Abstract
First identified in Drosophila, the Crumbs (Crb) proteins are important in epithelial polarity, apical membrane formation, and tight junction (TJ) assembly. The conserved Crb intracellular region includes a FERM (band 4.1/ezrin/radixin/moesin) binding domain (FBD) whose mammalian binding partners are not well understood and a PDZ binding motif that interacts with mammalian Pals1 (protein associated with lin seven) (also known as MPP5). Pals1 binds Patj (Pals1-associated tight-junction protein), a multi-PDZ-domain protein that associates with many tight junction proteins. The Crb complex also binds the conserved Par3/Par6/atypical protein kinase C (aPKC) polarity cassette that restricts migration of basolateral proteins through phosphorylation. Here, we describe a Crb3 knockout mouse that demonstrates extensive defects in epithelial morphogenesis. The mice die shortly after birth, with cystic kidneys and proteinaceous debris throughout the lungs. The intestines display villus fusion, apical membrane blebs, and disrupted microvilli. These intestinal defects phenocopy those of Ezrin knockout mice, and we demonstrate an interaction between Crumbs3 and ezrin. Taken together, our data indicate that Crumbs3 is crucial for epithelial morphogenesis and plays a role in linking the apical membrane to the underlying ezrin-containing cytoskeleton.
INTRODUCTION
Studies over the last 20 years have identified proteins that are essential for the formation and maintenance of epithelial polarity (1–4). One of the key proteins in this process is the evolutionarily conserved Crumbs (Crb) family of proteins (2, 4). Crb was first identified in Drosophila, where it has been shown to be important in epithelial polarity, apical membrane formation, and tissue morphogenesis (5). Crb family members have a conserved intracellular domain that contains a FERM (band 4.1/ezrin/radixin/moesin) binding domain (FBD) and PDZ binding domain. Targets for the PDZ domain have been well described. One mammalian partner is protein associated with lin seven (Pals1, also known as MPP5), which is an ortholog of Drosophila Stardust. Pals1 is a scaffold protein that interacts with the multi-PDZ-domain protein Pals1-associated tight junction (TJ) protein (Patj). Patj can in turn bind to a large number of tight junction proteins, including claudin-1, zonula occludens (ZO), and angiomotin proteins (2). In addition, the Crb complex can bind to the evolutionarily conserved complex consisting of Par3, Par6, and atypical protein kinase C (aPKC) (2). In turn, this interaction can localize aPKC to the apical surface, where it phosphorylates lateral targets and confines them to the basolateral membrane (6, 7). Thus, the Crb complex is felt to play an important role in both apical identity and tight junction formation in mammalian cells.
FERM domain-containing-proteins that bind Crb have been identified in Drosophila, but mammalian binding partners are less well understood (4). One group of binding partners includes the ERM family of proteins consisting of ezrin, radixin, and moesin. In Drosophila, Crb has been shown to interact with DMoesin and beta heavy spectrin connecting the apical membrane with the underlying cytoskeleton (8). Another well-described Crb interactor is the FERM domain-containing protein called Yurt in Drosophila (9, 10) or Mosaic Eyes (Moe) in zebrafish (11). This protein localizes to the lateral membrane and negatively regulates the Crb protein. The mammalian homologs of Yurt known as YMO1, EHM2, and EPB41L have been proposed to have similar functions. The FERM binding domain of Crb also intersects the Hippo tumor suppressor pathway in Drosophila (12, 13). The Crb proteins can interact via the band 4.1 proteins Merlin and Expanded to activate the Hippo pathway and negatively regulate the Yap transcription factor. In this manner, apically localized Crb can control cell proliferation.
There are three mammalian paralogs of Crb. All of these Crb proteins have conserved intracellular domains with FBD and PDZ binding motifs. Mutations in Crb1 lead to retinitis pigmentosa (14). Crb2 has relatively limited expression in adult tissues, including brain, eye, and kidney glomerulus, but Crb2 knockout mice die at gastrulation (15). Crb3 is the most widely expressed isoform in epithelial tissues, and it is markedly upregulated in the mesenchyme-to-epithelium transition (16) and downregulated in the epithelium-to-mesenchyme transition (17, 18). Knockdown or overexpression of Crb3 in MDCK cells leads to disorganized epithelial structures (19, 20). In this article, we describe a Crumbs3 knockout mouse which dies shortly after birth with defects in epithelial tissue morphogenesis.
MATERIALS AND METHODS
Construction of the targeting vector.
Crb3 genomic regions were PCR amplified from Mus musculus (Linnaeus) 129X1/SvJ DNA (DNA resource no. 00691; Jackson Laboratories, Bar Harbor, ME). The 5′ arm of homology comprised an ∼3.5-kb fragment encoding exon 1 and exon 2 and was subcloned into the SwaI site of pLoxPFlpNeo (a gift from James Shayman [21], University of Michigan). An ∼450-bp endogenous region of Crb3 encompassing targeted exon 3 was manipulated to add monomeric green fluorescent protein (mGFP) in-frame to the coding region (similar to the GFP-Crb3 cDNA described previously [20]), yielding an ∼1.2-kb fragment that was subcloned into the BamHI site of the vector backbone. Finally, the 3′ arm of homology contained an ∼3.4-kb region encoding exon 4 or exon 4/5 (Crb3a versus Crb3b splice variants, respectively) and was subcloned into the XhoI site of the vector. The construct was verified by DNA sequencing. Two FLP recombination target (FRT) sites flank the “Neo” cassette, and two LoxP sites flank targeted exon 3.
Generation of Crumbs3 mice.
The targeting vector was linearized and electroporated into R1 embryonic stem (ES) cells (a gift from Andras Nagy, Reka Nagy, and Wanda Abramow-Newerly [22], Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada). Clones resistant to G418 were analyzed for homologous recombination by Southern blotting using 5′ and 3′ probes containing genomic sequences outside the targeting vector. Targeted clones 11G, 12A, and 12C were injected into blastocysts from the breeding of C57BL/6NCrl female mice with (C57BL/6J × DBA/2J) F1 male mice and transferred to surrogate mothers (Transgenic Animal Model Core, University of Michigan).
Male chimeras were mated with Pgk1-FlpO females [catalog no. 11065; B6(C3)-Tg(Pgk1-FLPo)10Sykr/J; Jackson Laboratories] for transmission of the Neo allele through the germ line and for removal of the neomycin cassette, producing the “floxed” allele. Deletion of exon 3 and creation of the null allele were performed by crossing Crb3+/flox males with germ line Cre-deleter females, EIIa-Cre [catalog no. 3724; B6.FVB-Tg(EIIa-cre)C5379Lmgd/J; Jackson Laboratories], to produce Crb3+/− animals.
Heterozygous Crb3+/− mice arising from two independent ES cell clones (11G and 12A) were then backcrossed onto C57BL/6J (catalog no. 664; Jackson Laboratories) for up to six more generations (backcrossing is still ongoing in the laboratory) and intercrossed in timed matings to obtain Crb3+/+, Crb3+/−, and Crb3−/− animals. Littermates were used as controls.
Preterm pups from timed matings were dissected at embryonic day 12.5 (E12.5) to E18.5. Pups intentionally delivered alive at E18.5 were placed on moist paper towels on a 37°C warmer and stimulated to breathe with gentle prodding. The lifetime of each of the null pups was calculated as the difference between the time of delivery and its last movement, as judged by video recording of the animals for ∼3 h following delivery.
All experiments involving mice were approved by the University Committee on Use and Care of Animals at the University of Michigan and were in compliance with NIH guidelines outlined in the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Genotyping.
Tail biopsy specimens were digested in DirectPCR lysis reagent (Viagen, Los Angeles, CA) and subjected to PCR genotyping using Platinum Taq (Invitrogen, Carlsbad, CA).
The primer sequences were designed using Perlprimer (23) as follows: P1, 5′-CTGGTGAGTTGGGAAGGTGTAA-3′; P2, 5′-GTTTCTCTGTGTAGCCCTGT-3′; P3, 5′-AGGATCTCCTGTCATCTCACCTTGCTCCTG-3′; P4, 5′-AAGAACTCGTCAAGAAGGCGATAGAAGGCG-3′; P5, 5′-ACCTTCCCTCAGTCTCTAGTTCC-3′; P6, 5′-AAACGCCTTCCAAAGTCTCC-3′; P7, 5′-CTTCCCTCAGTCTCTAGTTCC-3′; and P8, 5′-GTTTCTCTGTGTAGCCCTGTA-3′.
Tail biopsy specimens were also sent to Transnetyx, Inc. (Cordova, TN) for analysis.
Antibodies.
The primary antibodies used were acetylated tubulin (Sigma, St. Louis, MO; catalog no. T7451), aPKC (R&D, Minneapolis, MN; catalog no. AF4465), Aquaporin 1 (Aqp1) (Santa Cruz, Santa Cruz, CA; catalog no. SC-25287), Aqp2 (Santa Cruz; catalog no. SC-9882), Aqp5 (Abcam, Cambridge, MA; catalog no. AB78486), α-tubulin (Sigma; catalog no. T6199), β-actin (Sigma; catalog no. A2228), β-catenin (BD Bioscience, San Jose, CA; catalog no. 610154), CC10 (Santa Cruz; catalog no. SC-9772), chemokine C-C motif ligand 2 (CCL2) (R&D; catalog no. AF479-NA), claudin-4 (Santa Cruz; catalog no. SC-17664), cleaved caspase-3 (Cell Signaling, Danvers, MA; catalog no. 9661), Crumbs3a (Custom [24]), E-cadherin (R&D; catalog no. AF748), ERM (Cell Signaling; catalog no. 3142), ezrin (Sigma; catalog no. E8897), GFP-agarose (MBL, Woburn, MA; catalog no. D153-8), hemagglutinin (HA) (Santa Cruz; catalog no. SC-805), Hes1 (MBL; catalog no. D134-3), Ki67 (Abcam; catalog no. AB16667), mucin 1 (Muc1) (Novus Biologicals, Littleton, CO; catalog no. NB120-15481), Nkx2.1 (Epitomics, Burlingame, CA; catalog no. 2044-1), occludin (Invitrogen; catalog no. 71-1500), Pals1 for immunoblotting (IB) (Santa Cruz; catalog no. SC-365411), Pals1 for immunohistochemistry (IHC) (Custom [25]), Par3 (Novus Biologicals; catalog no. NBP1-88861), Patj (Abcam; catalog no. AB102113), phospho-ERM (Cell Signaling; catalog no. 3141), phospho-Yap1 (Cell Signaling; catalog no. 4911), surfactant protein B (SPB) (EMD Millipore, Billerica, MA; catalog no. 07-614), surfactant protein C (SPC) (Seven Hills Bioreagents, Cincinnati, OH; catalog no. WRAB-9337), Yap1 (Epitomics; catalog no. EP1674Y), and ZO1 (Invitrogen; catalog no. 33-9100).
