Abstract
Type IV pili (T4P) are ubiquitous bacterial cell surface structures that undergo cycles of extension, adhesion, and retraction. T4P function depends on a highly conserved envelope-spanning macromolecular machinery consisting of 10 proteins that localizes polarly in Myxococcus xanthus. Using this localization, we investigated the entire T4P machinery assembly pathway by systematically profiling the stability of all and the localization of eight of these proteins in the absence of other T4P machinery proteins as well as by mapping direct protein-protein interactions. Our experiments uncovered a sequential, outside-in pathway starting with the outer membrane (OM) PilQ secretin ring. PilQ recruits a subcomplex consisting of the inner membrane (IM) lipoprotein PilP and the integral IM proteins PilN and PilO by direct interaction with the periplasmic domain of PilP. The PilP/PilN/PilO subcomplex recruits the cytoplasmic PilM protein, by direct interaction between PilN and PilM, and the integral IM protein PilC. The PilB/PilT ATPases that power extension/retraction localize independently of other T4P machinery proteins. Thus, assembly of the T4P machinery initiates with formation of the OM secretin ring and continues inwards over the periplasm and IM to the cytoplasm.
INTRODUCTION
Type IV pili (T4P) are important virulence factors in bacterial pathogens, such as Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa, and Vibrio cholerae, and are among the most widespread filamentous cell surface structures in bacteria (1). T4P are involved in a variety of processes including attachment to eukaryotic host cells during pathogenesis (2), biofilm formation (3, 4), cellular motility (5), protein secretion (6), and DNA uptake (7). Key to this versatility, and a distinguishing feature of T4P compared to other bacterial surface structures, is their ability to undergo cycles of extension and retraction (8, 9). Therefore, the assembly and function of T4P represent potential targets for novel antibacterial drugs (10). T4P are several micrometers in length, have a diameter of 5 to 8 nm, and are composed of the PilA pilin subunit and in some cases also of minor pilins (11). The extended T4P may adhere to an appropriate surface, and due to this adherence, retractions pull a cell forward (8, 9). During retractions, forces of up to 150 pN per pilus are generated (12, 13), making the T4P machinery the strongest molecular motor characterized.
T4P can be divided into the T4aP and T4bP subgroups, which are distinguished based on pilin size and assembly systems (1, 14). Here, we focus on T4aP and refer to these as T4P. T4P function depends on a set of 11 highly conserved proteins that localize to the cytoplasm, inner membrane (IM), periplasm, and outer membrane (OM) (1). All these proteins, with the exception of PilT, share sequence and/or structural similarity to proteins of type II secretion systems (T2SS), which are involved in the translocation of proteins from the periplasm over the OM to the extracellular environment, suggesting that the two machineries share functional characteristics (15–18). Consistently, T2SS have been hypothesized to depend on the formation of pseudopili that would function as a piston to “push” secretory proteins over the OM (18–20). In the case of T4P, PilA is the T4P pilin subunit and is synthesized as a preprotein with an N-terminal type III signal peptide and secreted via the Sec system (21, 22). The signal peptide is removed by the prepilin peptidase PilD (23), leaving the mature pilin anchored in the IM by an N-terminal hydrophobic α-helix (24). During T4P extension, PilA is incorporated at the base of a T4P from this IM reservoir; T4P retraction involves the removal of PilA subunits from the base of the T4P and their reinsertion into the IM (25). The remaining nine proteins can be divided into three subcomplexes based on localization, function, and protein-protein interactions (Fig. 1A) (from here on, these nine proteins and PilA are collectively referred to as the T4P system [T4PS]). In the OM, the secretin PilQ assembles into a highly stable oligomer suggested to consist of 12 to 15 subunits, which form a channel that serves as a conduit for T4P (26–30). Generally, insertion of PilQ monomers into the OM and possibly also oligomer formation depend on an OM lipoprotein, referred to as a pilotin (31–34). Based on analyses mostly of T4PS in P. aeruginosa and N. meningitidis, the PilM, -N, -O, -P, and -Q proteins are thought to form a complex that connects from the cytoplasm to PilQ in the OM. Based on bacterial two-hybrid system analyses, direct protein-protein interaction analyses, and protein structure determination, the bitopic IM proteins PilN and PilO interact directly by means of their periplasmic domains, possibly forming a heterodimer (35–37). PilN/PilO, in turn, interacts with the periplasmic domain of the IM lipoprotein PilP (38, 39). The periplasmic domain of PilP also interacts directly with the periplasmic domains of PilQ (37, 40–42). The short cytoplasmic N terminus of PilN interacts with the cytoplasmic actin-like protein PilM (35, 40, 43). Accordingly, the PilM/N/O/P complex has been suggested to align motor complexes (see below) in the IM/cytoplasm with the PilQ secretin channel in the OM and to regulate gating of the PilQ secretin channel (40, 44).
FIG 1.
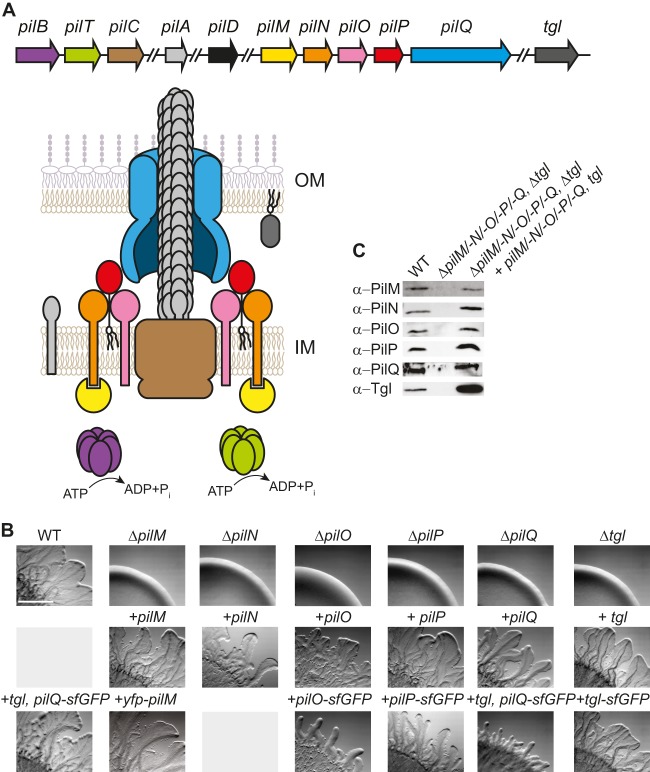
PilN, PilO, PilP, and Tgl are required for T4P-dependent motility in M. xanthus. (A) Model of the T4P machinery. The color code for the proteins is similar to the one used for the genes in the genetic map. PilD is not included in the model of the machinery. Genes and proteins are not drawn to scale. OM, outer membrane; IM, inner membrane. All the pil genes are clustered at the same locus, but not all pil genes are shown (49). tgl is not part of the pil cluster. (B) Motility phenotypes of the indicated mutants and complementation strains. Strains in the same column all contain the same in-frame deletion. Strains were incubated at 32°C for 24 h on 0.5% agar–0.5% CTT. Scale bar, 1 mm. (C) Immunoblots of T4PS protein accumulation. Total cell lysates from exponentially growing cultures were separated by SDS-PAGE (proteins from 7 × 107 cells loaded per lane) and analyzed by immunoblotting using specific antibodies as indicated. For PilQ, only the heat- and detergent-resistant oligomer is shown. The middle lane in each blot contains lysate from the relevant in-frame deletion mutant, and the last lane contains lysate from the relevant complementation strain.
The cytoplasmic secretion ATPases PilB and PilT provide the energy for T4P extension and retraction, respectively (45–47). Recently, two different cytoplasmic domains in the polytopic IM protein PilC were suggested to interact directly with PilB and PilT (48), forming motor complexes for extension and retraction of T4P, respectively (30). Moreover, PilC and PilO have been suggested to interact based on bacterial two-hybrid system analyses (35). Therefore, in the current model, the T4PS forms a macromolecular complex that spans all subcellular compartments. With the exception of PilT, all proteins of the T4PS are involved in T4P extension, whereas PilT is specifically required for T4P retraction (8). T2SS contain only one ATPase, which is a homolog of PilB (15, 17).
The rod-shaped deltaproteobacterium Myxococcus xanthus has emerged as a model system to study the function and assembly of T4P. M. xanthus contains all the conserved T4PS proteins (49). In M. xanthus, T4P power a type of surface motility equivalent to twitching motility in P. aeruginosa and Neisseria spp. (5, 50). Cells of M. xanthus assemble 5 to 10 T4P at the leading cell pole, which generate cell movement in the direction of the long axis of cells (51). Occasionally, cells stop and then reverse their direction of movement (52). During a reversal, the T4P disassemble at the old leading pole and reassemble at the new leading cell pole (53–55). The mechanism underlying the switch in polarity of T4P during reversals is relatively well understood. Specifically, PilC, PilM, and PilQ localize in clusters at both cell poles and do not change localization during reversals (33, 54), and PilA is localized along the entire length of cells (56). In contrast, the ATPases PilB and PilT primarily localize to the leading and lagging cell poles, respectively, and PilT only occasionally accumulates at the leading cell pole (54). Based on the localization pattern of PilB and PilT, it has been argued that the spatial separation of PilB and PilT allows the temporal separation of T4P extension and retraction and that the occasional accumulation of PilT at the leading cell pole coincides with T4P retractions (54). During a reversal, PilB and PilT dissociate from their respective poles and then associate with the new leading and lagging poles, respectively, thus laying the foundation for the assembly of T4P at the new leading pole (54). Based on these observations, it has been suggested that several T4PS proteins form preassembled complexes at both poles that are complemented by the dynamically localized proteins PilB or PilT.
