Abstract
In osteoclasts (OCs) podosomes are organized in a belt, a feature critical for bone resorption. Although microtubules (MTs) promote the formation and stability of the belt, the MT and/or podosome molecules that mediate the interaction of the two systems are not identified. Because the growing “plus” ends of MTs point toward the podosome belt, plus-end tracking proteins (+TIPs) might regulate podosome patterning. Among the +TIPs, EB1 increased as OCs matured and was enriched in the podosome belt, and EB1-positive MTs targeted podosomes. Suppression of MT dynamic instability, displacement of EB1 from MT ends, or EB1 depletion resulted in the loss of the podosome belt. We identified cortactin as an Src-dependent interacting partner of EB1. Cortactin-deficient OCs presented a defective MT targeting to, and patterning of, podosomes and reduced bone resorption. Suppression of MT dynamic instability or EB1 depletion increased cortactin phosphorylation, decreasing its acetylation and affecting its interaction with EB1. Thus, dynamic MTs and podosomes interact to control bone resorption.
INTRODUCTION
The homeostasis of the skeleton depends on the balanced action of bone-resorbing osteoclasts (OCs) and bone-forming osteoblasts. To resorb bone, OCs attach to the bone surface and seal off an extracellular compartment into which they transport H+ and Cl− through their ruffled-border apical membrane, lowering the pH and dissolving the mineral phase of the bone matrix. OCs also transport vectorially and secrete lysosomal enzymes, in particular cathepsin K, to degrade the organic phase of the bone matrix (1–3). OCs and other cells derived from monocytic lineage precursors adhere and migrate using specific actin-rich structures called podosomes (also termed invadosomes) (4–7).
Podosomes are dynamic structures that rapidly assemble and disassemble, undergoing fusion, fission, or sliding during their short life span (approximately 2 to 4 min) (8). Podosomes consist of an F-actin-rich core that contains a subset of actin-regulatory proteins, like Wasp, Arp2/Arp3 (Arp2/3) complex, and cortactin, and of a surrounding web of actin filaments, called the cloud, associated with proteins such as integrins, adaptors (Cbl and paxillin), kinases (Src and Pyk2), and Rho GTPases (6). Both the core and the cloud are sites of constant actin polymerization (8–10). This rapid podosome turnover is required for the mobility of the cells and for efficient sealing of the bone-resorbing compartment during cell migration along the bone surface.
Podosomes form early during OC differentiation and are initially organized in clusters which radially evolve into dynamic rings that merge and form a peripheral belt, ultimately circumscribing the ruffled border and forming the sealing zone (8, 10, 11). The reversible transition between podosome clusters and the podosome belt/sealing zone is a unique feature of the OC (11, 12).
The formation of podosomes is initiated by contact with the bone matrix through integrin receptors, in particular, the αvβ3 integrin (vitronectin receptor, VnR), which is highly expressed in OCs (13–16). Attachment-induced activation of the VnR in OCs sequentially activates tyrosine kinase signaling via both Pyk2 and Src, inducing the formation of a complex that recruits the E3 ubiquitin ligase Cbl (15, 17). Deletion of Src leads to osteopetrosis due to the failure of the OCs to resorb bone (18). This defect in Src−/− OCs appears to be linked to defective podosome patterning (15) as they are able to form clusters but fail to expand these clusters into podosome belts (17, 19), a transition that requires the kinase activity of Src (17, 20). Pyk2 is also required for the transition from clusters to the podosome belt and for bone resorption (21, 22), but, in contrast with Src, this appears to be largely independent of Pyk2 catalytic activity (17, 21). Cbl, the third component of the Pyk2-Src-Cbl complex, also promotes podosome belt formation; depleting both c-Cbl and Cbl-b, the two Cbl proteins expressed in OCs, prevents the formation of podosome belts (23).
Several studies pointed to a critical role of microtubules (MTs) in podosome biology and in particular in the expansion of podosome clusters into a belt. Indeed, disruption of the MT network leads to the loss of the podosome belt (8, 24). Pretreating macrophages with nocodazole, a tubulin polymerization inhibitor, inhibits podosome formation (25) although the rate of actin polymerization, measured by fluorescence recovery after photobleaching (FRAP), is not affected by MT depolymerization or stabilization (26). In macrophages and OCs, podosomes partially colocalize with MTs (26–29), and podosomes are targeted by MTs, leading to fusion, fission, or disappearance of the podosome (29). Finally, both Pyk2 and Cbl proteins affect MT acetylation and stability (21, 23).
Despite the detailed molecular information gained from these studies, the mechanism by which MTs are linked to actin polymerization and podosome dynamics is not yet elucidated. For the most part, MT growth occurs at one end, the plus end, which switches continuously between phases of disassembly/shrinkage and growth (catastrophe and recovery). A diverse group of multidomain proteins, the plus-end tracking proteins (+TIPs), regulate MT dynamics both spatially and temporally. The +TIPs accumulate at the growing MT plus ends, binding directly to tubulin or to another +TIP via specific interactions, and dissociate when the MT shrinks (30). The +TIPs also regulate the actin cytoskeleton (31). Among the plus-end proteins, the related +TIPs EB1, EB2, and EB3 have recently emerged as key regulators; they bind directly to tubulin via their N-terminal calponin domains (32) and recruit other +TIP proteins, such as CLIP170, to the MT growing end through their C-terminal domains (33), effectively acting as adaptor proteins mediating the interactions of the other +TIP binding partners at growing MT ends (34, 35).
Since the OC-specific transition from podosome clusters to belt appears to be dependent on MTs (21, 23, 24), we have investigated the mechanisms by which MTs interact with podosomes, in particular, during this transition. We tested the hypothesis that proteins enriched at plus ends could interact with proteins present in podosomes, allowing the physical interaction of podosomes and MTs to control podosome behavior. We show that indeed OCs express various +TIPs and that EB1 has a critical role in podosome belt formation. We identified cortactin as an Src-dependent interacting partner of EB1 in the podosomes and observe that, accordingly, cortactin also plays a role in podosome belt formation. Deletion of cortactin leads to a defect in the formation of peripheral belts, preventing effective bone resorption. Mechanistically, dynamic MT plus ends regulate cortactin phosphorylation by Src, and cortactin phosphorylation and acetylation are inversely and dynamically related. Thus, EB1-decorated MTs interact with podosomes through cortactin, and this is regulated by phosphorylation and acetylation events.
MATERIALS AND METHODS
Animals.
Generation of CTTN−/− mice (36, 37) and c-Src−/− mice (18) was described previously. Animals were handled in accordance with the guidelines of the Harvard University Standing Committee on Animals.
Antibodies and reagents.
The following primary and secondary antibodies were used in this study: mouse anti-α tubulin, mouse anti-acetylated tubulin, rabbit anti-EB1, mouse antivinculin, mouse anti-FLAG (Sigma), mouse anti-EB1, mouse antiactin, mouse anticortactin, rabbit anti-acetyl cortactin, mouse anti-phosphotyrosine 4G10 (Millipore), mouse anti-green fluorescent protein (anti-GFP), rabbit anti-dsRed (Clontech), rabbit anticortactin, rabbit antiphosphocortactin (Tyr421), mouse anti-Src (Cell Signaling), rabbit anti-EB1 (Abcam), rabbit anticortactin (Santa Cruz), rabbit anti-mouse IgG (Invitrogen), goat anti-mouse and anti-rabbit IgG–horseradish peroxidase (HRP) conjugate (Promega), and mouse TrueBlot Ultra Ig HRP (eBioscience). Rhodamine phalloidin was from Invitrogen. Reagents used include nocodazole, paclitaxel (originally named taxol) (Sigma), trichostatin A (TSA; Calbiochem), and tubacin (provided by Ralph Mazitschek and Stuart Schreiber, The Broad Institute and Harvard University). Minimum essential medium with the alpha modification (α-MEM) and fetal bovine serum (FBS) were purchased from Invitrogen.
Plasmids.
The following mammalian expression plasmids have been described previously: actin-enhanced GFP (eGFP) (Clontech), EB1-GFP (plasmid 17234; Addgene), cortactin-eGFP (gift from Marko Kaksonen, EMBL), cortactin-FLAG, and FLAG-tagged cortactin mutants where 9 lysines are replaced by glutamine or arginine (cortactin 9KQ-FLAG or cortactin 9KR-FLAG, respectively) (gift from Yingtao Zhang, University of Florida). Actin-mCherry was generated by cloning the actin coding sequence from actin-eGFP into pmCherry-C1 (Clontech). Cortactin-tdTomato (tdTom) was generated by cloning the cortactin coding sequence from cortactin-eGFP into ptdTomato-N1 (Clontech). EB1C-GFP (amino acids 190 to 269) was generated from EB1-GFP using a BamHI site located between the coding sequence for eGFP and EB1 and by creating an additional BamHI site upstream of amino acid 190, using a site-directed mutagenesis kit (QuikChange II; Agilent Technologies) and digesting the mutant to delete residues 1 to 189 of EB1. EB1Res, an EB1 mutant resistant to siRNA, was generated by silent mutations of at least four nucleotides from the target sequences of EB1 using a site-directed mutagenesis kit (QuikChange II).
siRNA.
Two sets of small interfering RNA (siRNA) duplexes (MISSION; Sigma) against each target gene (mouse EB1) were used. The MISSION siRNA universal negative control, whose sequence has no homology to any mature and predicted mRNA sequences was used as a control. The functionality of the siRNAs was tested by transfecting RAW 264.7 cells using N-TER Nanoparticle siRNA transfection system (Sigma) with a final concentration of siRNAs of 20 nM. Once validated, the siRNAs were microinjected into primary OCs (final concentration, 5 μM in microinjection buffer consisting of 5 mM phosphate sodium buffer and 100 mM KCl) along with actin-GFP (25 ng/μl).
Osteoclast primary cultures, TRAP staining, and pit formation assay.