Secondary antibodies conjugated to Alexa Fluor 488, 594, and 647 for immunostaining and optimized for multiple labeling were purchased from Jackson ImmunoResearch (West Grove, PA). Secondary antibodies conjugated to horseradish peroxidase (HRP) for immunoblotting were purchased from GE Healthcare (Livonia, MI).
Southern blotting.
Nonradioactive Southern blotting was performed using digoxigenin (DIG) reagents from Roche Applied Science (Basel, Switzerland) and closely followed the manufacturer's protocol and instructions in the DIG Application Manual for Filter Hybridization.
Preparation of tissue lysates.
Mouse organs were homogenized on ice using a PowerGen 125 handheld homogenizer with a 5- by 95-mm microtip (ThermoFisher, Waltham, MA) in a nondenaturing buffer containing 50 mM HEPES (pH 7.4), 1 mM EDTA, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 50 mM NaF, with protease and phosphatase inhibitors. Lysates were incubated on a shaker for 1 h at 4°C and then processed with a microcentrifuge at 20,000 × g for 40 min at 4°C. Supernatants were recovered for analysis.
Western blotting.
Immunoblotting was performed essentially as described previously (18). Samples typically contained 50 μg protein per lane.
Immunoprecipitation.
Tissue lysates were prepared, and 1 mg protein–1 ml total buffer volume was mixed with 10 μl GFP-agarose beads for 2 h overnight with rotation at 4°C. Immune complexes were washed three times with nondenaturing buffer (above) and once in Tris-EDTA (TE) buffer. Samples were mixed in 2× lithium dodecyl sulfate (LDS) buffer (Invitrogen), incubated for 1 h at 37°C, and loaded on a 10% Bis-Tris acrylamide gel for immunoblotting analysis.
Histology.
Whole-mount organs were dissected using a Leica MZFL III stereo dissecting microscope and images captured using an Olympus DP-70 camera.
Individual embryonic organs were fixed for 1 to 2 h in 4% paraformaldehyde (PFA)–phosphate-buffered saline (PBS) at 4°C. For frozen sections, tissue was transferred through a sucrose gradient to 30% sucrose, embedded in optimal cutting temperature (OCT) compound (Tissue-Tek, Torrance, CA), and then cryosectioned onto slides. For paraffin-embedded sections, tissue was dehydrated through an ethanol (EtOH) gradient to 70% EtOH, placed in cassettes, paraffin processed, paraffin embedded, and sectioned onto slides. Sections were typically 5 μm thick.
Paraffin sections were prepared for classic IHC staining by deparaffinizing in xylenes and rehydrating through an ethanol series to distilled water. Antigen retrieval was performed in antigen unmasking buffer (Citra buffer; catalog no. H-3300, Vector Laboratories, Burlingame, CA) (pH 6.0), and tissues were permeabilized for 10 min in 0.5% Triton X-100 (TX-100)–PBS. Staining was carried out using an ImmPress peroxidase polymer kit (mouse on mouse, rabbit, or goat; Vector Laboratories) and following the manufacturer's instructions. Endogenous peroxidase activity was blocked with Bloxall (Vector Laboratories), washes were performed with 0.05% Tween 20–PBS, ImmPact DAB detection reagent (Vector Laboratories) was used as the peroxidase substrate, tissues were counterstained with hematoxylin QS (Vector Laboratories), and slides were dehydrated again through an ethanol gradient to xylenes before mounting in Vectamount (Vector Laboratories).
Paraffin sections were prepared for fluorescent staining by deparaffinizing in xylenes and rehydrating through an ethanol series as described above before antigen retrieval. Frozen sections were prepared for fluorescent staining by warming to room temperature for 10 min and rehydrating in PBS before antigen retrieval. Antigen retrieval and permeabilization were performed as described above. Slides were then blocked for ≥15 min in 2% donkey serum with 0.05% Tween 20–PBS, incubated with primary antibodies for ≥2 h in blocking buffer, washed in 0.05% Tween 20–PBS, incubated with donkey fluorophore-conjugated secondary antibodies for 1 h in blocking buffer, washed in 0.05% Tween 20–PBS, stained with DAPI (4′,6-diamidino-2-phenylindole), and mounted with Prolong Gold (Invitrogen).
Hematoxylin and eosin (H&E) staining was carried out using standard procedures.
H&E and conventional IHC imaging was performed on an Olympus BX-51 upright microscope with an Olympus DP-70 high-resolution digital camera (Tokyo, Japan).
Fluorescent imaging was performed on a Nikon Eclipse TE2000-U inverted microscope (Nikon, Tokyo, Japan) with a Roper Scientific CoolSnap digital camera (Roper, Sarasota, FL) and MetaMorph software (Molecular Devices, Sunnyvale, CA) for epifluorescence or on an Olympus IX-71 inverted microscope attached to an Olympus Fluoview 500 confocal imaging system.
Electron microscopy.
For transmission electron microscopy (TEM) and scanning electron microscopy (SEM), embryonic organs were fixed overnight at 4°C in 2.5% glutaraldehyde–0.1 M Sorensen's phosphate buffer (pH 7.4), rinsed 3 times for 15 min each time in 0.1 M Sorensen's buffer, postfixed for 1 h at 4°C in 1% osmium tetroxide–0.1 M Sorensen's buffer, and rinsed 3 times for 15 min in 0.1 M Sorensen's buffer.
For TEM, samples were then dehydrated through an EtOH gradient, infiltrated with Epon resin, allowed to polymerize for 24 h at 60°C, cut into 70-nm-thick sections, placed on grids, and stained with uranyl acetate and lead citrate. Electron micrographs were collected on a Philips CM-100 transmission electron microscope (FEI, Hillsboro, OR).
For SEM, samples were then dehydrated through an EtOH series, cleared in hexamethyldisilazane 3 times for 15 min each time, air-dried, mounted on an SEM stub with sticky tape and colloidal graphite, dried overnight, and coated with gold in a Polaron E5100 sputter coater (Bio-Rad, West Sussex, United Kingdom). Electron micrographs were captured using an Amray 1910 field emission scanning electron microscope (KLA-Tencor/Amray Division, Bedford, MA).
Immunoelectron microscopy.
MDCK cells expressing inducible green fluorescent protein (iGFP)-Crb3 were grown on transwell filters as described previously (26). Labeling was performed using Crb3a antibody (20 μg/ml) followed by incubation with a mixture of 6 nm and 10 nm immunogold-conjugated goat anti-rabbit IgG (Aurion, Wageningen, Netherlands), adhering to the protocol as described previously (27). Images were collected on a Philips CM-100 transmission electron microscope.
GST-Crb3 pulldown assay.
The GST-Crb3 pulldown assay was performed as described previously (25). Briefly, glutathione S-transferase (GST) or GST-Crb3a (intracellular tail) was expressed in bacteria and purified. A human HA-ezrin FERM mammalian expression construct was transfected into 293 cells using Fugene (Roche Applied Science). Lysates from the 293 cells were prepared, incubated with purified GST or GST-Crb3a immobilized on glutathione beads, washed, resolved by polyacrylamide gel electrophoresis, and immunoblotted using HA antibodies.
PAS-diastase assay.
Reagents were purchased from PolyScientific (Bay Shore, NY). Periodic acid-Schiff (PAS)-diastase assay staining was performed using conventional protocols (28).
Image preparation and statistical analysis.
Image analysis was performed using ImageJ (29).
Figures were prepared in Adobe Photoshop and Adobe Illustrator (Adobe, San Jose, CA). Statistical analyses were performed in GraphPad Prism (GraphPad, La Jolla, CA), Microsoft Excel (Microsoft, Redmond, WA), and SPSS (IBM, Armonk, NY). Analysis of variance (ANOVA), Student's t test (independent samples, equal variance, two tailed), and a χ2 test for goodness of fit were evaluated, where a P value of less than 0.05 was considered statistically significant.
RESULTS
Creation of Crumbs3 targeted mice.