Although the components of the T4PS are known and the functions of several of these proteins have been investigated, the precise mechanisms of how and in which order the individual components are assembled to generate a macromolecular machinery that spans from the cytoplasm to the outside remain to be elucidated. Here, we took advantage of cell biology tools for studying T4PS in M. xanthus and analyzed the assembly process of the T4PS. By systematically profiling the stability and localization of T4PS proteins in the absence of other T4PS proteins in combination with mapping of direct protein-protein interactions, we uncovered that the T4PS assembles in an outside-in manner starting with the PilQ secretin ring in the OM. PilQ provides an assembly platform for a periplasmic/IM subcomplex composed of PilP, PilO, and PilN by direct interactions between PilP and PilQ as well as between PilO and PilN. The PilP/PilO/PilN subcomplex, in turn, recruits the cytoplasmic protein PilM, by direct interaction between PilN and PilM, and the IM PilC protein, likely by direct interaction between PilO and PilC. Moreover, we demonstrate that assembly of this complex does not require PilB and PilT and that PilB and PilT localize independently of all other T4PS proteins to the cell poles.
MATERIALS AND METHODS
Cell growth and construction of strains.
Strain DK1622 was used as the wild-type (WT) M. xanthus strain throughout, and all M. xanthus strains used are derivatives of DK1622. The M. xanthus strains used are listed in Table 1. Plasmids are listed in Table S1 in the supplemental material, and primers used in this work are listed in Table S2 in the supplemental material. Plasmid constructions are described in Materials and Methods in the supplemental material. All plasmids were verified by sequencing. M. xanthus strains were grown at 32°C in 1% CTT broth (57) and on CTT agar plates supplemented with 1.5% agar. Kanamycin (50 μg/ml) or oxytetracycline (10 μg/ml) was added when appropriate. Plasmids were introduced into M. xanthus by electroporation. Site-specific integration of plasmids at the Mx8 attB site on the chromosome was confirmed by PCR. In-frame deletions were generated as described previously (58).
TABLE 1.
M. xanthus strains used in this work
| M. xanthus strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| DK1622 | Wild type | 51 |
| DK8615 | ΔpilQ | 81 |
| DK10409 | ΔpilT | 82 |
| DK10410 | ΔpilA | 83 |
| SA3002 | ΔpilM | 54 |
| DK10416 | ΔpilB | 82 |
| DK10417 | ΔpilC | 82 |
| DK10405 | Tcr::Δtgl | 33 |
| SA3046 | ΔpilM/PpilA-yfp-pilM (pSC8) | 54 |
| SA3045 | ΔpilT yfp pilT (pIB75) | 54 |
| SA3001 | ΔpilO | This work |
| SA3005 | ΔpilP | This work |
| SA3006 | ΔpilQ/PpilA-pilQ (pIB55) | This work |
| SA3007 | Δtgl/PpilA-tgl (pIB54) | This work |
| SA3044 | ΔpilN | This work |
| SA3053 | ΔpilM/PpilA-pilM (pSC2) | This work |
| SA4028 | ΔpilO/PpilA-pilO (pSC38) | This work |
| SA4029 | ΔpilP/PpilA-pilP (pSC39) | This work |
| SA4041 | ΔpilM/PpilA-yfp-pilMR388A (pSC56) | This work |
| SA4058 | ΔpilM/PpilA-yfp-pilMD204A (pSC97) | This work |
| SA4059 | ΔpilM/PpilA-yfp-pilMV204D (pSC98) | This work |
| SA4060 | ΔpilMNOPQ | This work |
| SA4061 | ΔpilN/PpilA-piN (pSC37) | This work |
| SA4067 | WT/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4070 | ΔpilP/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4076 | Δtgl/ PpilA-tgl-sfGFP (pSC104) | This work |
| SA4077 | ΔpilA/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4078 | ΔpilB/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4079 | ΔpilC/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4080 | ΔpilM/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4081 | ΔpilN/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4082 | ΔpilO/PpilA-pilP-sfGFP(pSC102) | This work |
| SA4083 | ΔpilT/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4086 | WT/PpilA-pilO-sfGFP (pSC106) | This work |
| SA4088 | ΔpilO/PpilA-pilO-sfGFP (pSC106) | This work |
| SA4094 | ΔpilQ/PpilA-pilP-sfGFP (pSC102) | This work |
| SA4095 | ΔpilM/PpilA-pilO-sfGFP (pSC106) | This work |
| SA4096 | ΔpilN/PpilA-pilO-sfGFP (pSC106) | This work |
| SA4097 | ΔpilP/PpilA-pilO-sfGFP (pSC106) | This work |
| SA4098 | ΔpilQ/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6001 | ΔpilA/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6002 | ΔpilB/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6003 | ΔpilC/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6005 | ΔpilT/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6024 | ΔpilBTCMNOPQ | This work |
| SA6025 | ΔpilBTCMNOPQ/PpilA-yfp-pilT (pIB75) | This work |
| SA4073 | ΔpilTCMNOPQ | This work |
| SA6026 | WT/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6027 | ΔpilQ/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6028 | ΔpilA/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6029 | ΔpilB/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6030 | ΔpilN/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6031 | ΔpilP/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6033 | ΔpilC/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6034 | ΔpilM/PpilA-tgl pilQ-sfGF P (pSC120) | This work |
| SA6035 | ΔpilO/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6036 | ΔpilT/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6053 | Δtgl | This work |
| SA6056 | Δtgl/PpilA-pilP-sfGFP (pSC102) | This work |
| SA6057 | Δtgl/PpilA-pilO-sfGFP (pSC106) | This work |
| SA6058 | Δtgl/PpilA-yfp-pilT (pIB75) | This work |
| SA6060 | ΔaglZ/PpilA-tgl pilQ-sfGFP (pSC120) | This work |
| SA6061 | Δtgl/PpilA-pilQ-sfGFP (pSC110) | This work |
In constructs with PpilA, genes were transcribed from the pilA promoter. Plasmids containing the indicated expression constructs are in parentheses and were integrated at the Mx8 attB site.
Motility assays.
Cells from exponentially growing cultures were harvested and resuspended in 1% CTT to a density of 7 × 109 cells/ml. Five-microliter volumes were spotted on 0.5% agar supplemented with 0.5% CTT and incubated at 32°C for 24 h. Colony edges were documented using a Leica MZ75 stereomicroscope equipped with a Leica DFC280 camera.
Immunoblot analysis.
For immunoblot analysis, cells from exponentially growing cultures were harvested and resuspended in SDS lysis buffer, and proteins from 7 × 107 cells were separated by SDS-PAGE. Immunoblotting was done using standard procedures (59) with polyclonal anti-PilA (60), anti-PilT, anti-PilB (47), anti-PilC, anti-PilM, anti-PilQ (54), anti-PilN, anti-PilO, anti-PilP, anti-Tgl, and anti-mCherry (Roche) antibodies and secondary anti-rabbit immunoglobulin G peroxidase conjugate (Sigma). For detection of yellow fluorescent protein (YFP)- or green fluorescent protein (GFP)-tagged proteins, monoclonal anti-GFP mouse antibodies (Roche) and peroxidase-conjugated rabbit anti-mouse immunoglobulin G secondary antibodies (DakoCytomation) were used. Blots were developed using Luminata Western HRP Substrate (Merck Millipore).
Protein purification for pulldown experiments.
For overexpression and purification of PilNΔ42-His6, PilOΔ37-His6, PilPΔ20-His6, PilQ20–656-His6, MalE-PilNΔ42, MalE-PilOΔ37, and MalE-PilPΔ20, the plasmids pSC5, pSC6, pSC43, pSC108, pSC44, pSC45, and pSC46 were separately transformed into Escherichia coli Rosetta 2 [F− ompT hsdSB(rB− mB−) gal dcm pRARE2] (Novagen). Cells were grown in 500 ml LB medium with appropriate antibiotics at 37°C to an optical density at 550 nm (OD550) of 0.6. Overexpression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were shifted to 18°C and grown overnight. Cells were harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) containing protease inhibitors (Roche), and disrupted by sonication. Cell debris was removed by centrifugation at 4,000 × g for 10 min at 4°C. Subsequently, the lysate was centrifuged at 160,000 × g for 1 h at 4°C. All proteins used for in vitro analyses were purified under native conditions except for PilOΔ37-His6. His-tagged proteins were purified on an Ni2+-nitrilotriacetic acid (NTA)-agarose column as described by the manufacturer (Qiagen) using lysis buffer and elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). PilOΔ37-His6 was purified under denaturing conditions using lysis buffer B (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 8.0) and elution buffers D and E (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 4.5). Elution fractions were dialyzed stepwise against lysis buffer. MalE-tagged proteins were purified on an amylose matrix (New England BioLabs) using buffer CB1 (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM EDTA). Bound proteins were eluted using buffer CB2 (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM DTT, 1 mM EDTA, 10 mM maltose).
Pulldown experiments.
