Abstract
Hypoxia-inducible factor 1 (HIF-1) plays a key role in the cellular adaptation to hypoxia. Although HIF-1 is usually strongly suppressed by posttranslational mechanisms during normoxia, HIF-1 is active and enhances tumorigenicity in malignant tumor cells that express the membrane protease MT1-MMP. The cytoplasmic tail of MT1-MMP, which can bind a HIF-1 suppressor protein called factor inhibiting HIF-1 (FIH-1), promotes inhibition of FIH-1 by Mint3 during normoxia. To explore possible links between HIF-1 activation by MT1-MMP/Mint3 and tumor growth signals, we surveyed a panel of 252 signaling inhibitors. The mTOR inhibitor rapamycin was identified as a possible modulator, and it inhibited the mTOR-dependent phosphorylation of Mint3 that is required for FIH-1 inhibition. A mutant Mint3 protein that cannot be phosphorylated exhibited a reduced ability to inhibit FIH-1 and promoted tumor formation in mice. These data suggest a novel molecular link between the important hub proteins MT1-MMP and mTOR that contributes to tumor malignancy.
INTRODUCTION
Energy is vitally required for cell activities. Oxidative phosphorylation (OXPHOS) in mitochondria is the major source of ATP production in aerobic organisms. However, tissues are frequently exposed to hypoxia accidentally or under pathological conditions. Under hypoxic conditions, OXPHOS is inactivated and anaerobic glycolytic activity increases to produce ATP. Hypoxia-inducible factor 1 (HIF-1), a transcription factor that regulates multiple hypoxia stress response genes, plays a major role in the adaptation to hypoxia (1–3). In addition to HIF-1, HIF-2 and HIF-3 are also known to regulate the response to hypoxia, although their expression and action are more cell specific than those of HIF-1 and their target genes differ. For example, the glycolytic pathway is more preferentially regulated by HIF-1 (4). HIF-1 induces the expression of glycolysis-related genes, such as GLUT1, HK2, PGK1, LDHA, and PDK1, and shuts off the supply of pyruvate for OXPHOS by using PDK1. HIF-1 also regulates angiogenesis by inducing VEGF expression and promotes cell motility and invasion to allow escape from a hostile environment by affecting the expression of a variety of genes (5, 6). Thus, HIF-1 has been studied extensively, with a particular focus on its regulation and roles during hypoxia. In contrast, the roles of HIF-1 during normoxia have been studied to a lesser extent, largely because HIF-1 activity is believed to be negligible during normoxia. However, accumulating evidence has indicated that HIF-1 plays pivotal roles even during normoxia in some cell types, such as macrophages and tumor cells. Therefore, understanding the mechanisms by which HIF-1 is activated under such nonhypoxic conditions is of particular interest.
HIF-1 consists of a regulatory α subunit and a constitutively active β subunit. The α subunit is encoded by HIF1A, which is constitutively transcribed and translated in most cell types. However, during normoxia, the expression of the α subunit protein is maintained at a low level because of constitutive proteasomal degradation following oxygen-dependent hydroxylation of proline residues by HIF prolyl hydroxylases (PHDs) (7, 8). The ability of HIF-1α to promote the transcription of HIF-1 target genes is also suppressed by factor inhibiting HIF-1 (FIH-1), another oxygen-sensitive enzyme. FIH-1 hydroxylates an asparagine residue of HIF-1α, which prevents HIF-1α from binding to the transcriptional coactivator p300/CBP (9, 10). Thus, HIF-1 activity is strongly suppressed during normoxia by two different types of posttranslational mechanisms. Hypoxia inactivates PHDs and FIH-1 and relieves the suppression of HIF-1 activity.
However, certain circumstances can activate HIF-1 during normoxia. For example, aggressive tumor cells often exhibit a higher level of glucose consumption than noninvasive ones during normoxia; this phenomenon is known as the Warburg effect (11, 12). HIF-1 is constitutively active in such cells. Although the exact mechanism underlying this effect is not fully understood, a shift from OXPHOS to aerobic glycolysis occurs, which renders the cells addicted to glucose for growth and survival. This increased glycolysis rate not only supplies energy but also generates a variety of glucose metabolites that are used as building blocks for the cellular constituents that must be created to support cell division (13). We have recently reported that the suppression of HIF-1α by FIH-1 is abrogated in malignant tumor cells because of FIH-1 inactivation (14). However, other mechanisms of HIF-1α inactivation that are mediated by PHDs remain active in such cells. Although HIF-1α protein levels can be suppressed by PHD activity during normoxia, the residual HIF-1α protein is sufficient to promote the expression of target genes after release of HIF-1α from suppression by FIH-1. Indeed, knockdown of FIH-1 expression in nontransformed cells significantly activates HIF-1 activity, enhances the expression of HIF-1 target genes, and boosts the glycolytic activity of the cells (14).