Mouse osteoclasts (OCs) were generated by two methods. (i) For differentiation and resorption assays, gene expression analysis, microscopy, and all biochemical analyses, murine bone marrow macrophages (BMMs) were cultured on untreated tissue culture plastic (Corning) in α-MEM for 3 days with M-CSF (30 ng/ml), lifted with EDTA (0.5 mM), replated and cultured on FBS-coated glass coverslips, glass bottom dishes (MatTek Corporation) or plastic dishes for an additional 3 to 4 days in a medium with macrophage colony-stimulating factor (M-CSF) (30 ng/ml; R&D Systems), and RANKL (10 ng/ml; R&D Systems). (ii) For differentiation assays and resorption assays, OCs were also generated by coculturing BMMs with primary calvarial osteoblasts in α-MEM containing 10 nM 1,25-dihydroxyvitamin D3 and 1 μM PGE2 on plastic. Staining for TRAP in OCs was performed using a leukocyte acid phosphatase labeling kit (Sigma), according to the manufacturer's instructions. For pit formation assays, OCs generated on dentin slices and cultured for 4 days were fixed and counted following TRAP staining. The dentin slices were then washed, incubated in 1 N ammonium hydroxide for 5 min, and sonicated for 10 s to remove cells. The slices were then stained with rhodamine. Three-dimensional profiles of resorbed pits were generated using the fluorescence mode of a Zeiss LSM 510 confocal microscope (Plan-Neofluar 40×/1.3 numerical aperture [NA] objective lens). Resorbed pit depth was quantified using ImageJ software.
RNA extraction and qPCR.
RNA from primary cells (BMM-derived OCs and osteoblasts) stopped at different time points was isolated using an RNeasy minikit, including on-column RNase-free DNase 1 digestion, according to the manufacturer's instructions. The middiaphysis of the left femora was ground in liquid nitrogen, and this step was followed by RNA extraction using TRIzol reagent (Invitrogen) and RNA cleanup using an RNeasy minikit (Qiagen) according to the manufacturer's protocols. RNA was quantified spectrophotometrically. cDNA was synthesized by reverse transcription using a SuperScript VILO cDNA synthesis kit (Invitrogen). Real-time quantitative PCR (RT-qPCR) expression analysis was performed using a Bio-Rad iCycler iQ and iQ SYBR Green mix (Bio-Rad Laboratories). Aldolase A and 18S were used for normalization. Sets of primers were designed from the online mouse library probes of Roche Diagnostics for specific detection of the following genes: aldolase A, 18S, TRAP, CTSK, CLIP1, CLIP2, APC, CLASP1, CLASP2, MACF1, STIM1, EB1, EB2, and EB3.
Transient transfections.
HEK293VnR cells (HEK293 cells stably expressing the αvβ3 integrin, the major vitronectin receptor) (15) were transiently transfected with cortactin-FLAG, cortactin 9KQ-FLAG, or cortactin 9KR-FLAG using FuGENE HD (Roche Applied Sciences) according to the manufacturer's specifications. Total DNA (5 μg per 100-mm2 tissue culture plate) was kept constant by the addition of empty pcDNA3 plasmid. Transfected cells were maintained in α-MEM containing 10% fetal calf serum (FCS) for 24 or 36 h after transfection, lysed, and used for coimmunoprecipitation (co-IP) assays. Cells treated with epidermal growth factor (EGF; 100 ng/ml) were serum starved for the last 12 h.
Coimmunoprecipitation and immunoblot analysis.
HEK293VnR cells and primary OCs generated from BMMs were lysed in modified radioimmunoprecipitation assay (mRIPA) buffer (50 mM Tris-Cl, 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate) containing protease and phosphatase inhibitors (Complete Mini and Phosphostop; Roche). Lysates were incubated for 20 min at 4°C, and the supernatant was clarified by centrifugation. Cell lysate (500 μg) was precleared by incubation with Dynabeads protein G (Invitrogen) and 1 μg of appropriate control IgG for 30 min. The supernatant was then incubated with 2 μg of primary antibody at 4°C for 2 h, with rotation. Immunoprecipitation was performed using Dynabeads protein G (Invitrogen) for 1 h. Nonspecific proteins were removed by three washes in mRIPA buffer. Samples were boiled in SDS-containing sample buffer under reducing conditions, and proteins were resolved by 4 to 12% Bis-Tris Nu-PAGE gel (Invitrogen) and transferred to Immun-Blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked by incubation with 5% bovine serum albumin (BSA) in Tris-buffered saline–Tween 20 (TBST). Membranes were washed in TBST and incubated with primary antibodies. Membranes were then washed in TBST and incubated with either anti-rabbit or anti-mouse IgG horseradish peroxidase-conjugated secondary antibody diluted in TBST. Membranes were washed, incubated for 1 min with enhanced chemiluminescence (ECL; GE Healthcare), and exposed to film (Kodak).
Microinjection.
Intranuclear and cytoplasmic microinjection of cDNAs (25 ng/μl) and siRNA (5 μM) into OCs plated on glass-bottom dishes or coverslips was performed at room temperature on an inverted microscope (model IX 71; Olympus) equipped with a heated stage and a CO2 chamber using an InjectMan NI2 micromanipulator and a FemtoJet Microinjector (Eppendorf). After microinjection, cells were maintained at 37°C and 5% CO2 for at least 12 h in differentiation medium before imaging.
Time-lapse and confocal microscopy.
The glass-bottom dishes were placed on a 37°C heated stage in a 5% CO2 chamber, and confocal images were collected using 488-nm laser illumination with a Yokogawa CSU-X1 spinning-disk confocal unit (Andor Technology, South Windsor, CT) mounted on a Nikon Ti-E inverted microscope (Nikon Instruments, Melville, NY). Images were acquired using a 60× Plan Apo objective lens with an Orca-ER charge-coupled-device (CCD) camera (Hamamatsu Photonics, Bridgewater, NJ). Acquisition parameters, shutters, filter positions, and focus were controlled by Andor iQ software (Andor Technology, South Windsor, CT). Extracted images from stacks were processed with Image J (http://rsb.info.nih.gov/ij/). At least 25 cells in three to five independent experiments were analyzed for each condition. Comet tracking was performed manually using the tracking function of the MTrackJ plug-in of ImageJ software, following comets appearing in at least three consecutive sections.
For immunofluorescence, cells plated on FBS-coated coverslips were fixed with 3.7% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), pH 7.4, for 10 min. Cells were permeabilized in 0.05% saponin for 30 min. Coverslips for actin labeling were incubated in a 1:40 dilution of rhodamine phalloidin stock solution for 1 h, washed with PBS, and mounted in FluorSave (Calbiochem, San Diego, CA). All coverslips were blocked in 5% normal goat serum (Roche, Indianapolis, IN) for 30 min and incubated in appropriate primary antibodies, washed, incubated with fluorescent secondary antibody (Alexa Fluor 488, green; Alexa Fluor 568, red; Alexa Fluor 647, blue), washed again, and mounted in FluorSave. Nuclei were labeled with TO-PRO-3 (1:,1000) in the secondary antibody solution. Fixed cells were imaged with a Zeiss LSM 510 laser scanning confocal microscope (software AIM, version 4.2) using a Zeiss Plan-Neofluar 40×/1.3 NA objective. To ensure that only one fluorochrome was detected at any one time, each channel was imaged sequentially using a multitrack recording module before images were merged, using the same settings to allow comparisons. For actin belt analysis, the fluorescence intensity of rhodamine phalloidin-stained F-actin podosomes and associated actin clouds was quantified using the ImageJ Plot Profile plug-in program, which expresses pixel intensity as height. For all fixed-cell experiments, all coverslips were stored in PBS and imaged the following day after fixation.
Statistical analysis.
Statistical differences were assessed with an unpaired Student's t test. Calculations were performed using StatView, version 4.1, statistic analysis software (Statistical Analysis System Institute). Data are presented as means ± standard deviations (SD), and P values smaller than 0.05 were considered statistically significant.
RESULTS
EB1 expression increases in mature osteoclasts, and EB1 localizes around the podosomes.
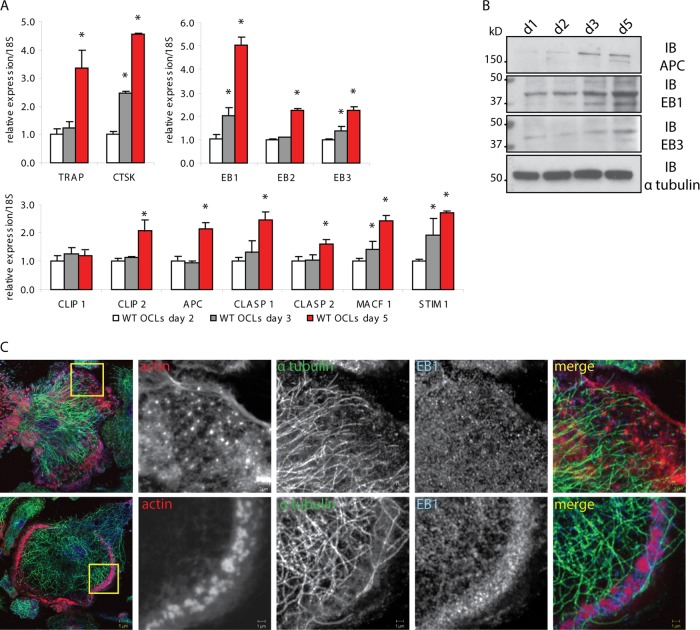
To identify the +TIPs involved in OCs, we first analyzed by real-time quantitative PCR (RT-qPCR) the expression levels of several +TIPs, including EB1, EB2, EB3, CLIP1, CLIP2, APC, CLASP1, CLASP2, MACF1, and STIM1, during OC differentiation. In wild-type (WT) cell cultures, most cells are mononucleated at day 2, multinucleated with clusters of podosomes at day 3, and multinucleated with podosome belts at day 5. As expected, the expression of TRAP and cathepsin K, two OC differentiation markers, increased 3- and 4-fold, respectively, during this period of time. Of all the +TIPs tested, the expression of EB1 stood out with a 5-fold increase by day 5 while most of the others increased only approximately 2-fold during OC differentiation (Fig. 1A). These findings were confirmed at the protein level by Western blot analysis of APC, EB1, and EB3 (Fig. 1B).