After purification of individual proteins, they were split and dialyzed against lysis buffer for Ni2+-NTA chromatography or CB1 buffer for amylose chromatography. In the pulldown experiments, proteins in the same buffer were added to the same final equimolar concentration, incubated for 1 to 2 h at 4°C, and applied to an Ni2+-NTA-agarose (Qiagen) or an amylose column (New England BioLabs). Equivalent volumes of the flowthrough, wash, and elution fractions were separated by SDS-PAGE and visualized by Coomassie blue G-250 staining.
Fluorescence microscopy.
Phase-contrast, differential inference contrast (DIC), and fluorescence microscopy was done as described previously (61). Briefly, cells from exponentially growing cultures were transferred to a thin 1.5% agar pad with A50 buffer (10 mM MOPS [morpholinepropanesulfonic acid] [pH 7.2], 10 mM CaCl2, 10 mM MgCl2, 50 mM NaCl) on a glass slide and covered with a coverslip. Cells were immediately visualized with a Leica DM6000B microscope using a Leica Plan Apo 100×/numerical aperture (NA) 1.40 phase-contrast oil objective and imaged with a Roper Photometrics Cascade II 1024 camera or a Leica DFC350FX camera. Images were recorded and processed with Metamorph (Molecular Devices). For time-lapse recordings, cells were applied directly on a 0.1% CTT agarose pad on a coverslip at 32°C, images were recorded every 20 min using a temperature-controlled Leica DMI6000B microscope with an adaptive focus control and a motorized stage and a Hamamatsu Flash 4.0 camera, and images were processed using the Leica MM AF software package. Immunofluorescence microscopy was done essentially as described previously (55). Briefly, exponentially growing M. xanthus cells were fixed with 3.2% (anti-PilC, anti-PilM, anti-PilN, and anti-PilP) or 1.6% (anti-PilB and anti-Tgl) paraformaldehyde and 0.008% glutaraldehyde. Fixed cells were spotted on a poly-l-lysine-treated 12-well diagnostic slide (Thermo Scientific) and permeabilized with glucose-Tris-EDTA (GTE) buffer (50 mM glucose, 20 mM Tris-HCl, 10 mM EDTA, pH 7.5) for 5 min. After blocking with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) for 20 min, primary antibody was added in 2% BSA–PBS overnight at 4°C. After washing with PBS, secondary antibody Alexa Fluor 594 goat anti-rabbit IgG (Molecular Probes, Life Technologies) was added for 2 h. After washing with PBS, Slow Fade Anti Fade Reagent (Molecular Probes, Life Technologies) was added to each well, and cells were covered with a coverslip. Cells were imaged as described above. For each protein, polar fluorescent clusters were defined in the WT, assessing the signal-to-noise ratio, and all other strains were compared to the WT.
Operon mapping.
Total RNA was isolated using a hot phenol extraction method (62). RNA was treated with DNase I (Ambion) and purified with the RNeasy kit (Qiagen). PCR analysis was used to confirm that the RNA was DNA free. One microgram of RNA was used to synthesize cDNA with the High capacity cDNA Archive kit (Applied Biosystems) using random hexamer primers. For the operon mapping, different primer combinations were used to amplify intragenic and intergenic regions. Genomic DNA was used to confirm that all primer pairs worked. RNA was used as a negative control to exclude DNA contamination of the RNA used for cDNA synthesis. The following primer pairs were used to amplify internal or intergenic regions (see Table S2 in the supplemental material): intragenic pilM (opilM-6, opilM-7), intergenic pilM-pilN (opilM-8, opilN-5-o), intragenic pilN (opilN-6-o, opilN-7-o), intergenic pilN-pilO (opilN-8-o, opilO-5-o), intragenic pilO (opilO-6-o, opilO-7-o), intergenic pilO-pilP (opilO-8-o, opilP-5-o), intragenic pilP (opilP-6-o, opilP-7-o), intergenic pilP-pilQ (opilP-8-o, opilQ-12), and intragenic pilQ (opilQ-13, opilQ-14).
RESULTS
PilN, PilO, PilP, and Tgl are required for T4P-dependent motility.
pilN, pilO, pilP, and tgl have been suggested to be important for T4P-dependent motility in M. xanthus based on analysis of transposon insertions and point mutations (33, 49, 63). pilN, pilO, and pilP are part of an operon and cotranscribed with pilM and pilQ (Fig. 1A; see Fig. S1 in the supplemental material). Tgl is an OM lipoprotein and was suggested to function as a pilotin to stimulate PilQ OM insertion and, in this way, PilQ oligomer formation in the OM (33, 63). As a starting point, we sought to confirm these observations and to rule out polar effects of previously analyzed mutants. Therefore, in-frame deletions of pilN, pilO, pilP, and tgl were generated and characterized. For assessment of T4P-dependent motility in the four in-frame deletion mutants, cells were spotted on 0.5% agar, which is favorable to T4P-dependent motility (64). The WT strain DK1622 formed the long flares characteristic of T4P-dependent motility, whereas the ΔpilM and ΔpilQ control strains, which are unable to assemble T4P (54), generated smooth colony edges without flares (Fig. 1B). Similarly, all four new in-frame deletion mutants were unable to move by T4P-dependent motility (Fig. 1B). Importantly, the defect in T4P-dependent motility in all six mutants was complemented by ectopic expression of the relevant WT gene from the constitutively active pilA promoter at the Mx8 attB site (Fig. 1B). Immunoblots using specific antibodies confirmed that the proteins accumulated at levels comparable to those in WT in the complementation strains (Fig. 1C), with the exception of Tgl, which accumulated at increased levels. Moreover, in the absence of Tgl, only PilQ monomers accumulated whereas PilQ oligomers were undetectable (Fig. 2A), confirming that Tgl stimulates the formation of PilQ oligomers in the OM.
FIG 2.
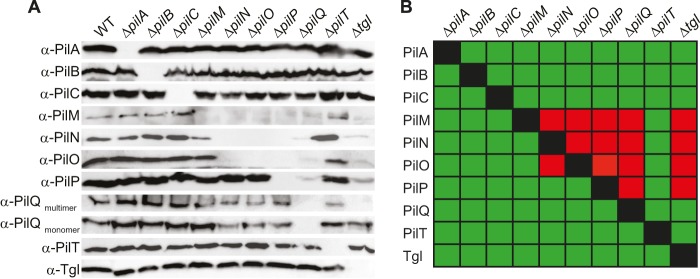
T4PS protein accumulation in in-frame deletion mutants. (A) Cells were grown and analyzed as described for Fig. 1C using specific antibodies as indicated. (B) Summary of observed effects on protein stability. Green and red indicate no effect or negative effect, respectively.
We also aimed at testing the importance of the PilD prepilin peptidase for T4P motility. However, all attempts to generate an in-frame deletion or an insertion mutant in pilD were unsuccessful. pilD is the only M. xanthus gene that encodes a full-length prepilin peptidase, whereas MXAN_3105 encodes a truncated prepilin peptidase (65). Because none of the other T4PS proteins are essential, we speculate that PilD is involved in processing other proteins with a type III signal sequence that are either required for survival or are toxic to the cells when they accumulate as unprocessed proteins. Because we were unable to generate a pilD mutant, we did not include pilD in further analyses.
Stability of T4PS proteins in the absence of each individual other T4PS protein.
To begin to understand how the T4PS proteins interact, we systematically determined the accumulation of every T4PS protein in the absence of each individual other T4PS protein using immunoblot analysis (Fig. 2A and B). If the stability of one protein depends on the presence of another protein, this is an indication that the two proteins interact directly or indirectly as part of a complex. This approach has been successfully used in the case of proteins of the T4PS in P. aeruginosa (44).
PilQ and Tgl accumulated at WT levels in all nine relevant deletion mutants, and only the absence of Tgl blocked the formation of PilQ oligomers. In the absence of Tgl or PilQ, PilM, PilN, PilO, and PilP either were not detectable or accumulated at strongly reduced levels. In the absence of the IM lipoprotein PilP, accumulation of PilM, PilN, and PilO was strongly reduced. PilN and PilO were mutually dependent for accumulation and both essential for PilM accumulation. In contrast, lack of PilM did not affect the accumulation of any of the analyzed proteins. PilB, PilT, PilC, and PilA accumulated in all nine relevant in-frame deletion mutants. Also, lack of PilB, PilT, PilC, or PilA did not interfere with the accumulation of any other T4PS protein. These effects on protein accumulation are not due to polar effects, because the motility defect in each of the in-frame deletion mutants is complemented by the ectopic expression of the relevant WT gene (Fig. 1B) (47, 54).
PilN, PilO, and PilP localize in a bipolar pattern.
PilQ, PilM, and PilC localize in a bipolar, symmetric pattern (33, 54; see also Fig. S4 in the supplemental material), and PilA localizes dispersedly in the IM (56). Moreover, it has been suggested that Tgl localizes in a unipolar pattern (33). To visualize the remaining components of the T4PS, we performed immunofluorescence microscopy with specific antibodies against PilN, PilO, and PilP. In the case of PilN and PilP, weak nonpolar signals were detected in the WT as well as in the ΔpilN and ΔpilP mutants; importantly, bipolar clusters were observed only in the WT, suggesting that the bipolar signals are specific (Fig. 3). In contrast, specific PilO signals could not be detected (data not shown). To be able to follow PilN, PilO, and PilP by live-cell microscopy, C-terminal fluorescent fusions to superfolder GFP (sfGFP), which folds correctly in the periplasm when exported via the Sec system (66, 67), were generated for PilN, PilO, and PilP. Ectopic expression of the fusion proteins from the pilA promoter restored the motility defects in the ΔpilO and ΔpilP mutants (Fig. 1B), whereas the PilN-sfGFP was unable to complement the ΔpilN mutant. Therefore, the PilN-sfGFP protein was not considered further. PilO-sfGFP and PilP-sfGFP accumulated at levels comparable to those of the native proteins in the WT and with some degradation in the case of PilP-sfGFP (see Fig. S2A and B in the supplemental material), suggesting that the fusion proteins are active. As shown in Fig. 3, PilO-sfGFP and PilP-sfGFP formed clusters that localized symmetrically to both cell poles. In total, PilN localizes in a bipolar symmetric pattern as determined using immunofluorescence microscopy, PilO localizes in a bipolar symmetric pattern as determined using the PilO-sfGFP fusion, and PilP localizes in a bipolar symmetric pattern as determined using immunofluorescence microscopy as well as the PilP-sfGFP fusion.