Macrophages uniquely use aerobic glycolysis as a major source of ATP during normoxia, and HIF-1 is constitutively active in these cells (15, 16). Mint3/APBA3 has been identified as an FIH-1 inhibitory factor in macrophages (16). Mint3 is a member of the X11 protein family that is characterized by the presence of common C-terminal PTB and PDZ domains, but Mint3 has a unique N-terminal domain (17, 18). Mint3 binds membrane proteins via its C-terminal domain, whereas the unique N-terminal domain binds FIH-1. Mint3 localizes to the trans-Golgi network by interacting with membrane proteins, such as amyloid precursor protein (APP) and furin (19, 20). Mint3 is expressed in most cell types (17, 18). Although Mint3 can bind and inhibit FIH-1, this interaction also requires the FIH-1-binding cytoplasmic tail of the membrane protease MT1-MMP for inhibition of FIH-1 by neighboring Mint3. Indeed, the colocalization of MT1-MMP with Mint3 has been observed mostly at the Golgi compartment (21). Cooperation between the two FIH-1-binding proteins MT1-MMP cytoplasmic tail and Mint3 thus efficiently recruits cytoplasmic FIH-1 to the membrane, where FIH-1 is then inhibited by Mint3. This MT1-MMP/Mint3-dependent inactivation of FIH-1 occurs in both macrophages and tumor cells (14, 16, 21). Depletion of MT1-MMP in macrophages restores FIH-1-mediated suppression of HIF-1α and thereby reduces ATP production by glycolysis to nearly 40% of the level observed in wild-type cells (21). Mint3-deficient mice are viable but exhibit defects in macrophage function because of reduced HIF-1 activity and ATP production, which is similar to the phenotype in mice lacking MT1-MMP (21–23). Although MT1-MMP is a potent proinvasive protease that is expressed in a variety of malignant tumor cells and macrophages (24), the ability of MT1-MMP to activate HIF-1 is dependent upon its cytoplasmic tail and does not require protease activity (14, 21, 25). Given the ability of MT1-MMP/Mint3 to induce the activation of HIF-1 in tumor cells, this pathway contributes to the Warburg effect (MT1-MMP-dependent Warburg effect). Disruption of the MT1-MMP-dependent Warburg effect by knocking down Mint3 expression in human breast carcinoma MDA-MB-231 cells suppresses tumor growth in mice (14). We have observed enhanced HIF-1 activity due to inhibition of FIH-1 activity in all of the MT1-MMP-expressing tumor cells tested to date, which indicates that the expression of MT1-MMP in tumor cells can activate HIF-1; however, the degree of activation varies between different tumor cell types (14).
Because MT1-MMP and HIF-1 play diverse roles during tumor growth, the mechanism by which the MT1-MMP/Mint3 pathway-mediated regulation of HIF-1 activity coordinates with tumor growth signals is of particular interest. To address this question, we examined the effects of 252 well-characterized inhibitors of specific cellular signaling pathways on MT1-MMP/Mint3-mediated HIF-1α activation. Five inhibitors suppressed HIF-1α activation, and three of these inhibitors (including rapamycin, an mTOR inhibitor) did so in an MT1-MMP/Mint3-dependent manner. Here, we demonstrate that rapamycin inhibits the phosphorylation of Mint3 by mTOR and that mTOR cooperates with MT1-MMP in the Mint3-mediated inhibition of FIH-1. We have thus uncovered a critical mechanism of cross talk between the protumorigenic membrane protease MT1-MMP and the signal integrator mTOR to activate HIF-1 during normoxia.
MATERIALS AND METHODS
Cell culture.
HT1080 fibrosarcoma, HEK293 human embryonic kidney, and MDA-MB-231 breast cancer cells were purchased from the American Type Culture Collection. 293FT cells (HEK293 cells expressing the simian virus 40 large T antigen) were purchased from Invitrogen. HT1080 and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with low glucose (Invitrogen), 10% fetal bovine serum (FBS), and penicillin-streptomycin. HEK293 and 293FT cells were cultured in DMEM with high glucose, 10% FBS, and penicillin-streptomycin. For experiments conducted under hypoxic conditions, the cells were cultured with 1% O2 and 5% CO2 in a model 9200 incubator (Wakenyaku).
Plasmids.
Mint3, MT1-MMP, and FIH-1 cDNAs were cloned into the pENTR vector (Invitrogen) and inserted into the pcDNA or pLenti6 vector (Invitrogen) as previously described (16, 21, 26). N-terminally His6- and/or C-terminally FLAG-tagged Mint3 and mutant proteins with alanine substitutions were prepared by PCR. pcDNA FLAG-rat mTOR and pcDNA AUI-rat mTOR were kind gifts from Tatsuhiro Sato (Kitasato University) (27). For the short hairpin RNA (shRNA) sequences used in the shRNA vectors, see Table S1 in the supplemental material. The deoxyribose versions of the targeted gene sequences were subcloned into pENTR/U6 TOPO (Invitrogen) before being transferred via recombination into the lentiviral vector pLenti6 BLOCKiT (Invitrogen) as previously described (16, 21, 26).