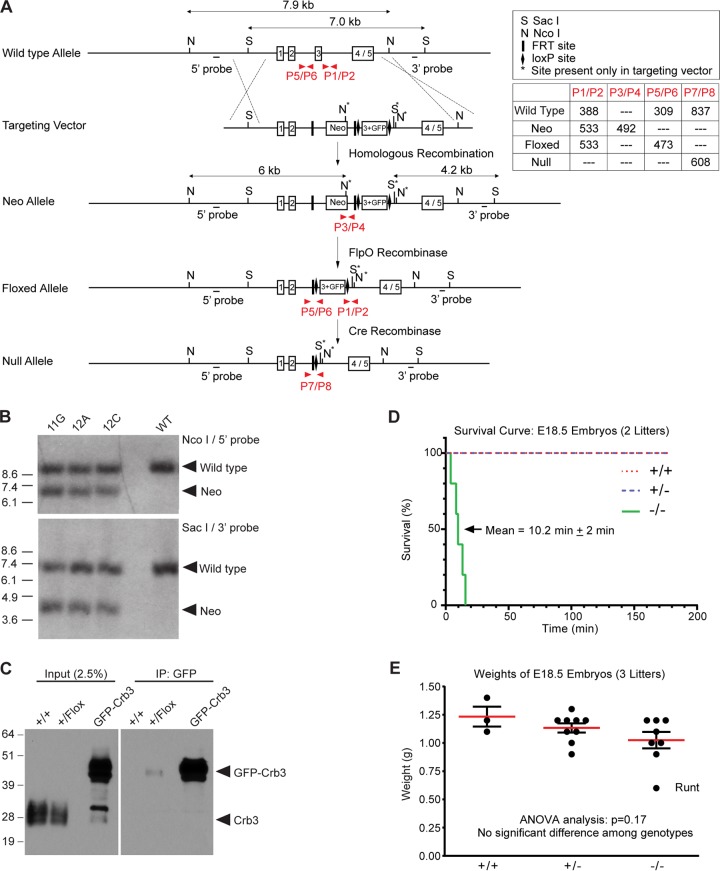
To establish the in vivo role of Crb3 in epithelial morphogenesis and recapitulate our studies in MDCK cells (20), we used gene-targeting techniques to target the Crb3 locus. The targeting construct contained an in-frame fusion of EGFP in exon 3 and a neomycin cassette in intron 2 surrounded by FRT sites. LoxP sites were placed on either side of this exon 3/GFP cassette to allow Crb3 gene deletion (Fig. 1A). We confirmed homologous recombination of the Neo allele in three embryonic stem (ES) cell clones by Southern blotting (Fig. 1B). The three ES cell clones were injected into blastocysts, yielding chimeras of each independent line, all of which successfully accomplished germ line transmission. We chose to carry two independent lines, 11G and 12A, forward and crossed them to a FlpO-deleter mouse to remove the Neo cassette, producing the Crb3 floxed allele that would express GFP-Crb3. We recovered Crb3+/+ and Crb3+/flox weanlings but not Crb3flox/flox animals (data not shown; evaluated by χ2 test for goodness of fit to expected Mendelian distribution; P < 0.001) (genotyping strategy shown in Fig. 1A). Immunoblotting analysis of lung tissue from Crb3+/+ and Crb3+/flox adults demonstrated that the GFP-Crb3 fusion protein was expressed at extremely low levels (Fig. 1C). We concluded that further analysis of this GFP-Crb3 allele would not be feasible.
FIG 1.
Generation of Crumbs3-deficient mice. (A) The diagram depicts the gene-targeting strategy and PCR genotyping scheme. The upper box shows the genomic elements; the lower box displays the amplicon sizes for each primer pair (shown in red) which distinguish the alleles. (B) Southern blot analysis of three targeted clones (11G, 12A, and 12C) and wild-type R1 ES cells confirms homologous recombination. (C) Western blot analysis of +/+ and +/flox embryonic lung lysates (embryonic day 18.5 [E18.5]) demonstrates that the GFP-Crb3 protein, a product of the floxed allele, is present in very small quantities in the Crb3+/flox animals. Both panels are taken from the same gel and same exposure time, indicating that GFP-Crb3 is expressed at a tiny fraction of the level of endogenous Crb3 (left panel). The GFP-Crb3 fusion protein is barely detectable following specific immunoprecipitation (IP) of 1 mg lung lysate (right panel). The lanes marked “GFP-Crb3” are positive-control lanes using MDCK cell lysates expressing GFP-Crb3. (D) A survival curve was plotted after videotaping the delivery of E18.5 embryos. The mean lifetime for Crb3−/− pups ± standard error of the mean is indicated. (E) Weights of E18.5 embryos do not appear significantly different among the genotypes by ANOVA.
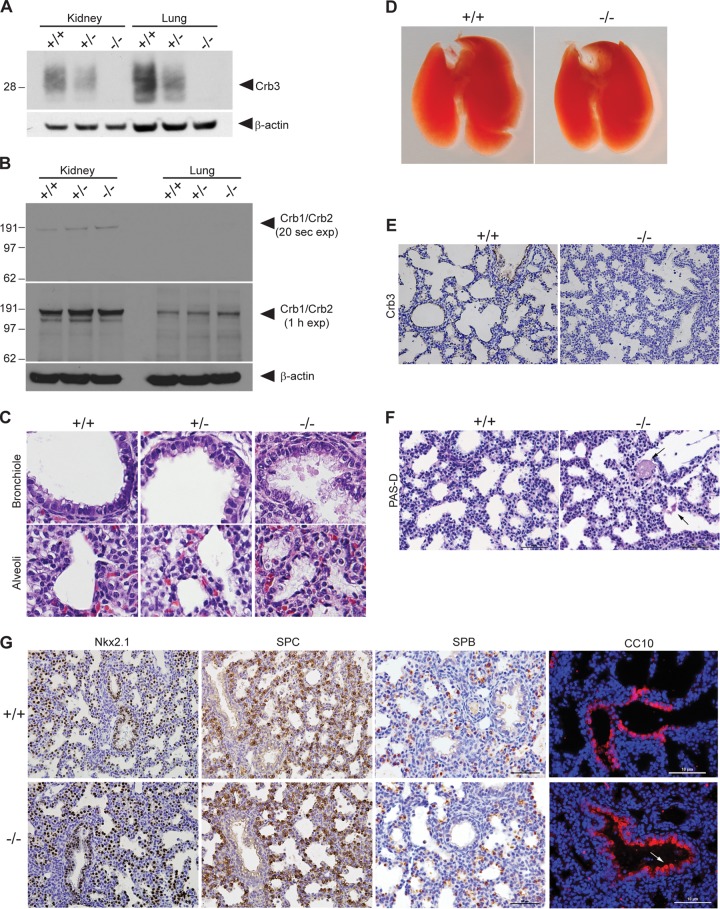
Therefore, we generated a germ line Crb3 knockout mouse by crossing the Crb3+/flox animals to the germ line Cre-deleter, EIIa-Cre (30). We recovered Crb3+/+ and Crb3+/− weanlings but not Crb3−/− animals (P < 0.0001) (Table 1), suggesting that Crb3 must be crucial for embryonic development. Thus, we performed timed matings by intercrossing Crb3 heterozygous animals and discovered Crb3−/− pups in utero at E12.5 and E17.5 (data not shown). We dissected pregnant females at E18.5, 1 day before the expected parturition (P0), and were able to deliver Crb3+/+, Crb3+/−, and Crb3−/− pups alive; however, Crb3−/− pups expired ∼10 min after delivery due to apparent respiratory distress (Fig. 1D). The animals looked well developed and well matched in size (Fig. 1E). Western blot analysis revealed a true half-dosage in Crb3 protein levels in Crb3+/− animals and its complete absence in Crb3−/− animals (Fig. 2A). Compensatory upregulation of higher-molecular-weight proteins Crumbs1 and Crumbs2, whose carboxy-terminal tails share a significant degree of identity and similarity to that of Crumbs3, was not detected by immunoblotting with the Crumbs3 antibody in the null animals (Fig. 2B). H&E analysis of E18.5 lungs of animals stimulated to breathe revealed normal-appearing bronchioles and primitive alveolar saccules with open air spaces in Crb3+/+ and Crb3+/− genotypes (Fig. 2C). However, the alveolar walls of Crb3−/− appeared thickened and hypercellular, and debris filled the airways of the Crb3−/− animals. The air spaces appeared to contain alveolar macrophages and eosinophilic fibrillar material, representing protein-rich fluid. Air space expansion was decreased in the knockout animals, but there did not appear to be a difference in the numbers of proximal airways or sacculations per section. Furthermore, immunohistochemical analysis for altered cell proliferation (Ki67) or apoptosis (cleaved caspase-3) did not show differences between the wild-type and null lungs (data not shown). Genotypic analysis of E17.5 to P0 pups revealed Crb3+/+ (18.4%), Crb3+/− (43.9%), and Crb3−/− (37.7%) (Table 2). Thus, Crb3 is not required for embryonic development but is essential for viability immediately after birth. We also note that half-dosage of Crb3 is sufficient: the Crb3+/− animals breed well and appear healthy even as they mature past 1 year of age (E. L. Whiteman and B. Margolis, unpublished results).
TABLE 1.
Genotypic distribution of weanlings from heterozygous matingsa
| Age | Strain | No. (%) of weanlings with indicated genotype |
P valueb | Litter no. | ||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | ||||
| P14–P21 | 11G | 23 (36.5) | 40 (63.5) | 0 (0) | <0.0001 | 14 |
| P14–P21 | 12A | 15 (36.6) | 26 (63.4) | 0 (0) | 0.0009 | 9 |
| Total | 38 (36.5) | 66 (63.5) | 0 (0) | <0.0001 | 23 | |
Data evaluated by χ2 test for goodness of fit to expected Mendelian distribution.
Absence of Crb3−/− weanlings is statistically significant.
FIG 2.
Analysis of Crumbs3-deficient mice and examination of Crumbs3 knockout lungs. (A) Western blot analysis of kidney and lung lysates (E18.5) confirms Crb3 loss. (B) Upregulation of highly similar proteins Crb1 and Crb2 is not detected in Crb3−/− kidneys or lungs. E18.5 kidney or lung homogenates (100 μg) were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF), probed using an antibody to the Crumbs3 carboxy-terminal tail, and detected using the very sensitive Bio-Rad Clarity substrate for short and long exposures. We speculate that the positive bands in the 20-s kidney exposure may represent Crb2, whose cross-reactivity with the Crb3 antibody is evident when staining kidney glomeruli (Fig. 6A). We did not observe any enhancement in higher-molecular-mass bands potentially representing Crb1 (>151 kDa) or Crb2 (>137 kDa) in the Crb3−/− animals. (C) Paraffin-embedded lungs from E18.5 pups stimulated to breathe were stained for H&E (×400). (D) Whole-mount images of +/+ and −/− mouse lungs at E18.5. (E) Paraffin sections from E18.5 lungs were stained for Crb3 (brown) and hematoxylin (blue) (×400). (F) Paraffin sections from E18.5 +/+ and −/− mouse lungs were subjected to a PAS-diastase assay. Arrows indicate residual, patchy PAS-diastase-resistant material in the knockout lung alveoli. Scale bar = 50 μm. (G) Paraffin sections from E18.5 +/+ and −/− mouse lungs were stained for Nkx2.1 (brown, ×400), SPC (brown, ×400), SPB (brown; scale bar = 50 μm), and CC10 (red; scale bar = 10 μm [the arrow indicates blebbing of Clara cells]). Nuclei (blue) were stained with hematoxylin or DAPI.
