for cell association and the kinetics of its accumulation in the medium. Aliquots from exponentially growing cultures were washed and resuspended in the same volume of fresh growth medium. Samples were then prepared from cells and from the medium at specific time intervals, and CwlO levels were determined by Western analysis using a CwlO-specific antibody. At 30 min after resuspension, the level of each CwlO variant protein that is cell associated and that has accumulated in the medium is similar to that observed for the wild-type protein (Fig. 5D). The cell-associated CwlOΔN1 protein variant is processed differently from the other proteins, with the unique cell-associated processed species not detected in the medium, suggesting that it either is secreted or is unstable in the medium. We conclude that despite being nonfunctional, the CwlOΔN1 and CwlOΔN3 variants associate with the cell and accumulate in the medium similarly to wild-type CwlO protein. Thus, an intact N-terminal domain is essential for CwlO function in vivo but does not significantly influence cell association or secretion into the medium.
Phenotypic analysis of strains mutated in cwlO regulation.
The extent to which CwlO expression is regulated shows the importance of controlling its cellular level and activity. Therefore, we sought to assess the contribution of transcript destabilization and antisense RNA expression to CwlO cellular function. Our results show that the growth profile and cellular morphology of strains are unaffected by transcript stabilization (DN3000, leaderless cwlO), by abolition of the cwlO antisense RNA (LS047, ΔPsigB) or when both of these features are combined (data not shown). Furthermore, no significant phenotype was observed when each of these mutations was combined with deletion of lytE.
Since the sense and antisense cwlO RNA transcripts are normally expressed during exponential growth and stationary phase, respectively, we tested whether expression of the cwlO antisense RNA affects the transition between these two growth phases. Overnight cultures (∼16 h of growth in LB at 37°C with shaking) of wild-type strain 168 and strain LS047 (with the SigB promoter deleted) were used to inoculate fresh medium, and growth was monitored. We consistently observe that strain LS047 begins exponential growth earlier than wild-type strain 168, suggesting that the cwlO antisense transcript influences exit from the stationary-phase state (see Fig. S1 in the supplemental material).
In view of the role played by CwlO and LytE in cell wall metabolism, we tested the susceptibility to five cell wall-acting antibiotics (bacitracin, vancomycin, penicillin G, fosfomycin, and d-cycloserine) and to lysozyme of strains (i) expressing a stabilized transcript, (ii) unable to express the cwlO antisense RNA alone, and (iii) with each feature in combination with deletion of ΔlytE as described in Materials and Methods. Neither cwlO regulatory mutation significantly affected susceptibility to any of the antibiotics or to lysozyme. However, all strains harboring a ΔlytE mutation (i.e., that express only the CwlO autolysin) are more susceptible to bacitracin and fosfomycin, the two antibiotics that inhibit the intracellular steps of lipid II synthesis. This suggests that cwlO expression, and hence WalRK activation, is sensitive to the intracellular levels of peptidoglycan metabolic intermediates (data not shown).
In summary, removal of the cwlO leader sequence and of the capability to synthesize a cwlO antisense transcript has little phenotypic impact.
DISCUSSION
cwlO message instability makes cellular CwlO protein levels sensitive to growth conditions.
We report that the cwlO transcript is very unstable, with a half-life of approximately 1 min, in exponentially growing cells. The instability is caused by RNase Y cleavage at an ACCA/UAAA site within the 187-nucleotide leader sequence (Fig. 1B and 4). The RNase Y endoribonuclease is a global regulator of RNA stability that influences the abundance of ∼1,000 transcripts and generally increases half-lives approximately 4-fold (34, 35). Deletion of the cwlO leader sequence or of the rny gene stabilizes the message and increases the amount of CwlO protein expressed. This is manifest in increased levels of CwlO protein both cell associated (Fig. 5A and B) and accumulated in the medium (Fig. 5C). We consistently observe CwlO protein associated with phosphate-limited cells for longer periods when the RNA transcript is stabilized. This suggests that by lowering the level of CwlO protein, transcript destabilization may function to restrict CwlO endopeptidase activity to actively dividing cells.