FIG 1.
EB1 expression increases in mature osteoclasts and localizes around the podosomes. (A) The expression of OC differentiation markers and +TIPs during OC differentiation was measured by RT-qPCR in WT OCs, generated from bone marrow macrophages cultured with M-CSF and RANKL and stopped at the indicated time points. TRAP and cathepsin K (CTSK) were used as OC differentiation markers. The expression of several +TIPs, including EB1, increased during the differentiation in mature cells. (B) Expression of several +TIPs was detected by immunoblotting (IB) using WT OC lysates and specific antibodies against APC, EB1, and EB3. α-Tubulin was used to control equal loading. d, day. (C) Immunolocalization of actin, α-tubulin, and EB1 was analyzed by confocal microscopy in WT OCs fixed at day 3 (upper row) and day 5 (lower row). In differentiating OCs with clusters of individual podosomes, EB1 was present on MTs and frequently accumulated around individual podosomes. In mature OCs with podosome belts, EB1 colocalized with densely packed podosomes in the belt. Data are presented as means ± SD. *, P < 0.05, significant difference from the control (day 2). OCL, osteoclast lysate.
EB family members, including EB1, accumulate autonomously at growing MT ends (38) and regulate dynamic +TIP interaction networks (35). We thus examined the cellular localization of EB1 in OCs, comparing differentiating OCs with clusters of podosomes and mature OCs with a podosome belt. As expected, EB1 labeling appeared as dots or dashes, frequently colocalizing with α-tubulin and at the junction between MTs and actin in podosomes (Fig. 1C). EB1 frequently localized around the cores of individual podosomes both in clusters (day 3) and at the podosome belt (day 5), which was highly enriched in EB1. Taken together, these results suggested a functional role for EB1 in podosomes in OCs. Furthermore, live-cell imaging showed that EB1-positive MT tracks are targeted to the belt and stop growing when they reach podosomes (see Fig. S1 and Movies S1A and B in the supplemental material).
Depletion of EB1 reduces podosome belt formation.
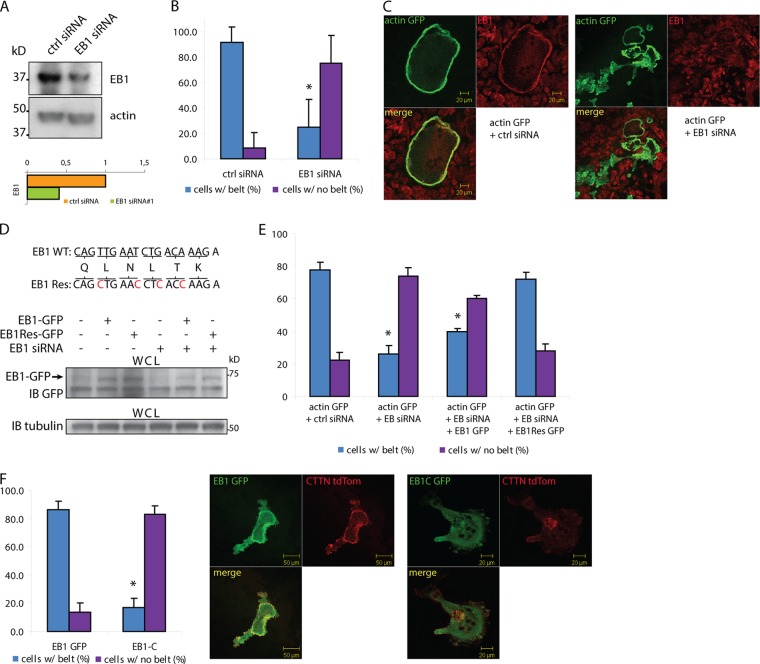
These findings suggested a role for EB1 in the formation and/or stability of the podosome belt. To explore this possibility, we analyzed the effect of EB1 depletion on podosome patterning. We first used siRNAs delivered directly to the cytoplasm via microinjection (39). The functionality of several EB1 siRNAs was first tested and validated in the murine macrophage cell line RAW264.7, transfected either with the targeted siRNA or with a universal negative control. The most efficient EB1 siRNA induced a 60% knockdown of EB1 protein (Fig. 2A). The selected siRNA was then microinjected along with an actin-GFP expression vector in differentiating OCs, prior to podosome belt formation. EB1 siRNA decreased markedly the percentage of actin-GFP-positive cells with belts after 24 h (Fig. 2B and C). Immunostaining of EB1 confirmed the depletion in injected cells (Fig. 2C). These results were confirmed using a second set of EB1 siRNAs that target a different sequence of EB1 mRNA (data not shown). To exclude a possible off-target effect of EB1 siRNA, we designed an EB1 mutant with silent mutations in the siRNA-targeted sequence (EB1Res). Resistance of this EB1 mutant to siRNA was first validated in RAW264.7 cells (Fig. 2D); the siRNA partially lost its efficacy in the presence of EB1Res. When the EB1Res expression vector was microinjected along with EB1 siRNA and actin-GFP, it abrogated the effect of the siRNA on podosome belt formation, in contrast with WT EB1 that only weakly increased the percentage of microinjected cells with belts (Fig. 2E).
FIG 2.
Depletion of EB1 strongly reduces podosome belt formation. (A) The effect of EB1 deletion on podosome belt formation was investigated by knocking down EB1 using siRNA. The functionality of the siRNA was first tested by transfecting RAW264.7 cells, and EB1 expression levels were assessed by Western blotting after 24 h. (B) The siRNA was microinjected along with a plasmid encoding actin-GFP in differentiating OCs that do not have a podosome belt. MISSION siRNA universal negative control was used as a control. The cells were allowed to differentiate for 24 h and fixed. The number of actin-GFP cells with podosome belts was quantified. More than 65 GFP-positive cells were counted for the presence or absence of podosome belts. (C) OCs were microinjected as described for panel B, fixed after 24 h, and stained for EB1 to assess the reduced expression of EB1 in cells microinjected with EB1 siRNA. (D) A mutant EB1-GFP was designed (EB1Res-GFP) that bears silent point mutations in the siRNA target sequence, preventing siRNA binding to the mRNA. Resistance of EB1Res to the siRNA was confirmed following transfection in RAW264.7 cells (compare lanes 2 and 5 with lanes 3 and 6). WCL, whole-cell lysate. (E) Microinjection of EB1Res-GFP along with EB1 siRNA strongly reduced the siRNA effect, whereas the siRNA still affected podosome belt formation when injected with EB1-GFP. More than 200 GFP-positive cells were counted for the presence or absence of a podosome belt. (F) OCs were microinjected with either EB1-GFP or EB1C-GFP, a mutant construct that lacks the N-terminal half necessary for MT binding. The number of actin-GFP cells with a podosome belt was counted 24 h after microinjection. EB1C-GFP induced a strong reduction in podosome belt formation. More than 65 GFP-positive cells were counted for the presence or absence of a podosome belt. Data are presented as means ± SD. *, P < 0.05, significant difference from control.
We further explored the role of EB1 in podosome belt formation by expressing another EB1 mutant (EB1C) that lacks the N-terminal half necessary for MT binding but still retains its ability to homo- and heterodimerize via the C-terminal half with endogenous EB family members, acting as a dominant negative mutant (40). Microinjection of an EB1C expression vector into differentiating OCs reduced significantly the percentage of cells with belts (Fig. 2F). Thus, EB1 is required for the formation and/or stabilization of the podosome belt in OCs.
Microtubule dynamic instability and the presence of EB1 at microtubule plus ends are critical for the podosome belt.