TABLE 2.
Genotypic distribution of late embryos and neonates from heterozygous matingsa
| Strain (age) | No. (%) of late embryos or neonates with indicated genotype |
P valueb | Litter no. | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| 11G (E17.5–P0) | 11 (20.0) | 23 (41.8) | 21 (38.2) | 0.08 | 9 |
| 12A (E17.5–P0) | 7 (16.3) | 20 (46.5) | 16 (37.2) | 0.14 | 6 |
| Total | 18 (18.4) | 43 (43.9) | 37 (37.7) | 0.01 | 15 |
Data evaluated by χ2 test for goodness of fit to expected Mendelian distribution.
All genotypes, including Crb3−/−, were present during late embryonic development and birth.
Crumbs3−/− lungs display membrane blebs and are filled with proteinaceous material.
To assess the role of Crb3 in lung development, we dissected E18.5 lungs. Lungs appear grossly normal (Fig. 2D), and Crb3 stained the apical membranes of bronchioles and alveoli in Crb3+/+ animals and was absent in Crb3−/− lungs (Fig. 2E). The fibrillar material seen within the air spaces was periodic acid-Schiff (PAS) positive and diastase resistant, indicative of the presence of glycoproteins or glycolipids, though we have not been able to identify its precise nature (Fig. 2F). The usual pulmonary cell types were represented in the wild-type and null lungs, as judged by staining for Nkx2.1 (which marks all respiratory epithelial cells), surfactant proteins B and C (SPB and SPC; expressed by alveolar type II pneumocytes), and Clara cell secretory protein (CCSP/CC10, expressed by bronchiolar Clara cells) (Fig. 2G). Surfactant proteins were not aberrantly detected. However, the debris in the Crb3−/− alveoli stained positively for SPB, indicating that focal proteinosis is present. We note some Clara cell blebbing in the Crb3−/− lungs which may indicate membrane instability or abnormal secretion.
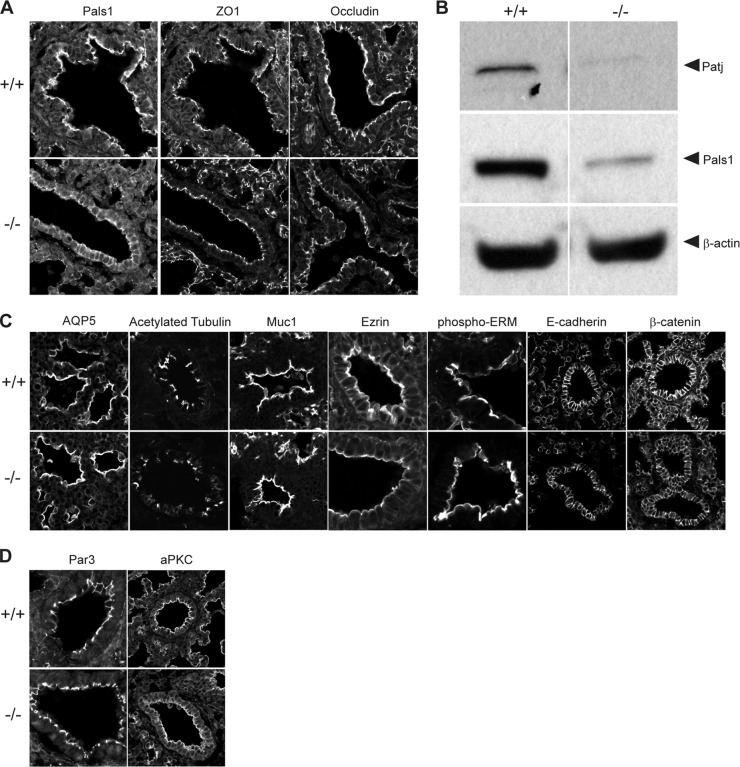
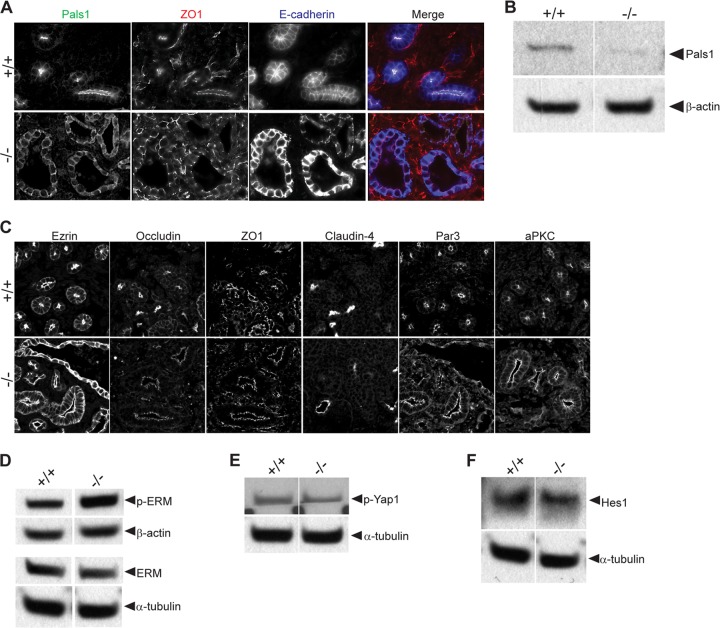
Next, we assayed the lungs for epithelial polarity and junctions. Pals1, the direct binding partner for Crb3, was strictly confined to the apical membranes in wild-type lungs but was localized at both apical and basolateral membranes in the knockout lungs; Pals1 expression levels were reduced in the absence of Crb3 (Fig. 3A and B). The third member of the Crb3 protein complex, Patj, was almost undetectable by immunoblotting analysis in Crb3 knockout lungs (Fig. 3B). Our previous studies have demonstrated that the individual stabilities of Crb3/Pals1/Patj are linked (18, 31, 32). Surprisingly, TJs were present in the null lungs as judged by ZO1 and occludin staining (Fig. 3A). Further evaluation of the knockout lung by light microscopy failed to reveal additional abnormalities: aquaporin 5 (Aqp5, marking predominantly alveolar type I pneumocytes), mucin 1 (Muc1; expressed by airway and alveolar epithelial cells), Ezr, and phospho-ezrin/radixin/moesin (p-ERM) remained apically localized; acetylated tubulin marked the presence of motile cilia, and E-cadherin and β-catenin appropriately illuminated adherens junctions (AJ) (Fig. 3C). Finally, Par3 and aPKC stained the apical membrane/TJ of null lungs, demonstrating that the Par3 polarity signaling cassette was still correctly localized (Fig. 3D).
FIG 3.
Evaluation of epithelial polarity in Crumbs3 knockout lungs. (A) Frozen sections from E18.5 lungs were stained for Pals1, ZO1, and occludin (×600; zoom, 1.5). (B) Western blot of Crb complex proteins Pals1 and Patj in E18.5 lungs. β-Actin was used as a loading control. Samples were run on nonadjacent lanes of the same gel. (C) Frozen sections from E18.5 +/+ and −/− mouse lungs were stained for AQP5 (alveolar type I cells) (×600; zoom, 2), acetylated tubulin (cilia) (zoom, 1.5), Muc1 (apical surface) (zoom, 1.5), ezrin and phospho-ERM (cytoskeleton) (zoom, 3), E-cadherin and β-catenin (adherens junction/basolateral surface) (zoom, 1.5). (D) Frozen sections from E18.5 +/+ and −/− mouse lungs were stained for Par3 (×600) (zoom, 3) and aPKC (apical polarity complex) (zoom, 1.5).
To further define changes in lung morphology in Crb3−/− mice, we next acquired ultrastructural information. Transmission electron microscopy (TEM) confirms the presence of TJs, motile cilia, and lamellar bodies (storing pulmonary surfactants in alveolar type II pneumocytes) in null lungs (Fig. 4A). However, we note lamellar bodies and proteinaceous material (presenting as a gray haze) that persisted in the air space outside the cell boundaries of Crb3−/− lungs. In addition, we find dramatic membrane blebs extending from the surface of some bronchioles and interrupting the motile cilia (Fig. 4B). SEM also reveals striking membrane blebs interrupting the arrangement of motile cilia in Crb3−/− lungs and membrane protrusions in the cells lining the bronchioles (Fig. 4C). To further assay defects in the actin cytoskeleton that might account for this blebbing, we assayed levels of p-ERM and ERM proteins by immunoblot analysis. The protein levels are indistinguishable between wild-type and null animals (Fig. 5A). SEM images of the alveoli show smooth, catacomb-like structures in the wild-type lungs and a more collapsed arrangement with puckered cells in the null lungs (Fig. 4C).
FIG 4.