Inhibitors.
SCADS inhibitor kits I to III were kindly provided by the Screening Committee of Anticancer Drugs in Japan (28, 29). For further characterization after screening, rapamycin (WAKO), KT5823 (WAKO), Akt Inhibitor VIII (Merck), actinomycin D (WAKO), and trichostatin A (WAKO) were used.
Reporter assay.
The reporter plasmid contains the firefly luciferase gene under the control of a transcriptional regulatory unit comprising four Gal4-binding elements and the thymidine kinase (TK)-derived TATA box. A pRL vector expressing Renilla luciferase (Promega) served as an internal control. For the screenings with SCADS kits I to III, HT1080 cells (2.5 × 105) were seeded into a 90-mm dish and cotransfected with the reporter plasmid (1 μg), the internal control vector (100 ng), and a Gal4 binding domain (Gal4BD)–C-terminal activation domain (CAD) plasmid (500 ng). Twenty-four hours after transfection, the cells (2.5 × 103/well) were seeded into the wells of 96-well plates and treated with the SCADS chemicals (10 μM) for 24 h. Luciferase activity was measured with the Dual-Glo luciferase assay system (Promega) in accordance with the manufacturer's instructions. Luminescence was measured in a FLUOstar OPTIMA luminometer (BMG LABTECH). For experiments other than the SCADS kit screen, HT1080 cells (1.25 × 104/well) or HEK293 cells (2.5 × 104/well) were seeded into 24-well plates and cotransfected with the reporter plasmid (100 ng), the internal control vector (10 ng), a Gal4BD-CAD plasmid (50 ng), and other plasmids (200 ng) that expressed the vector alone, Mint3, MT1-MMP, or mTOR. Transfections were performed with Lipofectamine 2000 (Invitrogen). Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Luminescence was measured in a TD20/20 luminometer (Promega).
Immunoblot analyses.
The cells were lysed with lysis buffer and centrifuged at 20,000 × g for 15 min at 4°C. The supernatants were collected, and total protein levels were quantified with the Bradford assay (Bio-Rad). Nuclear lysates were collected with the Nuclear Extract kit (Active Motif). The lysates were separated by SDS-PAGE, transferred to membrane filters, and analyzed by immunoblotting with a mouse anti-MT1-MMP antibody (Daiichi Fine Chemical), a mouse anti-Mint3 antibody (BD Biosciences), a goat anti-FIH-1 antibody (Santa Cruz Biotechnology), a mouse antiactin antibody (Millipore), a mouse anti-mTOR antibody (Millipore), a mouse anti-HIF-1α antibody (BD Biosciences), a mouse anti-lamin A/C antibody (Santa Cruz Biotechnology), or a rabbit anti-FLAG antibody (Sigma).
RNA isolation, RT, and real-time PCR.
Total RNA was isolated from cells with TRIzol reagent (Invitrogen) and subjected to reverse transcription (RT) with SuperScript II (Invitrogen) and random primers. The RT products were then subjected to real-time PCR in a Smart Cycler II System (Cepheid) with SYBR green I (BioWhittaker Molecular Applications) and specific primers for each gene (see Table S2 in the supplemental material). The PCR products were sequenced, and their homogeneity was confirmed by monitoring the dissociation temperature of SYBR green I fluorescence.
Two-dimensional (2D) electrophoresis.
The cells were lysed in buffer containing 1% SDS and 50 mM Tris (pH 7.4). The lysates were purified with a 2-D cleanup kit and dissolved in DeStreak rehydration solution (GE). The proteins in the cell lysates were separated on the basis of their isoelectric points with the Immobiline DryStrip and Ettan IPGphor system (GE), followed by SDS-PAGE and immunoblot analysis. The intensity of each spot was quantified with ImageJ software (NIH).
Phosphatase treatment.
The cell lysates were incubated with the phosphatase PP1 (5 U; NEB) in reaction buffer (50 mM HEPES [pH 7.0], 100 mM NaCl, 2 mM dithiothreitol, 0.01% Brij 35, 1 mM MnCl2) at 30°C for 60 min. The resulting lysates were subjected to 2D electrophoresis.
Immunoprecipitation.
The cells were lysed with lysis buffer (1% NP-40, 50 mM Tris [pH 8.0], 150 mM NaCl) and centrifuged at 20,000 ×g for 15 min at 4°C. The supernatants were collected and incubated with anti-FLAG M2 antibody-conjugated beads (Sigma). The beads were washed, and bound proteins were eluted with FLAG peptides and detected by immunoblot analysis.
Identification of Mint3 posttranslational modifications by mass spectrometry.
The lysates from HT1080 cells expressing N-terminally His6- and C-terminally FLAG-tagged Mint3 (HF-Mint3) were subjected to immunoprecipitation as described above. The eluates were analyzed with a, LTQ-Orbitrap Velos (Thermo Fisher Scientific) (30).
