Abstract
Helicobacter pylori uses natural competence and homologous recombination to adapt to the dynamic environment of the stomach mucosa and maintain chronic colonization. Although H. pylori competence is constitutive, its rate of transformation is variable, and little is known about factors that influence it. To examine this, we first determined the transformation efficiency of H. pylori strains under low O2 (5% O2, 7.6% CO2, 7.6% H2) and high O2 (15% O2, 2.9% CO2, 2.9% H2) conditions using DNA containing an antibiotic resistance marker. H. pylori transformation efficiency was 6- to 32-fold greater under high O2 tension, which was robust across different H. pylori strains, genetic loci, and bacterial growth phases. Since changing the O2 concentration for these initial experiments also changed the concentrations of CO2 and H2, transformations were repeated under conditions where O2, CO2, and H2 were each varied individually. The results showed that the increase in transformation efficiency under high O2 was largely due to a decrease in CO2. An increase in pH similar to that caused by low CO2 was also sufficient to increase transformation efficiency. These results have implications for the physiology of H. pylori in the gastric environment, and they provide optimized conditions for the laboratory construction of H. pylori mutants using natural transformation.
INTRODUCTION
H. pylori is a naturally competent (1), Gram-negative bacterium that colonizes the human gastric mucosa and causes peptic ulcer or gastric cancer in about 10% of those infected (2). Strains isolated from different individuals demonstrate extraordinary genomic diversity (3). Like other organisms with small genomes (4), H. pylori has relatively few regulatory networks (5) and instead exploits natural competence and genetic changes to adapt to changing conditions (6). Since infection is chronic over the lifetime of the host, sometimes with more than one resident strain (7–9), frequent genetic exchange enables H. pylori to better adapt to the changing environment of the host stomach (10, 11) and to acquire antibiotic resistance (12). Thus, natural competence is essential to the lifestyle of H. pylori.
H. pylori natural competence is mediated by a two-step process: uptake of double-stranded DNA across the outer membrane followed by transport of single-stranded DNA through the inner membrane (13). DNA is transported across the H. pylori outer membrane and into the periplasm via a ComB type IV secretion system (T4SS) similar to the Vir system of Agrobacterium tumefaciens. ComB components are expressed from two operons and have been named according to homology with Vir proteins. Several have been shown to be essential for H. pylori natural competence. For example, ComB9, homologous to VirB9, is thought to be an important structural component, tightly anchored to the outer membrane within the periplasmic space (1), and ComB4 is an ATPase putatively responsible for function of the ComB structure (13). Both proteins are thought to be involved in the biogenesis of the ComB apparatus (1). Once inside the periplasm, double-stranded DNA is processed to single-stranded DNA and transported across the bacterial inner membrane by a ComEC channel (14). Single-stranded DNA can then become incorporated into the bacterial chromosome via RecA-mediated homologous recombination (13).
Transformation efficiency in H. pylori is variable and can be affected by the length and similarity of DNA sequences and restriction-modification systems (15–17). Variation also occurs during H. pylori growth phases, with peaks at mid-log- and late-stationary-phase growth (18). Since natural transformation is an important mechanism for bacterial adaptation to the gastric environment, certain conditions of stress may increase H. pylori competence. For example, induction of double-strand breaks by treatment with ciprofloxacin can increase expression of comB genes and enhance competence (19). Taken together, these studies suggest that H. pylori has evolved to increase natural competence when this is beneficial for its survival.
Another stress condition that might alter H. pylori competence is a change in the partial pressure of O2 or CO2. CO2 is highly prevalent in the gastric environment, while O2 tension is low. In the live mouse, for example, the partial O2 pressure (pO2) in the stomach is estimated at 58 ± 15 torr (∼7.6%) (20). O2 and CO2 concentrations are variable within the gastric environment and can be affected by disease state, food consumption, and location within the gastric mucosa (21, 22). For example, the CO2 concentration tends to be low nearest the epithelium (where the pH is high) and higher toward the gastric lumen (where the pH is low). Although, H. pylori is generally regarded as a microaerophile (2), it expresses high levels of catalase and superoxide dismutase, which facilitate the elimination of toxic oxygen intermediates (23, 24). More recent studies have shown that many strains can tolerate and even thrive in the presence of high O2 levels as long as CO2 levels are high (25). These studies indicate that H. pylori is a capnophile whose growth is promoted by atmospheric O2 as long as sufficient CO2 is present.
Here we show that low levels of CO2 markedly increase H. pylori transformation efficiency and induce expression of the ComB T4SS. This change in competence seems to be a result of CO2 influence on the pH of the bacterial culture medium. These results are important for understanding the physiology of H. pylori in the gastric environment and also have practical implications for using natural transformation to generate H. pylori mutants in the laboratory.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Helicobacter pylori strains J166 and SS1 were cultured on brucella agar (Becton, Dickinson and Company, Sparks, MD) supplemented with 5% heat-inactivated newborn calf serum (Gibco, Grand Island, NY), 5 μg/ml trimethoprim, 10 μg/ml vancomycin, 2.5 units/ml polymyxin B, and 2.5 μg/ml amphotericin B (Sigma-Aldrich, Inc., St. Louis, MO). Cultures were grown at 37°C using the Anoxomat system (Advanced Instruments, Inc., Norwood, MA), which is a sealed jar connected to a pressurized tank that contained one of several defined gas mixtures (Table 1). Specific gas conditions in the sealed jar were achieved by removing a defined volume of ambient air and replacing it with the appropriate defined gas. Transformation efficiency was initially studied under 5% O2 (7.6% CO2, 7.6 H2) or 15% O2 (2.9% CO2, 2.9% H2), where O2 was manipulated by changing the volume of a single defined gas mixture, allowing the other gases to vary. Separate defined gas mixtures were then used to manipulate O2, CO2, and H2 individually, using N2 as a replacement gas (Table 1).
TABLE 1.
