Abstract
We recently demonstrated that Pseudomonas aeruginosa PAO1 undergoes a pronounced phenotypic change when introduced into the intestines of rats during surgical injury. Recovered strains displayed a specific phenotype (termed the P2 phenotype) characterized by altered pyocyanin production, high collagenase activity, high swarming motility, low resistance to chloramphenicol, and increased killing of Caenorhabditis elegans compared to the inoculating strain (termed the P1 phenotype). The aims of this study were to characterize the differences between the P. aeruginosa P1 and P2 phenotypes in quorum sensing and competitiveness. We then determined the presence of the P2 phenotype among PAO1 strains from various laboratories. Results demonstrated that P2 cells display accelerated growth during early exponential phase and early activation of quorum-sensing systems and overcome the growth of P1 cells in a mixed population. Among eight PAO1 strains obtained from different laboratories, four exhibited the P2 phenotype. Of 27 mutants analyzed from the P. aeruginosa MPAO1 transposon library, 25 displayed P2 phenotypes. The P2 phenotype in both cases correlated with a lack of expression of mexE or mexF due to mutations in mexT and mexF genes. In summary, strains possessing the P2 phenotype are distributed among PAO1 strains commonly used across a variety of research laboratories. Genetically, they are characterized by various mutations in mexT or mexF.
INTRODUCTION
Stable genetic mutations that change phenotypes can arise as microbes adapt to their environment to maximize propagation. This ecologically dependent shift in phenotype has been documented in many bacterial species and in diverse contexts (1, 2). Recently, we demonstrated that the MPAO1 strain of Pseudomonas aeruginosa became transformed to (or selected for) a more virulent phenotype when present in the colon of rats subjected to preoperative radiation followed by colon resection and anastomosis (3). The strain recovered from the anastomotic tissue (termed MPAO1-P2, i.e., having the P2 phenotype) exhibited high collagenase activity, swarming ability, increased pyocyanin production in liquid medium, and increased tissue-destroying capacity compared to the initial strain (termed MPAO1-P1, having the P1 phenotype). Comparative genomic sequencing analysis revealed that MPAO1-P2 harbored a single-nucleotide polymorphism (SNP) in the mexT gene that results in a truncated and nonfunctional MexT protein. Transformation of mexT-P1 into MPAO1-P2 reverted the strain back to the P1 phenotype. The phenotypes of P. aeruginosa that are similar to P1 and P2 have been previously described (4–7). In the cited studies, the wild-type PAO1 strain harbored an 8-bp insertion in mexT that results in a nonfunctional MexT protein and a phenotype similar to P2. Its derivative mutant that was selected by norfloxacin (referred to as a nfxC-type mutant) had a functional MexT protein and was characterized by attenuated pyocyanin production (5), high resistance to chloramphenicol (6), and absence of swarming motility (8) similar to the P1 strains in our study (3). mexT has been described as a mutational “hot spot,” where mutations can contribute to global phenotypic changes in P. aeruginosa (6, 9–11). MexT is a transcriptional regulator of the MexEF-OprN efflux pump, which has been extensively characterized in the context of antimicrobial agents (12–15). It has also been suggested that the MexEF-OprN pump is involved in the efflux of quorum-sensing (QS) signaling molecules, which could account for the attenuated virulence observed in strains with a functional MexT protein (5, 16, 17).
In this study, we performed detailed analyses of intra- and extracellular levels of three QS signaling molecules—C4-homoserine lactone (C4-HSL), 3OC12-HSL, and Pseudomonas quinolone signal (PQS)—in strains MPAO1-P1 and MPAO1-P2 during the early stationary phase prior to visible pyocyanin production in MPAO1-P2. MPAO1-P2 demonstrated accumulation of intra- and extracellular C4-HSL, 3OC12-HSL, and PQS and significantly higher activation of RhlI at the transition to stationary phase. We further observed that MPAO1-P2 had greater competitiveness in mixed cultures with MPAO1-P1, suggesting that strains with mutations in genes that affect the functionality of the MexEF-OprN efflux pump possessing the P2 phenotype will predominate. We then analyzed eight strains of P. aeruginosa PAO1 obtained from several laboratories and found that they exhibited either the P1 or P2 phenotype. Strains with the P2 phenotype harbored mutations in mexT and mexF. Additionally, we analyzed mutant strains from the P. aeruginosa transposon library (18), which had been constructed from the parent strain MPAO1-P1, and discovered that many strains harbor the P2 phenotype. Our data demonstrate that the broad distribution of the P2 phenotype among the various PAO1 strains may be due to mutations affecting the MexEF-OprN pump.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa strain MPAO1 (referred to as MPAO1-P1) and the 27 mutants (Table 1) were obtained from the University of Washington transposon mutant library (18). MPAO1-P2 is the transformed MPAO1-P1 strain isolated from leaking rat anastomoses as previously described (3). W-PAO1 was obtained from Klaus Winzer, and D-PAO1 was obtained from Steve Diggle, University of Nottingham, United Kingdom. Although they both came from the laboratory of Paul Williams, these strains were obtained 10 years apart. H-PAO1 was obtained from Tong Chuan He, University of Chicago, Chicago, IL. R-PAO1 was obtained from Cornelia Reimmann, University of Lausanne, Switzerland. C-PAO1 was obtained from Pierre Cornelis, Vrije University, Brussels, Belgium. Ig-PAO1 was obtained from Barbara Iglewski, University of Rochester, Rochester, NY. L-PAO1 was obtained from Linda McCarter, University of Iowa, Iowa City, IA. Ma-PAO1 was obtained from Colin Manoil, University of Washington, Seattle, WA. All strains were stored in 10% glycerol stock at −80°C. Only cells freshly plated from stock were used in experiments. Cells from stock were plated onto tryptic soy broth (TSB) plates and grown overnight at 37°C and were further used as designed.
TABLE 1.