Measurement of lactic acid levels and ATP concentrations.
Lactic acid levels and ATP concentrations were determined with the l-lactic acid kit (R-Biopharm) or ATP bioluminescence assay kit CLS II (Roche Applied Science), respectively, as previously described (14, 22).
Measurement of glucose consumption.
The cells were seeded into the wells of 24-well plates (1 × 105/well) in triplicate. Conditioned medium was collected after 8 h, and glucose was measured with a Glucose (GO) Assay kit (Sigma).
Immunostaining.
The cells were fixed with 4% paraformaldehyde and permeabilized with 0.01% Triton X-100 for 10 min. After blocking in phosphate-buffered saline (PBS) containing 5% goat serum and 3% bovine serum albumin, the cells were incubated with a mouse anti-Mint3 antibody (BD Biosciences) and a rabbit anti-GM130 antibody (Novus Biologicals) for 1 h, washed three times, and then incubated for 1 h with Alexa Fluor 488-conjugated anti-mouse IgG or Alexa Fluor 594-conjugated anti-rabbit antibody (Invitrogen). The cells were counterstained with Hoechst 33342, washed five times with PBS, mounted, and observed by confocal microscopy.
Cell growth assay.
The cells (1 × 104) were seeded onto a plastic dish and cultured at 37°C in a humidified CO2 incubator. The cells were counted periodically with a Coulter counter (Beckman).
Tumor growth assay.
The tumorigenicity of the cells was examined by using 6-week-old female BALB/c nude mice (Clea, Japan). Briefly, 1 × 106 cells were injected subcutaneously into the dorsal side of a mouse and the volumes of the implanted tumors were measured with a caliper and calculated with the formula V = (L × W2)/2, where V is volume (mm3), L is the largest diameter (mm), and W is the smallest diameter (mm). The experiments were performed according to the institutional ethical guidelines for animal experiments and the safety guidelines for gene manipulation experiments (The Institute of Medical Science, University of Tokyo).
Statistical analyses.
Two-subject groups were compared by using the two-sided t test or the Mann-Whitney U test.
RESULTS
Screening for inhibitors of MT1-MMP/Mint3-mediated posttranslational activation of HIF-1α.
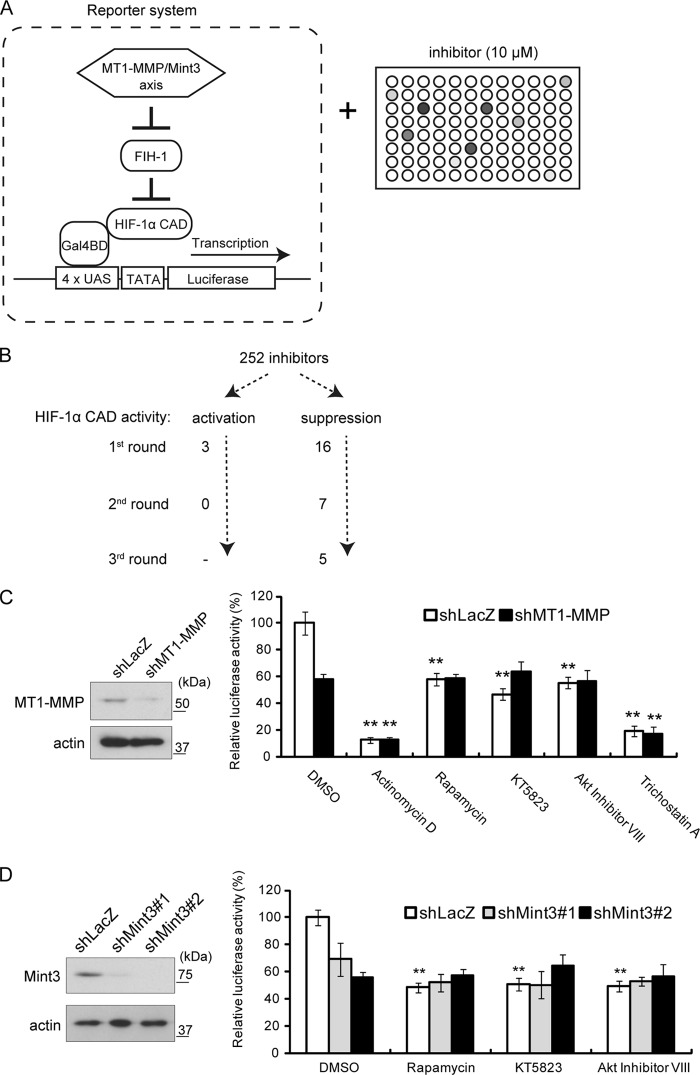
To identify signaling factors that potentially contribute to the regulation of MT1-MMP/Mint3-dependent HIF-1 activation, we screened a panel of 252 well-characterized signaling inhibitors (28, 29) by using a luciferase reporter assay to monitor the FIH-1-dependent HIF-1α activity in HT1080 cells (Fig. 1A). HIF-1-dependent transcription of target genes is mediated by the HIF-1α CAD, which contains an arginine residue that can be hydroxylated by FIH-1, leading to inhibition of transcription. Therefore, we can specifically monitor FIH-1-regulated HIF-1α activity by using the reporter construct shown in Fig. 1A. Because HT1080 cells express MT1-MMP, FIH-1-mediated suppression of HIF-1α CAD is constitutively abrogated in a Mint3-dependent manner (21). Three repeated screenings of the inhibitors shown in Fig. 1B revealed that actinomycin D, rapamycin, KT5823, Akt inhibitor VIII, and trichostatin A suppressed reporter activity in HT1080 cells. We next examined whether the suppression by the five inhibitors was mediated by the MT1-MMP/Mint3 axis by knocking