Transformation conditions and defined gas mixtures
| Transformation conditions (%, vol/vol) |
Defined gas mixture (%, vol/vol) |
||||
|---|---|---|---|---|---|
| Oxygen | Carbon dioxide | Hydrogen | Carbon dioxide | Hydrogen | Nitrogen |
| 5.0 | 7.6 | 7.6 | 10.0 | 10.0 | 80.0 |
| 15.0 | 2.9 | 2.9 | 10.0 | 10.0 | 80.0 |
| 15.0 | 7.7 | 7.7 | 27.0 | 27.0 | 46.0 |
| 5.0 | 2.9 | 7.6 | 3.8 | 10.0 | 86.2 |
| 5.0 | 7.6 | 2.9 | 10.0 | 3.8 | 86.2 |
Plasmid construction.
Two plasmids were used to measure transformation efficiency, one that deleted an outer membrane protein (pOMP) and a second that deleted an intergenic region (pIG) between oppositely oriented genes and did not disrupt a known open reading frame. A third plasmid (pRecA) was developed to disrupt the recA gene to determine the influence of RecA expression on natural competence genes under relevant experimental conditions. Construction of the plasmids is shown schematically in Fig. 1; primer sequences are given in Table 2. To construct pOMP for strain J166 (pOMPJ166), a chloramphenicol resistance cassette (26) was amplified and ligated to DNA sequences from upstream and downstream of the promoter region in the babA gene (designated HP1243 in H. pylori 26695), which encodes the blood group antigen binding protein (27). A similar plasmid was constructed for strain SS1 (pOMPSS1), except that upstream and downstream sequences were amplified so as to delete the entire HP1243 locus, which in SS1 carries a babA paralog, babC. To construct pIG, a nonpolar kanamycin cassette (aphA3) was amplified from pUC18K (28) and ligated between HP0512 and HP0519 amplified from strain J166 (pIGJ166) or SS1 (pIGSS1), which in both strains are transcribed in opposite directions and contain no intervening open reading frames. These features, along with the insertion of a nonpolar cassette, minimized potential effects on the strain phenotype. Construction of pRecA was the same as for pIG except that aphA3 was ligated into the recA open reading frame of H. pylori SS1. PCR-amplified fragments were digested with the appropriate enzymes (New England BioLabs, Ipswich, MA), ligated into NotI/XhoI-digested pBluescript SK− vector (Stratagene, La Jolla, CA) using T4 DNA ligase (New England BioLabs, Ipswich, MA) according to the manufacturer's instructions, and cloned into Escherichia coli Top10 (Invitrogen, Carlsbad CA) with selection on chloramphenicol (5 μg/liter) or kanamycin (25 μg/liter) as appropriate.
FIG 1.
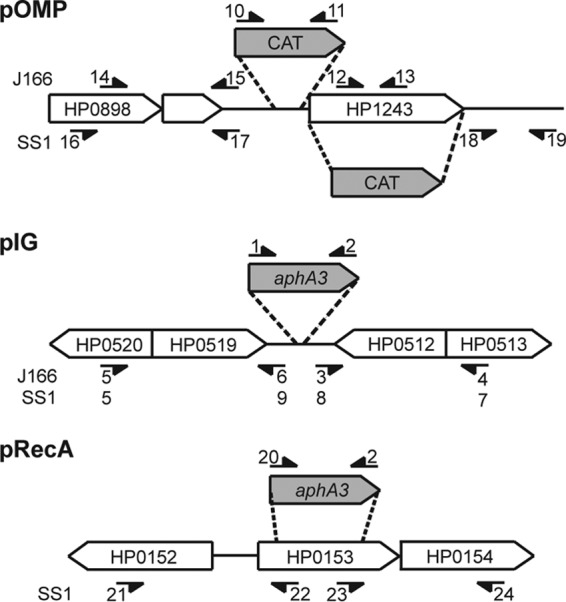
Schematic of plasmid construction. pOMP was constructed by inserting a chloramphenicol resistance cassette (CAT) into the promoter region (H. pylori J166) or the open reading frame of babA (HP1234) (H. pylori SS1). pIG was constructed by insertion of a kanamycin resistance cassette (aphA3) into an intergenic region between HP0519 and HP0512 in H. pylori J166 and SS1. pRecA was constructed by insertion of aphA3 into the recA gene in H. pylori SS1. The primers used for amplification of plasmid components for each strain are represented by small numbered arrows and correspond to the primers in Table 2. Gene designations are for H. pylori 26695.
TABLE 2.
Primers used for plasmid construction and RT-PCR
| Number | Primer name | Restriction site | Sequence (5′ → 3′)a |
|---|---|---|---|
| 1 | aphA3_F | SacI | AAC GAGCTC GGT ACC CGG GTG AC |
| 2 | aphA3_R | HincII | AAC GTCGAC TCT AGA GGA TCC CC |
| 3 | J166_IG_F1 | HincII | AAT GTCGAC AAA TCT TAT TAT CGA CAC TCT ATT AGA A |
| 4 | J166_IG_R1 | XhoI | AAT CTCGAG CCC TTT GTT GGC CGT CAA |
| 5 | J166_IG_F2 | NotI | AAT GCGGCCGC TAG CAC CGC TCA AGA CAC AA |
| 6 | J166_IG_R2 | SacI | AAT GAGCTC CCA AGT GAT AGT AAA GAT TAG GTA A |
| 7 | SS1_IG_F1 | XhoI | AAT CTCGAG ATC ATA CCC CTA TTT CTA GGA |
| 8 | SS1_IG_R1 | HincII | AAT GTCGAC AAA TCT TAT CAT CGA CAC TCT ATT AAA G |
| 9 | SS1_IG_R2 | SacI | AAT GAGCTC TCA AGT GAT AGT AAA GAT TAG GTA G |
| 10 | cat_F | SacI | AAC GAGCTC GAT GCT TTA TAA CTA TGG ATT AAA CAC |
| 11 | cat_R | BamHI | AAC GGATCC TTA TCA GTG CGA CAA ACT GGG AT |
| 12 | J166_OMP_F1 | BamHI | AAC GGATCC TTG CTC CAC GCT GAA GAC |
| 13 | J166_OMP_R1 | XhoI | AAC CTCGAG GAC GCT CGT TTG ATT GAC CA |
| 14 | J166_OMP_F2 | NotI | AAC GCGGCCGC AGC CAC AAA ACC TCT AAA GA |
| 15 | J166_OMP_R2 | SacI | AAC GAGCTC GGG GTA TTT TGA AAT AAC TCT C |
| 16 | SS1_OMP_F1 | XhoI | AAC CTCGAG TTT TGA GCC GGT GGA TAT ATT AG |
| 17 | SS1_OMP_R1 | SacI | AAC GAGCTC GCA AGC TCT TTT ATT ATT TAT CTT A |
| 18 | SS1_OMP_F2 | BamHI | AAC GGATCC CGT TAA AAC CTT TTG TGA AAC T |
| 19 | SS1_OMP_R2 | NotI | AAA GCGGCCGC TGT GGA TCT AGC GGT TCT T |