A second consequence of a highly unstable transcript is that it makes cwlO expression highly responsive to the conditions that activate the WalRK two-component signal transduction system, its only known transcriptional regulator (9, 14). When WalRK is active, cwlO transcript is expressed at a high level. However, the level of cwlO mRNA will be reduced to a low cellular level within a very short period of WalRK deactivation due to its high instability. This is evident in the very rapid loss of cell-associated CwlO protein when cells become phosphate limited, a condition of greatly reduced WalRK activity. Thus, RNase Y-mediated initiation of cwlO transcript degradation is not regulatory in the sense that message stability varies under different growth or environmental conditions; it is not thought that cellular RNase Y activity varies significantly (34). However, extreme transcript instability makes protein production highly responsive to transcriptional initiation, in this instance making CwlO protein production highly responsive to the conditions that (de)activate the WalRK two-component system. Thus, transcript instability contributes to the overall regulation of CwlO and its cellular function, as outlined below (5, 6, 9).
It is interesting that RNase Y-mediated control of cwlO transcript stability was not reported in any of three recent global studies (35, 36, 37). However, a close examination of the global transcript profiles obtained using tiled arrays (see Fig. S1 in reference 37) shows that upon RNase Y depletion, the signal at the 5′ end of the cwlO transcript increases while the signal at the 3′ end of the transcript is unaffected. This suggests that under normal conditions, RNase Y cleavage of the cwlO transcript generates two transcripts, with the released fragment of the leader sequence being particularly unstable (38). In support of this interpretation, RNase Y also appears to have a differential effect on degradation of the 5′ and 3′ ends of the srfAA single-cistron transcript (see Fig. S1 in reference 37). RNase Y depletion results in an increased signal for a short region at the 5′ end of the srfAA transcript without affecting the signal for the remainder of this very long transcript. Crucially, Nicolas et al. (18) have reported that srfAA (like cwlO) has a 300-nucleotide 5′ leader sequence (named S126) with the capability to form secondary structure. We posit that the S126 region of srfAA may also be a target for RNase Y cleavage, similar to that seen for cwlO. Thus, the failure to identify RNase Y processing of the cwlO and srfAA transcripts in these global studies may be due to RNase Y cleavage of some transcripts leading to the generation of RNA fragments that are differentially sensitive to further degradation. Thus, the difference in the total signal for each cistron in RNase Y-replete and -depleted cells may fall below the significance threshold applied (35, 36, 37). These observations suggest that additional details on the cellular role of RNase Y remain to be established.
CwlO d,l-endopeptidase: a highly regulated enzyme.
Autolysins have the potential to disrupt the integrity of the cell wall, with potentially lethal consequences. For example, unregulated cwlO transcription, generated by a constitutively active WalR response regulator, compromises the integrity of the cell wall to the extent that protoplasts emerge from the sacculus in isotonic medium (39, 40). The degree of control required to allow normal autolysin function without compromising cell viability is strikingly illustrated by the multiplicity of controls on CwlO expression and enzyme activity. Transcription of cwlO is activated by the WalRK two-component signal transduction system, its only know transcriptional regulator, under conditions conducive to cell growth (9). The cwlO transcript is highly unstable, ensuring that transcript is present and CwlO protein is produced only when WalRK is activated (this study). Transcript stability and/or translation may also be regulated by production of an antisense cwlO transcript under stress conditions (this study). We were unable to achieve clear and reproducible results by producing the cwlO sense and antisense transcripts in exponentially growing cells concomitantly (data not shown). However, cells unable to produce an antisense transcript exit the lag phase and resume exponential growth more rapidly, suggesting that it may play a regulatory role under specific growth conditions (see Fig. S1 in the supplemental material). CwlO is rapidly secreted from the cell, as shown by the rapid disappearance of cell-associated protein upon the onset of phosphate limitation (this study). Furthermore, CwlO is enzymatically active only when associated with the FtsEX ATP-like transporter, which is positioned within the membrane and requires ATP hydrolysis (5, 6). These requirements not only restrict CwlO enzymatic activity to the membrane-proximal layer of peptidoglycan but also ensure that the secreted enzyme is inactive (D. Noone and K. M. Devine, unpublished results). Moreover, a limiting cellular level of FtsEX may explain why increasing the cell-associated CwlO has little phenotypic consequence, with excess protein being secreted from the cell without being activated. While the CwlO N-terminal domain is important for function and required for interaction with FtsEX, it does not appear to play a role in retaining CwlO in the cell. Finally, the cellular location of CwlO enzymatic activity is restricted by its dependence on FtsEX and the actin-like protein Mbl (5, 6). This multiplicity of control mechanisms ensures that the CwlO d,l-endopeptidase enzyme is active only at specific cellular locations and under specific growth conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ciaran Condon for the gift of strains and for helpful discussion on RNases.