down the expression of MT1-MMP (Fig. 1B) with an shRNA sequence from a previous study (31). Actinomycin D and trichostatin A continued to suppress HIF-1α activity in MT1-MMP knockdown cells (Fig. 1C, shLacZ versus shMT1-MMP). In contrast, rapamycin, KT5823, and Akt inhibitor VIII suppressed reporter activity only in the presence of MT1-MMP (Fig. 1C, shLacZ versus shMT1-MMP). Mint3 knockdown produced results similar to those observed upon the treatment of MT1-MMP knockdown cells with the three inhibitors (Fig. 1D). These results indicate that rapamycin, KT5823, and Akt inhibitor VIII specifically abrogate MT1-MMP/Mint3-dependent HIF-1α activation.
FIG 1.
Screening of 252 compounds for inhibitors of HIF-1α transcriptional activity with a reporter assay. (A) Schematic illustration of the reporter screening with SCADS kits. (B) Summary of the inhibitor screenings. (C) Reporter assays performed with MT1-MMP knockdown cells. (Left) Immunoblot analysis performed with MT1-MMP knockdown cells. (Right) Results of the reporter assay. Note that rapamycin, KT5823, and Akt inhibitor VIII did not decrease HIF-1α CAD activity further in MT1-MMP knockdown cells. (D) Reporter assays were performed with Mint3 knockdown cells. (Left) Immunoblot analysis performed with MT1-MMP knockdown cells. (Right) Results of the reporter assay. The data in panels C and D were analyzed with the Student t test. **, P < 0.01. DMSO, dimethyl sulfoxide.
mTORC1 promotes HIF-1 activity.
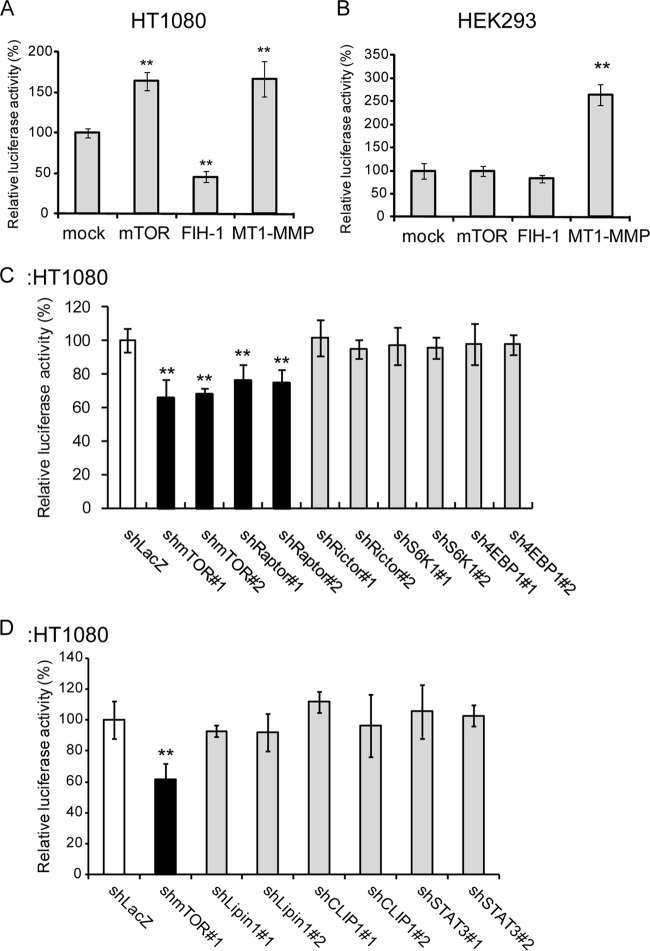
Rapamycin inhibits mTOR (mTORC1, specifically), KT5823 inhibits PKG, and Akt inhibitor VIII inhibits Akt1/2. Because Akt activates mTOR (32, 33), rapamycin and the Akt inhibitor are thought to affect the same pathway. The Akt/mTOR pathway is a major signaling pathway that acts downstream of receptor TKs, such as epidermal growth factor receptor and insulin-like growth factor I receptor (IGF-IR), and is frequently constitutively activated in cells harboring mutations in pathway components (32, 33). Therefore, we focused our study on HIF-1α regulation by mTOR. The MT1-MMP/Mint3 regulatory axis increased reporter expression in HT1080 cells; overexpression of FIH-1 suppressed reporter activity (Fig. 2A, FIH-1), whereas overexpression of MT1-MMP enhanced reporter activity (Fig. 2A, MT1-MMP), as previously described (16, 21). mTOR overexpression increased reporter expression (Fig. 2A, mTOR), indicating that the effect of rapamycin on reporter expression was indeed mediated by mTOR inhibition. Unlike HT1080 cells, which constitutively express MT1-MMP, HEK293 cells do not express MT1-MMP and therefore Mint3 is unable to inhibit the FIH-1-mediated suppression of HIF-1α (16, 21). Ectopic overexpression of FIH-1 in HEK293 cells did not further affect reporter activity (Fig. 2B, FIH-1), although expression of MT1-MMP markedly enhanced the reporter activity (Fig. 2B, MT1-MMP). Interestingly, overexpression of mTOR did not increase reporter activity in HEK293 cells (Fig. 2B, mTOR). Coexpression of MT1-MMP and mTOR strikingly boosted HIF-1α reporter activity which was repressed by rapamycin in HEK293 cells (see Fig. S1 in the supplemental material). These results are consistent with the specific effect of rapamycin on MT1-MMP-mediated activation of HIF-1α.