Mutants displaying P1 and P2 phenotypesa
| Mutant no. | P. aeruginosa IDb | Mutant IDc | Function | Pyocyanin production |
Chloramphenicol resistance | Collagenase activity | Swarming motility | |
|---|---|---|---|---|---|---|---|---|
| Agar | Liquid | |||||||
| 1 | PA1003 | ID13375 | MvfR | − | − | + | − | − |
| 2 | PA5361 | ID30430 | PhoR | + | − | + | − | − |
| 3 | PA5394 | PW10101 | Cardiolipin synthase | − | + | − | + | + |
| 4 | PA0452 | PW1826 | Flotilin | − | + | − | + | + |
| 5 | PA2389 | ID32408 | PvdR | − | + | − | + | + |
| 6 | PA2391 | ID9422 | OpmQ | − | + | − | + | + |
| 7 | PA0685 | ID39358 | Probable type II secretion protein | − | + | − | + | + |
| 8 | PA0690 | ID18158 | Hypothetical protein | − | + | − | + | + |
| 9 | PA0691 | ID4721 | Hypothetical protein | − | + | − | + | + |
| 10 | PA0696 | ID11082 | Hypothetical protein | − | + | − | + | + |
| 11 | PA0051 | ID39650 | PhzH | − | + | − | + | + |
| 12 | PA0674 | ID21628 | VreA | − | + | − | + | + |
| 13 | PA0675 | ID9556 | ECF sigma factor - VreI | − | + | − | + | + |
| 14 | PA0676 | ID16317 | Sigma factor regulator - VreR | − | + | − | + | + |
| 15 | PA0678 | ID42074 | HxcU putative pseudopilin | − | + | − | + | + |
| 16 | PA0677 | ID19639 | HxcW putative pseudopilin | − | + | − | + | + |
| 17 | PA0678 | ID54030 | HxcU putative pseudopilin | − | + | − | + | + |
| 18 | PA0679 | ID20782 | Hypothetical protein | − | + | − | + | + |
| 19 | PA0680 | ID35854 | HxcV putative pseudopilin | − | + | − | + | + |
| 20 | PA0682 | ID33753 | HxcX atypical pseudopilin | − | + | − | + | + |
| 21 | PA5170 | ID31482 | ArcD | − | + | − | + | + |
| 22 | PA0688 | ID12226 | Probable binding protein component of ABC transporter | − | + | − | + | + |
| 23 | PA0681 | ID5144 | HxcT pseudopilin | − | + | − | + | + |
| 24 | PA4221 | ID48243 | FptA | − | + | − | + | + |
| 25 | PA2388 | ID6650 | FpvR | − | + | − | + | + |
| 26 | PA5360 | ID40588 | PhoB | − | + | − | + | + |
| 27 | PA2426 | PW5085 | Sigma factor, PvdS | − | + | − | + | + |
Chloramphenicol resistance is defined as growth on 500 ng/μl chloramphenicol (n = 2). Collagenase activity is defined as production of collagenase at 4 h of growth in TSB at levels at least 5 times higher than that of P1 (negatives had lower collagenase than P1) (n = 2). Mutant 1 (MvfR) does not produce pyocyanin because of a lack of quinolone production.
As reported in the Pseudomonas Genome Database at http://www.pseudomonas.com/.
As reported in the P. aeruginosa PAO1 transposon mutant library at http://www.gs.washington.edu/labs/manoil/libraryindex.htm.
Construction of MPAO1-P1/RFP (P1/RFP) expressing red fluorescence.
P. aeruginosa MPAO1 was transformed with our constructed plasmid pUCP24/RFP to create the red fluorescent strain P1/RFP. To construct the pUCP24/RFP plasmid, we amplified the gene encoding DS-Red Express 2 protein from plasmid pQE60NA-Express 2 (19) using primers XbaI-Ds-Red and Ds-Red-HindIII (see Table S1 in the supplemental material) and cloned it in the Escherichia coli-P. aeruginosa shuttle vector pUCP24 (20) via XbaI-HindIII cleavage sites to create the plasmid pUCP24/RFP. The plasmid was electroporated into competent cells of MPAO1 to get strain P1/RFP, which was selected on Pseudomonas isolation agar plates containing 100 μg/ml gentamicin. The selected clones were verified for production of red fluorescence with confocal microscopy.
Construction of MPAO1-P2/GFP (P2/GFP) expressing green fluorescence.
P. aeruginosa MPAO1-P2 was chromosomally tagged using a mini-Tn7 insertion that constitutively expressed green fluorescent protein (GFP) (21). Uptake of pBK-miniTn7-gfp2 plasmid DNA in the presence of a pRK600 mobilization plasmid and pUX-BF13 conjugation helper plasmid into the MPAO1-P2 recipient was achieved by conjugational mating where transformants were selected on 100 μg/ml gentamicin (22, 23).
Collagenase assay.
Collagenase activity was assessed by the rate of degradation of fluorescein-conjugated gelatin using the EnzChek gelatinase/collagenase assay kit (Molecular Probes). To each well of a 96-well black clear-bottom plate, 180 μl of TSB, 10 μl of overnight P. aeruginosa culture in TSB, and 10 μl of gelatin substrate (1.0 mg/ml; DQ gelatin [catalog no. D12054; Molecular Probes] from pig skin, fluorescein conjugate) were added. The negative control consisted of 190 μl TSB and 10 μl fluorescein-labeled gelatin. Fluorescence due to gelatin degradation by bacteria was measured after 3 h of incubation at 37°C and 160 rpm. Values from the negative control were subtracted from values of each strain. Fluorescence values were normalized to cell density at 600 nm. Experiments were performed in triplicate.
Swarming assay.
Swarming medium consisted of 20 mM NH4Cl, 12 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 1 mM MgSO4, 1 mM CaCl2, 11 mM dextrose, 0.5% Casamino Acids, and 0.5% Difco Bacto-agar as described previously (24). Prepared plates were dried overnight at room temperature and then were inoculated with P. aeruginosa colonies picked with a sterile pipette tip from overnight Pseudomonas isolation agar (PIA) plates. Swarming plates were incubated at 30°C overnight.
Caenorhabditis elegans killing assay.
Wild-type C. elegans N2 provided by the Caenorhabditis Genetic Center (CGC), University of Minnesota, was used in all experiments. Culturing, cleaning, egg preparation for synchronization, and transferring were performed accordingly to reference 25. Nematodes were prepared for the experiment as previously described (26). The killing assay was performed as previously described (26) with small modifications. Briefly, prepared nematodes were transferred from kanamycin (Km)-containing plates into 1.3 ml P. aeruginosa culture grown for 5 h in 0.1× TY (1 g/liter tryptone, 0.5 g/liter yeast extract, and 0.1 mM potassium phosphate buffer [pH 6.0]). Worms were incubated at room temperature without shaking. Fifteen worms per plate and three plates per experiments were used for each variant. Nematodes were considered dead if they did not respond to the touch of a platinum picker.
Complementation of ΔhxcT with hxcT.
Complementation of ΔhxcT with hxcT was performed using the plasmid pUCP24/hxcT harboring the hxcT gene. To construct pUCP24/hxcT, the hxcT gene was amplified from MPAO1 genome DNA using primers HxcT-XbaI and HxcT-HindIII (see Table S1 in the supplemental material) and cloned into pUCP24 (20) via restriction sites XbaI and HindIII. The complemented clone was selected on Pseudomonas isolation agar plates containing 100 μg/ml gentamicin and PCR verified to have the intact region where the mutant had the transposon inserted. Primers used were designed with Primer3 software (see Table S1 in the supplemental material).