FIG 3.
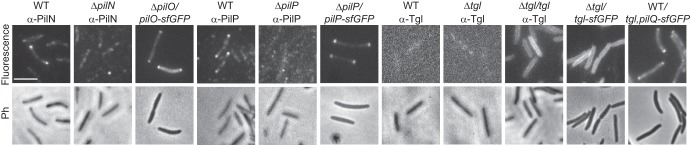
Localization of PilN, PilO, PilP, PilQ, and Tgl. Cells from exponentially growing cultures were transferred to a 1.5% agar pad on a microscope slide and imaged by fluorescence microscopy in the case of the fluorescent fusion proteins of PilO, PilP, PilQ, and Tgl. For native PilN, PilP, and Tgl, cells were fixed and imaged by immunofluorescence microscopy using anti-PilN, anti-PilP, and anti-Tgl antibodies, respectively. Upper and lower rows show fluorescence and phase-contrast (Ph) microscopy images, respectively. Scale bar, 5 μm.
To determine the localization of Tgl, immunofluorescence microscopy using anti-Tgl antibodies as described previously (33) was performed. Using this method, we were unable to detect a specific Tgl signal in the WT (Fig. 3); however, in the Δtgl/tgl strain, which overexpresses Tgl (Fig. 1C), we observed a fluorescence signal along the entire cell circumference, suggesting that Tgl in the OM is localized along the entire cell length. Because this observation is at odds with the suggested unipolar Tgl localization, we analyzed the localization of a Tgl-sfGFP fusion protein. Ectopic expression of the fusion protein from the pilA promoter in a Δtgl mutant resulted in Tgl-sfGFP accumulation at a slightly higher level than in the native protein in the WT, and moreover, Tgl-sfGFP was partially degraded, resulting in the accumulation of a protein with the size of Tgl (see Fig. S2C in the supplemental material). Expression of Tgl-sfGFP restored the motility defect in the Δtgl mutant (Fig. 1B), suggesting that the fusion is active. Importantly, Tgl-sfGFP also showed a uniform localization to the cell periphery and along the length of cells (Fig. 3). Nudleman et al. detected polar Tgl localization in approximately one-third of the cells, whereas the remaining cells had a uniform localization of Tgl along the cell periphery. It has been reported that overexpression of proteins of the T2SS may result in their polar localization whereas the same proteins expressed at native levels are localized at the cell periphery along the length of cells (68, 69). Despite our use of strains overexpressing Tgl or Tgl-sfGFP, we observed only the circumferential localization pattern. The unipolar localization suggested for Tgl implies that the protein relocates between the poles during cellular reversals as reported for cytoplasmic motility proteins (54, 55). Cellular reversals occur on a time scale of 15 to 30 s; we find it unlikely that an OM lipoprotein would be able to relocate from one pole to the other within 15 to 30 s. Alternatively, a unipolar localization of Tgl could be accomplished by a mechanism in which newly synthesized Tgl specifically localizes to one cell pole and is then fully degraded within one cell cycle. To test this idea, we determined the half-life (t1/2) of Tgl by blocking protein synthesis in WT M. xanthus cells using chloramphenicol and then monitored Tgl levels by immunoblotting. This experiment showed that Tgl has a t1/2 of >360 min (see Fig. S3 in the supplemental material). Because one M. xanthus cell cycle is approximately 330 min, we conclude that this hypothesis cannot explain a unipolar localization of Tgl. Therefore, we suggest that Tgl localizes uniformly throughout the OM.
The PilQ secretin was previously shown to localize in a bipolar, symmetric pattern using immunofluorescence microscopy (33, 54). To be able to follow PilQ localization over time, we took advantage of the approach that was used to determine the localization of the PulD secretin of the T2SS in Escherichia coli (70). Therefore, we expressed a PilQ-sfGFP fusion together with its pilotin Tgl from the pilA promoter. PilQ-sfGFP accumulated at a reduced level compared to native PilQ in the WT (see Fig. S2D in the supplemental material). In motility assays, expression of PilQ-sfGFP together with Tgl in the ΔpilQ mutant partially restored motility, suggesting that the fusion protein is partially active (Fig. 1B). Expression of PilQ-sfGFP together with Tgl did not have a dominant negative effect on motility in the WT (Fig. 1B). Moreover, PilQ-sfGFP formed monomers as well SDS-resistant oligomers when expressed in an otherwise WT strain (see Fig. S2D and E in the supplemental material), suggesting that the fusion protein formed mixed multimers with native PilQ and that the fusion protein is efficiently incorporated into functional secretin complexes. Consistently, we observed that PilQ-sfGFP localized in a bipolar, symmetric pattern in an otherwise WT strain (Fig. 3) as reported for native PilQ using immunofluorescence microscopy.
From these and previous analyses (33, 54, 56), we conclude that eight T4PS proteins (PilQ, PilP, PilN, PilO, PilM, PilC, PilB, and PilT) (see also Fig. S4 in the supplemental material) localize to the cell poles and that the T4P subunit PilA and likely also the pilotin Tgl localize uniformly in the cell envelope along the cell length. Because M. xanthus only assembles T4P at one pole at a time (51) and the pole at which T4P are assembled switches within 15 to 30 s during cellular reversals, we suggest that the fluorescent clusters observed at the poles correspond to preassembled T4PS complexes and that the specific localization of PilB and PilT determines at which pole T4P are assembled.
Assembly of the T4PS starts from multimeric PilQ in the OM and proceeds inwards.
The observation that eight of the T4PS proteins localize to clusters at the cell poles and likely correspond to preassembled T4PS complexes offered the opportunity to decipher the assembly pathway of the T4PS. Therefore, we systematically investigated the localization of all eight polarly localized T4PS protein in the WT as well as in the absence of each individual other T4PS protein using the array of mutants containing nonpolar in-frame deletions.
PilQ localization was investigated using PilQ-sfGFP coexpressed with Tgl. In WT as well as in nine in-frame deletion mutants, PilQ-sfGFP accumulated (see Fig. S2D and E in the supplemental material) and displayed bipolar localization indistinguishable from that in the WT in all these strains (Fig. 4A; see Fig. S4 in the supplemental material). When PilQ-sfGFP was expressed in the absence of extra Tgl in a Δtgl mutant, the polar localization was abolished and PilQ-sfGFP localized throughout the cells (Fig. 4A; see Fig. S4 in the supplemental material). Because Tgl is the only protein required for PilQ oligomer formation in the OM, we conclude that only oligomeric PilQ in the OM is polarly localized. Because Tgl likely localizes throughout the OM, these data also suggest that Tgl is not a polar targeting determinant for PilQ but functions to stimulate OM insertion and in this way PilQ oligomer formation and polar localization.
FIG 4.
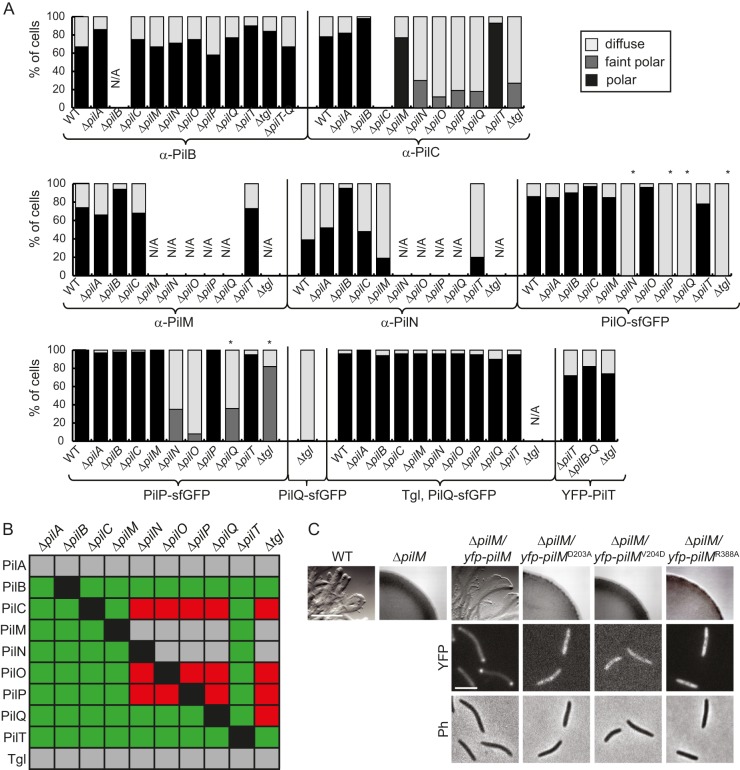
Localization of proteins of the T4PS. (A) Cells were analyzed as indicated for Fig. 3. Immunofluorescence microscopy was used to analyze the localization of PilB, PilC, PilM, and PilN. PilO, PilP, PilQ, and PilT were localized using the indicated sfGFP/YFP-tagged fusion proteins. Patterns of fluorescence signals were grouped into three categories. The histograms illustrate the distribution of localization patterns of the indicated proteins in the indicated in-frame deletion mutants (n = 57 to 317). *, in-frame deletion mutants in which the relevant native protein does not accumulate (compare with Fig. 2). Strains marked N/A (not applicable) were not analyzed. (B) Summary of observed effects on protein localization. Green and red indicate no effect or negative effect, respectively. Gray combinations were not analyzed. (C) Motility assays of the indicated pilM mutants and localization of the indicated PilM proteins. Upper row, motility assay of the indicated strains; scale bar, 1 mm. Cells were treated as indicated for Fig. 1B. Middle and lower rows, fluorescence (YFP) and phase-contrast (Ph) microscopy images of the indicated strains; scale bar, 2 μm. Cells were treated as indicated for Fig. 3.