FIG 2.
mTORC1 promotes HIF-1 transcriptional activity in HT1080 cells. (A and B) mTOR promoted HIF-1α CAD activity in HT1080 cells (A) but not in HEK293 cells (B). (C) Knockdown of raptor but not rictor decreased HIF-1α CAD activity. (D) Knockdown of known mTOR target genes did not affect HIF-1α CAD activity in HT1080 cells. The data in panels A to D were analyzed with the Student t test. **, P < 0.01.
mTOR forms two functionally distinct complexes, mTORC1 and mTORC2 (32). To determine which complex regulates HIF-1α, we knocked down components of each mTOR complex and examined the effects on reporter expression (Fig. 2C). Knockdown of the expression of either mTOR or the mTORC1-specific component raptor (34, 35) significantly decreased reporter activity in HT1080 cells (Fig. 2C, shmTOR#1 and shmTOR#2 and shRaptor#1 and shRaptor#2). However, knockdown of the mTORC2-specific protein rictor (36) did not affect reporter activity (Fig. 2C, shRictor#1 and shRictor#2). These results indicate that mTORC1, but not mTORC2, regulates HIF-1α activity, which is consistent with the greater specificity of rapamycin for mTORC1 (35, 36). mTORC1 has been reported to enhance the translation of HIF-1α mRNA via phosphorylation of S6K or 4EBP1 (32, 37, 38). However, knockdown of S6K and 4EBP1 did not affect MT1-MMP/Mint3-sensitive reporter expression in HT1080 cells (Fig. 2C, S6K, 4EBP1), indicating that mTORC1 appears to regulate the HIF-1α CAD in a manner that is independent of its effects on translation. We further examined the effects of other reported mTOR target genes, LIPIN1 (39, 40), CLIP1 (41), and STAT3 (42), on reporter expression. Knockdown of these genes also did not affect reporter activity in HT1080 cells (Fig. 2D).
mTOR induces Mint3 modifications.
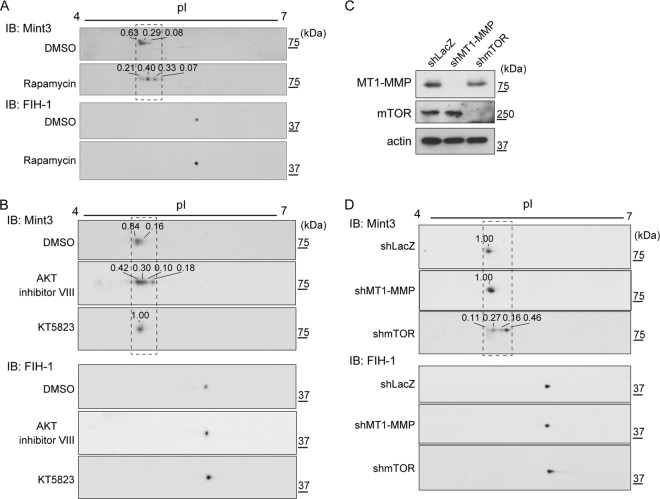
Rapamycin suppressed HIF-1α CAD activity in a MT1-MMP/Mint3 axis-dependent manner. Because mTOR has serine/threonine kinase activity, we analyzed the phosphorylation of Mint3 and FIH-1 in HT1080 cells. The cells were exposed to rapamycin or the vehicle, and cell lysates were prepared and separated by 2D electrophoresis, followed by immunoblot analysis. Rapamycin induced a shift in the isoelectric point of Mint3 protein (Fig. 3A, Mint3; for an enlarged picture and densitometry analysis, see Fig. S2 in the supplemental material). In contrast, rapamycin treatment did not affect the isoelectric point of FIH-1 (Fig. 3A, FIH-1). Thus, mTOR may regulate Mint3 phosphorylation. Akt inhibitor VIII (but not KT5823) also increased the isoelectric point of Mint3 (Fig. 3B; see Fig. S2). These results indicate that Akt inhibitor VIII and rapamycin inhibit Mint3 modification through an mTOR-dependent mechanism, whereas KT5823 appears to act through a different mechanism.