This work was supported by Science Foundation Ireland principal investigator 08/1N.1/B1859 award to K.M.D.
Footnotes
Published ahead of print 25 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00986-13.
REFERENCES
- 1.Vollmer W, Joris B, Charlier P, Foster SJ. 2008. Bacterial peptidoglycan (murein) hydroylases. FEMS Microbiol. Rev. 32:259–286. 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- 2.Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262 [DOI] [PubMed] [Google Scholar]
- 3.Sekiguchi J, Yamamoto H. 2012. Cell wall structure of E. coli and B. subtilis, p 115–148 In Sadaie Y, Matsumoto K. (ed), Escherichia coli and Bacillus subtilis: the frontiers of molecular microbiology revisited. Research Signpost, Kerala, India [Google Scholar]
- 4.Yamaguchi H, Furuhata K, Fukushima T, Yamamoto H, Sekiguchi J. 2004. Charaacterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J. Biosci. Bioeng. 98:174–181. 10.1016/S1389-1723(04)00262-2 [DOI] [PubMed] [Google Scholar]
- 5.Domínguez-Cuevas P, Porcelli I, Daniel RA, Errington J. 2013. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol. Microbiol. 89:1084–1098. 10.1111/mmi.12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 89:1069–1083. 10.1111/mmi.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa S, Hara Y, Ohnishi R, Sekiguchi J. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847. 10.1111/j.1365-2958.2008.06211.x [DOI] [PubMed] [Google Scholar]
- 9.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180–200. 10.1111/j.1365-2958.2007.05782.x [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto M, Ooiwa S, Sekiguchi J. 2012. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of d,l-endopeptidase activity at the lateral cell wall. J. Bacteriol. 194:796–803. 10.1128/JB.05569-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto H, Kurosawa S, Sekiguchi J. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185:6666–6677. 10.1128/JB.185.22.6666-6677.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzberg LI, Powell L, Hokamp K, Botella E, Noone D, Devine KM. 2013. The WalRK (YycFG) and σ(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol. Microbiol. 87:180–195. 10.1111/mmi.12092 [DOI] [PubMed] [Google Scholar]
- 13.Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell 11:399–409. 10.1016/j.devcel.2006.07.017 [DOI] [PubMed] [Google Scholar]
- 14.Bisicchia P, Lioliou E, Noone D, Salzberg LI, Botella E, Hübner S, Devine KM. 2010. Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate-limited Bacillus subtilis cells. Mol. Microbiol. 75:972–989. 10.1111/j.1365-2958.2009.07036.x [DOI] [PubMed] [Google Scholar]
- 15.Botella E, Hübner S, Hokamp K, Hansen A, Bisicchia P, Noone D, Powell L, Salzberg LI, Devine KM. 2011. Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology 157:2470–2484. 10.1099/mic.0.049205-0 [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen S, Nielsen HB, Jarmer H. 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73:1043–1057. 10.1111/j.1365-2958.2009.06830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irnov I, Sharma CM, Vogel J, Winkler WC. 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38:6637–6651. 10.1093/nar/gkq454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller JP, An Z, Merad T, Hancock IC, Harwood CR. 1997. Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143:947–956. 10.1099/00221287-143-3-947 [DOI] [PubMed] [Google Scholar]
- 22.Botella E, Fogg M, Jules M, Piersma S, Doherty G, Hansen A, Denham EL, Le Chat L, Veiga P, Bailey K, Lewis PJ, van Dijl JM, Aymerich S, Wilkinson AJ, Devine KM. 2010. pBaSysBioII: an integrative plasmid generating gfp transcriptional fusions for high-throughput analysis of gene expression in Bacillus subtilis. Microbiology 156:1600–1608. 10.1099/mic.0.035758-0 [DOI] [PubMed] [Google Scholar]