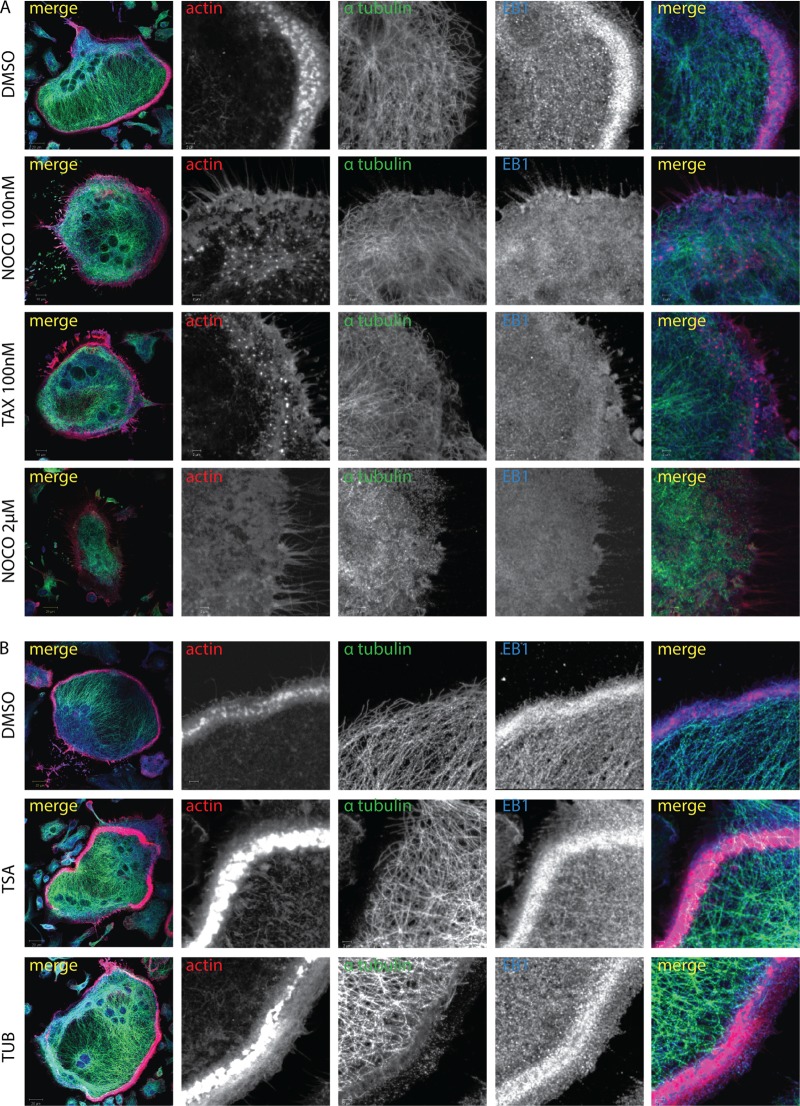
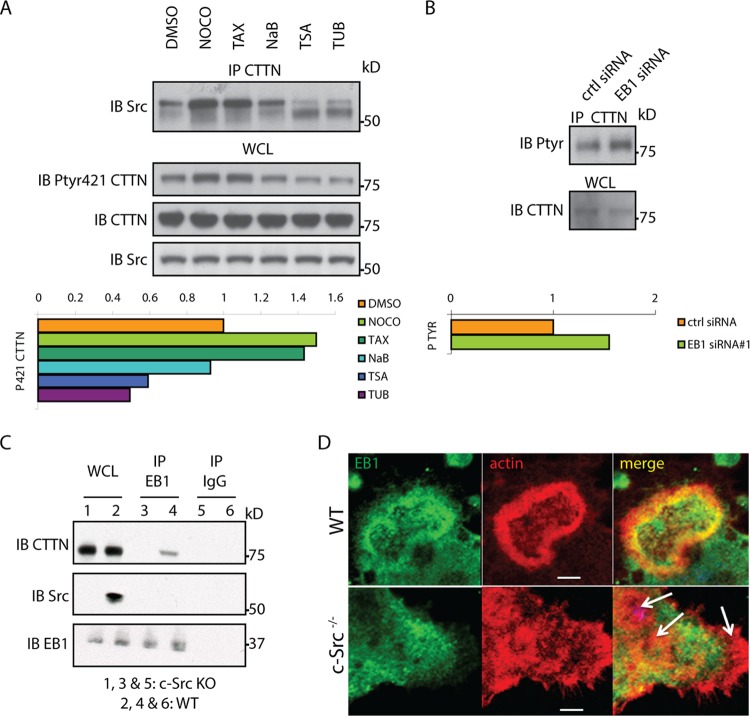
EB1 functions at plus ends to maintain MT dynamic instability. To determine if the effect of EB1 depletion on the podosome belt was related to this function, we used low concentrations of nocodazole or paclitaxel and short treatment periods to block MT dynamic instability. This protocol affects the switch between MT growth and shrinkage, reducing rates of growth and shortening and decreasing the time spent in growth. This results in the loss of +TIPs from MTs, without disrupting the MT network or changing the total content of +TIPs in the cell (41–44). Mature OCs with podosome belts were treated with dimethyl sulfoxide (DMSO), nocodazole (100 nM, 30 min), or paclitaxel (100 nM, 30 min). As expected, treatment with nocodazole or paclitaxel eliminated EB1-positive dots while not inducing the collapse of the entire MT network (Fig. 3A), in contrast with a higher concentration of nocodazole (2 μM, 30 min). Both nocodazole and paclitaxel suppressed the accumulation of EB1 in the belt and reduced the number of podosomes present at the cell periphery, resulting in the disappearance of the belt (Fig. 3A; see also Fig. S2A in the supplemental material).
FIG 3.
Suppression of microtubule dynamic instability and displacement of EB1 from microtubule ends disrupts the podosome belt. (A) Mature OCs generated from WT BMMs with RANKL/M-CSF were treated with DMSO, nocodazole (Noco; 100 nM or 2 μM for 30 min), or paclitaxel (Tax; 100 nM for 30 min) and fixed, and the immunolocalization of actin, tubulin (Tub), and EB1 was analyzed by confocal microscopy. Treatments with low doses of nocodazole (100 nM) and paclitaxel induced the loss of the podosome belt and EB1 concentration at the belt without inducing the collapse of the entire MT network, in contrast to results with a high dose of nocodazole (2 μM). (B) Mature OCs were generated as described for panel A and treated with DMSO, TSA (500 nM, 1 h), or tubacin (2 μM, 1 h). Inhibition of HDAC6 induced a wider and denser podosome belt, where EB1 accumulated.
Histone deacetylase 6 (HDAC6) inhibition can also reduce the velocity of both the growth and shortening of the MTs, similar to low concentrations of nocodazole or paclitaxel, but without affecting the duration spent in growth and, therefore, without altering the presence of EB1 on dynamic MTs (45). Highly acetylated MTs observed after HDAC6 inhibition exhibit delayed drug-induced depolymerization, suggesting that α-tubulin acetylation enhances the stability of MTs (44, 46). We therefore used HDAC6 inhibition to test whether the presence or absence of EB1 at the MT plus end affected podosome organization. Mature OCs were treated with two different inhibitors of HDAC6: the class I and II inhibitor trichostatin A (TSA) (44–46) or tubacin, a more specific inhibitor of HDAC6 (45, 47). DMSO or sodium butyrate (NaB), a deacetylase inhibitor that does not affect α-tubulin acetylation (48), was used as a control. Immunostaining and immunoblotting confirmed that both inhibitors increased the amount of acetylated α-tubulin (see Fig. S2B and C in the supplemental material). HDAC6 inhibition maintained EB1 at the plus end of MTs and induced an accumulation of podosomes in the belt (Fig. 3B; see also Fig. S2D). Altogether, these experiments demonstrate that MT dynamic instability plays a critical role in the regulation of the podosome belt and that EB1 functions as a +TIP critical for podosome patterning. Since EB1 can localize to the podosomes (Fig. 1C), we next determined which podosome protein(s) interacts with EB1 and establishes the link with dynamic MTs.
EB1 and cortactin form a molecular complex in osteoclasts, and deletion of cortactin alters the dynamics of podosomes and bone resorption.
In neurons, EB3, a homolog of EB1 (49), regulates dendritic spine morphology and actin polymerization, forming a molecular complex with the actin-regulatory protein cortactin and the adaptor p140Cap (43). Cortactin is enriched in the podosome core and is involved in podosome formation (50, 51), making it a candidate for EB1 interaction in OCs. EB1 and cortactin are both highly expressed in mature OCs and are frequently colocalized, with EB1 preferentially localized around the core of podosomes where cortactin is enriched (Fig. 4A). Live-cell imaging of OCs microinjected with expression vectors for EB1-GFP and cortactin-tdTomato confirmed that a subset of EB1-positive tracks targeted cortactin-positive podosome cores (see Movie S2 in the supplemental material).
FIG 4.
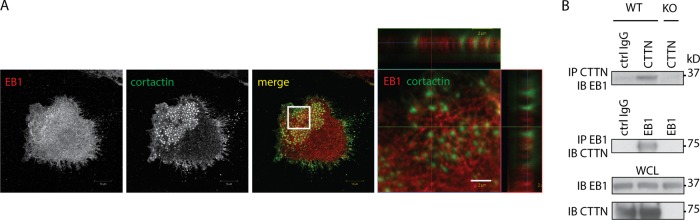
EB1 and cortactin form a molecular complex in osteoclasts. (A) Colocalization of cortactin with EB1 in podosome clusters from WT OCs and an x-z series image of individual podosomes. (B) Co-IP experiments performed using lysates of OCs derived from BMMs of WT mice showed an interaction between EB1 and cortactin. The same experiment using irrelevant antibody or lysates from CTTN−/− OCs did not show any interaction.
Furthermore, coimmunoprecipitation of endogenous proteins from WT OCs confirmed the presence of EB1 and cortactin in the same molecular complexes (Fig. 4B). EB1 was detected in anticortactin immunoprecipitates of WT but not from cortactin knockout (KO; CTTN−/−) (36, 37) OC lysates, demonstrating that the presence of EB1 in the immunoprecipitate is due to its interaction with cortactin and not to a nonspecific interaction with the antibodies.
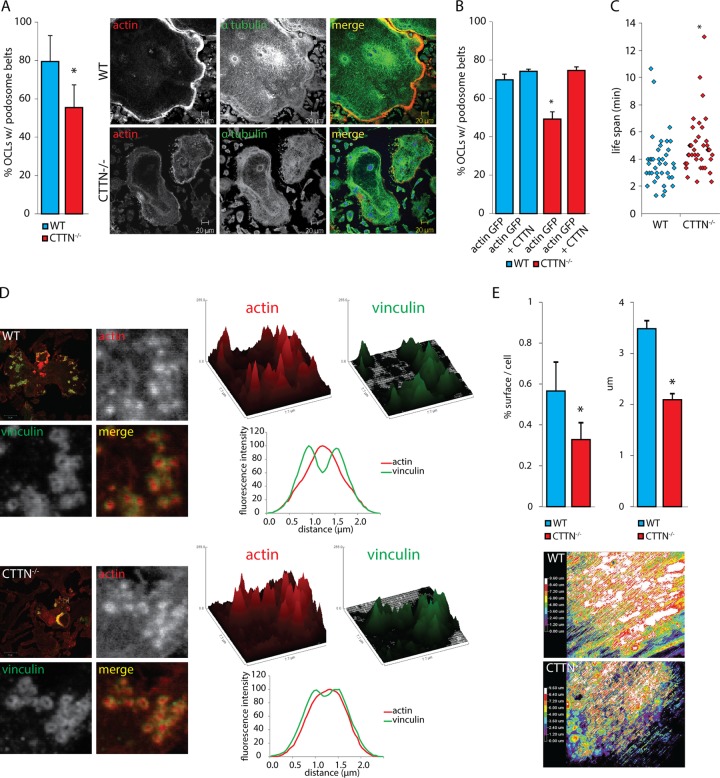
To assess the role of cortactin in podosome dynamics, we used cortactin-deficient OCs generated from the bone marrow of cortactin knockout mice. Cortactin is highly expressed in mature OCs (52, 53; also our unpublished observations), and we confirmed its deletion in CTTN−/− OCs (see Fig. S3A in the supplemental material). Cortactin deletion impaired belt formation (Fig. 5A; see also Fig. S3B), and this phenotype was rescued by microinjecting cortactin-tdTomato along with actin-GFP plasmids (Fig. 5B). Deletion of cortactin also affected podosome dynamics and the podosome life span, which increased by 30% (P ≤ 0.05) in the absence of cortactin (Fig. 5C). Furthermore, the architecture of single podosomes was also altered in the absence of cortactin, showing an enlargement of the actin core and a disorganization of the vinculin cloud (Fig. 5D).