C4-HSL/3OC12-HSL/PQS assay.
P. aeruginosa strains were grown overnight in TSB, diluted to an optical density at 600 nm (OD600) of 0.03 with fresh TSB, and continued to grow at 37°C and 160 rpm before bacterial cell culture collection at 6 h. Bacterial cultures were centrifuged at 5,000 rpm for 5 min to separate pellet and culture supernatant. The supernatant was tested for extracellular quorum-sensing (QS) molecules. Briefly, supernatant was filtered through a 0.22-μm filter and used as a medium for luminescent reporter strains. Luminescence was recorded after 2 h of reporter strain incubation with supernatants and normalized to P. aeruginosa cell density at the time of collection (6 h). The bacterial pellet was tested for intracellular QS molecules. Briefly, bacterial pellets were washed twice with 1 ml TSB per wash, and subsequently, 100 μl of 0.1 mm zirconia beads and 500 μl TSB were added to the pellet. Cells were lysed using the vortex adaptor for Genie 2 (Ambion). After the cells had been lysed by vortexing at maximum speed for 10 min, 700 μl fresh TSB was added and the supernatant was filtered through a 0.22-μm filter. Protein concentrations were measured with the Qubit 2.0 fluorometer using the Qubit protein assay kit (Invitrogen) with the measured value from fresh TSB subtracted, and supernatants were used as media for luminescent reporter strains. Luminescence measured after 2 h of reporter strain incubation was normalized to protein concentration. The following reporter strains were used: for C4-HSL, E. coli harboring the reporter plasmid pSB536 (27); for 3OC12-HSL, E. coli harboring the reporter plasmid pSB1705 (27); and for PQS, P. aeruginosa ΔPqsA/pqsA::luxCDABE (28).
Competitiveness assay.
To assess competitiveness, red fluorescent MPAO1-P1 (P1/RFP) and green fluorescent MPAO1-P2 (P2/GFP) were incubated either separately or in mixed cultures in 2 ml TSB. Each pure culture at 0 h had an OD600 of 0.03, while mixed cultures at 0 h contained equal amounts of each strain for a total OD600 of 0.06. The TSB cultures were incubated for 6 h, after which cells were pelleted by centrifugation at 5,000 rpm and resuspended in 500 μl phosphate-buffered saline (PBS). The cell suspension was analyzed by fluorescence measured on a Tecan Safire 2 multidetection microplate reader at 560-nm/580-nm excitation/emission for red fluorescence and at 344-nm/434-nm excitation/emission for green fluorescence. Background red fluorescence displayed by MPAO1-P2/GFP (P2/GFP) was subtracted from red fluorescent values of pure MPAO1-P1/RFP (P1/RFP) and mixed cultures assigned to measure P1/RFP. Similarly, background green fluorescence displayed by P1/RFP was subtracted from green fluorescence values of pure P2/GFP and mixed cultures assigned to measure P2/GFP. Lastly, all fluorescence values were normalized so that the average fluorescence of all pure cultures equaled 1,000 relative fluorescence units. Direct comparisons were made between fluorescence of a mixed culture and that of a pure culture for a particular color of fluorescence.
To measure P1/P2 competitiveness during coincubation with intestinal epithelial cells, HT-29 monolayers were first prepared as follows. HT-29 cells between passages 19 and 34 were grown in Dulbecco's minimal essential medium (DMEM; Gibco, Long Island, NY) supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine (Gibco), and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin). Cells were cultured in a water-saturated atmosphere of 95% air/5% CO2 to form monolayers. P1/RFP and P2/GFP solutions were prepared in DMEM at an OD600 of 0.03, while the mixed solution was prepared in DMEM at an OD600 of 0.06 with equal amounts of each strain. The original medium of HT-29 monolayers was replaced by 1 ml of either pure or mixed cultures. The coculture with HT-29 monolayers was incubated for 5 h, after which the DMEM was aspirated and centrifuged at 5,000 rpm to pellet the planktonic cells. The cells were resuspended in 500 μl PBS in which the red and green fluorescence was measured. To examine the P. aeruginosa adhered to the HT-29 monolayer, 1 ml lysis buffer consisting of 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 1% Triton X-100 was added to the cell dish. Cells were then scraped and used for fluorescence analysis. Fluorescence values were normalized to background fluorescence from either pure PBS or lysis buffer. Additionally, background red fluorescence values from pure P2/GFP cultures and background green fluorescence values from pure P1/RFP cultures were subtracted from normalized fluorescence values before the mean fluorescence of the pure cultures was adjusted to 1,000 relative fluorescence units as described above. Direct comparisons were made between fluorescence of a mixed culture and that of a pure culture for a particular color of fluorescence.
Measurement of pyocyanin.
Pyocyanin was measured as previously described (26, 29) from 1.8 ml of bacterial culture using 500 μl of chloroform for the extraction of pyocyanin from conditioned medium followed by reextraction into 250 μl of 0.2 M HCl. A 100-μl portion of the aqueous layer containing pyocyanin was used for OD520 measurements by a PowerWave XS microplate spectrophotometer (Bio-Tek).
Growth of P1 in P1/P2 conditioned medium.
To prepare the conditioned medium, we grew P1 and P2 in TSB for 8 h at 37°C and 160 rpm. Cells were removed via centrifugation, and the conditioned medium was passed through a 0.22-μm filter.
Overnight P1 cells were diluted 1:100 into the cell-free medium for a total volume of 200 μl in a 96-well plate, which was incubated at 37°C and 160 rpm. Optical density at 600 nm was tracked dynamically from 0 to 18 h.
Quantitative reverse transcriptase PCR (qRT-PCR) analysis.
Three individual colonies of P1 or P2 were picked from freshly prepared plates, inoculated into 2 ml TSB, and grown overnight. Overnight cultures were diluted in 2 ml fresh TSB to an OD600 of 0.03 and grown for 3.5 or 6 h. Two volumes of RNA Protect bacterial reagent (Qiagen) was added per 2 ml of culture before isolation of RNA using the UltraClean microbial RNA isolation kit (Mo Bio). DNA was removed using the Turbo DNA-free kit (Invitrogen). For cDNA synthesis, 10 ng of RNA was converted to cDNA using the high-capacity RNA-to-cDNA kit (Applied Biosystems). For qPCRs, a total volume of 10 μl was used that included 1 μl of cDNA, 0.2 μl 10 mM each forward and reverse primer (see Table S1 in the supplemental material), and 5 μl Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). qPCR was performed using the 7900HT fast real-time PCR system (Applied Biosystems). The program for amplification had an initial heating step at 50°C for 2 min, followed by the denaturation step at 95°C for 15 s and then 40 cycles of 95°C for 15 s and 60°C for 1 min. Each sample was run in triplicate. The RT-negative control was run to ensure complete DNA removal. Gene expression was normalized to expression of the housekeeping gene PA4748 as previously described (30). Primers used were designed with Primer3 software.