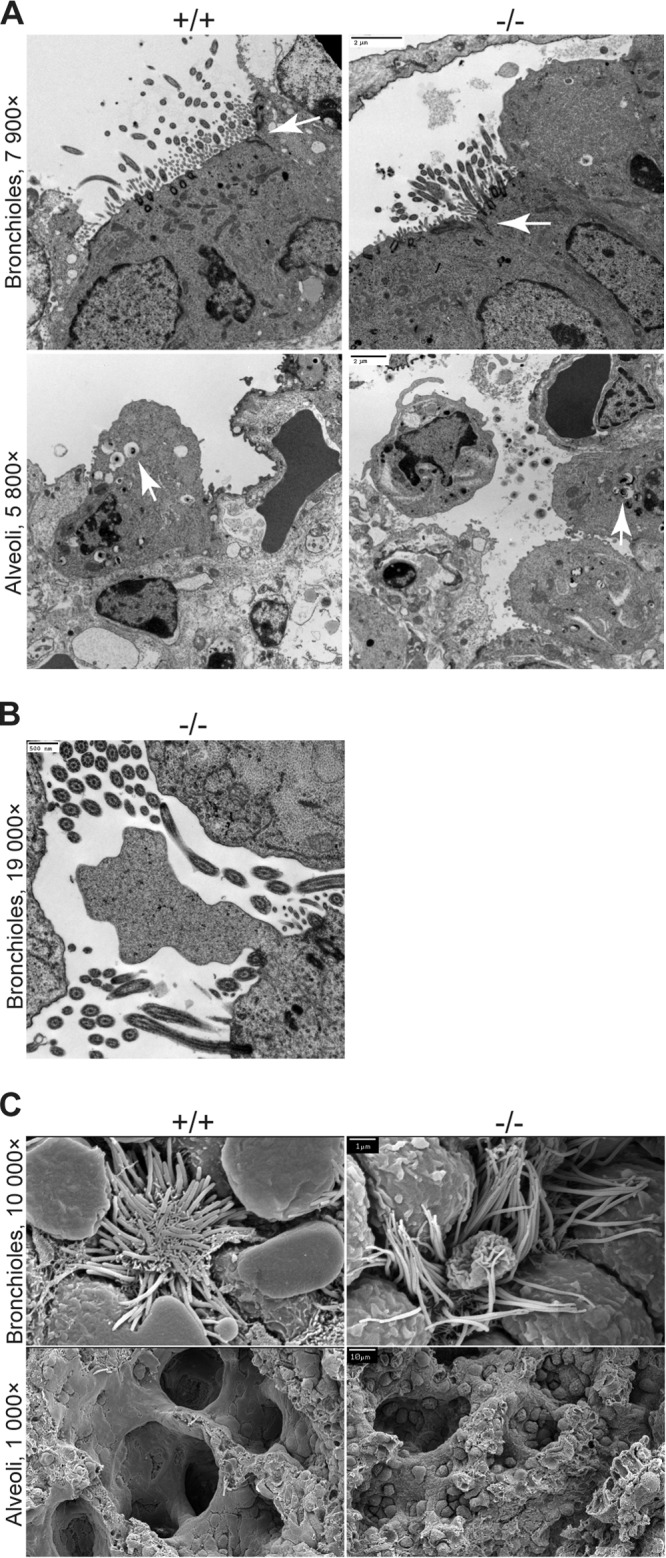
Electron micrographs of lungs from Crumbs3 knockout mice. (A) Transmission electron micrographs of bronchioles and alveoli from E18.5 lungs (scale bars = 2 μm). Arrows indicate tight junctions (bronchioles) or lamellar bodies (alveoli). (B) Transmission electron micrograph of membrane bleb in Crb3−/− bronchioles from E18.5 lungs (scale bar = 500 nm). (C) Scanning electron micrographs of bronchioles and alveoli from E18.5 lungs (scale bars = 1 μm [top] and 10 μm [bottom]).
FIG 5.
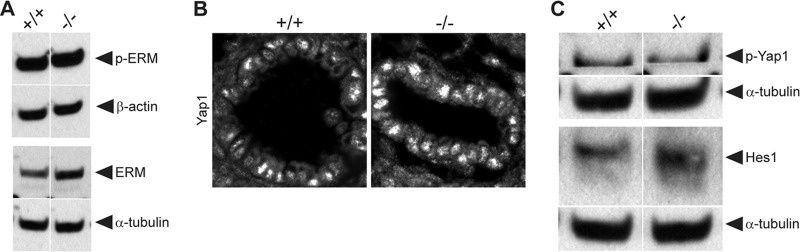
Investigation of signaling pathways in Crumbs3 knockout lungs. (A) Western blot of p-ERM and ERM protein levels among +/+ and −/− lungs at E18.5. β-Actin and α-tubulin are shown as loading controls. Samples were run on nonadjacent lanes of the same gel. (B) Frozen sections from E18.5 +/+ and −/− mouse lungs were stained for Yap1 (transcription factor downstream of Hippo pathway) (×600; zoom, 3). (C) Western blot of p-Yap1 (Hippo pathway activation) and Hes1 (Notch pathway activation) among +/+ and −/− lungs at E18.5. α-Tubulin is shown as the loading control. Samples were run on nonadjacent lanes of the same gel.
Finally, we assayed the lungs for more subtle defects in cellular signaling pathways, since Crb proteins positively regulate the Hippo tumor suppressor signaling pathway (33–35) and negatively regulate the Notch signaling cascade (36–38). According to the models developed in lower organisms and newly emerging evidence in mammalian systems, we would predict that absence of Crb3 might lead to dephosphorylation of Yap1 and its resulting nuclear localization (a readout of Hippo pathway inactivity) and an enhancement in the levels of the transcription factor Hes1 (an indication of Notch pathway activation). However, in wild-type and null lungs alike, Yap1 was similarly localized to the nucleus, and Yap1 phosphorylation appeared unchanged and Hes1 protein levels were also very comparable (Fig. 5B and C). In all experiments described here, heterozygous and wild-type lungs were indistinguishable (data not shown).
Crumbs3−/− kidneys reveal cysts and abnormal brush borders.
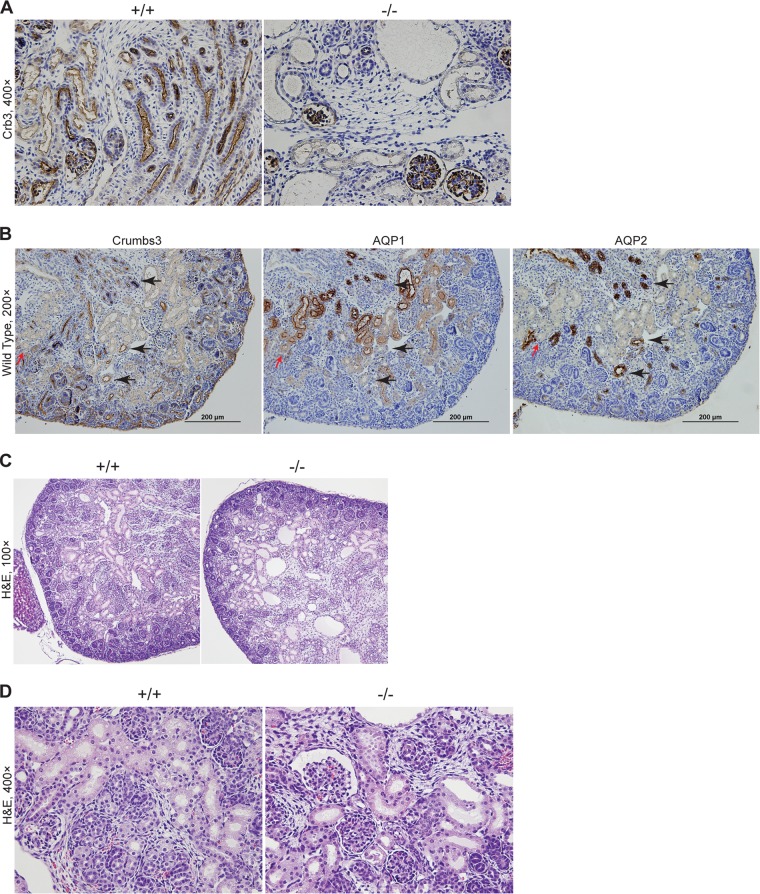
To evaluate the role of Crb3 in kidney development, where Crb3 is enriched during mesenchyme-to-epithelium transition, we isolated kidneys from E17.5 to E18.5 animals. We show that Crb3 staining was absent from tubules in Crb3−/− kidneys, though we do see residual staining of glomeruli (Fig. 6A). This staining likely represents cross-reactivity with Crb2, a highly conserved isoform also expressed in kidney (15). However, we were unable to detect a compensatory upregulation of Crb2 in Crb3−/− kidneys by immunoblotting (Fig. 2B). In the wild-type kidneys, Crb3 was expressed at lower levels in proximal tubules and was more highly enriched in distal tubules, as judged by staining with Aqp1 (proximal tubules) and Aqp2 (collecting ducts) in serial sections (Fig. 6B). Inspection of H&E-stained sections revealed the presence of cysts and dilated tubules in the null kidneys (Fig. 6C) but normal-appearing glomeruli (Fig. 6D). We observe an ∼13% increase in the luminal diameter of null tubules compared to wild-type tubules at E18.5 (wild type, 28.0 μm ± 8.3 μm; null, 31.6 μm ± 13.4 μm [P < 0.001 by t test]).
FIG 6.
Examination of Crumbs3 knockout kidney. (A) Paraffin sections from E18.5 kidneys were stained for Crb3 (brown) and hematoxylin (blue). (B) Paraffin-embedded kidneys from wild-type E18.5 mice were stained for Crb3, Aqp1, and Aqp2 (brown) using serial sections. Red arrows indicate areas where weak Crb3-positive tubules were overlaid with Aqp1. Black arrows indicate the overlap of strongly Crb3-positive tubules with Aqp2. Nuclei were counterstained with hematoxylin (blue). (C) Paraffin sections from E18.5 kidneys were stained with H&E. (D) Paraffin-embedded kidneys from E18.5 mice were stained for H&E.