FIG 3.
mTOR regulates the posttranslational modification of Mint3. (A) Rapamycin treatment increased the isoelectric point of the Mint3 protein. (B) Treatment with Akt inhibitor VIII but not KT5823 decreased Mint3 acidity. (C and D) Knockdown of mTOR decreased Mint3 acidity, but knockdown of MT1-MMP did not. Knockdown of mTOR or MT1-MMP was confirmed by immunoblot (IB) analysis (C), and the acidity of Mint3 in control (shLacZ), mTOR, or Mint3 knockdown cells was analyzed by 2D electrophoresis, followed by immunoblot analysis (D). In panels A, B, and D, the relative intensity of each Mint3 signal is shown above the spot. For an analysis of the boxed areas, see Fig. S2 in the supplemental material. DMSO, dimethyl sulfoxide.
MT1-MMP cooperates with Mint3 to abrogate FIH-1-mediated suppression of HIF-1α (14, 21). Therefore, we next examined whether knockdown of MT1-MMP affects the posttranslational modification of Mint3. MT1-MMP knockdown did not affect the isoelectric point of Mint3 (Fig. 3C and D; see Fig. S2 in the supplemental material). In contrast, knockdown of mTOR increased the isoelectric point of Mint3, similar to the effects of exposure to rapamycin (Fig. 3C and D; see Fig. S2). Thus, the mTOR-mediated modification of Mint3 occurs in an MT1-MMP-independent manner; in contrast, MT1-MMP is essential for mTOR-dependent activation of Mint3-dependent HIF-1 activation.
mTOR phosphorylates Mint3.
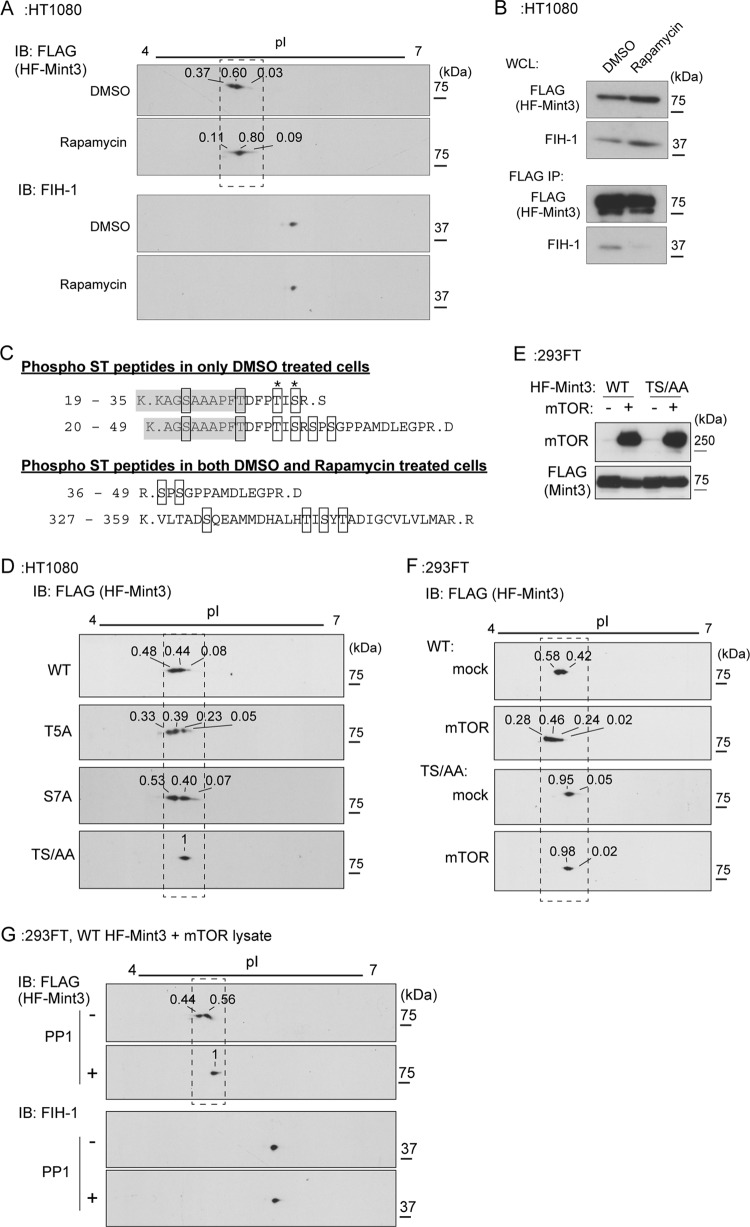
To analyze the mechanism by which the isoelectric point of Mint3 is regulated by mTOR, we expressed recombinant Mint3 with a His6 tag fused to its N terminus and a FLAG tag fused to its C terminus (HF-Mint3) in HT1080 cells. HF-Mint3-expressing cells were exposed to rapamycin or the vehicle, and the protein was analyzed by 2D electrophoresis of cell lysates, followed by immunoblot analysis. In vehicle-treated cells, HF-Mint3 appeared in the form of two polypeptides with distinct migratory properties, whereas only the less acidic form was observed in the rapamycin-treated cells (Fig. 4A; see Fig. S3 in the supplemental material). The changes in the effective migration of the second species were similar for both proteins and hence likely due to similar modifications (Fig. 3A; see Fig. S3). Simultaneously, rapamycin treatment also greatly reduced the binding of HF-Mint3 to FIH-1 (Fig. 4B). Because Mint3 binding to FIH-1 is necessary for FIH-1 inhibition and HIF-1α activation (14, 16), these results provide a plausible mechanism for rapamycin-mediated inhibition of HIF-1α CAD activity.