| 20 | aphA3_F2 | PstI | AAT CTGCAG GGT ACC CGG GTG AC |
| 21 | SS1_RecA_F1 | NotI | AAT GCGGCCGC TCG TTA CTG CCC TTA ATG AGC TC |
| 22 | SS1_RecA_R1 | PstI | AAT CTGCAG CAA TTT GTT TGA TCG CTA AAG AAA TCG C |
| 23 | SS1_RecA_F2 | HincII | AAT GTCGAC AAT GAA GAG ATC ATG CCC TTA CCC |
| 24 | SS1_RecA_R1 | XhoI | AAT CTCGAG AAAAGACAATCAGGGAGCTATGGC |
| 25 | comB4_F1 | TGT TAG AAA AGC TTT TAA GCG C | |
| 26 | comB4_R1 | TTG CTC TGT GCT TAA ATG GGT G | |
| 27 | comB9_F | AAG CCG ATG ATT TTT TAG AAG AAG C | |
| 28 | comB9_R | AAA GCT CCC TTG AAT GGC GTT TAA | |
| 29 | recA_F | GTG CGC CTT GGG GAT AAG CA | |
| 30 | recA_R | CTT AGA GTG GTC TTC CCG CTT GAC | |
| 31 | 16S_F | GGA GTA CGG TCG CAA GAT TAA A | |
| 32 | 16S_F | CTA GCG GAT TCT CTC AAT GTC AA |
Underlining indicates a restriction site.
Transformations.
Transformations were performed as previously described (29). Briefly, overnight (18- to 24-h) H. pylori cultures were harvested from agar plates using a sterile swab and suspended in brucella broth to an optical density at 600 nm (OD600) of 5, and 10 μl was spread evenly in a 1.5-cm-diameter circle on a fresh agar plate. Five replicate circles of these bacterial cells were then incubated at 37°C in an Anoxomat jar adjusted to contain the appropriate atmospheric conditions. After 3 to 6 h, 0.3 to 3 μg of plasmid or genomic DNA suspended in deionized water was mixed with each of the circles using a sterile loop. The cells with DNA were then returned to 37°C in the same gas mixture with which they had been conditioned. After overnight incubation, the bacteria were removed from the 1.5-cm circle using a sterile loop and suspended in 300 μl brucella broth. Dilutions were plated on nonselective and selective plates with 5 μg/ml chloramphenicol or 25 μg/ml kanamycin. Transformation efficiency was calculated as the number of CFU present on selective medium divided by the number of CFU on nonselective medium divided by the amount (μg) of DNA used for transformation.
Effects of growth phase on competence.
H. pylori SS1 harvested from an overnight agar culture was plated on brucella agar in 1.5-cm-diameter circles as described for transformations. Plates were incubated under 5% or 15% O2 conditions, and at 6-h intervals from 0 to 30 h, 3.0 μg of pIGSS1 was added to three replicate circles and mixed with a sterile loop. After 6 h of additional incubation with DNA, bacteria were removed from the circles with a sterile loop, suspended in brucella broth, and then plated and analyzed for both CFU and transformation efficiency.
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR).
A 100-μl suspension in brucella broth (OD600 = 5) was harvested from an overnight brucella agar culture of H. pylori SS1, spread evenly on a brucella agar plate, and incubated at 37°C with 5% or 15% O2 for 18 to 24 h. Bacterial cells were suspended in 1 ml TRIzol reagent (Ambion, Carlsbad, CA), and total RNA was isolated and then treated with DNase I (Roche Applied Science, Mannheim, Germany) according to the manufacturer's directions. RNA was purified using an RNeasy Minikit (Qiagen Sciences, Germantown, MD). Gene-specific primers (Table 2) were used to synthesize cDNA with the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A 1-μl aliquot of a 1:5 dilution of cDNA was used to perform quantitative PCR with the same primers using SYBR green PCR master mix with standard cycling parameters (Applied Biosystems, Warrington, WA) in 20-μl reaction mixtures on an ABI 7900HT 96 well block.
Determination of CO2 effects on brucella medium pH.
The pH of 25 ml sterile brucella broth (Becton, Dickinson and Company, Sparks, MD) was measured using a pH meter (Beckman Coulter) after overnight incubation under 7.6% CO2 (5% O2) or 2.9% CO2 (15% O2) at 37°C. Brucella agar plates, supplemented with 11 mg/liter phenol red (Matheson, Coleman & Bell, Norwood, OH), were incubated under 7.6% CO2 (5% O2) or 2.9% CO2 (15% O2) at 37°C overnight. The pH of brucella agar plates from each condition was visually estimated by comparison to a standard curve, prepared by supplementing 25-ml aliquots of brucella broth with 11 mg/liter phenol red and adjusting the pH in increments of 0.2 from 5.8 to 6.8 using 6 N HCl or NaOH. The agar pHs of two to four brucella agar plates from each condition were independently estimated by four observers, and the average values are reported.
Statistical analysis.
Pairwise comparisons were performed using the Student t test with Welch's correction on GraphPad Prism software. Data involving bacterial counts were log transformed prior to statistical analysis. Error bars in all figures represent standard error.