Sequencing of mexT and mexF.
For sequencing of mexT and mexF, a colony from the overnight TSB agar plate was collected with a sterile pipette tip and placed into 10 μl distilled water, of which 1 μl was used as the DNA template for the PCR. Each PCR mixture (total volume, 20 μl) additionally contained 0.5 μl of each forward and reverse 10 mM primer (see Table S1 in the supplemental material) and 18 μl Platinum PCR high-fidelity SuperMix (Invitrogen). PCR products were verified through gel electrophoresis in 0.8% agar and cleaned with ExoSAP-IT (Affymetrix) before being submitted for sequencing at the University of Chicago DNA Sequencing & Genotyping Facility (Chicago, IL). Primers used were designed with Primer3 software.
Statistical analysis.
Statistical significance was determined by the Student t test (P < 0.05). Standard errors are displayed in all figures.
RESULTS
MPAO1-P2 demonstrates early activation of quorum sensing.
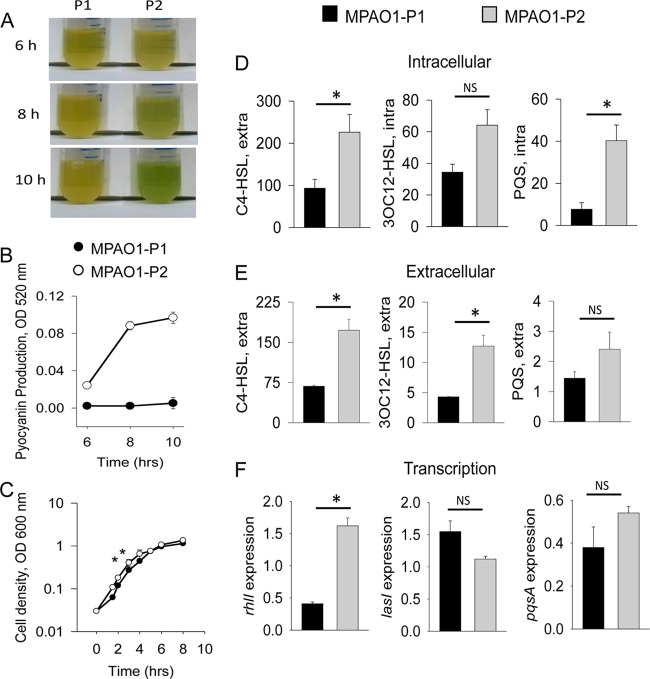
We have previously shown that liquid cultures of MPAO1-P2 (P2) produce large amounts of pyocyanin compared to MPAO1-P1 (P1), which produces a negligible amount of pyocyanin after overnight growth in TSB (3). Here, we performed dynamic tracking of pyocyanin-specific pigmentation in P1 and P2 and found that green pigmentation in P2 appears at as early as 8 h (Fig. 1A). OD520 measurements of pyocyanin extracted from culture supernatants confirmed increased pyocyanin levels in P2 at 8 h and above (Fig. 1B). Since pyocyanin biosynthesis is regulated by quorum sensing (31), the production of pyocyanin at this time point suggests earlier activation of quorum sensing in the P2 strain. Therefore, we next measured the accumulation of QS signaling molecules in P1 and P2 during growth in TSB. Specifically, we examined the accumulation of C4-HSL, 3OC12-HSL, and PQS both intracellularly and extracellularly at the transition to stationary phase (6 h) just before visible pigmentation in P2. Cell-free conditioned media and cell lysates were analyzed for QS molecule production using E. coli (27) and P. aeruginosa (28) reporter strains for C4-HSL, 3OC12-HSL, and PQS activity. The data were normalized to either cell density (for extracellular measurements) or protein concentration (for intracellular measurements). Results demonstrated that at 6 h of growth, the P2 strain showed significantly higher intracellular levels of C4-HSL and PQS and also higher extracellular levels of C4-HSL and 3OC12-HSL (Fig. 1D and E). Intracellular 3OC12-HSL and extracellular PQS also seemed to be higher in P2, although the differences did not reach statistical significance (Fig. 1D and E).
FIG 1.
MPAO1-P2 demonstrates early activation of quorum sensing and accumulation of C4-HSL and 3OC12-HSL -P2 compared to MPAO1-P1 during growth in TSB. (A) Greenish pigmentation suggesting higher pyocyanin production by P2. (B) Pyocyanin measured from 6 h to 10 h. (C) Growth curves of MPAO1-P1 and MPAO1-P2. n = 3; *, P < 0.001. (D and E) Intracellular (D) and extracellular (E) levels of C4-HSL, 3OC12-HSL, and PQS at 6 h. n = 3; *, P < 0.05. (F) Expression of rhlI, lasI, and pqsA at 6 h. n = 3; *, P < 0.01. Black bars represent MPAO1-P1 and gray bars represent MPAO1-P2.
We noticed that P2 growth was consistently 50 to 70% higher during the early exponential phase than that of P1, yet both strains entered the stationary phase at the same cell density (Fig. 1C). The differences in growth were most likely affected by a shorter transitioning time of P2 from lag to log phase (see Fig. S1 in the supplemental material). Accelerated growth of P2 could potentially contribute to faster accumulation of QS molecules, so that they reach threshold concentrations at earlier time points that activate quorum sensing, which was evident at 8 h.
To confirm earlier activation of the QS system in P2 strains, we next performed transcriptional analysis of genes involved in the biosynthesis of quorum-sensing molecules: rhlI (C4-HSL synthase), lasI (3OC12-HSL synthase), and pqsA (biosynthesis of PQS precursor). Results demonstrated significantly higher expression of rhlI and pqsA at 6 h in P2 (Fig. 1F). The expression of lasI seemed to be higher in P1 than P2, although this was not statistically significant.
We next used the nutrient-limited medium 0.1× TY, in which P1 and P2 strains have equally low growth rates (see Fig. S2 in the supplemental material), barely reaching a cell density of 0.2 at OD600, similar to our previously published data on MPAO1 growth (26). MPAO1 (P1 phenotype) grown in this medium never reached the point where pyocyanin became visible. In contrast, we observed that P2 displayed visible pyocyanin production at 12 h (Fig. 2A), suggesting that quorum sensing, and hence virulence, is activated in the P2 strain even under nutrient-poor conditions. OD520 measurements of pyocyanin from culture supernatant confirmed increased pyocyanin levels in P2 at 14 h (Fig. 2B). To confirm a more virulent phenotype in the P2 strain, we performed C. elegans mortality assays in which nematodes were transferred into liquid cultures of P1 and P2 grown in 0.1× TY. The mortality rate caused by P2 was almost 2-fold higher than P1 (Fig. 2C).