To determine PilP localization, we expressed the active PilP-sfGFP fusion in the WT and in the 10 in-frame deletion mutants. Although native PilP is unstable in the Δtgl and ΔpilQ mutants, PilP-sfGFP accumulated in both mutants (see Fig. S2B and F in the supplemental material) suggesting that the C-terminal sfGFP tag protects PilP from proteolytic degradation in the absence of Tgl and PilQ and that the fusion protein can be used to follow PilP localization in the absence of Tgl and PilQ. PilP-sfGFP formed faint polar clusters in the absence of Tgl and showed diffuse localization or faint polar clusters in a few cells in the ΔpilQ, ΔpilN, and ΔpilO mutants. In all other mutants, PilP localized in a symmetric bipolar pattern (Fig. 4A; see Fig. S4 in the supplemental material). These data suggest that polar localization of PilP-sfGFP and, by implication, incorporation into the T4PS complex depend on Tgl, PilQ, PilN, and PilO.
Immunoblot analyses revealed that active PilO-sfGFP accumulated in the WT as well as in all in-frame deletion mutants, although accumulation was slightly lower in the ΔpilN and ΔpilP mutants (see Fig. S2A and G in the supplemental material), suggesting that the C-terminal sfGFP tag stabilizes PilO in the Δtgl, ΔpilQ, ΔpilP, and ΔpilN mutants and that the fusion protein can be used to follow PilO localization in the absence of Tgl, PilQ, PilO, and PilN. Despite its accumulation, PilO-sfGFP did not form polar clusters in the Δtgl, ΔpilQ, ΔpilP, and ΔpilN mutants (Fig. 4A; see Fig. S4 in the supplemental material). Bipolar PilO-sfGFP clusters were detected in all other mutants as in the WT (Fig. 4A; see Fig. S4). These data suggest that PilO-sfGFP incorporation into the T4PS complex depends on Tgl, PilQ, PilP, and PilN.
Because we were unable to generate an active fluorescently tagged PilN protein, we determined the localization of PilN using immunofluorescence microscopy. PilN does not accumulate in the ΔpilO, ΔpilP, ΔpilQ, and Δtgl mutants (Fig. 2A and B); therefore, we determined PilN localization only in the remaining in-frame deletion mutants. In all these mutants, PilN formed bipolar clusters (Fig. 4A; see Fig. S4 in the supplemental material). We conclude that incorporation of PilN into the T4PS complex is independent of PilB, PilT, PilC, PilM, and PilA.
To localize PilM, we performed immunofluorescence microscopy. PilM does not accumulate in the ΔpilN, ΔpilO, ΔpilP, ΔpilQ, and Δtgl mutants (Fig. 2A and B). Therefore, these mutants were excluded from the localization experiment. In the remaining four mutants, polar PilM localization was observed as in the WT (Fig. 4A; see Fig. S4 in the supplemental material). We conclude that PilB, PilT, PilC, and PilA are dispensable for incorporation of PilM into the T4PS complex.
PilC localization was investigated by immunofluorescence microscopy. Because PilC accumulates in all mutants (Fig. 2), PilC localization was determined in the WT as well as in all the in-frame deletion mutants. Polar PilC clusters were observed in the absence of PilB, PilT, PilA, and PilM (Fig. 4A; see Fig. S4 in the supplemental material). Interestingly, although PilC was stable in the absence of Tgl, PilQ, PilP, PilO, and PilN, lack of any one of these five proteins resulted in a decreased number of cells with polar clusters and the clusters formed were fainter than those formed in the WT (Fig. 4A; see Fig. S4 in the supplemental material). We conclude that incorporation of PilC into the T4PS complex depends on Tgl, PilQ, PilP, PilO, and PilN.
PilB localization was investigated using immunofluorescence microscopy as previously described (54). Weak nonpolar signals were detected in all strains tested; however, in the single-deletion mutants, polar PilB signals indistinguishable from those in the WT were observed only in pilB+ strains (Fig. 4A; see Fig. S4 in the supplemental material), suggesting that none of the T4PS proteins are essential for polar PilB localization. Consistently, polar PilB clusters were also observed in a mutant lacking PilT, PilC, PilM, PilN, PilO, PilP, and PilQ (Fig. 4A; see Fig. S4).
To investigate the localization of PilT, a functional YFP-PilT fusion expressed at native levels (54) was used. YFP-PilT accumulated (see Fig. S2H in the supplemental material) and localized polarly in the ΔpilT mutant as well as in a strain lacking PilB, PilT, PilC, PilM, PilN, PilO, PilP, and PilQ (Fig. 4A; see Fig. S4 in the supplemental material). Moreover, YFP-PilT localized polarly independently of Tgl (Fig. 4A; see Fig. S4). These data suggest that YFP-PilT, similarly to PilB, localizes independently of all other T4PS proteins. A summary of the observed effects on protein localization is shown in Fig. 4B.
PilN recruits PilM to the T4P machinery.
To establish a clear connection between PilM and the remaining T4PS complex, we took advantage of the recently solved X-ray structure of PilM of Thermus thermophilus (43). In this structure, PilM binds a 15-amino-acid peptide that corresponds to the conserved, cytoplasmic N terminus of PilN. Similarly, PilM of P. aeruginosa binds the conserved N terminus of PilN (40). The N terminus of PilN of T. thermophilus and P. aeruginosa is conserved in PilN of M. xanthus (see Fig. S5A in the supplemental material). Similarly, the amino acid residues in PilM of T. thermophilus involved in binding the PilN peptide are conserved in PilM of M. xanthus (see Fig. S5B), suggesting that PilM in M. xanthus also interacts with the N terminus of PilN. To test in vivo whether the interaction between the cytoplasmic N terminus of PilN is important for recruiting PilM to the T4PS in M. xanthus, three single-amino-acid substitutions (D203A, V204D, and R388A [see Fig. S5B]) of residues involved in PilN peptide binding by PilM of T. thermophilus (43) were separately introduced into a functional YFP-PilM fusion that accumulates at native levels (54). All three mutant variants accumulated similarly to the WT YFP-PilM protein (see Fig. S2I in the supplemental material). As opposed to YFP-PilM, none of the three mutant proteins were able to restore motility in the ΔpilM strain (Fig. 4C). Furthermore, all three mutant variants displayed diffuse localization as opposed to the bipolar localization of the YFP-PilM WT protein (Fig. 4C). On the basis of these data, we suggest that the interaction between PilM and the cytoplasmic N terminus of PilN results in the recruitment of PilM to the T4PS complex.
PilN and PilO as well as PilP and PilQ interact directly.
Our data suggest that T4PS assembly initiates with formation of the PilQ oligomer in the OM and then proceeds inwards over the periplasm and the IM to the cytoplasm. To corroborate this assembly pathway, we tested for direct protein-protein interactions among T4PS proteins with a special focus on PilN, PilO, PilP, and PilQ. Because all four proteins are membrane proteins, only the periplasmic parts were purified as His6-tagged proteins corresponding to PilNΔ42-His6, PilOΔ37-His6, PilPΔ20-His6, andPilQ20–656-His6 (see Fig. S6A in the supplemental material). In size exclusion chromatography, PilNΔ42-His6 and PilPΔ20-His6 eluted in two peaks with masses corresponding to dimers and tetramers. PilOΔ37-His6 eluted in a single peak corresponding by mass to tetramers, while PilQ20–656-His6 eluted with a mass corresponding to dimers (see Fig. S6B, C, D, and E in the supplemental material). In comparison, the periplasmic domain of PilO of P. aeruginosa forms a dimer in solution while the periplasmic domain of PilN of P. aeruginosa is insoluble (36). To our knowledge, multimeric forms of PilP and PilQ of T4PS have not been reported previously. The periplasmic domain of the XcpD secretin of the P. aeruginosa T2SS was reported to form a dimer (71).
To test for direct protein interactions, we purified N-terminally MalE-tagged PilN (MalE-PilNΔ42), PilO (MalE-PilOΔ37), and PilP (MalE-PilPΔ20) (see Fig. S6A in the supplemental material). As shown in Fig. 5A, MalE-PilOΔ37 bound to an amylose matrix interacted with PilNΔ42-His6. Similarly, PilNΔ42-His6 bound to an Ni2+-NTA-agarose matrix interacted with MalE-PilOΔ37. In control experiments, PilNΔ42-His6 and MalE-PilOΔ37 did not separately bind to the amylose matrix and the Ni2+-NTA-agarose matrix, respectively. Using a similar experimental approach, we observed that MalE-PilPΔ20 and PilQ20–656-His6 interacted (Fig. 5B). Because purified PilQ20–656-His6 contains an additional protein with a size similar to that of MalE-PilPΔ20 (see Fig. S6A in the supplemental material), we performed immunoblot analyses using PilP-specific antibodies on the PilQ20–656-His6/MalE-PilPΔ20 elutions. These analyses confirmed that MalE-PilPΔ20 coelutes with PilQ20–656-His6 (see Fig. S6F).