RESULTS
H. pylori transformation efficiency under 5% and 15% O2.
To determine the transformation efficiency of H. pylori under different atmospheric conditions, we first incubated H. pylori SS1 with plasmid DNA directed to a babA paralog (pOMPSS1) and compared the results under 5% O2 to 15% O2 using a single defined gas mixture (Table 1). The transformation efficiency was increased approximately 6-fold in 15% O2 (Fig. 2A). To determine if these results were strain specific, we repeated the experiment with H. pylori J166 using the corresponding plasmid, pOMPJ166. This resulted in approximately 8-fold-greater transformation efficiency in 15% O2 than in 5% O2 (Fig. 2B). The transformation efficiency could be increased under high O2 conditions because deletion of an OMP increases H. pylori fitness under high O2, though it seems unlikely since two different OMPs were targeted (babA in J166 and babC in SS1). In other words, the effect of O2 on transformation efficiency could be specific to one genetic locus. To test this, we next transformed H. pylori SS1 and J166 with plasmid DNA containing a kanamycin resistance cassette directed to an intergenic region (pIGSS1 and pIGJ166) that would not be expected to disrupt an open reading frame. Under 15% O2, the transformation efficiencies in strains SS1 and J166 increased 32-fold and 10-fold, respectively (Fig. 2C and D). These results show that a change in atmospheric conditions can cause a robust change in H. pylori natural competence that is independent of strain or genetic locus.
FIG 2.
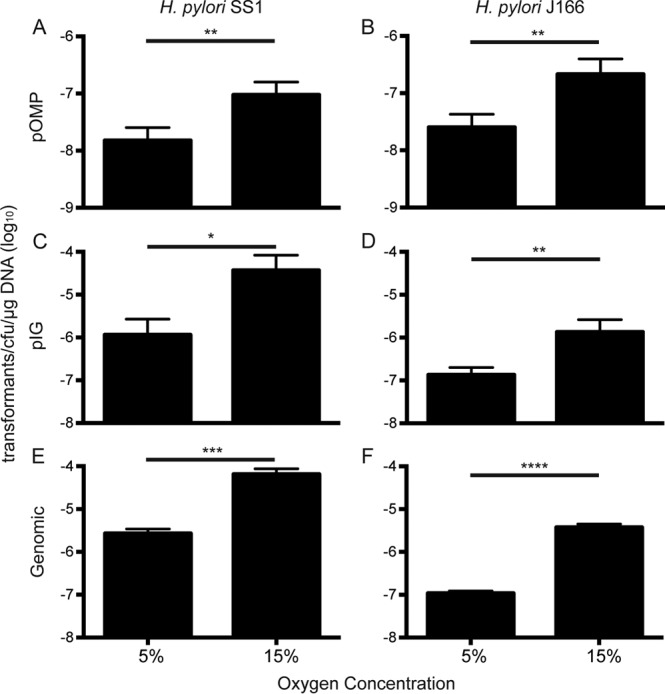
Transformation efficiency in H. pylori is increased under conditions of high O2 and low CO2. H. pylori SS1 and J166, as indicated, were transformed with pOMPSS1 (A), pOMPJ166 (B), pIGSS1 (C), pIGJ166 (D), genomic DNA from pIGSS1-transformed H. pylori SS1 (E) and genomic DNA from pIGJ166-transformed H. pylori J166 (F) at 5% O2 (7.6% CO2, 7.6% H2) or 15% O2 (2.9% CO2, 2.9% H2). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To ensure that this phenomenon was not specific to plasmid DNA, genomic DNA was isolated from H. pylori SS1 and J166 which had been transformed with pIGSS1 and pIGJ166, respectively. This DNA was then used to perform transformation efficiency experiments as described above. Under 15% O2, the transformation efficiency increased 24-fold and 34-fold in strains SS1 and J166, respectively (Fig. 2E and F), illustrating that both plasmid and genomic DNAs are taken up by the bacteria with increased efficiency under 15% O2 conditions.
Atmospheric effects on transformation efficiency are independent of growth phase.
High O2 conditions that increased transformation efficiency (Fig. 2) also resulted in a trend of decreased bacterial growth (Fig. 3). Since it has previously been shown that transformation efficiency is variable throughout H. pylori growth phases (18), we considered the possibility that the effects of changing atmospheric conditions on competence were due to altered growth. To address this, we measured the transformation efficiency of H. pylori SS1 at different growth phases under both 5% and 15% O2. pIGSS1 DNA was added at 6-h intervals under 5% and 15% O2, and the transformation efficiency was determined 6 h later. As expected, 15% O2 caused a trend of decreased bacterial growth (Fig. 4A). No transformants were detected at 0 h, but when DNA was added after 6 to 30 h of growth (mid-log to stationary phase), the transformation efficiency was markedly higher at 15% O2 at each point in the growth curve (Fig. 4B). Thus, although 15% O2 moderately impaired H. pylori growth, it also increased competence at every point in the growth curve, indicating that growth phase is not responsible for the increase in natural competence at 15% O2 (Fig. 4).
FIG 3.
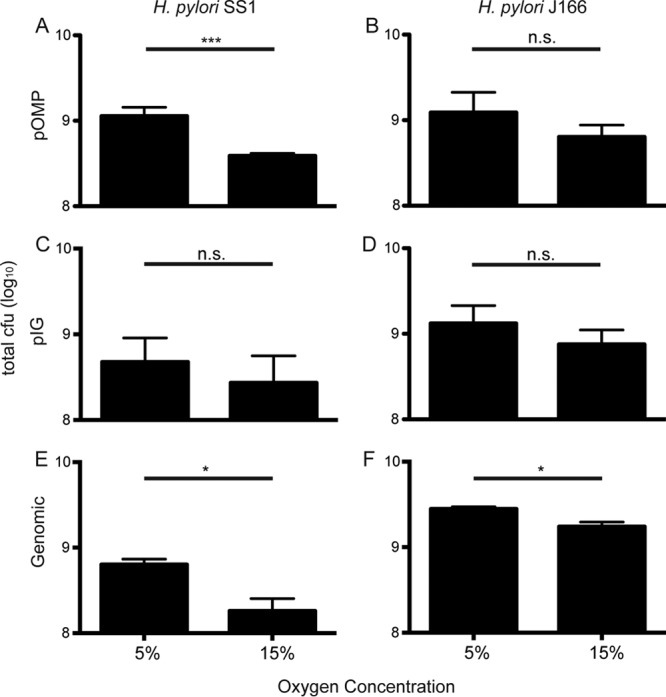
A trend of reduced H. pylori growth is observed under conditions of high O2 and low CO2. CFU for H. pylori SS1 (A, C, and E) and J166 (B, D, and F) under high and low O2 conditions are shown. The data correspond to the transformation efficiency results shown in Fig. 2. *, P < 0.05; ***, P < 0.001; n.s., not significant.