FIG 5.
Cortactin deletion alters the dynamics of podosomes. (A) Mature OCs (day 5) generated from BMMs of WT and CTTN−/− mice were fixed and stained for actin and α-tubulin, and the number of cells with a podosome belt was counted. More than 500 OCs per genotype were counted for the presence or absence of a podosome belt. (B) Differentiating OCs (day 4) generated from BMMs of WT and CTTN−/− mice were microinjected with actin-GFP alone or together with CTTN-tdTomato. After 24 h, cells were fixed, and the number of actin-GFP-positive OCs with a podosome belt was counted. More than 200 GFP-positive cells were counted. (C) Differentiating WT and CTTN−/− OCs were microinjected with expression vector for GFP-actin and observed by time-lapse microscopy. Individual podosomes in clusters were followed, and their life span (the overall time in which a fluorescently labeled podosome exists) was calculated and plotted. Eighty individual podosomes were analyzed. (D) WT and CTTN−/− OCs were fixed and stained for vinculin (green) and F-actin (red). Analysis of individual podosomes by processing the actin and vinculin images with the a three-dimensional surface plot plug-in revealed a larger actin cloud and disorganized vinculin ring. Fluorescence intensity profiles for actin and vinculin in individual podosomes from WT and CTTN−/− OCs were calculated from measurements of over 200 podosomes. (E) The average resorbed area was measured after toluidine blue staining of dentin slices plated with WT and CTTN−/− OCs and was normalized by the number of OCs on the slices, identified by TRAP staining. Pit depth was measured by confocal microscopy. OCs were plated on dentin slices and allowed to resorb for 12 h. OCs were then removed from the slices that were stained with rhodamine. A z series was acquired with a confocal microscope, and color-coded images of the depth of the pits were generated with ImageJ. Data are presented as means ± SD. *, P < 0.05, significant difference from control.
Since defects in podosome patterning or signaling often lead to a decrease in OC activity (54, 55), we next determined whether CTTN−/− OCs were impaired in their ability to resorb bone in vitro. The average resorbed area produced on dentin slices per individual OC was reduced in the absence of cortactin, and the average pit depth showed a similar pattern (Fig. 5E). Thus, in the absence of cortactin, OCs show a cell-autonomous decrease in bone-resorbing activity. In addition, the ability of CTTN−/− bone marrow macrophages (BMMs) to form multinucleated OCs was reduced (see Fig. S3C in the supplemental material), possibly also as a consequence of podosome disruption.
Microtubule targeting of podosomes is altered in CTTN−/− osteoclasts.
Since EB1 and cortactin interact and since both play a role in podosome patterning, this interaction may contribute to the connection between MTs and podosomes. To answer this question, we compared the length, duration, and speed of EB1-positive tracks in WT and CTTN−/− OCs following expression of actin-mCherry and EB1-GFP. Length and speed of growing MTs were increased when cortactin was absent (Fig. 6A; see also Movies S3A and B in the supplemental material). Since MT growth stops when MTs reach podosomes, these results suggest that the targeting of EB1-decorated MT plus ends to podosomes was altered in the absence of cortactin. We reasoned that if this was true, then the interaction of EB1 with other components of the podosome could also be disturbed in the absence of cortactin. We found that vinculin, another protein enriched in podosomes, was present in EB1 immunoprecipitates and that this interaction was also decreased after treatment with low doses of nocodazole (Fig. 6B). Interestingly, the interaction between EB1 and vinculin was also reduced in the absence of cortactin, supporting the idea that deletion of cortactin reduces the targeting of podosomes by EB1-decorated MTs.
FIG 6.
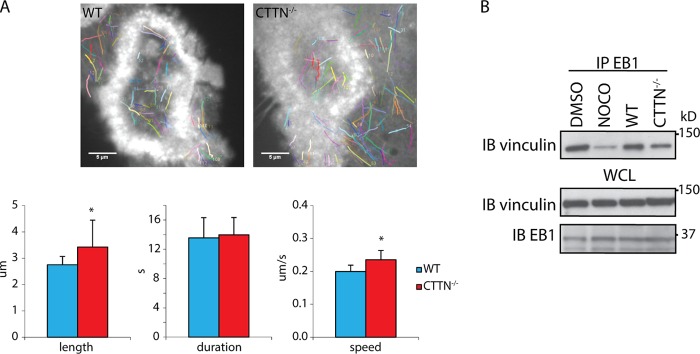
Microtubule targeting of podosomes is altered in CTTN−/− osteoclasts. (A) GFP-positive structures were manually tracked with the ImageJ plug-in MtrackJ in WT and CTTN−/− OCs microinjected with GFP-EB1 and mCherry-actin. The duration, length, and speed of the EB1-positive tracks were calculated. A total of 10 cells were analyzed for MT dynamics. (B) EB1 interaction with vinculin, a marker of podosomes, was assessed by co-IP in WT OCs treated with DMSO or nocodazole (100 nm, 30 min) and in nontreated CTTN−/− OCs.
Microtubules and EB1 regulate cortactin phosphorylation.
Cortactin's activity is controlled in part by phosphorylation, including its ability to promote actin polymerization (56) and podosome belt formation (10). Having shown that cortactin interacts with EB1, we then asked whether alterations in MT stability could regulate cortactin phosphorylation status. Suppression of dynamic instability of MTs with nocodazole or paclitaxel led to an increase in the interaction of cortactin with c-Src and in the phosphorylation of cortactin at tyrosine 421 (Fig. 7A) (57). In contrast, cortactin phosphorylation and its interaction with c-Src were decreased when MTs were stabilized following HDAC6 inhibition with TSA or tubacin (Fig. 7A). Thus, the Src-dependent phosphorylation of cortactin is at least in part dependent upon MT stability. EB1 could therefore be involved in the regulation of cortactin phosphorylation. Because enough material for biochemical analysis cannot be obtained from microinjected OCs, to test this possibility, we used RAW264.7 cells, a cell line that can be induced to differentiate into OCs by RANKL, and transfected them with EB1 siRNA. Tyrosine phosphorylation of cortactin was increased in EB1-depleted cells (Fig. 7B), similar to the changes observed after disruption of dynamic MTs. Thus, the tyrosine phosphorylation of cortactin is in part downregulated by EB1 and MT dynamic instability. Interestingly, the interaction between EB1 and cortactin is Src dependent, and deletion of c-Src prevented the interaction, as shown by co-IP experiments using WT and c-Src KO OCs (Fig. 7C). In agreement with this finding, confocal microscopy demonstrated that in contrast to WT OCs, EB1 fails to efficiently colocalize with podosomes in Src KO OCs, where podosomes remain in clusters and cannot form peripheral belts (15) (Fig. 7D). Thus, phosphorylation of cortactin by Src is required for EB1-cortactin interaction, and EB1 and MT instability decrease the phosphorylation of cortactin. As discussed below, this may be linked to an MT-dependent switch to acetylation of cortactin.
FIG 7.
Microtubules and EB1 regulate cortactin phosphorylation. (A) WT OCs with podosome belts were treated as described previously with DMSO, nocodazole, paclitaxel, TSA, or tubacin, and the level of cortactin phosphorylation was measured by Western blotting with a specific anticortactin phospho-Tyr421 antibody. The membrane was then stripped and reprobed with an anticortactin antibody. The ratio of phosphorylated to total cortactin was determined by densitometry using ImageJ. Cortactin interaction with Src was detected by co-IP in the same lysates. (B) Cortactin phosphorylation was measured in RAW264.7 cells following EB1 depletion by siRNA by immunoprecipitating cortactin and probing with a pan-phosphotyrosine antibody (4G10). The ratio of phosphorylated to total cortactin was determined by densitometry using ImageJ. (C) WT osteoclasts or c-Src-KO OCs were lysed at day 4, and equal amounts of proteins were immunoprecipitated with either IgG (control) or EB1 antibody. The immunoprecipitated protein was blotted with CTTN, c-Src, and EB1 antibodies. (D) OCs from WT (upper panel) and c-Src−/− (lower panel) mice were generated and cultured on dentin. At day 5, the cells were fixed and stained for EB1 and actin. Arrows indicate clusters of podosomes in c-Src KO OCs. Scale bar, 5 μm.
Alternate and dynamic acetylation/phosphorylation of cortactin is required for podosome belt formation.
Cortactin is also a substrate of HDAC6, and its acetylation affects its activity, including its actin binding ability (58–61). The amount of acetylated cortactin in OCs was increased after treatment with the HDAC6 inhibitor TSA or tubacin (see Fig. S4A in the supplemental material), indicating that cortactin acetylation does occur endogenously in OCs. Cross-regulation between acetylation and other posttranslational modifications, including phosphorylation, occurs not only for histones but also for cytoplasmic proteins (62), and it has been suggested that lysine acetylation of cortactin could compete with phosphorylation of selected tyrosines (58, 63). We therefore determined whether cortactin acetylation affects cortactin phosphorylation. To test this possibility, we used HEK293VnR cells, a model we have extensively used to verify phosphorylation events and in which EGF induces a robust induction of cortactin phosphorylation (64, 65), and two cortactin mutants where 9 lysines are replaced by glutamine to mimic the acetylated state (cortactin 9KQ) or by arginine to mimic the nonacetylated state (cortactin 9KR) (58). HEK293VnR cells transfected with WT cortactin or the cortactin mutant 9KQ or 9KR and treated with EGF (100 ng/ml, 5 min) showed that the cortactin mutant that mimics the acetylated state (9KQ) was hypophosphorylated compared to WT cortactin, and, in contrast, the mutant that mimics the nonacetylated state (9KR) was hyperphosphorylated (see Fig. S4D in the supplemental material). Most importantly, the interaction of cortactin with EB1 was modified by the acetylation/phosphorylation status of cortactin: EGF-induced phosphorylation of WT cortactin or cortactin 9KR decreased markedly the interaction, whereas the cortactin acetylated-state mutant (9KQ) showed an increased interaction with EB1.