FIG 5.
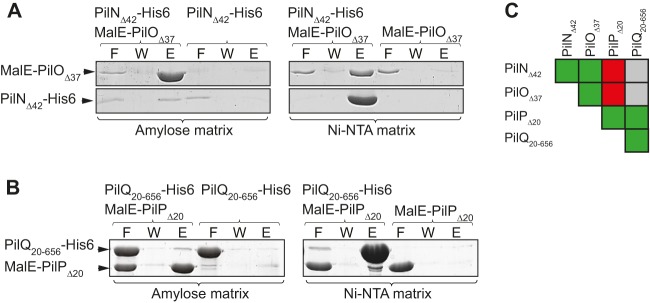
PilP and PilQ as well as PilN and PilO interact directly. (A) PilN and PilO interact directly. The indicated proteins were mixed and applied to amylose and Ni-NTA matrices as indicated. Shown are relevant sections of Coomassie blue G-250-stained polyacrylamide gels of the flowthrough (F), wash (W), and elution (E) fractions with PilNΔ42-His6 (23 kDa) and MalE-PilOΔ37 (62 kDa). (B) PilP and PilQ interact directly. The experiments were carried out and are presented as for panel A, except that gel sections with MalE-PilPΔ20 (60 kDa) and PilQ20–656-His6 (69 kDa) are shown. (C) Summary of protein-protein interactions analyzed. Green, combinations of one or two proteins in which interactions were observed; red, combinations of two proteins in which no interactions were observed; gray, combinations were not analyzed.
We did not detect interactions between MalE-PilNΔ42 and PilP-His6Δ20 or PilNΔ42-His6 and MalE-PilPΔ20. Similarly, we did not detect interactions between MalE-PilOΔ37 and PilPΔ20-His6 or PilOΔ37-His6 and MalE-PilPΔ20 (see Fig. S6G in the supplemental material). We conclude that the periplasmic parts of PilN and PilO, as well as the periplasmic parts of PilP and PilQ, interact directly. A summary of the protein-protein interactions observed is shown in Fig. 5C.
Timing of polar PilQ localization.
Because our data suggest that assembly of the T4PS complex begins with oligomeric PilQ in the OM, we investigated when PilQ becomes polarly localized using time-lapse fluorescence microscopy on M. xanthus cells expressing PilQ-sfGFP and extra Tgl and with images captured every 20 min. As shown in Fig. 6 and in Fig. S7 in the supplemental material, PilQ-sfGFP was present at the old cell poles and accumulated at the division site in cells with a deep constriction or immediately after cell division. We conclude that oligomeric PilQ-sfGFP becomes polarly localized late during or immediately after cell division.
FIG 6.
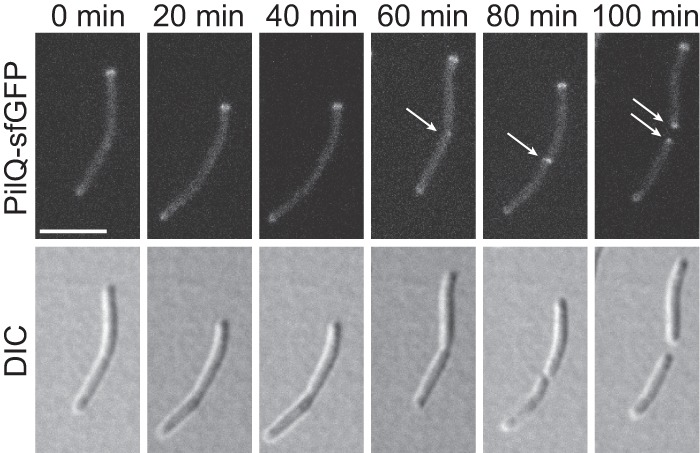
PilQ localizes polarly late during or immediately after cell division. M. xanthus cells from an exponentially growing culture were applied directly on a coverslip with a 0.1% CTT agarose pad and imaged by fluorescence and DIC microscopy at 20-min intervals. Arrows indicate newly formed polar clusters. Strain used, SA6060 (ΔaglZ/PpilA-tgl pilQ-sfGFP). The ΔaglZ mutation blocks gliding motility. Scale bar, 5 μm.
DISCUSSION
The dynamic cycles of T4P extension and retraction depend on a molecular machine that includes at least 10 proteins and that spans from the cytoplasm, over the IM and the periplasm, and to the OM. Here, we report that this nanomachine is assembled in an outside-in manner and that multimeric PilQ in the OM functions as an assembly platform for this machine and with additional components being added sequentially from the outside in. Additionally, the two ATPases PilB and PilT, which provide the energy for T4P extension and retraction, respectively, localize independently of all other T4PS proteins.
To gain insights into functional interactions between T4PS proteins and the assembly process, we initially profiled in vivo the stability of the 10 T4PS proteins in the absence of each individual other T4PS protein and we tested in vitro for direct interactions between PilN, PilO, PilP, and PilQ in various binary combinations. The OM pilotin lipoprotein Tgl, the secretin PilQ, the integral IM protein PilC, the cytoplasmic ATPases PilB and PilT, and the pilin subunit PilA stably accumulate independently of all other T4PS proteins. Moreover, Tgl is the only analyzed protein required for PilQ oligomer formation in the OM. On the other hand, the stability of PilM, PilN, PilO, and PilP depend on other T4PS proteins. Specifically, Tgl and PilQ are required for the stability of all of these four proteins. PilP, in turn, is required for the stability of PilM, PilN, and PilO. Finally, PilN and PilO mutually stabilize each other as well as PilM. The observation that Tgl and PilQ in the OM affect the stability of PilM in the cytoplasm demonstrates that this experimental approach does not distinguish between direct and indirect effects. Lack of Tgl or PilQ has similar effects on the stability of other T4PS proteins. Because Tgl functions as a pilotin for PilQ, we suggest that the effects of lack of Tgl are indirect and caused by the lack of the PilQ secretin oligomer in the OM. In total, the protein stability profiling experiments suggest the existence of a complex consisting of PilQ, PilP, PilN, PilO, and PilM. Consistently, we observed that the periplasmic domain of PilQ interacts with the periplasmic domain of PilP and that the periplasmic domains of PilN and PilO interact. These observations are in agreement with observations from N. meningitidis and P. aeruginosa in assays using purified proteins, i.e., the periplasmic domains of PilN and PilO interact directly to form heterodimers in P. aeruginosa (36), and the periplasmic domains of PilQ and PilP interact directly in both organisms (40–42). We did not observe an interaction between PilP and PilN or PilO. In P. aeruginosa, the periplasmic domain of PilP was found to interact with a PilN/PilO complex (38). Of note, Li et al. (37) recently provided evidence using yeast two-hybrid system analyses that PilO and PilP of M. xanthus interact directly. We speculate that PilP in M. xanthus may also interact with the PilN/PilO complex and that we did not observe this interaction because we tested only for binary interactions.
Our observation that PilM accumulation depends on Tgl, PilQ, PilP, PilN, and PilO is in agreement with the observation that PilM interacts with the cytoplasmic N terminus of PilN (35, 40, 43). In total, these data are consistent with a model in which PilP connects multimeric PilQ in the OM and PilN/PilO in the IM and that PilN interacts with PilM in the cytoplasm, as also suggested for the T4PS in P. aeruginosa and N. meningitidis (35, 36, 40, 44).
To elucidate the T4PS assembly pathway, we determined the localization of all T4PS proteins using immunofluorescence microscopy and/or fluorescent fusion proteins. Based on previous data and the data reported here, oligomeric PilQ, PilP, PilN, PilO, PilM, and PilC localize to both cell poles. On the other hand, PilA localizes dispersedly in the IM (56) and our data suggest that Tgl localizes dispersedly in the OM. Finally, PilB and PilT localize mostly unipolarly (54). Because the dependency for polar localization of T4PS proteins largely (but with some informative exceptions) parallels their stability dependency as well as direct interactions, and because M. xanthus assembles T4P at one pole at a time (51) and the pole at which T4P are assembled can switch within 15 to 30 s, we suggest that the protein clusters observed at both poles correspond to preassembled T4PS complexes consisting of oligomeric PilQ, PilP, PilN, PilO, PilM, and PilC and that the mostly unipolar localization of PilB and PilT to opposite cell poles determines at which pole T4P are assembled.