FIG 4.
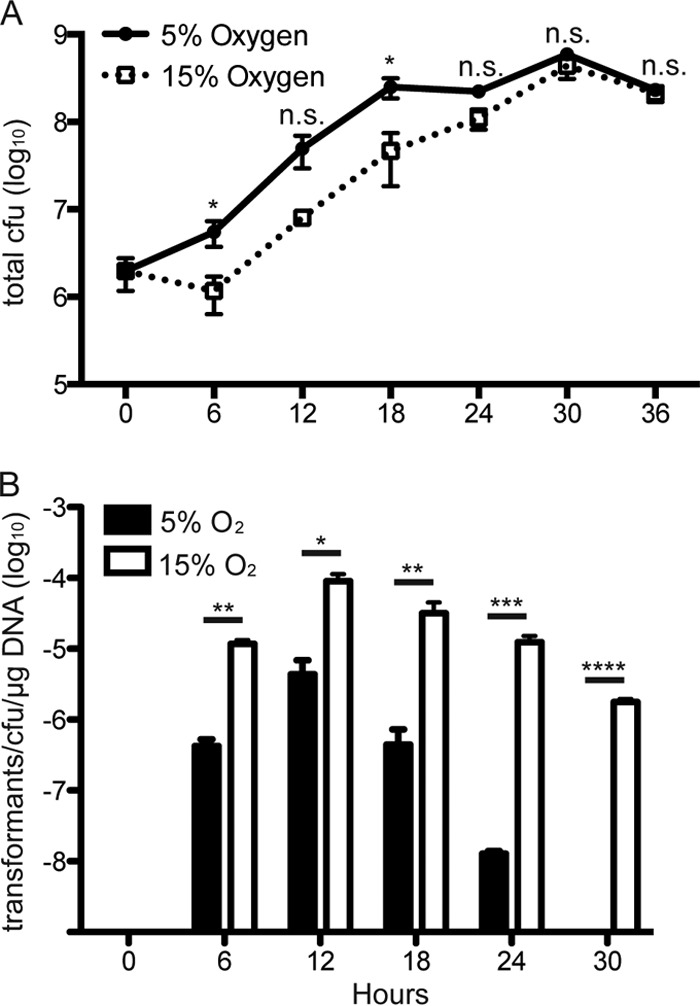
H. pylori growth and transformation efficiency are affected by atmospheric conditions throughout the growth curve. (A) Total CFU present under 5% O2 (solid line) or 15% O2 (dotted line) when transformation efficiency was measured for each time point shown. (B) Transformation efficiency of H. pylori SS1 under 5% O2 (filled bars) or 15% O2 (open bars), as measured 6 h after the addition of pIGSS1, which was added at the time points shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001, n.s. = not significant.
The ComB T4SS is induced under 15% O2 conditions independent of RecA.
H. pylori natural competence occurs primarily through expression of the ComB T4SS apparatus. RecA is also an important protein involved in homologous recombination following uptake of exogenous DNA. Therefore, we examined expression of the comB and recA genes under both high and low O2 conditions. Since the ComB apparatus is composed of elements present on two operons, comB2-4 and comB6-10 (30), we examined expression of one comB gene from each operon (comB4 and comB9). qRT-PCR analysis of RNA isolated from H. pylori SS1 incubated under either high or low O2 conditions showed significantly increased expression of both comB4 and comB9 under 15% O2 conditions (Fig. 5A). No changes in expression of recA were seen. This result is in agreement with previous studies (19) indicating that recA expression was not increased during induction of other natural competence genes. 16S rRNA, which served as a loading control, was also unchanged. These results suggest that increased H. pylori competence under high O2 is mediated by the ComB T4SS.
FIG 5.

ComB expression is induced by conditions of high O2 and low CO2 independent of RecA. H. pylori SS1 (A) or SS1 ΔrecA (B) mRNA expression levels of the comB4, comB9, recA, and 16S rRNA genes after overnight incubation with 5% O2 (filled bars) or 15% O2 (open bars) are shown. In both SS1 and SS1 ΔrecA, comB4 and comB9 were significantly induced under 15% O2. Induction of recA did not occur. 16S rRNA was used as a loading control. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s. = not significant.
It has previously been shown that RecA is required for H. pylori induction of comB genes in response to DNA double-strand breaks (19). To determine whether the induction of comB genes in response to increased O2 was also dependent on RecA, we developed a recA deletion mutant of H. pylori SS1 (SS1 ΔrecA) using pRecA. qRT-PCR was performed on RNAs from three individual SS1 ΔrecA clones incubated under either 5% or 15% O2 conditions. Significant induction of both comB4 and comB9 was detected under 15% O2 (Fig. 5B). These data indicate that induction of the ComB apparatus under 15% O2 conditions occurs by a novel mechanism, independent of the response to double-strand break induction.
H. pylori natural competence is induced in response to low CO2.
In all of the above-described experiments in which a single defined gas mixture was used, CO2 and H2 concentrations were decreased to accommodate an increased O2 concentration. Because recent work shows that H. pylori is a capnophile (25, 31), we asked whether the change in CO2, rather than O2, was responsible for the increase in transformation efficiency. Transformation efficiency in H. pylori SS1 was examined using pIGSS1 under a series of conditions (Table 1) in which we individually manipulated O2, H2, or CO2, leaving the other gases at the same levels found under low O2 conditions. Nitrogen was used as a replacement gas. When O2 was increased to 15% with no change in CO2 or H2, a small (1.8-fold) increase in competence was observed. In contrast, reducing CO2 without changing O2 or H2 caused a marked (40-fold) increase in transformation efficiency (Fig. 6A). Reduction of H2 alone had no effect. Additionally, the least amount of growth was measured under conditions of low CO2, which resulted in the highest transformation efficiency (Fig. 6B). These results indicate that the increase in H. pylori transformation efficiency under different atmospheric conditions is predominantly a result of a decreased CO2 concentration.
FIG 6.