Thus, there is an inverse relationship between the Src-dependent phosphorylation and the HDAC6-dependent acetylation status of cortactin. Since we have shown that this balance is regulated in part by the instability of MTs and determines the level of interaction of EB1 with cortactin, this balance may regulate the interaction between MTs and podosomes and the formation and/or stability of the podosome belt in OCs.
DISCUSSION
MTs play a central role in processes essential for OC activity and bone resorption: polarization of the cells (66), vesicular traffic (67–72), motility (68), and podosome belt formation (21, 23, 24, 70, 73–75). Formation of the podosome belt has been the focus of this study because if other cell types use podosomes/invadosomes to migrate, adhere, and degrade the extracellular matrix (7), OCs are the only cells where podosomes follow a precise patterning such that clusters form rings that fuse and expand to form a characteristic peripheral belt (6, 7, 12), establishing the sealing zone when OCs are plated on bone or dentin (11). Our understanding of the molecular mechanisms responsible for this specificity is limited, and little is known about the molecular interactions between MTs and podosomes in OCs. Here, we show that MT plus ends and their associated +TIPs are essential regulators of podosome patterning in OCs.
Radial MTs are oriented toward the cell periphery where podosomes form the belt (68, 76), and growing MTs target podosomes (29) and invadopodia (77). We found that a subset of growing MTs targets podosomes. The direction of growing MTs was skewed toward podosomes, and MT growth stopped when they reached individual podosomes. Suppression of MT growth with low doses of nocodazole or paclitaxel rapidly disrupted the podosome belt. However, and in contrast with the effects of complete suppression of the entire MT network by high doses of these agents, podosomes were still present. Thus, the interaction between growing MTs and podosomes is not required for the formation or maintenance of podosomes but for their patterning to form and/or stabilize the belt. This suggested that molecular events at the plus ends of MTs, including dynamic instability, were crucial to establish this connection.
We found that the presence of EB1 at the growing ends of MTs is required for podosome belt formation. First, analysis of +TIPs during OC differentiation showed a marked increase in EB1 expression, and EB1 accumulates in the belt and around individual podosomes. Depletion of EB1 in OCs prevented podosome belt formation, similar to the effects of preventing MT growth. EB1 selectively binds to growing MT plus ends (41), where it forms a platform that recruits several other +TIPs, establishing the link with actin or membranes (33). EB1 is consequently involved in the majority of MT-dependent processes. Several molecular mechanisms are responsible for these properties, including a direct autonomous accumulation at growing MT ends through the N-terminal domain of EB1 and interaction with other +TIPs via its C-terminal domain (33–35, 40, 78). Consequently, overexpression of the EB1 C-terminal domain mutant suppresses EB1 binding to MTs while maintaining its interaction with other +TIPs (40, 79). The fact that we observe similar alterations of the podosome belt after MT growth inhibition, which displaces EB1 from MTs, after depletion of EB1, or after overexpression of EB1C confirms that the patterning of podosomes into a belt in OCs is dependent upon interactions of MT plus ends with EB1 and the subsequent recruitment of other +TIPs. Of note, EB1C also displaces other +TIPs from growing MTs and sequesters other EB family members through dimer formation (40). This additional perturbation could explain the strong effect of EB1C on podosome belt formation compared to siRNA-mediated depletion of EB1 alone.
The next question was to identify the component(s) of the podosome and actin-regulatory machinery which directly or indirectly interacts with EB1 to establish the link between MTs and podosomes. In neuronal dendritic spines, dynamic MTs regulate the actin cytoskeleton via a molecular complex formed of EB3, p140Cap, and cortactin (43). We assessed the expression of p140Cap in OCs but could not unequivocally detect its mRNA or protein. In contrast, cortactin is highly expressed in mature OCs (80) and localizes to individual podosomes and in the podosome belt (8, 10, 52, 53, 81, 82). In OCs, EB1 and cortactin belong to a similar molecular complex, as shown by immunoprecipitation, even though a direct interaction between the proteins cannot be ascertained. Furthermore, cortactin is a prominent Src substrate (83), and Src activity is essential in OCs to ensure proper podosome dynamics and patterning (15, 17, 20), possibly in part due to changes in cortactin phosphorylation.
Cortactin is present in podosomes and invadopodia and plays a central role in their formation and function (84). siRNA-mediated knockdown of cortactin in fibroblasts (85), smooth muscle cells (86), tumor cells (51, 87–90), or OCs (52, 53) prevents the formation of podosome and invadopodia. To determine which role cortactin plays in podosome patterning and belt formation, we used OCs derived from cortactin knockout mice (36). In contrast to previous reports, we found that cortactin is dispensable for podosome formation in OCs. In contrast, it is important for podosome patterning and belt formation, similar to EB1, and regulates podosome life span. These findings suggest that cortactin functions as a scaffolding protein linking and integrating the podosome and MT machineries. This hypothesis is further supported by our finding of reduced interaction of vinculin with EB1 and altered MT dynamics in the absence of cortactin. Furthermore, the track length of MTs is increased in the absence of cortactin, indicating a defect in MT targeting to podosomes, which could explain the increased life span of podosomes in the absence of cortactin (29). The detection of podosomes after 20 min of dynamic instability suppression, when the average podosome life span is under 5 min (8, 20, 21; this study), supports the view that defective targeting decreases the rate at which podosomes disappear. Taken together these data suggest that the targeting of the plus ends of MTs to podosomes is dependent upon the presence of EB1 at the growing MT end and of cortactin in the podosome.
How is this interaction regulating the life span and patterning of podosomes? Cortactin phosphorylation is involved in the regulation of podosome and invadopodium dynamics (10) and is correlated with the invasiveness of invadopodia (91). Stepwise models detailing the events involved in invadopodium formation and stability reveal that cortactin serves as an adaptor to form a complex with N-WASP, cofilin, and Arp2/3 (51, 87). We show here that tyrosine phosphorylation of cortactin is regulated by growing MTs, thus influencing the stability and maturation of podosomes. When MT instability was suppressed, it led to the loss of EB1 at the plus ends of MTs and to increased interaction of Src with cortactin, resulting in increased phosphorylation. Similarly, depletion of EB1 led to the disruption of the interaction between MTs and podosomes and an increase in cortactin phosphorylation. Interestingly, cortactin is highly phosphorylated in clustered podosomes while podosome belt-associated cortactin is not, and overexpression of constitutively active Src results in abnormal podosome belt formation (10). Furthermore, c-Src-deficient OCs are incapable of organizing podosomes into a peripheral belt (15, 20), and we show here that the interaction between cortactin and EB1 and the association of EB1 with podosomes are lost in these cells. These findings illustrate the fact that phosphorylation of cortactin through Src activity and its subsequent EB1-dependent dephosphorylation need to be tightly regulated during the life cycle of podosomes, possibly explaining the need of nonactive Src in v-Src transformed cells to form invadopodia (92). Taken together, these results indicate that EB1-decorated MT plus ends target and interact with podosomes through a cortactin-dependent mechanism and that this interaction results in a decrease in Src-dependent tyrosine phosphorylation of cortactin, an event expected to stabilize podosomes (51).
In contrast to suppression of MT dynamic instability, which increased cortactin phosphorylation, HDAC6 inhibition decreased the phosphorylation of cortactin, leading to an increase in the actin content in the podosome belt. Thus, increased acetylation of substrates in the OC led to inhibition of cortactin phosphorylation and promoted podosome patterning. HDAC6 most likely deacetylates numerous substrates in OCs (93), with α-tubulin among them, and increased MT acetylation is associated with podosome belt stabilization, through mDia2 and HDAC6 (24). Deletion of Pyk2 (21), Cbl (23), Capn6 (73), or Akt (75) leads to decreased tubulin acetylation and absence of the podosome belt. Despite all of these concordant observations indicating a link between tubulin acetylation and the podosome belt, whether tubulin acetylation is responsible for these podosome alterations or merely a consequence of changes in the acetylation of other substrates is unclear. Most relevant to this paper is the fact that cortactin can be acetylated at nine different lysines by PCAF and ATAT1 (94), inhibiting binding to F-actin. Deacetylation of these same lysines by HDAC6 reverses this process (58). Using HEK293VnR cells, a model system we have used extensively to dissect molecular signaling events linked to OC adhesion (15, 21, 23, 55, 95), we observed (i) that phosphorylation and acetylation of cortactin are inversely and dynamically related and (ii) that acetylated (and thereby dephosphorylated) cortactin exhibits enhanced association with EB1. In OCs, both increased acetylation and decreased phosphorylation of cortactin are associated with stabilization of the podosome belt. Conversely, when the belt was disrupted following suppression of MT instability, cortactin acetylation was markedly decreased, and phosphorylation increased. Thus, tyrosine phosphorylation and lysine acetylation of cortactin in OCs are inversely and dynamically related, as recently reported in fibroblasts (63), and reflect two functional states of cortactin. These findings suggest that events that modulate cortactin phosphorylation and/or acetylation could determine how podosomes are linked to and respond to MTs. This may also apply to invadopodium-mediated invasiveness of tumor cells, as silencing of either HDAC6 (61) or ATAT1 (94) impair the cell's invasive ability.