We observed that oligomeric PilQ in the OM is polarly localized whereas monomeric PilQ is not. Insertion and oligomer formation by PilQ in the OM depends on Tgl (33). Because Tgl is likely localized throughout the OM, we suggest that Tgl does not recruit PilQ to the cell poles but functions in the formation of PilQ oligomers, which then localize polarly. In this scenario, the PilQ secretin complex localizes polarly independently of all other T4PS proteins. PilP and PilO localization is mutually dependent and also depends on Tgl, PilQ, and PilN. These observations together with the protein stability profiling experiments and direct protein-protein interaction analyses suggest that PilP, PilN, and PilO form a subcomplex that is incorporated into the T4P machinery by interaction with oligomeric PilQ. Interestingly, PilP interacts directly with PilQ; however, this interaction is not sufficient to incorporate PilP into the T4P machinery. PilP incorporation also depends on PilN and PilO. We speculate that the PilQ-PilP interaction is transient or of low affinity, precluding stable complex formation. In the presence of the integral IM proteins PilN and PilO and with the formation of the PilP/PilN/PilO subcomplex, PilP dynamics could be restrained in this way, allowing stable complex formation with PilQ and incorporation into the T4P machinery. Our data also suggest that PilM incorporation into the T4P machinery depends on a direct interaction with the cytoplasmic N terminus of PilN. Finally, despite PilC accumulating independently of all other T4PS proteins, PilC incorporation into the T4P machinery depends on Tgl, PilQ, PilP, PilO, and PilN. In N. meningitidis, PilG (the PilC homolog in N. meningitidis) was suggested to interact directly with PilO based on bacterial two-hybrid analyses (35). Therefore, we hypothesize that PilO is the direct interaction partner of PilC and that all other effects on PilC localization are indirect by affecting the stability of PilO.
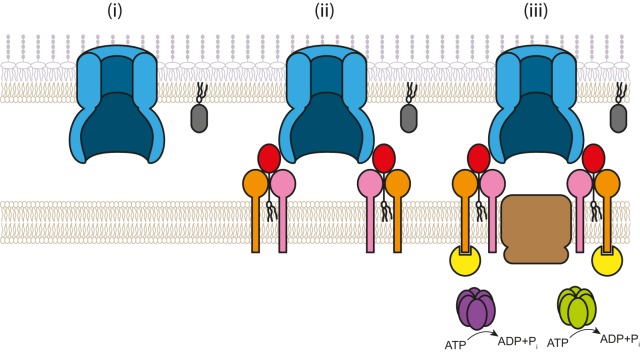
Combining the observations from the stability and localization profiling experiments with observations on direct interactions, we suggest that the assembly pathway for the T4P machinery follows an outside-in route (Fig. 7). Assembly is initiated by formation of the oligomeric PilQ secretin ring in the OM (Fig. 7i). Oligomeric PilQ, by directly interacting with PilP, recruits a subcomplex consisting of the IM lipoprotein PilP and the integral IM proteins PilN and PilO; in this subcomplex, PilN and PilO interact directly and PilP interacts with PilN, PilO, or both (Fig. 7ii). PilN by direct interactions recruits PilM in the cytoplasm; and PilO, likely by direct interactions, recruits the integral IM protein PilC (Fig. 7iii). An outside-in assembly pathway in which components are added sequentially has at least two advantages: (i) the individual parts of the machinery are automatically connected and aligned; and (ii) because T4P extension depends on PilC and PilM, T4P assembly is not initiated until the T4P machinery assembly process has been completed, thus avoiding the formation of T4P that would be unable to pass the OM.
FIG 7.
Model of assembly of the M. xanthus T4P machinery. See the text for details. The color code is as described for Fig. 1A.
In addition to being part of the T4P machinery, secretins also make up an OM ring structure in T2SS and type III secretion systems (T3SS) serving as OM conduits for the pseudopilus and injection needle, respectively (72). Interestingly, the assembly of the T3SS in Yersinia enterocolitica also initiates with the formation of the OM secretin ring and proceeds with the sequential addition of components in an outside-in direction (73). Similarly, in V. cholerae, incorporation of EpsC, the structural homolog of PilP, and EpsM, the homolog of PilO, into the T2SS depends on the secretin homolog EpsD, suggesting that EpsD is crucial for assembly of the T2SS (68). Thus, an outside-in assembly pathway is emerging as a conserved feature in secretin-containing trans-envelope machines. Also, for the T2SS and T3SS, an outside-in assembly pathway would preclude the formation of the pseudopilus and needle until the trans-envelope machine has assembled. An outside-in assembly pathway is in stark contrast to the flagellum assembly pathway in bacteria, which proceeds in an inside-out direction (74). A notable difference between flagella and T4PS, T2SS, and T3SS is that flagellar gene expression is coupled to flagellar assembly (74). We speculate that in the case of the T4PS, T2SS, and T3SS, synthesis of T4P, pseudopili, and needle structures, respectively, is delayed until the relevant nanomachines have been assembled whereas in the case of the flagellar system, premature flagellum synthesis does not pose a significant problem because the relevant genes are not transcribed until the hook-basal body structure has been completed.
The PilB and PilT ATPases, which are the only two members of the T4P machinery that primarily localize unipolarly, localize and accumulate independently of all tested T4PS proteins. Clearly, PilB and PilT must interact with one or more of the T4PS proteins to fulfill their function. Consistently, evidence supporting the idea that PilB interacts with an N-terminal cytoplasmic domain of PilC and that PilT interacts with a more C-terminally located cytoplasmic domain of PilC in P. aeruginosa has been provided (48). Similarly, a bacterial two-hybrid analysis of N. meningitidis T4PS proteins suggests an interaction between PilT and PilG (the PilC homolog in N. meningitidis) (35). In agreement, in the T2SS of V. cholerae, ATPase activity of EpsE (the PilB homolog) is stimulated by the cytoplasmic domain of EpsL (the PilM and PilN homolog) (75), and the proteins GspE (the PilB homolog), GspF (the PilC homolog), GspL (the PilM and PilN homolog), and GspM (the PilO homolog) of the T2SS of Erwinia chrysanthemi interact to form a complex (76). These observations suggest that PilB and PilT functionally interact with other T4PS proteins to stimulate T4P extension and retraction. However, our data suggest that this interaction(s) is neither required (because both proteins localize polarly in the absence of other T4PS proteins) nor sufficient (see below) for their polar localization.
Recently, it was shown that the small Ras-like GTPase SofG and the bactofilin BacP are required for the polar localization of PilB and PilT even in the presence of all other T4PS proteins (77). SofG and filamentous BacP interact directly, and this interaction brings about the localization of PilB and PilT at the same pole. Subsequently, the small Ras-like GTPase MglA and its cognate GTPase-activating protein (GAP) MglB sorts PilB to the leading cell pole and PilT to the lagging cell pole (78–80). Given that the T4PS proteins analyzed here, with the exception of Tgl and PilA, localize to both poles, it is not surprising that none of them are required for the mostly unipolar PilB and PilT localization. We would predict that if PilB and PilT were recruited by one or more T4PS proteins, then they would be bipolarly localized. Also, this localization would be difficult to reconcile with the pole switching of PilB and PilT during reversals.
We have provided evidence that the T4PS in M. xanthus assembles in an outside-in manner starting with the secretin ring in the OM, similarly to T2SS in V. cholerae (68) and T3SS in Y. enterocolitica (73). However, these T2SS and T3SS are not polarly localized whereas the T4PS in M. xanthus is. Using time-lapse microscopy, we observed polarly localized PilQ late during cell division or immediately after cell division, suggesting that PilQ becomes incorporated in the OM during the late stages of cell division. Future work will be directed at understanding how polar localization of oligomeric PilQ is accomplished in M. xanthus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris van der Does for many helpful discussions.
This work was supported by the German Research Council within the framework of the Collaborative Research Center 987 “Microbial Diversity in Environmental Signal Response” and the Max Planck Society.
We have no conflict of interest.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01094-13.