Next, we assayed epithelial polarity and cell-cell junctions in the kidney. We found that, in similarity to the lung results, Pals1 was no longer restricted to the apical membrane in the absence of Crb3 and its protein level was reduced (Fig. 7A and B). (Patj was not detectable by immunoblot in embryonic kidneys.) However, ZO1 (TJ) and E-cadherin (AJ) were appropriately localized (Fig. 7A). Further staining with TJ markers occludin, ZO1, and claudin-4 shows appropriate localization in null kidneys, and the cytoskeleton-associated protein Ezr also was correctly localized, although its staining pattern was disorganized in cysts (Fig. 7C). Levels of p-ERM and total ERM in wild-type and null kidneys were comparable (Fig. 7D). The Par3 signaling cassette was also localized appropriately to apical membranes/TJs in the null kidneys, as measured by Par3 and aPKC staining (Fig. 7C). We did not detect any changes in the Hippo tumor suppressor pathway or the Notch signaling pathway, as judged by immunoblotting for phospho-Yap1 and Hes1 (Fig. 7E and F).
FIG 7.
Evaluation of epithelial polarity and signaling pathways in Crumbs3 knockout kidneys. (A) Frozen sections from E17.5 kidneys were stained for Pals1 (green), ZO1 (red), and E-cadherin (blue) (×400 magnification). (B) Western blot of Crb complex protein Pals1 in E18.5 kidneys. β-Actin was used as a loading control. Samples were run on nonadjacent lanes of the same gel. (C) Frozen sections of kidneys from E17.5 to E18.5 mice were stained for ezrin (cytoskeleton), occludin and ZO1 (tight junction), E-cadherin (adherens junction/basolateral surface), claudin-4 (tight junction), Par3, and aPKC (apical polarity complex) (×600; zoom, 1.5). (D) Western blot of p-ERM and ERM protein levels among +/+ and −/− kidneys at E18.5. β-Actin and α-tubulin are shown as loading controls. Samples were run on nonadjacent lanes of the same gel. (E and F) Western blots of p-Yap1 (Hippo pathway activation) and Hes1 (Notch pathway activation) among +/+ and −/− kidneys at E18.5. α-Tubulin is shown as the loading control. Samples were run on nonadjacent lanes of the same gel.
We next analyzed kidney organization at the ultrastructural level. TEM verified the existence of TJs and brush borders (microvillus [MV]-based structures) in wild-type and null kidneys; however, we report the presence of some tubules displaying partial loss of the brush border in Crb3−/− kidneys (Fig. 8A and B). Additionally, SEM shows well-formed, interdigitating podocyte foot processes in wild-type and null glomeruli (Fig. 8C). Cilia were detected in the Crb3−/− kidneys, but the apical surface of some renal cells appeared irregular (Fig. 8C).
FIG 8.
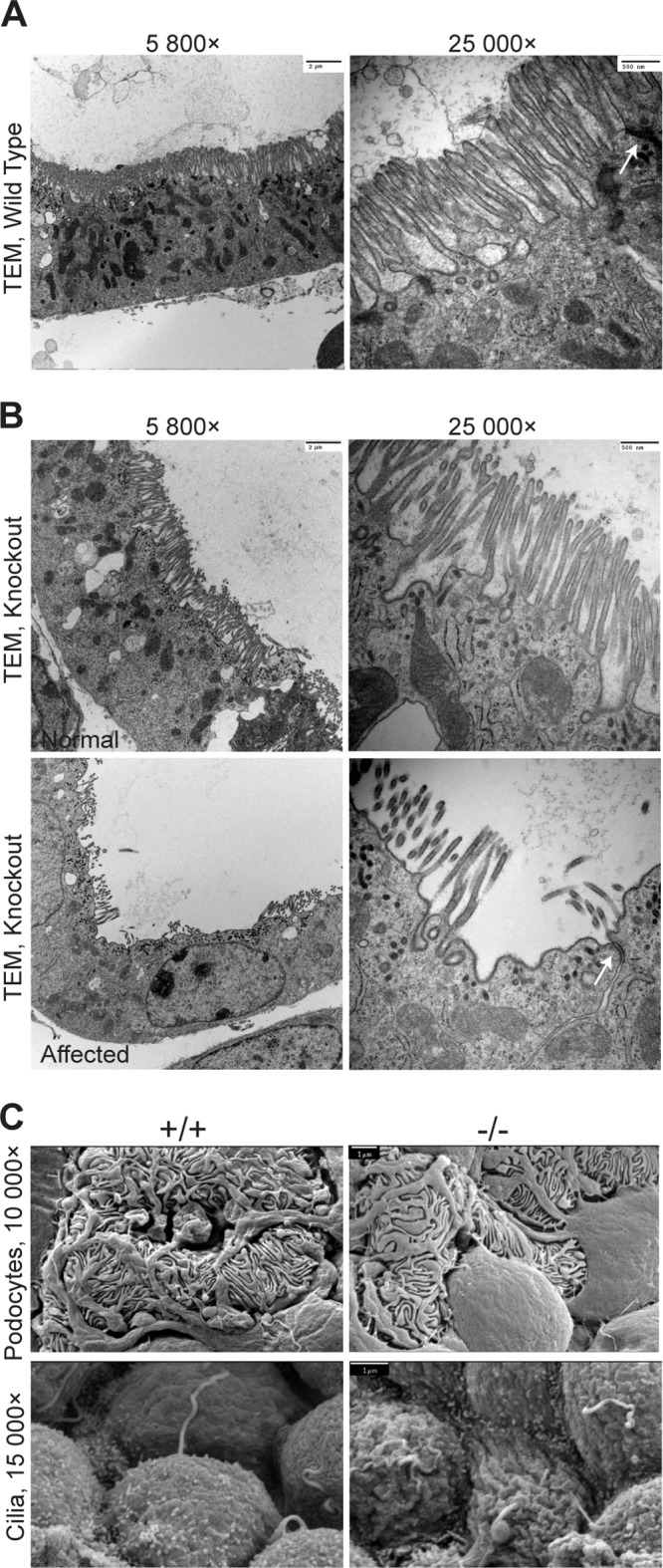
Electron micrographs of kidneys from Crumbs3-deficient mice. (A and B) Transmission electron micrographs of wild-type and knockout proximal tubules with brush borders from E18.5 kidneys (scale bars = 2 μm [left] and 500 nm [right]). Arrows indicate tight junctions. (C) Scanning electron micrographs of podocytes and cilia from E18.5 kidneys (scale bars = 1 μm).
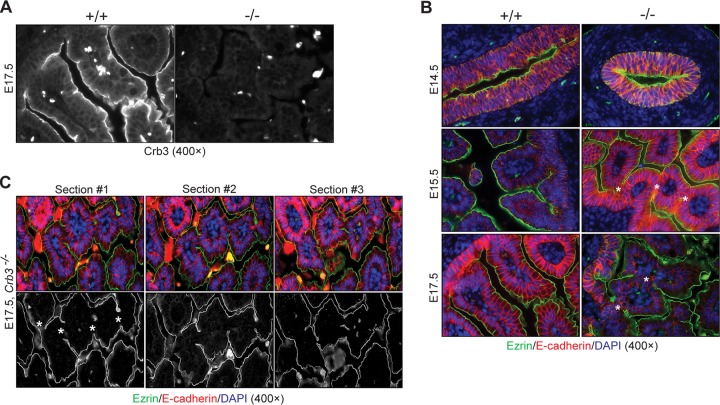
Crumbs3−/− intestines demonstrate villus fusion and irregular microvilli.
To examine the role of Crb3 in intestinal development, where dramatic cellular rearrangements lead to villus morphogenesis (39, 40), we isolated duodenum from embryonic mice. Crb3 staining verifies the absence of Crb3 in knockout intestine (Fig. 9A). Duodenum isolated and stained for Ezr (apical) and E-cadherin (basolateral) at various time points before (E14.5), during (E15.5), and after (E17.5) villus emergence shows normal organization of the epithelium at E14.5 but villus fusion at E15.5 and E17.5 in Crb3−/− animals (Fig. 9B). Fusion was also seen in Crb3−/− jejunum at E17.5 (data not shown). The persistence of fused villi was confirmed by analyzing serial sections of null tissue stained for Ezr and E-cadherin, even though the overall polarization of the villi was maintained (Fig. 9C). No villus fusions were observed in wild-type intestines. Heterozygous intestines also did not show evidence of villus fusion (data not shown).
FIG 9.
Evaluation of Crumbs3 knockout intestine. (A) Staining of E17.5 duodenum confirms the absence of Crumbs3. Bright spots represent autofluorescent blood cells. (B) Fluorescent staining of paraffin-embedded duodenum sections at different stages of villus development with ezrin (green), E-cadherin (red), and DAPI (blue, to mark nuclei) demonstrates the presence of bridges (marked by asterisks) between villi in Crb3−/− embryos. (C) Staining of Crb3−/− duodenum serial sections with ezrin (green), E-cadherin (red), and DAPI (blue) confirms the presence of bridges (asterisks).
The ultrastructure of the perinatal intestine is also disrupted. SEM shows irregular and fused villi in the Crb3−/− intestine at low magnification. At high magnification, we note that the MV are disorganized (Fig. 10A). TEM micrographs confirm the presence of TJs in Crb3+/+ and Crb3−/− intestine, although the MV were significantly shorter in the null animals and the gut lumen appears to have been filled with debris (Fig. 10B). This is consistent with the detection of Crb3 in microvilli in epithelial cells by immunoelectron microscopy (Fig. 10C).
FIG 10.