FIG 2.
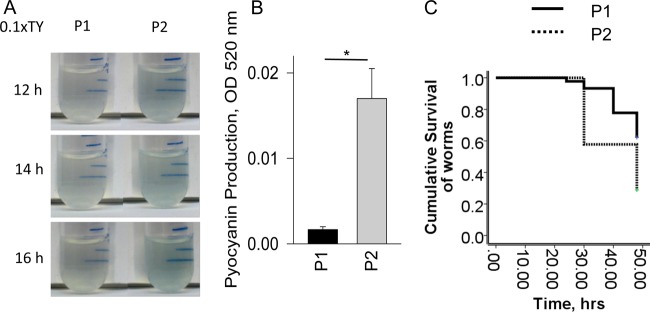
Enhanced virulence of P2 in low-nutrient medium. (A) Blue pigmentation suggesting higher pyocyanin production by P2. (B) Quantitative analysis of pyocyanin measured at 14 h. n = 3; *, P < 0.05. (C) Kaplan-Meier survival curves of C. elegans incubated with P2 versus P1 grown in 0.1× TY.
MPAO1-P2 outcompetes MPAO1-P1 in a mixed culture.
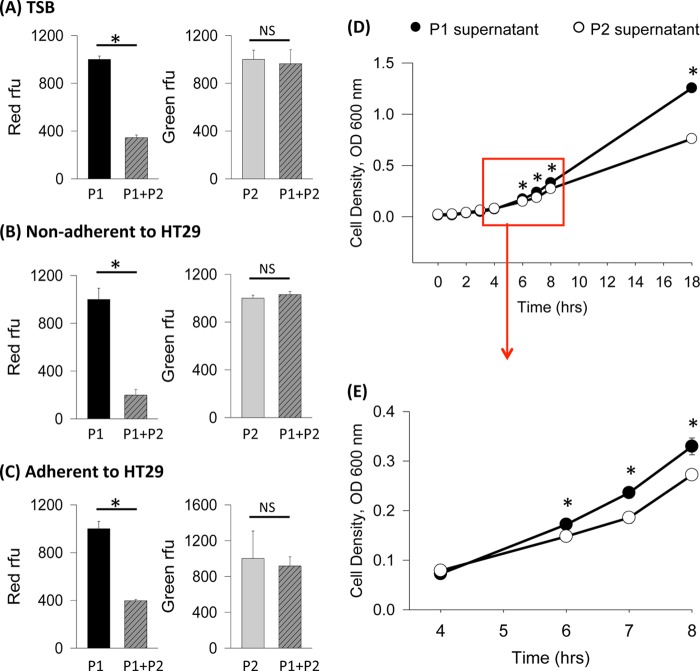
Early propagation and activation of quorum sensing may confer superior competitiveness to strains with the P2 phenotype. To test this hypothesis, we transformed MPAO1-P1 with the plasmid harboring red fluorescent protein and MPAO1-P2 with a plasmid harboring green fluorescent protein and measured red and green fluorescence to determine the relative abundance of each strain within a mixed culture. In Fig. 3A to C, the intensity of red fluorescence exhibited by the red fluorescent MPAO1-P1 strain in either the pure MPAO1-P1 culture or in the mixed MPAO1-P1/P2 culture reflects the growth of the MPAO1-P1 strain. Figure 3A to C also show the intensity of green fluorescence exhibited by the green fluorescent MPAO1-P2 strain in either the pure MPAO1-P2 culture or in the mixed MPAO1-P1/P2 culture, reflecting the growth of the MPAO1-P2 strain. We first performed the experiment in TSB-grown cultures and found that the relative intensity of red fluorescence in the mixed culture was 60 to 70% less than that of the pure MPAO1-P1 culture, suggesting that the growth of MPAO1-P1 is inhibited in the mixed cultures. However, the intensity of green fluorescence in the mixed culture was similar to that of the pure MPAO1-P2 culture (Fig. 3A), suggesting that MPAO1-P1 did not suppress MPAO1-P2 growth and that MPAO1-P2 growth in the mixed culture was equal to the growth of MPAO1-P2 in the individual cultures. Because the MPAO1 pure cultures and the mixed cultures had similar cell densities at the time of fluorescence analysis (data not shown), it is plausible that MPAO1-P2 remains in abundance at the cost of MPAO1-P1 growth inhibition in a coculture. We next examined if the phenomenon of MPAO1-P2 dominance over MPAO1-P1 persists in the context of host factors. MPAO1-P1 and MPAO1-P2 were incubated alone or as a mixed culture on intestinal epithelial monolayers of HT-29 cells. Fluorescence of planktonic and adherent bacterial cells was examined after 5 h of growth. In mixed cultures compared to pure MPAO1-P1 cultures, levels of MPAO1-P1 were ∼80% lower in both planktonic (Fig. 3B) and adherent cells (Fig. 3C). The amount of MPAO1-P2 in the mixed cultures did not differ significantly from the amount of pure MPAO1-P2 (Fig. 3B and C). These data suggest that MPAO1-P2 could have greater competitiveness than MPAO1-P1 when present at the intestinal epithelium. We next tested the hypothesis that MPAO1-P2, with its earlier induction of quorum sensing, secretes exoproducts that suppress the growth of MPAO1-P1. First, we obtained cell-free conditioned media by growing MPAO1-P1 and MPAO1-P2 in pure cultures for 8 h and removing the cells through centrifugation and filtration. Afterwards, overnight MPAO1-P1 cells were grown in either MPAO1-P1- or MPAO1-P2-conditioned media, and cell growth was tracked dynamically. The data demonstrated that growth of MPAO1-P1 cells in MPAO1-P2-conditioned medium was attenuated compared to growth in MPAO1-P1-conditioned media (Fig. 3D and E).
FIG 3.
MPAO1-P2 outcompetes MPAO1-P1 in mixed cultures. (A) Relative fluorescence exhibited by MPAO1-P1 (red relative fluorescence units [RFU]) and MPAO1-P2 (green RFU) after 6 h of growth in TSB in separate or mixed populations. n = 3; *, P < 0.001. (B and C) Relative fluorescence of MPAO1-P1 and MPAO1-P2 grown on intestinal epithelial monolayers of HT-29 cells. (B) Nonadherent bacterial cells (n = 3; *, P < 0.001); (C) adherent bacterial cells (n = 3; *, P < 0.01). (D and E) MPAO1-P1 growth in P1- versus P2-conditioned media. (D) All time periods from 0 to 18 h (n = 3; *, P < 0.05); (E) 4- to 8-h time points (n = 3; *, P < 0.05).