In conclusion, our data support a model where podosome patterning is dependent upon (i) the Src-dependent interaction between EB1 at the MT plus ends and cortactin within the podosomes and (ii) the dynamic instability of MTs and the posttranslational status of cortactin, where cortactin phosphorylation by Src and its deacetylation by HDAC6 favor interaction with actin, while cortactin acetylation favors interaction with EB1-decorated MTs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH, NIAMS (AR054450 and AR062054 to R.B.), and the Deutsche Forschungsgemeinschaft (RO2414/1-2 to K.R.) and fellowships from the Fondation Bettencourt-Schueller and the Fondation pour la Recherche Médicale (to M.B.D.).
We are very grateful to Francesca Gori for her help with this paper.
Footnotes
Published ahead of print 21 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00578-13.
REFERENCES
- 1.Baron R, Neff L, Brown W, Courtoy PJ, Louvard D, Farquhar MG. 1988. Polarized secretion of lysosomal enzymes: co-distribution of cation-independent mannose-6-phosphate receptors and lysosomal enzymes along the osteoclast exocytic pathway. J. Cell Biol. 106:1863–1872. 10.1083/jcb.106.6.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teitelbaum SL, Ross FP. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638–649. 10.1038/nrg1122 [DOI] [PubMed] [Google Scholar]
- 3.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. 1998. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 95:13453–13458. 10.1073/pnas.95.23.13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder S, Aepfelbacher M. 2003. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 13:376–385. 10.1016/S0962-8924(03)00128-4 [DOI] [PubMed] [Google Scholar]
- 5.Linder S, Kopp P. 2005. Podosomes at a glance. J. Cell Sci. 118:2079–2082. 10.1242/jcs.02390 [DOI] [PubMed] [Google Scholar]
- 6.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. 2009. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122:3037–3049. 10.1242/jcs.052704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy DA, Courtneidge SA. 2011. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. 2003. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell 14:407–416. 10.1091/mbc.E02-07-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochoa GC, Slepnev VI, Neff L, Ringstad N, Takei K, Daniell L, Kim W, Cao H, McNiven M, Baron R, De Camilli P. 2000. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150:377–389. 10.1083/jcb.150.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luxenburg C, Parsons JT, Addadi L, Geiger B. 2006. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J. Cell Sci. 119:4878–4888. 10.1242/jcs.03271 [DOI] [PubMed] [Google Scholar]
- 11.Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. 2007. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One 2:e179. 10.1371/journal.pone.0000179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurdic P, Saltel F, Chabadel A, Destaing O. 2006. Podosome and sealing zone: specificity of the osteoclast model. Eur. J. Cell Biol. 85:195–202. 10.1016/j.ejcb.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 13.Nakamura I, Lipfert L, Rodan GA, Le TD. 2001. Convergence of αvβ3 integrin- and macrophage colony stimulating factor-mediated signals on phospholipase Cγ in prefusion osteoclasts. J. Cell Biol. 152:361–373. 10.1083/jcb.152.2.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou W, Kitaura H, Reeve J, Long F, Tybulewicz VL, Shattil SJ, Ginsberg MH, Ross FP, Teitelbaum SL. 2007. Syk, c-Src, the αvβ3 integrin, and ITAM immunoreceptors, in concert, regulate osteoclastic bone resorption. J. Cell Biol. 176:877–888. 10.1083/jcb.200611083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. 2001. Cbl associates with Pyk2 and Src to regulate Src kinase activity, αvβ3 integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 152:181–195. 10.1083/jcb.152.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faccio R, Novack DV, Zallone A, Ross FP, Teitelbaum SL. 2003. Dynamic changes in the osteoclast cytoskeleton in response to growth factors and cell attachment are controlled by β3 integrin. J. Cell Biol. 162:499–509. 10.1083/jcb.200212082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne WC, Baron R. 2004. Src kinase activity is essential for osteoclast function. J. Biol. Chem. 279:17660–17666. 10.1074/jbc.M311032200 [DOI] [PubMed] [Google Scholar]
- 18.Soriano P, Montgomery C, Geske R, Bradley A. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693–702. 10.1016/0092-8674(91)90499-O [DOI] [PubMed] [Google Scholar]
- 19.Schwartzberg PL, Xing L, Hoffmann O, Lowell CA, Garrett L, Boyce BF, Varmus HE. 1997. Rescue of osteoclast function by transgenic expression of kinase-deficient Src in src−/− mutant mice. Genes Dev. 11:2835–2844. 10.1101/gad.11.21.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Destaing O, Sanjay A, Itzstein C, Horne WC, Toomre D, De Camilli P, Baron R. 2008. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol. Biol. Cell 19:394–404. 10.1091/mbc.E07-03-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil-Henn H, Destaing O, Sims NA, Aoki K, Alles N, Neff L, Sanjay A, Bruzzaniti A, De Camilli P, Baron R, Schlessinger J. 2007. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2−/− mice. J. Cell Biol. 178:1053–1064. 10.1083/jcb.200701148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duong LT, Nakamura I, Lakkakorpi PT, Lipfert L, Bett AJ, Rodan GA. 2001. Inhibition of osteoclast function by adenovirus expressing antisense protein-tyrosine kinase 2. J. Biol. Chem. 276:7484–7492. 10.1074/jbc.M008368200 [DOI] [PubMed] [Google Scholar]
- 23.Purev E, Neff L, Horne WC, Baron R. 2009. c-Cbl and Cbl-b act redundantly to protect osteoclasts from apoptosis and to displace HDAC6 from beta-tubulin, stabilizing microtubules and podosomes. Mol. Biol. Cell 20:4021–4030. 10.1091/mbc.E09-03-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. 2005. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J. Cell Sci. 118:2901–2911. 10.1242/jcs.02425 [DOI] [PubMed] [Google Scholar]
- 25.Linder S, Hufner K, Wintergerst U, Aepfelbacher M. 2000. Microtubule-dependent formation of podosomal adhesion structures in primary human macrophages. J. Cell Sci. 113:4165–4176 [DOI] [PubMed] [Google Scholar]
- 26.Evans JG, Correia I, Krasavina O, Watson N, Matsudaira P. 2003. Macrophage podosomes assemble at the leading lamella by growth and fragmentation. J. Cell Biol. 161:697–705. 10.1083/jcb.200212037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akisaka T, Yoshida H, Suzuki R, Takama K. 2008. Adhesion structures and their cytoskeleton-membrane interactions at podosomes of osteoclasts in culture. Cell Tissue Res. 331:625–641. 10.1007/s00441-007-0552-x [DOI] [PubMed] [Google Scholar]
- 28.Akisaka T, Yoshida H, Takigawa T. 2011. Differential distribution of posttranslationally modified microtubules in osteoclasts. J. Histochem. Cytochem. 59:630–638. 10.1369/0022155411405334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopp P, Lammers R, Aepfelbacher M, Woehlke G, Rudel T, Machuy N, Steffen W, Linder S. 2006. The kinesin KIF1C and microtubule plus ends regulate podosome dynamics in macrophages. Mol. Biol. Cell 17:2811–2823. 10.1091/mbc.E05-11-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galjart N. 2010. Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20:R528–R537. 10.1016/j.cub.2010.05.022 [DOI] [PubMed] [Google Scholar]
- 31.Basu R, Chang F. 2007. Shaping the actin cytoskeleton using microtubule tips. Curr. Opin. Cell Biol. 19:88–94. 10.1016/j.ceb.2006.12.012 [DOI] [PubMed] [Google Scholar]
- 32.Akhmanova A, Steinmetz MO. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- 33.Duellberg C, Fourniol FJ, Maurer SP, Roostalu J, Surrey T. 2013. End-binding proteins and Ase1/PRC1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell Biol. 23:54–63. 10.1016/j.tcb.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 34.Lansbergen G, Akhmanova A. 2006. Microtubule plus end: a hub of cellular activities. Traffic 7:499–507. 10.1111/j.1600-0854.2006.00400.x [DOI] [PubMed] [Google Scholar]
- 35.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, Jelesarov I, Winkler FK, Wuthrich K, Akhmanova A, Steinmetz MO. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- 36.Schnoor M, Lai FP, Zarbock A, Klaver R, Polaschegg C, Schulte D, Weich HA, Oelkers JM, Rottner K, Vestweber D. 2011. Cortactin deficiency is associated with reduced neutrophil recruitment but increased vascular permeability in vivo. J. Exp. Med. 208:1721–1735. 10.1084/jem.20101920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai FP, Szczodrak M, Oelkers JM, Ladwein M, Acconcia F, Benesch S, Auinger S, Faix J, Small JV, Polo S, Stradal TE, Rottner K. 2009. Cortactin promotes migration and platelet-derived growth factor-induced actin reorganization by signaling to Rho-GTPases. Mol. Biol. Cell 20:3209–3223. 10.1091/mbc.E08-12-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. 2012. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149:371–382. 10.1016/j.cell.2012.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. 2006. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 34:1369–1380. 10.1093/nar/gkl001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, Borisy GG, Steinmetz MO, Akhmanova A. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706. 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mimori-Kiyosue Y, Shiina N, Tsukita S. 2000. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148:505–518. 10.1083/jcb.148.3.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. 1997. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell 8:973–985. 10.1091/mbc.8.6.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC. 2009. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61:85–100. 10.1016/j.neuron.2008.11.013 [DOI] [PubMed] [Google Scholar]
- 44.Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 7:761–768. 10.1038/nmeth.1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zilberman Y, Ballestrem C, Carramusa L, Mazitschek R, Khochbin S, Bershadsky A. 2009. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J. Cell Sci. 122:3531–3541. 10.1242/jcs.046813 [DOI] [PubMed] [Google Scholar]