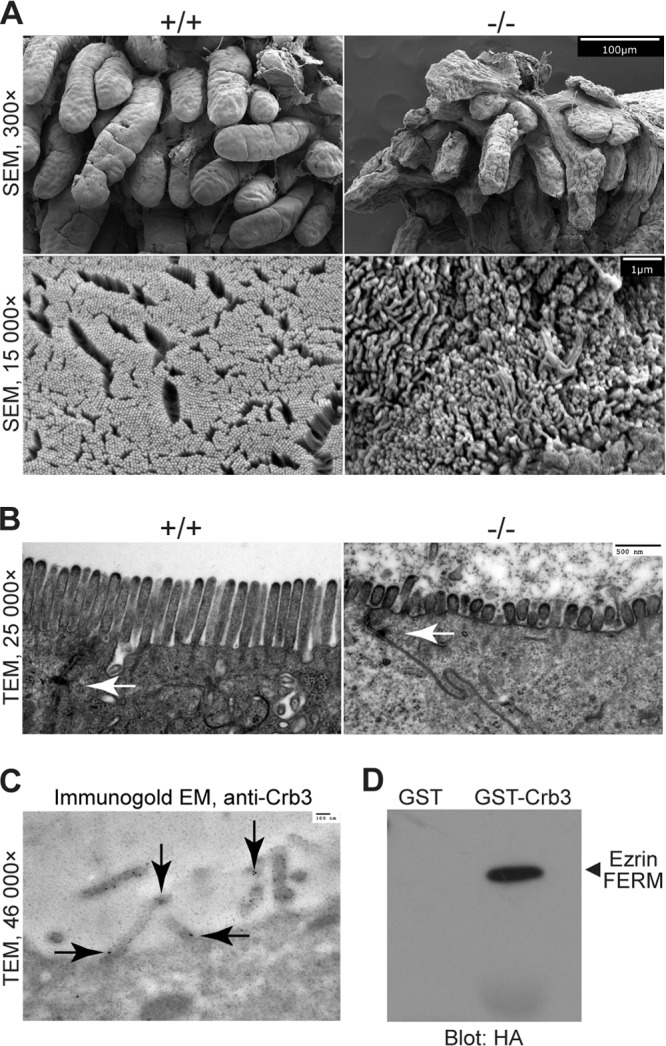
Electron micrographs of intestine from Crumbs3 knockout mice, Crb3 in microvilli (cell culture), and Crb3 interactions with ezrin. (A) Scanning electron micrographs of E18.5 wild-type and Crb3 null intestine (scale bars = 100 μm and 1 μm). (B) Transmission electron micrographs of E18.5 wild-type and Crb3 null intestine (scale bar = 500 nm). Arrows indicate tight junctions. (C) Transmission electron micrograph of immunogold-labeled Crb3 in MDCK cells (scale bar = 100 nm). Arrows indicate immunogold particles and localization of Crb3. (D) GST pulldown experiments assay the interaction between Crb3 intracellular domain and the HA-tagged ezrin FERM domain.
The presence of villus fusion, rough topology of the villi, and shortened MV are reminiscent of the Ezr−/− intestine (41, 42). In Drosophila, Crb has been shown to bind a member of the ERM family, DMoesin, in an interaction that depends on the Crb FBD (8). This interaction is thought to connect the apical membrane with the underlying cytoskeleton. To assay a possible similar interaction between Crb3 and Ezr, we performed GST pulldown assays and we found that Crb3 and Ezr can associate in vitro (Fig. 10D).
DISCUSSION
Crb3 knockout mice die shortly after birth, most likely due to respiratory distress. Prominent features in other organs are renal cysts and abnormal intestinal villi. Defects in the apical membrane were also detected, including brush border abnormalities and membrane blebbing.
Lumen formation and ciliogenesis.
Crb proteins have a key role in cell polarity and tight junction formation. Crb3 is upregulated in the mesenchyme-to-epithelium transition, suggesting that it might be crucial for lumen and tube formation (16). However, initial lumen formation appeared to proceed in lung, kidney, and intestine with normal localization of Par polarity complex proteins. Redundancy between the Crb polarity complex and the Par polarity complex has been described in Drosophila epithelia (43), and in some Drosophila tissues Par3 is upstream of Crb and does not require Crb for proper targeting (44, 45). It is thought that correct targeting of aPKC is essential for proper apicobasal polarity (7, 46–48), and this appears to occur in the Crb3−/− tissues we studied here. In addition, we detected TJs by TEM and proper localization of TJ proteins by light microscopy in Crb3−/− tissues. However, whether Crb3-deficient TJs function normally remains to be determined.
The lungs of Crb3−/− mice showed thickened, hypercellular alveolar walls and air spaces filled with alveolar macrophages and proteinaceous material. This phenotype resembles pulmonary alveolar proteinosis (PAP), a group of diseases marked by abnormal accumulation of PAS-positive glycolipids and glycoproteins in the alveoli, resulting in inadequate gas exchange and respiratory insufficiency (49). In the Crb3−/− mice, it may be caused by defective surfactant secretion, recycling, or degradation. Alternatively, the motile cilia, which are involved in the clearance of liquid from the newborn lung and the transition to air-filled neonatal lung, may malfunction in Crb3−/− lungs. Indeed, Crb3 is involved in ciliogenesis and localizes to primary cilia (50) as well as motile cilia (S. Fan and B. Margolis, unpublished results). We observed that CCL2/MCP-1 (chemokine C-C motif ligand 2/monocyte chemoattractant protein-1) staining appears to be enhanced in the null lungs compared to wild-type lungs, possibly providing a mechanism for macrophage recruitment (data not shown). CCL2 serves as a chemoattractant for macrophages and is thought to mediate the inflammatory response to alveolar hypoxia occurring in acute or chronic pulmonary diseases and also illnesses of high altitude (51). However, macrophage recruitment is a complex process, and there are many other factors that could be contributing to the enrichment of macrophages in the Crb3−/− lungs.
The cysts we detect in the Crb3 knockout kidneys could also be due to defective cilia (52). In zebrafish, Crb2 knockdown led to pronephros cysts that were attributed to abnormal cilia (53). We detected cilia in some kidney tubules of the Crb3 knockout mice, though we did not test their function. It is also possible that the presence of cysts and tubular dilations represents a polarity defect resulting in improperly oriented cell division or directed cell movement (54, 55). We noted that kidney tubule cross sections were larger in Crb3−/− animals, likely representing loss of control of luminal diameter or early cystic changes. Defects in polarity proteins result in renal cysts in other knockout mouse models. In particular, Lin-7c/Mals-3 deletion leads to a nephronophthisis-like disease with cysts and fibrosis (56).
Cytoskeleton.
Many Crb3−/− intestinal abnormalities phenocopied defects seen in the Ezr−/− mice described by McClatchey and coworkers. These defects include membrane blebbing and abnormal MV (41, 42). Ezr and other members of the ERM family are critical in the formation of the membrane cytoskeleton (57). Similarities between the Crb3 knockout and the Ezr knockout align well with the known ability of Crb to bind ERM proteins, including the Drosophila ERM, DMoesin (8). Furthermore, we have previously noted that expression of a Crb3 construct with a mutated FBD does not rescue TJ formation in MCF10A cells, indicating a strict requirement for this domain in cell culture (58). We have detected Crb3 throughout the apical membrane as well as in MV. Presumably, Crb3 can bind cytoskeletal proteins such as Ezr and connect the apical membrane to the cytoskeleton. Similarly, we find that loss of Crb3 leads to membrane blebbing. Blebbing is seen in the Ezr−/− mouse, and Ezr plays an important role in repairing membrane blebs (42, 59). Furthermore, the MV defect in the Ezr−/− model is thought to arise from irregularities in the actin terminal web which contains Ezr and provides an anchor for MV while infiltrating the apical junctional region (41). The defects observed in the kidney brush borders may be due to a similar loss of interaction with ERM proteins, or they may be caused by tubular defects related to the development of kidney cysts.
Also in striking similarity to the Ezr−/− mice, we see intestinal villus fusion in the Crb3−/− animals. The original model for embryonic gut development holds that stratified epithelial layers rearrange to form secondary lumina (39), and the villus fusion in the Ezr−/− mice has been ascribed to failure of secondary lumen expansion (41). In the absence of Crb3, secondary lumen formation may also be incomplete, leading to villus fusion. This may be similar to the multilumen phenotype seen in Crb3 knockdown cells (20). Data collected on villus regeneration in Ezr−/− adult gut propose that bridges can also be formed when adjacent cells fail to segregate as they emerge from the same crypt and remain in contact as they ascend two separate villi due to altered spindle orientation and defective apical junction remodeling, ultimately leading to fusion (42). Recent data indicate that secondary lumina do not form in embryonic gut epithelium. Rather, the new model suggests that an embryonic pseudostratified epithelium experiences interkinetic nuclear migration, apico-basal elongation, cell shape changes, and expansion of the apical surface resulting in villus formation (40). Therefore, it is possible that the fused villi observed in Ezr−/− and also Crb3−/− embryos were caused by randomized cell division due to altered spindle orientation. Finally, the possibility that ezrin affects junctional remodeling is also pertinent to understanding villus fusion in both the Ezr−/− and Crb3−/− animals. In this regard, Ezr interacts with Crb3 in vitro, and the two knockout models share an intestinal phenotype.
ACKNOWLEDGMENTS
This research was supported by NIH grants DK089119 (to J.L.H.), HL079339 (to M.B.H.), DK06914 and DK073722 (to G.R.D.), DK065850 and DK089933 (to D.L.G.), and DK069605 (to B.M.).
We thank members of our outstanding core facilities at the University of Michigan: Microscopy and Image Analysis Laboratory (especially S. Almburg, J. Harrison, and J. Poore), MDRTC Morphology and Image Analysis Core (S. Lentz), Transgenic Animal Model Core (especially E. Hughes, T. Saunders, M. Schmidt, and D. Vanheyningen), and the Unit for Laboratory Animal Medicine (Managed Breeding Colony, Technical Services, and Pathology Services, especially P. Arrowsmith, I. Bergin, R. McUmber, C. Schray, and B. Popoola). We also thank K. Bentley, M. Berger, M. Green, E. Hurd, J. Kushwaha, M. Nagy, S. Patel, B. Rockich, J. Spence, U. Sajjan, and C. Tsui for reagents, technical advice, and helpful discussions.
E.L.W., S.F., J.L.H., K.D.W., C.-J.L., A.S., V.C.F., and G.H.D. performed experiments and data analysis. E.L.W., M.B.H., G.R.D., G.H.D., D.L.G., and B.M. developed the concepts and experimental approaches. E.L.W. and B.M. wrote the manuscript.