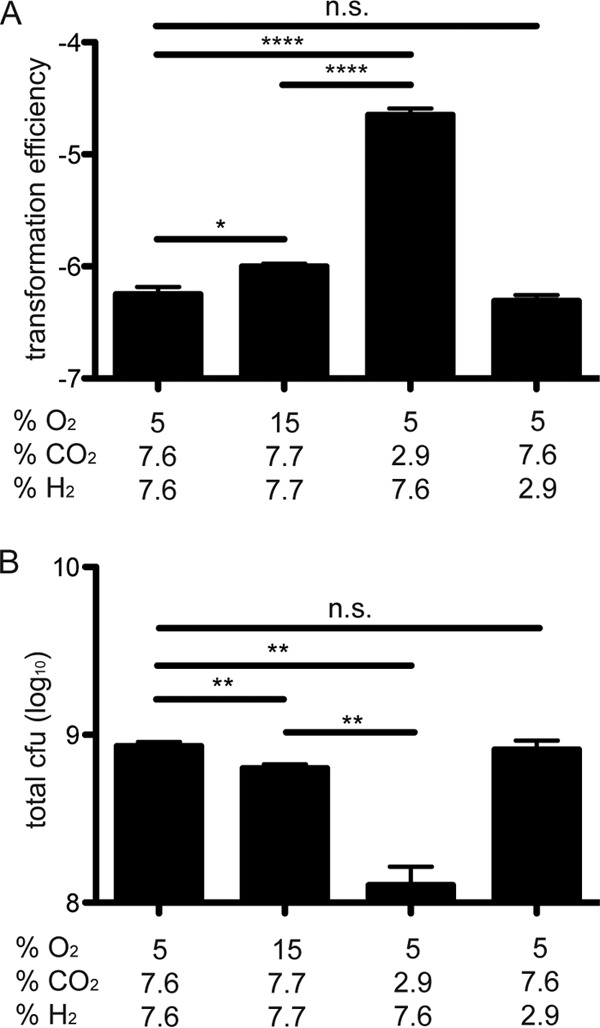
H. pylori transformation efficiency is induced by low CO2 rather than high O2 levels. Transformation efficiency as measured after the addition of pIGSS1 (A) and total CFU (B) of H. pylori SS1 under defined atmospheric conditions are shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; n.s. = not significant.
Increased pH mimics the increase in transformation efficiency caused by decreased CO2.
Since CO2 reacts with water to form carbonic acid, differences in CO2 concentration could affect the pH of the unbuffered culture medium. The pH of autoclaved brucella broth was 6.87 (±0.01), which decreased to 6.31 (±0.02) after overnight incubation under 7.6% CO2 (5% O2). Conditions of low (2.9%) CO2 resulted in a lesser decrease in pH, to 6.47 (±0.03), after overnight incubation. Since all transformations were done on agar-grown bacteria, phenol red was added to brucella agar medium prior to autoclaving in order to verify that a similar effect occurred in brucella agar plates. Brucella agar plates containing phenol red were incubated under 7.6% CO2 or 2.9% CO2 overnight. The color of the plates before and after incubation at each of the conditions was estimated by comparison to a phenol red standard curve. Under aerobic conditions, the pH of brucella agar was 6.76 (±0.04), and this decreased to 6.55 (±0.08) after incubation at 7.6% CO2. Low (2.9%) CO2 conditions again resulted in less of a decrease in pH, to 6.63 (±0.03). Thus, incubation under conditions of increased CO2 caused a decrease in the pH of unbuffered brucella medium that was relative to the amount of CO2 present.
To determine whether increased pH could be responsible for the increase in competence under low CO2 conditions, transformation efficiency was determined on buffered brucella agar plates adjusted to pH 6.5 or 7.0 with 20 mM phosphate buffer. After incubation at 7.6% CO2, the pH of the plates buffered at 6.5 or 7.0 decreased slightly to 6.46 (±0.06) and 6.69 (±0.02), respectively. H. pylori SS1 was transformed with pIGSS1 on these buffered plates under 7.6% CO2. The transformation efficiency of H. pylori SS1 was about 35-fold higher with pIGSS1 at pH 6.69 than at pH 6.46 under 7.6% CO2, which mimicked the increase in competence caused by conditions of low CO2 and high O2 (Fig. 7A). This was also accompanied by decreased growth (Fig. 7B). Incubation of H. pylori in liquid medium prepared in the same way as the plates described above showed that although the presence of H. pylori could moderately affect the absolute pH of the buffered medium, it did not significantly affect the difference in pH between the two buffers (see Fig. S1 in the supplemental material). Together these results show that increased pH is sufficient to increase transformation efficiency, and they suggest that this may explain the increased competence under conditions of low CO2.
FIG 7.
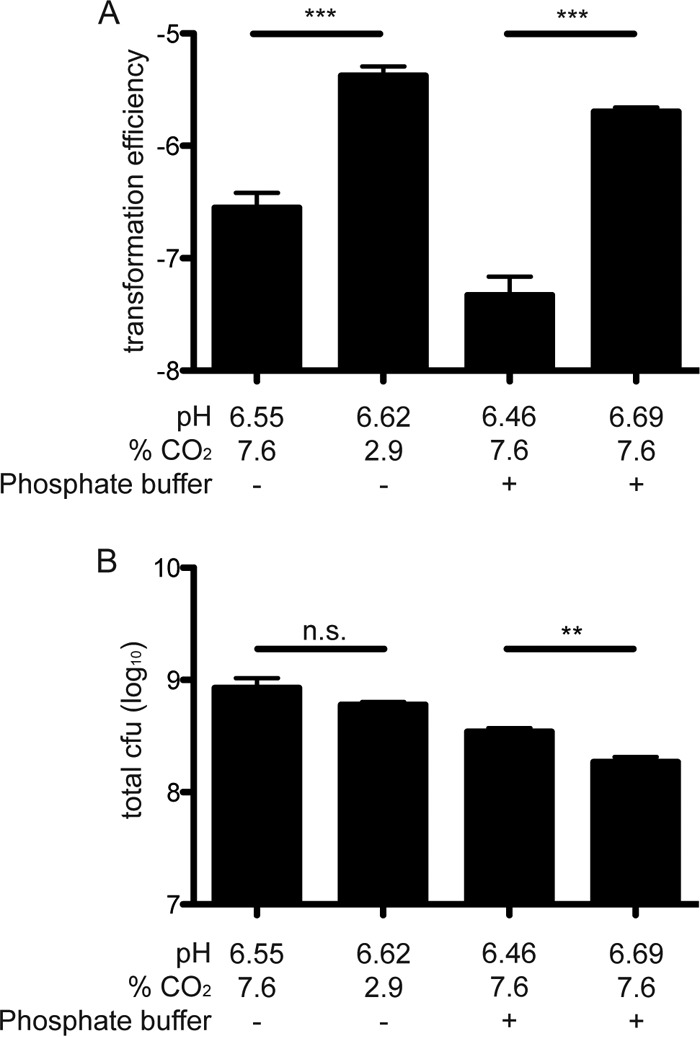
A change in pH is sufficient to reproduce the effects of CO2 on H. pylori transformation efficiency. (A) The transformation efficiency of H. pylori SS1 was increased both by a decrease in CO2 (2.9%) and by an increase in pH under buffered conditions (7.6% CO2). (B) Increased transformation efficiency was accompanied by small decreases in CFU. **, P < 0.01; ***, P < 0.001; n.s. = not significant.