REFERENCES
- 1.Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837. 10.1111/j.1365-2958.2008.06197.x [DOI] [PubMed] [Google Scholar]
- 2.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenecity. Nat. Rev. Microbiol. 2:363–378. 10.1038/nrmicro885 [DOI] [PubMed] [Google Scholar]
- 3.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61–68. 10.1046/j.1365-2958.2003.03677.x [DOI] [PubMed] [Google Scholar]
- 4.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304. 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- 6.Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227–237. 10.1111/j.1365-2958.2006.05365.x [DOI] [PubMed] [Google Scholar]
- 7.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249. 10.1038/nrmicro844 [DOI] [PubMed] [Google Scholar]
- 8.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. 10.1038/35024105 [DOI] [PubMed] [Google Scholar]
- 9.Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901–6904. 10.1073/pnas.121171698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron C. 2010. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13:100–105. 10.1016/j.mib.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 11.Craig L, Li J. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267–277. 10.1016/j.sbi.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U. S. A. 99:16012–16017. 10.1073/pnas.242523299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. 2009. High force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191:4633–4638. 10.1128/JB.00396-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strom MS, Lory S. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565–596. 10.1146/annurev.mi.47.100193.003025 [DOI] [PubMed] [Google Scholar]
- 15.Peabody CR, Chung YJ, Yen M-R, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072. 10.1099/mic.0.26364-0 [DOI] [PubMed] [Google Scholar]
- 16.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10:336–351. 10.1038/nrmicro2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U. S. A. 98:2503–2508. 10.1073/pnas.051436598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs M, Mattick JS. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233–243. 10.1111/j.1365-2958.1993.tb01949.x [DOI] [PubMed] [Google Scholar]
- 19.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221–2228. 10.1093/emboj/19.10.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchik VE, Robert-Baudouy J, Condemine G. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007–3016. 10.1093/emboj/16.11.3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189:1783–1793. 10.1128/JB.01230-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189:2069–2076. 10.1128/JB.01236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunn DN, Lory S. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U. S. A. 88:3281–3285. 10.1073/pnas.88.8.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom MS, Lory S. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656–1664 [PubMed] [Google Scholar]
- 25.Morand PC, Bille E, Morelle S, Eugene E, Beretti JL, Wolfgang M, Meyer TF, Koomey M, Nassif X. 2004. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 23:2009–2017. 10.1038/sj.emboj.7600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins RF, Davidsen L, Derrick JP, Ford RC, Tonjum T. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825–3832. 10.1128/JB.183.13.3825-3832.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins RF, Frye SA, Kitmitto A, Ford RC, Tonjum T, Derrick JP. 2004. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J. Biol. Chem. 279:39750–39756. 10.1074/jbc.M405971200 [DOI] [PubMed] [Google Scholar]
- 28.Jain S, Moscicka KB, Bos MP, Pachulec E, Stuart MC, Keegstra W, Boekema EJ, van der Does C. 2011. Structural characterization of outer membrane components of the type IV pili system in pathogenic Neisseria. PLoS One 6:e16624. 10.1371/journal.pone.0016624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkhardt J, Vonck J, Averhoff B. 2011. Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286:9977–9984. 10.1074/jbc.M110.212688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfgang M, van Putten JPM, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408–6418. 10.1093/emboj/19.23.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardie KR, Lory S, Pugsley AP. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978–988 [PMC free article] [PubMed] [Google Scholar]
- 32.Koo J, Tammam S, Ku SY, Sampaleanu LM, Burrows LL, Howell PL. 2008. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190:6961–6969. 10.1128/JB.00996-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nudleman E, Wall D, Kaiser D. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60:16–29. 10.1111/j.1365-2958.2006.05095.x [DOI] [PubMed] [Google Scholar]
- 34.Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64. 10.1111/j.1365-2958.2004.04364.x [DOI] [PubMed] [Google Scholar]
- 35.Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V. 2012. Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol. Microbiol. 84:857–873. 10.1111/j.1365-2958.2012.08062.x [DOI] [PubMed] [Google Scholar]
- 36.Sampaleanu LM, Bonanno JB, Ayers M, Koo J, Tammam S, Burley SK, Almo SC, Burrows LL, Howell PL. 2009. Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J. Mol. Biol. 394:143–159. 10.1016/j.jmb.2009.09.037 [DOI] [PubMed] [Google Scholar]
- 37.Li C, Wallace RA, Black WP, Li YZ, Yang Z. 2013. Type IV pilus proteins form an integrated structure extending from the cytoplasm to the outer membrane. PLoS One 8:e70144. 10.1371/journal.pone.0070144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tammam S, Sampaleanu LM, Koo J, Sundaram P, Ayers M, Andrew Chong P, Forman-Kay JD, Burrows LL, Howell PL. 2011. Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Mol. Microbiol. 82:1496–1514. 10.1111/j.1365-2958.2011.07903.x [DOI] [PubMed] [Google Scholar]
- 39.Drake SL, Sandstedt SA, Koomey M. 1997. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23:657–668. 10.1046/j.1365-2958.1997.2511618.x [DOI] [PubMed] [Google Scholar]
- 40.Tammam S, Sampaleanu LM, Koo J, Manoharan K, Daubaras M, Burrows LL, Howell PL. 2013. PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol. 195:2126–2135. 10.1128/JB.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balasingham SV, Collins RF, Assalkhou R, Homberset H, Frye SA, Derrick JP, Tonjum T. 2007. Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J. Bacteriol. 189:5716–5727. 10.1128/JB.00060-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berry J-L, Phelan MM, Collins RF, Adomavicius T, Tønjum T, Frye SA, Bird L, Owens R, Ford RC, Lian L-Y, Derrick JP. 2012. Structure and assembly of a trans-periplasmic channel for type IV pili in Neisseria meningitidis. PLoS Pathog. 8:e1002923. 10.1371/journal.ppat.1002923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karuppiah V, Derrick JP. 2011. Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J. Biol. Chem. 286:24434–24442. 10.1074/jbc.M111.243535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394:128–142. 10.1016/j.jmb.2009.09.034 [DOI] [PubMed] [Google Scholar]
- 45.Crowther LJ, Yamagata A, Craig L, Tainer JA, Donnenberg MS. 2005. The ATPase activity of BfpD is greatly enhanced by zinc and allosteric interactions with other Bfp proteins. J. Biol. Chem. 280:24839–24848. 10.1074/jbc.M500253200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Sakai D, Horiuchi T, Komano T. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 276:17968–17975. 10.1074/jbc.M010652200 [DOI] [PubMed] [Google Scholar]
- 47.Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pili function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421. 10.1128/JB.01793-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. 2013. The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288:9721–9728. 10.1074/jbc.M113.453506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10. 10.1046/j.1365-2958.1999.01339.x [DOI] [PubMed] [Google Scholar]
- 50.Wu SS, Kaiser D. 1995. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547–558. 10.1111/j.1365-2958.1995.mmi_18030547.x [DOI] [PubMed] [Google Scholar]
- 51.Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952–5956. 10.1073/pnas.76.11.5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackhart BD, Zusman DR. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. U. S. A. 82:8767–8770. 10.1073/pnas.82.24.8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H, Zusman DR, Shi W. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143–1146. 10.1016/S0960-9822(00)00705-3 [DOI] [PubMed] [Google Scholar]
- 54.Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Hone A, Maier B, Hoppert M, Søgaard-Andersen L. 2009. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol. 74:691–706. 10.1111/j.1365-2958.2009.06891.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mignot T, Merlie JP, Jr, Zusman DR. 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310:855–857. 10.1126/science.1119052 [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Lux R, Hu W, Hu C, Shi W. 2010. PilA localization affects extracellular polysaccharide production and fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 76:1500–1513. 10.1111/j.1365-2958.2010.07180.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodgkin J, Kaiser D. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938–2942. 10.1073/pnas.74.7.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi X, Wegener-Feldbrügge S, Huntley S, Hamann N, Hedderich R, Søgaard-Andersen L. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J. Bacteriol. 190:613–624. 10.1128/JB.01502-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60.Li Y, Lux R, Pelling AE, Gimzewski JK, Shi W. 2005. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology 151:353–360. 10.1099/mic.0.27614-0 [DOI] [PubMed] [Google Scholar]
- 61.Leonardy S, Freymark G, Hebener S, Ellehauge E, Søgaard-Andersen L. 2007. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 26:4433–4444. 10.1038/sj.emboj.7601877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Overgaard M, Wegener-Feldbrügge S, Søgaard-Andersen L. 2006. The orphan response regulator DigR is required for synthesis of extracellular matrix fibrils in Myxococcus xanthus. J. Bacteriol. 188:4384–4394. 10.1128/JB.00189-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Soto JP, Kaiser D. 1997. Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J. Bacteriol. 179:4372–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi W, Zusman DR. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. U. S. A. 90:3378–3382. 10.1073/pnas.90.8.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konovalova A, Petters T, Søgaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 34:89–106. 10.1111/j.1574-6976.2009.00194.x [DOI] [PubMed] [Google Scholar]
- 66.Dinh T, Bernhardt TG. 2011. Using Superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 193:4984–4987. 10.1128/JB.00315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aronson DE, Costantini LM, Snapp EL. 2011. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12:543–548. 10.1111/j.1600-0854.2011.01168.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. 2009. Docking and assembly of the type II secretion complex of Vibrio cholerae. J. Bacteriol. 191:3149–3161. 10.1128/JB.01701-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buddelmeijer N, Francetic O, Pugsley AP. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secreton components PulL and PulM. J. Bacteriol. 188:2928–2935. 10.1128/JB.188.8.2928-2935.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buddelmeijer N, Krehenbrink M, Pecorari F, Pugsley AP. 2009. Type II secretion system secretin PulD localizes in clusters in the Escherichia coli outer membrane. J. Bacteriol. 191:161–168. 10.1128/JB.01138-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Meeren R, Wen Y, Van Gelder P, Tommassen J, Devreese B, Savvides SN. 2013. New insights into the assembly of bacterial secretins: structural studies of the periplasmic domain of XcpQ from Pseudomonas aeruginosa. J. Biol. Chem. 288:1214–1225. 10.1074/jbc.M112.432096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korotkov KV, Gonen T, Hol WGJ. 2011. Secretins: dynamic channels for protein transport across membranes. TiBS 36:433–443. 10.1016/j.tibs.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29:1928–1940. 10.1038/emboj.2010.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465. 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WGJ, Sandkvist M. 2007. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 26:19–27. 10.1038/sj.emboj.7601481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Py B, Loiseau L, Barras F. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2:244–248. 10.1093/embo-reports/kve042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bulyha I, Lindow S, Lin L, Bolte K, Wuichet K, Kahnt J, van der Does C, Thanbichler M, Søgaard-Andersen L. 2013. Two small GTPases act in concert with the bactofilin cytoskeleton to regulate dynamic bacterial cell polarity. Dev. Cell 25:119–131. 10.1016/j.devcel.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 78.Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Søgaard-Andersen L. 2010. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 29:2276–2289. 10.1038/emboj.2010.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Franco M, Ducret A, Mignot T. 2010. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 8:e1000430. 10.1371/journal.pbio.1000430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, Leonardy S, Søgaard-Andersen L, Wittinghofer A. 2011. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J. 30:4185–4197. 10.1038/emboj.2011.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wall D, Kolenbrander PE, Kaiser D. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu SS, Wu J, Kaiser D. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109–121. 10.1046/j.1365-2958.1997.1791550.x [DOI] [PubMed] [Google Scholar]
- 83.Wu SS, Kaiser D. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.