- 46.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. 2002. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 21:6820–6831. 10.1093/emboj/cdf682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U. S. A. 100:4389–4394. 10.1073/pnas.0430973100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. 2003. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 22:1168–1179. 10.1093/emboj/cdg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y. 2000. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene 19:210–216. 10.1038/sj.onc.1203308 [DOI] [PubMed] [Google Scholar]
- 50.Webb BA, Eves R, Mak AS. 2006. Cortactin regulates podosome formation: roles of the protein interaction domains. Exp. Cell Res. 312:760–769. 10.1016/j.yexcr.2005.11.032 [DOI] [PubMed] [Google Scholar]
- 51.Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587. 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. 2006. Cortactin has an essential and specific role in osteoclast actin assembly. Mol. Biol. Cell 17:2882–2895. 10.1091/mbc.E06-03-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma T, Sadashivaiah K, Madayiputhiya N, Chellaiah MA. 2010. Regulation of sealing ring formation by l-plastin and cortactin in osteoclasts. J. Biol. Chem. 285:29911–29924. 10.1074/jbc.M109.099697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyauchi A, Hruska KA, Greenfield EM, Duncan R, Alvarez J, Barattolo R, Colucci S, Zambonin-Zallone A, Teitelbaum SL, Teti A. 1990. Osteoclast cytosolic calcium, regulated by voltage-gated calcium channels and extracellular calcium, controls podosome assembly and bone resorption. J. Cell Biol. 111:2543–2552. 10.1083/jcb.111.6.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruzzaniti A, Neff L, Sandoval A, Du L, Horne WC, Baron R. 2009. Dynamin reduces Pyk2 Y402 phosphorylation and SRC binding in osteoclasts. Mol. Cell Biol. 29:3644–3656. 10.1128/MCB.00851-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. 2007. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. U. S. A. 104:11933–11938. 10.1073/pnas.0701077104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Head JA, Jiang D, Li M, Zorn LJ, Schaefer EM, Parsons JT, Weed SA. 2003. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14:3216–3229. 10.1091/mbc.E02-11-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, Yao TP, Lane WS, Seto E. 2007. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27:197–213. 10.1016/j.molcel.2007.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, Nicosia SV, Zhang X. 2009. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28:445–460. 10.1038/onc.2008.388 [DOI] [PubMed] [Google Scholar]
- 60.Kaluza D, Kroll J, Gesierich S, Yao TP, Boon RA, Hergenreider E, Tjwa M, Rossig L, Seto E, Augustin HG, Zeiher AM, Dimmeler S, Urbich C. 2011. Class IIb HDAC6 regulates endothelial cell migration and angiogenesis by deacetylation of cortactin. EMBO J. 30:4142–4156. 10.1038/emboj.2011.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rey M, Irondelle M, Waharte F, Lizarraga F, Chavrier P. 2011. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur. J. Cell Biol. 90:128–135. 10.1016/j.ejcb.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 62.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23:607–618. 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 63.Meiler E, Nieto-Pelegrin E, Martinez-Quiles N. 2012. Cortactin tyrosine phosphorylation promotes its deacetylation and inhibits cell spreading. PLoS One 7:e33662. 10.1371/journal.pone.0033662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell DH, Sutherland RL, Daly RJ. 1999. Signaling pathways and structural domains required for phosphorylation of EMS1/cortactin. Cancer Res. 59:5376–5385 [PubMed] [Google Scholar]
- 65.Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ. 2003. A cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278:21805–21813. 10.1074/jbc.M211407200 [DOI] [PubMed] [Google Scholar]
- 66.Abu-Amer Y, Ross FP, Schlesinger P, Tondravi MM, Teitelbaum SL. 1997. Substrate recognition by osteoclast precursors induces C-src/microtubule association. J. Cell Biol. 137:247–258. 10.1083/jcb.137.1.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ng PY, Cheng TS, Zhao H, Ye S, Sm Ang E, Khor EC, Feng HT, Xu J, Zheng MH, Pavlos NJ. 2013. Disruption of the dynein-dynactin complex unveils motor-specific functions in osteoclast formation and bone resorption. J. Bone Miner. Res. 28:119–134. 10.1002/jbmr.1725 [DOI] [PubMed] [Google Scholar]
- 68.Ye S, Fowler TW, Pavlos NJ, Ng PY, Liang K, Feng Y, Zheng M, Kurten R, Manolagas SC, Zhao H. 2011. LIS1 regulates osteoclast formation and function through its interactions with dynein/dynactin and Plekhm1. PLoS One 6:e27285. 10.1371/journal.pone.0027285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pavlos NJ, Cheng TS, Qin A, Ng PY, Feng HT, Ang ES, Carrello A, Sung CH, Jahn R, Zheng MH, Xu J. 2011. Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption. Mol. Cell Biol. 31:1551–1564. 10.1128/MCB.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hazama R, Qu X, Yokoyama K, Tanaka C, Kinoshita E, He J, Takahashi S, Tohyama K, Yamamura H, Tohyama Y. 2009. ATP-induced osteoclast function: the formation of sealing-zone like structure and the secretion of lytic granules via microtubule-deacetylation under the control of Syk. Genes Cells 14:871–884. 10.1111/j.1365-2443.2009.01317.x [DOI] [PubMed] [Google Scholar]
- 71.Stenbeck G, Horton MA. 2004. Endocytic trafficking in actively resorbing osteoclasts. J. Cell Sci. 117:827–836. 10.1242/jcs.00935 [DOI] [PubMed] [Google Scholar]
- 72.Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada GH, Wada Y, Futai M. 2003. From lysosomes to the plasma membrane: localization of vacuolar-type H+-ATPase with the a3 isoform during osteoclast differentiation. J. Biol. Chem. 278:22023–22030. 10.1074/jbc.M302436200 [DOI] [PubMed] [Google Scholar]
- 73.Hong JM, Teitelbaum SL, Kim TH, Ross FP, Kim SY, Kim HJ. 2011. Calpain-6, a target molecule of glucocorticoids, regulates osteoclastic bone resorption via cytoskeletal organization and microtubule acetylation. J. Bone Miner. Res. 26:657–665. 10.1002/jbmr.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMichael BK, Cheney RE, Lee BS. 2010. Myosin X regulates sealing zone patterning in osteoclasts through linkage of podosomes and microtubules. J. Biol. Chem. 285:9506–9515. 10.1074/jbc.M109.017269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsumoto T, Nagase Y, Hirose J, Tokuyama N, Yasui T, Kadono Y, Ueki K, Kadowaki T, Nakamura K, Tanaka S. 2012. Regulation of bone-resorption and sealing zone formation in osteoclasts occurs through Akt-mediated microtubule stabilization. J. Bone Miner. Res. 28:1191–1202. 10.1002/jbmr.1844 [DOI] [PubMed] [Google Scholar]
- 76.Turksen K, Kanehisa J, Opas M, Heersche JN, Aubin JE. 1988. Adhesion patterns and cytoskeleton of rabbit osteoclasts on bone slices and glass. J. Bone Miner. Res. 3:389–400 [DOI] [PubMed] [Google Scholar]
- 77.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450:1100–1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- 79.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830. 10.1038/ncb1160 [DOI] [PubMed] [Google Scholar]
- 80.Hiura K, Lim SS, Little SP, Lin S, Sato M. 1995. Differentiation dependent expression of tensin and cortactin in chicken osteoclasts. Cell Motil. Cytoskeleton 30:272–284 [DOI] [PubMed] [Google Scholar]
- 81.Hurst IR, Zuo J, Jiang J, Holliday LS. 2004. Actin-related protein 2/3 complex is required for actin ring formation. J. Bone Miner. Res. 19:499–506. 10.1359/JBMR.0301238 [DOI] [PubMed] [Google Scholar]
- 82.Matsubara T, Myoui A, Ikeda F, Hata K, Yoshikawa H, Nishimura R, Yoneda T. 2006. Critical role of cortactin in actin ring formation and osteoclastic bone resorption. J. Bone Miner. Metab. 24:368–372. 10.1007/s00774-006-0701-4 [DOI] [PubMed] [Google Scholar]
- 83.Kanner SB, Reynolds AB, Vines RR, Parsons JT. 1990. Monoclonal antibodies to individual tyrosine-phosphorylated protein substrates of oncogene-encoded tyrosine kinases. Proc. Natl. Acad. Sci. U. S. A. 87:3328–3332. 10.1073/pnas.87.9.3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ammer AG, Weed SA. 2008. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton 65:687–707. 10.1359/JBMR.0301238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Webb BA, Jia L, Eves R, Mak AS. 2007. Dissecting the functional domain requirements of cortactin in invadopodia formation. Eur. J. Cell Biol. 86:189–206. 10.1016/j.ejcb.2007.01.003 [DOI] [PubMed] [Google Scholar]
- 86.Zhou S, Webb BA, Eves R, Mak AS. 2006. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 290:C463–C471. 10.1152/ajpcell.00350.2005 [DOI] [PubMed] [Google Scholar]
- 87.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043. 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- 88.Ayala I, Baldassarre M, Giacchetti G, Caldieri G, Tete S, Luini A, Buccione R. 2008. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J. Cell Sci. 121:369–378. 10.1242/jcs.008037 [DOI] [PubMed] [Google Scholar]
- 89.Clark ES, Whigham AS, Yarbrough WG, Weaver AM. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235. 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- 90.Desmarais V, Yamaguchi H, Oser M, Soon L, Mouneimne G, Sarmiento C, Eddy R, Condeelis J. 2009. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskeleton 66:303–316. 10.1002/cm.20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, Mueller SC. 2006. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp. Cell Res. 312:1240–1253. 10.1016/j.yexcr.2005.12.012 [DOI] [PubMed] [Google Scholar]
- 92.Kelley LC, Ammer AG, Hayes KE, Martin KH, Machida K, Jia L, Mayer BJ, Weed SA. 2010. Oncogenic Src requires a wild-type counterpart to regulate invadopodia maturation. J. Cell Sci. 123:3923–3932. 10.1242/jcs.075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 94.Castro-Castro A, Janke C, Montagnac G, Paul-Gilloteaux P, Chavrier P. 2012. ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of alpha-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur. J. Cell Biol. 91:950–960. 10.1016/j.ejcb.2012.07.001 [DOI] [PubMed] [Google Scholar]
- 95.Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R. 2001. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 276:35185–35193. 10.1074/jbc.M102219200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.