- 23.Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement for gram-positive bacteria. J. Bacteriol. 175:3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloor AE, Cranenburgh RM. 2006. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 72:2520–2525. 10.1128/AEM.72.4.2520-2525.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noone D, Howell A, Devine KM. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J. Bacteriol. 182:1592–1599. 10.1128/JB.182.6.1592-1599.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell A, Dubrac S, Noone D, Varughese KI, Devine KM. 2006. Interactions between the YycFG and PhoPR two-component systems in Bacillus subtilis: the PhoR kinase phosphorylates the non-cognate YycF response regulator upon phosphate limitation. Mol. Microbiol. 59:1199–1215. 10.1111/j.1365-2958.2005.05017.x [DOI] [PubMed] [Google Scholar]
- 27.King J, Laemmli UK. 1971. Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62:465–477. 10.1016/0022-2836(71)90148-3 [DOI] [PubMed] [Google Scholar]
- 28.Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484–1502. 10.1101/gr.182801 [DOI] [PubMed] [Google Scholar]
- 29.Chi BK, Gronau K, Mäder U, Hessling B, Becher D, Antelmann H. 2011. S-Bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics 10:M111.009506. 10.1074/mcp.M111.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Médigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775. 10.1099/mic.0.027839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecker M, Pané-Farré J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236. 10.1146/annurev.micro.61.080706.093445 [DOI] [PubMed] [Google Scholar]
- 33.Ohnishi R, Ishikawa S, Sekiguchi J. 1999. Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J. Bacteriol. 181:3178–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahbabian K, Jamalli A, Zig L, Putzer H. 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28:3523–3533. 10.1038/emboj.2009.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehnik-Habrink M, Schaffe M, Mäder U, Diethmaier C, Herzberg C, Stülke J. 2011. RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol. Microbiol. 81:1459–1473. 10.1111/j.1365-2958.2011.07777.x [DOI] [PubMed] [Google Scholar]
- 36.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. 2012. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 8:e1002520. 10.1371/journal.pgen.1002520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laalami S, Bessières P, Rocca A, Zig L, Nicolas P, Putzer H. 2013. Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays. PLoS One 8:e54062. 10.1371/journal.pone.0054062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Condon C. 2011. Regulated RNA stability in Gram positives. Curr. Opin. Microbiol. 14:148–154. 10.1016/j.mib.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez-Cuevas P, Mercier R, Leavera M, Kawai Y, Errington J. 2012. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol. Microbiol. 83:52–66. 10.1111/j.1365-2958.2011.07920.x [DOI] [PubMed] [Google Scholar]
- 40.Devine KM. 2012. Bacterial L-forms on tap: an improved methodology to generate Bacillus subtilis L-forms heralds a new era of research. Mol. Microbiol. 83:10–13. 10.1111/j.1365-2958.2011.07922.x [DOI] [PubMed] [Google Scholar]
- 41.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130. 10.1016/0022-2836(86)90385-2 [DOI] [PubMed] [Google Scholar]
- 42.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 63:127–138. 10.1111/j.1365-2958.2006.05499.x [DOI] [PubMed] [Google Scholar]
- 43.Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. 2013. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 195:2340–2348. 10.1128/JB.00164-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boylan SA, Redfield AR, Brody MS, Price CW. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931–7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. 10.1016/S0378-1119(96)00404-0 [DOI] [PubMed] [Google Scholar]
- 46.Maguin E, Duwat P, Hege T, Ehrlich SD, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.