DISCUSSION
Genomic diversity is a signature feature of H. pylori that arises from mutation, recombination, and importation of exogenous DNA via natural transformation, which allows H. pylori to adapt to the hostile environment of the human stomach. Variability in competence among different H. pylori strains is well documented (16, 32, 33), but little is known about conditions that affect transformation efficiency. This is important, not only for understanding the physiology of H. pylori in the gastric environment but also for optimizing methods that are commonly exploited in the laboratory to generate isogenic mutants. Here we systematically examined the effects of environmental growth conditions on H. pylori transformation efficiency.
Competence in naturally transformable bacteria is commonly induced by physiological stress (34). Since H. pylori is considered a microaerophile that is subject to toxic oxygen species under ambient O2 conditions (35), we first examined the impact of O2 concentration on natural competence. Culturing H. pylori under 15% versus 5% O2 decreased bacterial growth, increased transformation efficiency, and induced the comB operon, which is required for natural competence. These effects were robust across different H. pylori strains, genetic loci, and bacterial growth phases (Fig. 2 to 5). However, in these experiments manipulation of O2 was accomplished using a single defined gas mixture, so the increase in O2 was also accompanied by decreases in CO2 and H2. Since some studies have suggested that H. pylori is a capnophile (25, 31), we considered the possibility that low CO2 rather than high O2 was the relevant environmental variable responsible for increasing competence. Independent manipulation of O2 and CO2 revealed that in fact low CO2 was primarily responsible for the increased transformation efficiency, though there was also a small effect of increased O2, which can induce the comB operon (31). An increase in pH similar to that caused by incubating unbuffered agar plates under low CO2 also increased transformation efficiency. Thus, increased pH is itself sufficient to enhance H. pylori competence. Increased pH has also been observed to enhance competence in Campylobacter jejuni (36, 37) and Streptococcus pneumoniae (38), which suggests that this response may be a convergent evolutionary adaptation of naturally competent, opportunistic mucosal pathogens.
It may at first seem paradoxical that H. pylori transformation efficiency increased under high pH (low CO2), since the stomach is commonly thought of as a harsh, acidic environment in which the acidity might be expected to induce a stress response and increase competence. However, several lines of evidence are consistent with the observation that increased pH may induce stress in H. pylori. First, studies of gene expression in vitro and in vivo suggest that the natural habitat of H. pylori within the gastric mucosa is actually quite acidic, with a pH of ≤4.0 (39). In fact, H. pylori cannot survive neutral pH in the presence of physiological concentrations of urea because the pH rapidly rises (40). The stringent response is also required for H. pylori survival under high O2 conditions (41), which can be abrogated by addition of CO2 (25). Ex vivo measurements of guinea pig gastric tissue indicate that a pH gradient exists across the mucosa, with a pH of approximately 6.1 immediately adjacent to the epithelial cell surface that gradually decreases toward the gastric lumen (42) Therefore, in vivo it is likely that increased pH and reduced CO2 would be experienced immediately adjacent to the host epithelial cells (43), while H. pylori is thought to more commonly localize just offshore in the mucous layer (44). It may be that H. pylori undergoes a stress response when located close to the gastric epithelium, with consequent upregulation of ComB and increased competence.
We conclude that the increased transformation efficiency observed in vitro under elevated pH and decreased CO2 provides insight into H. pylori natural competence regulation within the gastric mucosa. Our results indicate that H. pylori competence is highly sensitive to subtle changes that could be experienced within microenvironments of the mucous layer. These conditions of increased pH and reduced CO2 can also be exploited to optimize transformation efficiency for generating isogenic mutants in the laboratory.
Supplementary Material
ACKNOWLEDGMENTS
The H. pylori SS1 sequence, used for the construction of pOMPSS1 and pIGSS1, was generously provided by Jenny Draper and Karen Ottemann. We also thank Lori Hansen for designing several primers.
This work was supported by grants R01 AI081037 and R01 AI070803 from the National Institutes of Health to Jay V. Solnick.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00633-13.