Footnotes
Published ahead of print 28 October 2013
REFERENCES
- 1.Dow LE, Humbert PO. 2007. Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int. Rev. Cytol. 262:253–302. 10.1016/S0074-7696(07)62006-3 [DOI] [PubMed] [Google Scholar]
- 2.Pieczynski J, Margolis B. 2011. Protein complexes that control renal epithelial polarity. Am. J. Physiol. Renal Physiol. 300:F589–F601. 10.1152/ajprenal.00615.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datta A, Bryant DM, Mostov KE. 2011. Molecular regulation of lumen morphogenesis. Curr. Biol. 21:R126–R136. 10.1016/j.cub.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tepass U. 2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28:655–685. 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- 5.Bulgakova N, Knust E. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122:2587–2596. 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- 6.Joberty G, Petersen C, Gao L, Macara IG. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- 7.Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, Yonemura S, Ohno S. 2009. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J. Cell Sci. 122:1595–1606. 10.1242/jcs.043174 [DOI] [PubMed] [Google Scholar]
- 8.Medina E, Williams J, Klipfell E, Zarnescu D, Thomas G, Le Bivic A. 2002. Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158:941–951. 10.1083/jcb.200203080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, Tepass U. 2006. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev. Cell 11:363–374. 10.1016/j.devcel.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, Beitel GJ, McGlade CJ, Tepass U. 2009. Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature 459:1141–1145. 10.1038/nature08067 [DOI] [PubMed] [Google Scholar]
- 11.Christensen A, Jensen A. 2008. Tissue-specific requirements for specific domains in the FERM protein Moe/Epb4.1l5 during early zebrafish development. BMC Dev. Biol. 8:3. 10.1186/1471-213X-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. 2010. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. U. S. A. 107:10532–10537. 10.1073/pnas.1004279107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grzeschik N, Parsons L, Allott M, Harvey K, Richardson H. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20:573–581. 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- 14.den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, Hoyng CB, Westerveld A, Brunner HG, Bleeker-Wagemakers EM, Deutman AF, Heckenlively JR, Cremers FP, Bergen AA. 1999. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat. Genet. 23:217–221. 10.1038/13848 [DOI] [PubMed] [Google Scholar]
- 15.Xiao Z, Patrakka J, Nukui M, Chi L, Niu D, Betsholtz C, Pikkarainen T, Vainio S, Tryggvason K. 2011. Deficiency in crumbs homolog 2 (Crb2) affects gastrulation and results in embryonic lethality in mice. Dev. Dyn. 240:2646–2656. 10.1002/dvdy.22778 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt-Ott K, Yang J, Chen X, Wang H, Paragas N, Mori K, Li J-Y, Lu B, Costantini F, Schiffer M, Bottinger E, Barasch J. 2005. Novel regulators of kidney development from the tips of the ureteric bud. J. Am. Soc. Nephrol. 16:1993–2002. 10.1681/ASN.2004121127 [DOI] [PubMed] [Google Scholar]
- 17.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. 2007. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26:6979–6988. 10.1038/sj.onc.1210508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whiteman EL, Liu CJ, Fearon ER, Margolis B. 2008. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 27:3875–3879. 10.1038/onc.2008.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torkko JM, Manninen A, Schuck S, Simons K. 2008. Depletion of apical transport proteins perturbs epithelial cyst formation and ciliogenesis. J. Cell Sci. 121:1193–1203. 10.1242/jcs.015495 [DOI] [PubMed] [Google Scholar]
- 20.Schlüter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S, Liu CJ, Margolis B. 2009. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell 20:4652–4663. 10.1091/mbc.E09-02-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiraoka M, Abe A, Lu Y, Yang K, Han X, Gross RW, Shayman JA. 2006. Lysosomal phospholipase A2 and phospholipidosis. Mol. Cell. Biol. 26:6139–6148. 10.1128/MCB.00627-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 90:8424–8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall OJ. 2004. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20:2471–2472. 10.1093/bioinformatics/bth254 [DOI] [PubMed] [Google Scholar]
- 24.Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B. 2003. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302:21–29. 10.1016/S0378111902010843 [DOI] [PubMed] [Google Scholar]
- 25.Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 157:161–172. 10.1083/jcb.200109010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harder JL, Whiteman EL, Pieczynski JN, Liu CJ, Margolis B. 2012. Snail destabilizes cell surface Crumbs3a. Traffic 13:1170–1185. 10.1111/j.1600-0854.2012.01376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheehan DC, Hrapchak BB. 1981. Theory and practice of histotechnology, 2nd ed, p 474 C. V. Mosby, St. Louis, MO [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93:5860–5865. 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straight SW, Shin K, Fogg VC, Fan S, Liu CJ, Roh M, Margolis B. 2004. Loss of PALS1 expression leads to tight junction and polarity defects. Mol. Biol. Cell 15:1981–1990. 10.1091/mbc.E03-08-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Margolis B. 2007. Apical junctional complexes and cell polarity. Kidney Int. 72:1448–1458. 10.1038/sj.ki.5002579 [DOI] [PubMed] [Google Scholar]
- 33.Robinson BS, Huang J, Hong Y, Moberg KH. 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 20:582–590. 10.1016/j.cub.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. 2010. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 107:15810–15815. 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. 2010. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev. Cell 19:831–844. 10.1016/j.devcel.2010.11.012 [DOI] [PubMed] [Google Scholar]
- 36.Herranz H, Stamataki E, Feiguin F, Milan M. 2006. Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of gamma-secretase activity. EMBO Rep. 7:297–302. 10.1038/sj.embor.7400617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsuishi Y, Hasegawa H, Matsuo A, Araki W, Suzuki T, Tagami S, Okochi M, Takeda M, Roepman R, Nishimura M. 2010. Human CRB2 inhibits gamma-secretase cleavage of amyloid precursor protein by binding to the presenilin complex. J. Biol. Chem. 285:14920–14931. 10.1074/jbc.M109.038760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson EC, Pichaud F. 2010. Crumbs is required to achieve proper organ size control during Drosophila head development. Development 137:641–650. 10.1242/dev.041913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathan M, Moxey PC, Trier JS. 1976. Morphogenesis of fetal rat duodenal villi. Am. J. Anat. 146:73–92. 10.1002/aja.1001460104 [DOI] [PubMed] [Google Scholar]
- 40.Grosse AS, Pressprich MF, Curley LB, Hamilton KL, Margolis B, Hildebrand JD, Gumucio DL. 2011. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development 138:4423–4432. 10.1242/dev.065789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saotome I, Curto M, McClatchey AI. 2004. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell 6:855–864. 10.1016/j.devcel.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 42.Casaletto JB, Saotome I, Curto M, McClatchey AI. 2011. Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc. Natl. Acad. Sci. U. S. A. 108:11924–11929. 10.1073/pnas.1103418108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanentzapf G, Tepass U. 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5:46–52. 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- 44.Franz A, Riechmann V. 2010. Stepwise polarisation of the Drosophila follicular epithelium. Dev. Biol. 338:136–147. 10.1016/j.ydbio.2009.11.027 [DOI] [PubMed] [Google Scholar]
- 45.McKinley RF, Yu CG, Harris TJ. 2012. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci. 125:1177–1190. 10.1242/jcs.091884 [DOI] [PubMed] [Google Scholar]
- 46.Wodarz A, Ramrath A, Grimm A, Knust E. 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150:1361–1374. 10.1083/jcb.150.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotillos S, Diaz-Meco MT, Caminero E, Moscat J, Campuzano S. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166:549–557. 10.1083/jcb.200311031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fletcher GC, Lucas EP, Brain R, Tournier A, Thompson BJ. 2012. Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 22:1116–1122. 10.1016/j.cub.2012.04.020 [DOI] [PubMed] [Google Scholar]
- 49.Carey B, Trapnell BC. 2010. The molecular basis of pulmonary alveolar proteinosis. Clin. Immunol. 135:223–235. 10.1016/j.clim.2010.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14:1451–1461. 10.1016/j.cub.2004.08.025 [DOI] [PubMed] [Google Scholar]
- 51.Chao J, Wood JG, Gonzalez NC. 2011. Alveolar macrophages initiate the systemic microvascular inflammatory response to alveolar hypoxia. Respir. Physiol. Neurobiol. 178:439–448. 10.1016/j.resp.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardenas-Rodriguez M, Badano JL. 2009. Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am. J. Med. Genet. C Semin. Med. Genet. 151C:263–280. 10.1002/ajmg.c.30227 [DOI] [PubMed] [Google Scholar]
- 53.Omori Y, Malicki J. 2006. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr. Biol. 16:945–957. 10.1016/j.cub.2006.03.058 [DOI] [PubMed] [Google Scholar]
- 54.Fedeles S, Gallagher AR. 16 November 2012. Cell polarity and cystic kidney disease. Pediatr. Nephrol. [Epub ahead of print.] 10.1007/s00467-012-2337-z [DOI] [PubMed] [Google Scholar]
- 55.Carroll TJ, Yu J. 2012. The kidney and planar cell polarity. Curr. Top. Dev. Biol. 101:185–212. 10.1016/B978-0-12-394592-1.00011-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olsen O, Funke L, Long JF, Fukata M, Kazuta T, Trinidad JC, Moore KA, Misawa H, Welling PA, Burlingame AL, Zhang M, Bredt DS. 2007. Renal defects associated with improper polarization of the CRB and DLG polarity complexes in MALS-3 knockout mice. J. Cell Biol. 179:151–164. 10.1083/jcb.200702054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fehon RG, McClatchey AI, Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11:276–287. 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fogg VC, Liu CJ, Margolis B. 2005. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J. Cell Sci. 118:2859–2869. 10.1242/jcs.02412 [DOI] [PubMed] [Google Scholar]
- 59.Charras GT, Hu CK, Coughlin M, Mitchison TJ. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490. 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]