PAO1 strains display the P1 or P2 phenotype.
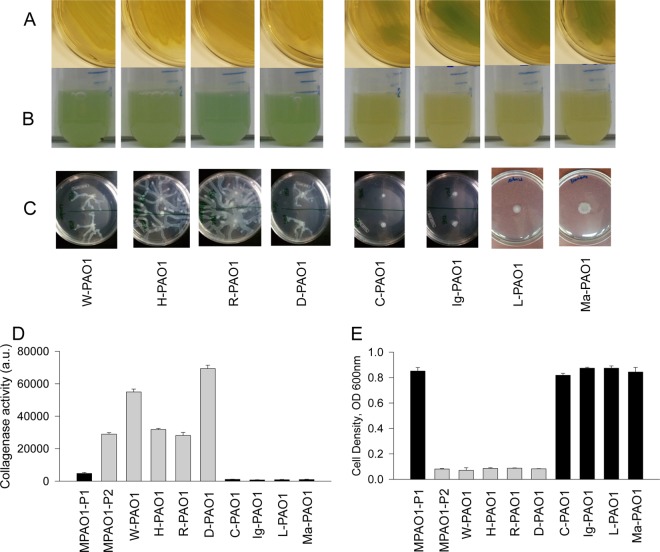
Increased competitiveness of P2 could allow emergent P2 cells to replace a P1 population. From experience working with P. aeruginosa PAO1 strains from various laboratories, we noticed pronounced differences in these strains in terms of altered pyocyanin production on solid agar and in liquid media that could be referred to as either P1- or P2-like. As described previously (3), the P2 phenotype is characterized by enhanced pyocyanin in liquid medium, attenuated pyocyanin on agarized medium, swarming motility, high collagenase activity, and low resistance to chloramphenicol. We assessed 8 PAO1 strains for these features and found that half (W-PAO1, H-PAO1, R-PAO1, and D-PAO1) possessed the P2 phenotype, while the other half (C-PAO1, Ig-PAO1, L-PAO1, and Ma-PAO1) possessed the P1 phenotype (Fig. 4).
FIG 4.
PAO1 strains exhibit the P1 or P2 phenotype. (A and B) Pyocyanin production on TSB agar (A) and liquid TSB (B). (C) Swarming motility. (D) Collagenase activity measured by degradation of fluorescein labeled gelatin. n = 3; P < 0.01 for all P2 strains compared to P1 strains. (E) Resistance to chloramphenicol. Cell density was measured in overnight cultures grown in TSB containing 500 μg/ml chloramphenicol. n = 3; P < 0.01 for all P2 strains compared to P1 strains. Black bars, P1 strains; gray bars, P2 strains.
PAO1 strains with P2 phenotype have mutations leading to dysfunction of MexT or MexF.
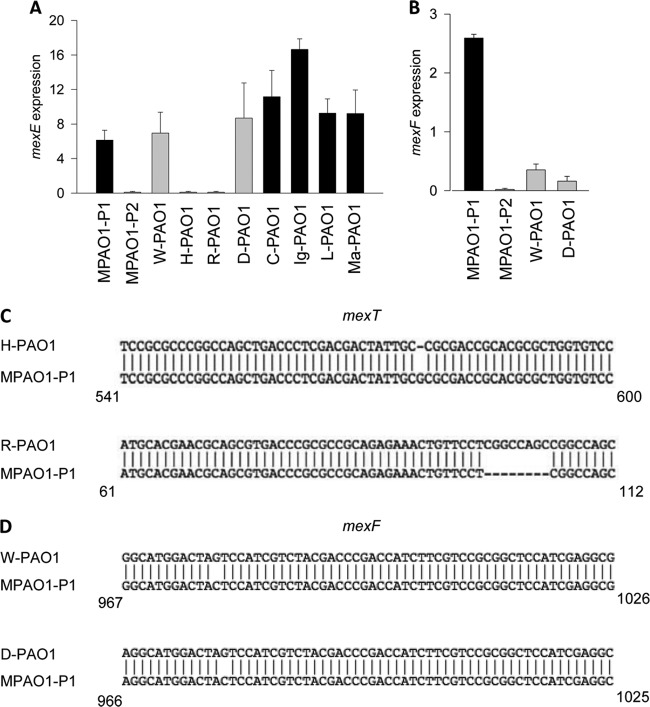
We next tested the hypothesis that P. aeruginosa PAO1 strains with the P2 phenotype had loss of MexEF-OprN efflux pump functionality. Due to the known implication of MexT in the switch from MPAO1-P1 to MPAO1-P2 (3), we first measured the expression of mexE, known to be positively regulated by MexT. The mexE expression was analyzed by qRT-PCR in cells collected after 3.5 h of growth in TSB. All P1 phenotype strains (C-PAO1, Ig-PAO1, L-PAO1, and Ma-PAO1) and two P2 phenotype strains (W-PAO1 and D-PAO1) demonstrated active transcription of mexE, while the remaining two P2 strains (H-PAO1 and R-PAO1) did not (Fig. 5A). Sequencing of mexT revealed that the strains with mexE transcription contained a mexT sequence identical to that of MPAO1-P1. H-PAO1 and R-PAO1, which did not transcribe mexE, contained frameshift mutations in mexT. H-PAO1 contained a 1-bp deletion (Fig. 5C), while R-PAO1 contained an 8-bp insertion (Fig. 5C) identical to that in the P. aeruginosa PAO1-UW strain.
FIG 5.
PAO1 strains with the P2 phenotype have mutations leading to dysfunction of MexT and MexF. (A and B) Expression of mexE (A) and mexF (B) determined by qRT-PCR. n = 3/group. (C) Sequence of mexT in H-PAO1 and R-PAO1, demonstrating frameshift mutations. (D) Sequence of mexF in W-PAO1 and D-PAO1, demonstrating SNP leading to a premature stop codon.
To elucidate the controversy between the P2 phenotype in strains W-PAO1 and D-PAO1 that have intact mexT and express mexE, we decided to examine the expression of mexF in these strains even though both are transcribed within the same operon. qRT-PCR revealed that both W-PAO1 and D-PAO1 expressed negligible amounts of mexF transcript (Fig. 5B) that were significantly lower than the levels of mexE transcript. Sequencing of mexF revealed that both strains harbored an identical C-to-G SNP at nucleotide 978, causing a shift from a tyrosine to a premature stop codon (Fig. 5D). These data demonstrate that PAO1 strains displaying the P2 phenotype have various mutations leading to dysfunction in the MexEF-OprN efflux pump.