REFERENCES
- 1.Hofreuter D, Odenbreit S, Haas R. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379–391. 10.1046/j.1365-2958.2001.02502.x [DOI] [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19:449–490. 10.1128/CMR.00054-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Megraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585. 10.1126/science.1080857 [DOI] [PubMed] [Google Scholar]
- 4.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182. 10.1128/JB.01887-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- 6.Baltrus DA, Guillemin K, Phillips PC. 2008. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62:39–49. 10.1111/j.1558-5646.2007.00271.x [DOI] [PubMed] [Google Scholar]
- 7.Taylor NS, Fox JG, Akopyants NS, Berg DE, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter FM, et al. 1995. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J. Clin. Microbiol. 33:918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales-Espinosa R, Castillo-Rojas G, Gonzalez-Valencia G, Ponce de Leon S, Cravioto A, Atherton JC, Lopez-Vidal Y. 1999. Colonization of Mexican patients by multiple Helicobacter pylori strains with different vacA and cagA genotypes. J. Clin. Microbiol. 37:3001–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patra R, Chattopadhyay S, De R, Ghosh P, Ganguly M, Chowdhury A, Ramamurthy T, Nair GB, Mukhopadhyay AK. 2012. Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PLoS One 7:e43370. 10.1371/journal.pone.0043370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. 2013. Natural competence promotes Helicobacter pylori chronic infection. Infect. Immun. 81:209–215. 10.1128/IAI.01042-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. 2008. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 69:994–1007. 10.1111/j.1365-2958.2008.06336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smeets LC, Arents NL, van Zwet AA, Vandenbroucke-Grauls CM, Verboom T, Bitter W, Kusters JG. 2003. Molecular patchwork: chromosomal recombination between two Helicobacter pylori strains during natural colonization. Infect. Immun. 71:2907–2910. 10.1128/IAI.71.5.2907-2910.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 107:1184–1189. 10.1073/pnas.0909955107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249. 10.1038/nrmicro844 [DOI] [PubMed] [Google Scholar]
- 15.Humbert O, Dorer MS, Salama NR. 2011. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol. Microbiol. 79:387–401. 10.1111/j.1365-2958.2010.07456.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine SM, Lin EA, Emara W, Kang J, DiBenedetto M, Ando T, Falush D, Blaser MJ. 2007. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 21:3458–3467. 10.1096/fj.07-8501com [DOI] [PubMed] [Google Scholar]
- 17.Kumar R, Mukhopadhyay AK, Ghosh P, Rao DN. 2012. Comparative transcriptomics of H. pylori strains AM5, SS1 and their hpyAVIBM deletion mutants: possible roles of cytosine methylation. PLoS One 7:e42303. 10.1371/journal.pone.0042303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baltrus DA, Guillemin K. 2006. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255:148–155. 10.1111/j.1574-6968.2005.00066.x [DOI] [PubMed] [Google Scholar]
- 19.Dorer MS, Fero J, Salama NR. 2010. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 6:e1001026. 10.1371/journal.ppat.1001026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U. S. A. 96:4586–4591. 10.1073/pnas.96.8.4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steer H. 2009. The source of carbon dioxide for gastric acid production. Anat. Rec. (Hoboken) 292:79–86. 10.1002/ar.20762 [DOI] [PubMed] [Google Scholar]
- 22.Taylor DE, Gutierrez G, Clark C, Hainley S. 1997. Measurement of gastric mucosal carbon dioxide tension by saline and air tonometry. J. Crit. Care 12:208–213. 10.1016/S0883-9441(97)90034-4 [DOI] [PubMed] [Google Scholar]
- 23.Krieg NR, Hoffman PS. 1986. Microaerophily and oxygen toxicity. Annu. Rev. Microbiol. 40:107–130. 10.1146/annurev.mi.40.100186.000543 [DOI] [PubMed] [Google Scholar]
- 24.Kelly DJ, Hughes NJ, Poole RK. 2001. Microaerobic physiology: aerobic respiration, anaerobic respiration, and carbon dioxide metabolism, p 113–124 In Mobley HLT, Mendz GL, Hazell SL. (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 25.Park SA, Ko A, Lee NG. 2011. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol. 11:96. 10.1186/1471-2180-11-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Styer CM, Hansen LM, Cooke CL, Gundersen AM, Choi SS, Berg DE, Benghezal M, Marshall BJ, Peek RM, Jr, Boren T, Solnick JV. 2010. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect. Immun. 78:1593–1600. 10.1128/IAI.01297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–377. 10.1126/science.279.5349.373 [DOI] [PubMed] [Google Scholar]
- 28.Menard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Roos KP, Taylor DE. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485–2493. 10.1099/00221287-139-10-2485 [DOI] [PubMed] [Google Scholar]
- 30.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. 2006. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188:882–893. 10.1128/JB.188.3.882-893.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SA, Lee NG. 2013. Global regulation of gene expression in the human gastric pathogen Helicobacter pylori in response to aerobic oxygen tension under a high carbon dioxide level. J. Microbiol. Biotechnol. 23:451–458. 10.4014/jmb.1209.09064 [DOI] [PubMed] [Google Scholar]
- 32.Donahue JP, Israel DA, Peek RM, Blaser MJ, Miller GG. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37:1066–1074. 10.1046/j.1365-2958.2000.02036.x [DOI] [PubMed] [Google Scholar]
- 33.Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Blaser MJ. 2009. Natural transformation of helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5:e1000337. 10.1371/journal.ppat.1000337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park AM, Li Q, Nagata K, Tamura T, Shimono K, Sato EF, Inoue M. 2004. Oxygen tension regulates reactive oxygen generation and mutation of Helicobacter pylori. Free Radic. Biol. Med. 36:1126–1133. 10.1016/j.freeradbiomed.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 36.Wilson DL, Bell JA, Young VB, Wilder SR, Mansfield LS, Linz JE. 2003. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149:3603–3615. 10.1099/mic.0.26531-0 [DOI] [PubMed] [Google Scholar]
- 37.Vegge CS, Brondsted L, Ligowska-Marzeta M, Ingmer H. 2012. Natural transformation of Campylobacter jejuni occurs beyond limits of growth. PLoS One 7:e45467. 10.1371/journal.pone.0045467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JD, Morrison DA. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J. Gen. Microbiol. 133:1959–1967 [DOI] [PubMed] [Google Scholar]
- 39.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. 2007. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc. Natl. Acad. Sci. U. S. A. 104:7235–7240. 10.1073/pnas.0702300104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clyne M, Labigne A, Drumm B. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mouery K, Rader BA, Gaynor EC, Guillemin K. 2006. The stringent response is required for Helicobacter pylori survival of stationary phase, exposure to acid, and aerobic shock. J. Bacteriol. 188:5494–5500. 10.1128/JB.00366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber S, Nguyen TH, Stuben M, Scheid P. 2000. Demonstration of a pH gradient in the gastric gland of the acid-secreting guinea pig mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G597–G604 [DOI] [PubMed] [Google Scholar]
- 43.Bury-Mone S, Mendz GL, Ball GE, Thibonnier M, Stingl K, Ecobichon C, Ave P, Huerre M, Labigne A, Thiberge JM, De Reuse H. 2008. Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect. Immun. 76:497–509. 10.1128/IAI.00993-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. U. S. A. 101:5024–5029. 10.1073/pnas.0308386101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