P2 phenotype is broadly distributed in the P. aeruginosa MPAO1 mutant strains of the transposon mutant library.
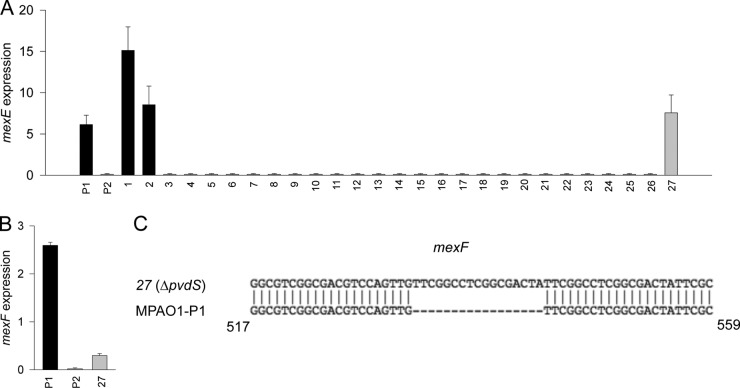
From our experience working with the P. aeruginosa MPAO1 transposon mutant library, we noticed the occasional appearance of high-pyocyanin-producing mutants that could not be explained by disruption of the gene. Given our current knowledge about the emergence of the P2 phenotype in MPAO1 populations, we hypothesized that pyocyanin production in the mutants could be due to the P2 phenotype. We randomly chose 66 mutants (verified by PCR to have a transposon insertion in the expected gene) and assessed them for swarming capacity, collagenase activity, resistance to chloramphenicol, and pyocyanin production. Among them, 27 mutants demonstrated either the P1 or P2 phenotype (Table 1) and were chosen for mexE expression analysis (Fig. 6A). In this analysis, the two P1 mutants showed transcription of mexE, while 24 of the 25 P2 mutants did not. The pvdS mutant (mutant 27) demonstrated active transcription of mexE despite its P2 phenotype. We then examined mexF transcription in the PvdS mutant and found it to be negligible mexF (Fig. 6B). Sequencing of the PvdS mutant mexF revealed a 17-bp insertion leading to a frameshift mutation and a downstream premature stop codon (Fig. 6C).
FIG 6.
MPAO1 mutant strains with the P2 phenotype have dysfunction of MexEF. (A and B) Expression of mexE (A) and mexF (B) determined by qRT-PCR. (C) Sequence of mexF in the ΔpvdS strain (no. 27 in Table 1), demonstrating a 17-base-pair insertion.
To confirm that the phenotype switch to P2 was not due to the transposon insertion in the mutant gene, we randomly chose one mutant with the P2 phenotype, the ΔhxcT mutant (no. 23 in Table 1), and complemented this mutant with a functional hxcT gene. The complemented ΔhxcT/hxcT mutant retained the P2 phenotype (see Fig. S3 in the supplemental material), confirming that the P2 switch in phenotype was not due to the effect of the disrupted gene.
DISCUSSION
PAO1 is one of the most widely used P. aeruginosa strains in scientific research by laboratories across the globe (32). Here we describe the emergence of the P2 phenotype in PAO1 populations that correlates with various mutations leading to the MexEF-OprN efflux pump dysfunction and global changes in PAO1 behavior.
We demonstrated higher intracellular levels of C4-HSL and PQS and higher extracellular levels of C4-HSL and 3OC12-HSL in P2 than P1. P2 also shows increased activation of the Rhl quorum-sensing subsystem, which may suggest that dysfunction of the MexEF-OprN pump mainly affects the Rhl/C4-HSL quorum-sensing pathway of P. aeruginosa. Although the MexEF-OprN efflux pump has been extensively characterized in the context of antibiotics, it has been suggested that the main purpose of the pump may not actually be antimicrobial extrusion (33) but may rather be metabolic function (34). For example, some isolates of P. aeruginosa from a mouse model of pneumonia express the MexEF-OprN pump without any exposure to antimicrobials (35). There is increasing evidence that the MexEF-OprN pump functions to extrude quorum-sensing molecules. For example, the MexEF-OprN efflux pump can export HHQ (4-hydroxy-2-heptylquinoline) (17), a precursor of PQS, which leads to decreased intracellular PQS synthesis. It has also been suggested that 3OC12-HSL may be secreted by the MexEF-OprN pump in cultures with very high cell densities (optical density, >4) (5). Intriguingly, the same study found that strains expressing the MexEF-OprN pump have an ∼60% lower extracellular concentration of C4-HSL. In contrast, others have suggested that the MexEF-OprN system may be involved in the export of C4-HSL from the cell (16). Consistent with others' findings (5, 16), we did observe intracellular accumulation of all three QS molecules in MPAO1-P2 involved in quorum sensing, which is known to regulate pyocyanin production. The transcriptional response demonstrated expression of the Rhl subsystem in P2, whereas there was no significant difference in lasI and pqsA expression. It could be speculated that there is competitive binding between C4-HSL and 3OC12-HSL for LasR, which may offset the increase of LasI overactivation predicted by the intracellular accumulation of 3OC12-HSL. RhlR has been shown to inhibit the expression of pqsA (36) and this may account for the discordance of the expected increase in pqsA expression in P2 at the transition to stationary phase. P2 cells therefore may shift toward an increased Rhl/C4-HSL subtype.
Precisely how P2 outcompetes P1 in nutrient-rich environments remains to be clarified. The increased competitiveness of P2 can be attributed to earlier activation of quorum sensing and accelerated growth at the early exponential phase. The finding that P2 can outcompete P1 led us to assess the prevalence of the P2 phenotype among PAO1 strains, including those in the MPAO1 transposon library (parental strain MPAO1 of the P1 phenotype) (18). Our results demonstrated that, among this group, P2 strains harbor various mutations in mexT and mexF, leading to dysfunction of MexEF-OprN.
Additionally, P2 phenotype strains seem to be quite stable over time. For example, W-PAO1 and D-PAO1 were obtained from the same laboratory but with a gap of almost 10 years. The fact that both contained the same SNP in mexF demonstrates the stability of the switch to the P2 phenotype. We have also demonstrated high stability in the P2 phenotype in strain MPAO1-P2, which was generated in our rat model of anastomotic leakage (3). The stability of the P2 phenotype and its appearance in multiple contexts suggest that certain environmental conditions may favor its emergence.
The emergence of the P2 phenotype within the transposon mutant library, which was constructed from a parental MPAO1 strain of the P1 phenotype, has several important implications. Other investigators have reported an unexpected increase in pyocyanin production and a universal hypersusceptibility of mutants to antimicrobials seen across a broad range of functionally unrelated mutants within this library (37). These reports did not provide a mechanism for this phenomenon but suggested that these mutants may have appeared as a result of an unknown but generalizable effect across the library. Because the P2 phenotype displays such significant phenotypic changes compared to the P1 phenotype independent of the disrupted gene in a given mutant, it could be argued that the P2 phenotype can mask the effect of the known mutation. Finally the finding that strains of P. aeruginosa PAO1 from various laboratories can express either the P1 or P2 phenotype may complicate comparisons between studies. Therefore, it may be advisable in future studies involving the use of PAO1 strains for them to be identified in terms of the expression of the P1 or P2 phenotype.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Biological Sciences Collegiate Division Research Endowments at the University of Chicago (P.M.L.) and NIH RO1 5R01GMO62344 (J.C.A.).
MPAO1-P1/RFP was constructed in collaboration with the laboratory of Deborah Nelson, Department of Neurobiology, Pharmacology & Physiology Lab, University of Chicago.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01050-13.
REFERENCES
- 1.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Hoiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119. 10.1099/mic.0.033993-0 [DOI] [PubMed] [Google Scholar]
- 2.Kalia A, Bessen DE. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J. Bacteriol. 186:110–121. 10.1128/JB.186.1.110-121.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivas AD, Shogan BD, Valuckaite V, Zaborin A, Belogortseva N, Musch M, Meyer F, Trimble WL, An G, Gilbert J, Zaborina O, Alverdy JC. 2012. Intestinal tissues induce an SNP mutation in Pseudomonas aeruginosa that enhances its virulence: possible role in anastomotic leak. PLoS One 7:e44326. 10.1371/journal.pone.0044326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, Van Delden C, Curty LK, Kohler T. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027–3033. 10.1128/JB.184.11.3027-3033.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213–5222. 10.1128/JB.183.18.5213-5222.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maseda H, Saito K, Nakajima A, Nakae T. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107–112. 10.1111/j.1574-6968.2000.tb09367.x [DOI] [PubMed] [Google Scholar]
- 7.Fukuda H, Hosaka M, Hirai K, Iyobe S. 1990. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob. Agents Chemother. 34:1757–1761. 10.1128/AAC.34.9.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian ZX, Mac Aogain M, O'Connor HF, Fargier E, Mooij MJ, Adams C, Wang YP, O'Gara F. 2009. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb. Pathog. 47:237–241. 10.1016/j.micpath.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 9.Zaoui C, Overhage J, Lons D, Zimmermann A, Musken M, Bielecki P, Pustelny C, Becker T, Nimtz M, Haussler S. 2012. An orphan sensor kinase controls quinolone signal production via MexT in Pseudomonas aeruginosa. Mol. Microbiol. 83:536–547. 10.1111/j.1365-2958.2011.07947.x [DOI] [PubMed] [Google Scholar]
- 10.Jin Y, Yang H, Qiao M, Jin S. 2011. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J. Bacteriol. 193:399–410. 10.1128/JB.01079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L, Tummler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192:1113–1121. 10.1128/JB.01515-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechere JC. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345–354. 10.1046/j.1365-2958.1997.2281594.x [DOI] [PubMed] [Google Scholar]
- 13.Tin S, Sakharkar KR, Lim CS, Sakharkar MK. 2009. Activity of chitosans in combination with antibiotics in Pseudomonas aeruginosa. Int. J. Biol. Sci. 5:153–160. 10.3923/ijb.2009.153.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llanes C, Kohler T, Patry I, Dehecq B, van Delden C, Plesiat P. 2011. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 55:5676–5684. 10.1128/AAC.00101-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maseda H, Yoneyama H, Nakae T. 2000. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:658–664. 10.1128/AAC.44.3.658-664.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1320–1328. 10.1128/AAC.48.4.1320-1328.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. 10.1371/journal.pone.0024310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS. 2008. A noncytotoxic DsRed variant for whole-cell labeling. Nat. Methods 5:955–957. 10.1038/nmeth.1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen RH, DeBusscher G, McCombie WR. 1982. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J. Bacteriol. 150:60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277. 10.1111/j.1365-2958.2006.05421.x [DOI] [PubMed] [Google Scholar]
- 22.Choi KH, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1:153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- 23.Koch B, Jensen LE, Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187–195. 10.1016/S0167-7012(01)00246-9 [DOI] [PubMed] [Google Scholar]
- 24.Tremblay J, Deziel E. 2008. Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 48:509–515. 10.1002/jobm.200800030 [DOI] [PubMed] [Google Scholar]
- 25.Stiernagle T. 2006. Maintenance of C. elegans. In Chalfie M. (ed), WormBook: the online review of C. elegans biology. http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html [DOI] [PMC free article] [PubMed]
- 26.Zaborin A, Gerdes S, Holbrook C, Liu DC, Zaborina OY, Alverdy JC. 2012. Pseudomonas aeruginosa overrides the virulence inducing effect of opioids when it senses an abundance of phosphate. PLoS One 7:e34883. 10.1371/journal.pone.0034883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185–192. 10.1111/j.1574-6968.1998.tb13044.x [DOI] [PubMed] [Google Scholar]
- 28.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14:87–96. 10.1016/j.chembiol.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 29.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR, Rahme LG, Chang E, Alverdy JC. 2007. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 3:e35. 10.1371/journal.ppat.0030035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U. S. A. 106:6327–6332. 10.1073/pnas.0813199106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333–343. 10.1111/j.1365-2958.1995.mmi_17020333.x [DOI] [PubMed] [Google Scholar]
- 32.Carilla-Latorre S, Calvo-Garrido J, Bloomfield G, Skelton J, Kay RR, Ivens A, Martinez JL, Escalante R. 2008. Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 8:109. 10.1186/1471-2180-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, Poole K. 2011. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob. Agents Chemother. 55:508–514. 10.1128/AAC.00830-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Schweizer HP. 2011. Evidence of MexT-independent overexpression of MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa in presence of metabolic stress. PLoS One 6:e26520. 10.1371/journal.pone.0026520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Join-Lambert OF, Michea-Hamzehpour M, Kohler T, Chau F, Faurisson F, Dautrey S, Vissuzaine C, Carbon C, Pechere J. 2001. Differential selection of multidrug efflux mutants by trovafloxacin and ciprofloxacin in an experimental model of Pseudomonas aeruginosa acute pneumonia in rats. Antimicrob. Agents Chemother. 45:571–576. 10.1128/AAC.45.2.571-576.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGrath S, Wade DS, Pesci EC. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27–34. 10.1016/S0378-1097(03)00849-8 [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Wang H, Zhao X. 2008. Antimicrobial studies with the Pseudomonas aeruginosa two-allele library require caution. Antimicrob. Agents Chemother. 52:3826–3827. 10.1128/AAC.00419-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.