FIG 4.
Mint3 is phosphorylated by mTOR. (A) Rapamycin treatment increased the isoelectric point of the HF-Mint3 protein. IB, immunoblot. (B) Rapamycin treatment decreased the binding of Mint3 to FIH-1. His6- and FLAG-tagged Mint3 (HF-Mint3) in vehicle- or rapamycin-treated HT1080 cells was precipitated with anti-FLAG antibody beads, and the eluate was subjected to immunoblot analysis. WCL, whole-cell lysates. (C) Phosphorylated peptides of HF-Mint3 were identified by mass spectrometry. The shaded amino acids were derived from the linker sequence between the His6 tag and Mint3. DMSO, dimethyl sulfoxide. (D) TS/AA mutant Mint3 migrated in the form of a single polypeptide in 2D electrophoresis. (E and F) The expression of mTOR decreased the isoelectric point of WT but not TS/AA mutant HF-Mint3 in 293FT cells. (E) Expression of mTOR and WT or TS/AA mutant Mint3 in 293FT cells was confirmed by immunoblot analysis. (F) 2D electrophoresis and immunoblot analysis were performed with the cell lysates assessed in panel E. (G) Phosphatase treatment increased the isoelectric point of WT HF-Mint3 coexpressed with mTOR. In panels A, D, F, and G, the relative intensity of each HF-Mint3 signal is shown above the spot. For enlargements of the boxed areas and a densitometry analysis, see Fig. S3 in the supplemental material.
We next affinity purified HF-Mint3 from cells treated with the vehicle or rapamycin and analyzed HF-Mint3 by mass spectrometry. In the vehicle-treated cells, four peptides were found to be possibly phosphorylated at serine and/or threonine residues, but two of these peptides were absent from the rapamycin-treated cells (Fig. 4C). The two peptides detected only in the vehicle-treated cells (Fig. 4C, amino acids 19 to 35 and 20 to 49) contained a linker sequence between the His6 tag and Mint3 (Fig. 4C, shaded sequences, amino acids 19 to 28 and 20 to 28). The last amino acid in the linker sequence, threonine, corresponds to the substituted translational initiation codon methionine. A comparison of the overlapping Mint3 sequences in the peptides revealed two potential phosphorylation sites of Mint3, the fifth amino acid counting from the initiation codon (threonine [T5]) and the seventh serine (S7).
We next prepared a mutant form of HF-Mint3 containing alanine substitutions at the candidate sites and analyzed the isoelectric points of the mutant proteins. Mutation of either site alone did not affect the isoelectric point of HF-Mint3 (Fig. 4D, wild-type [WT], T5A, and S7A; see Fig. S3 in the supplemental material). However, a mutant protein with alanine substitutions at both sites exhibited a single isoelectric point in 2D gel analysis (Fig. 4D, TS/AA; see Fig. S3), similar to the effects of exposure to rapamycin (Fig. 4C). To confirm that Mint3 T5 and S7 are phosphorylated by mTOR, WT or TS/AA mutant HF-Mint3 was transiently expressed either alone or together with mTOR in 293FT cells, and Mint3 modification was analyzed by 2D electrophoresis, followed by immunoblot analysis. The migration of WT HF-Mint3 shifted to acidic when mTOR was expressed (Fig. 4E and F; see Fig. S3). In contrast, mTOR expression did not affect the migration of TS/AA mutant HF-Mint3 (Fig. 4E and F; see Fig. S3). Treatment of the sample with the phosphatase PP1 shifted the isoelectric point of WT HF-Mint3 to that of the TS/AA mutant (Fig. 4G; see Fig. S3). Taken together, these results indicate that mTOR mediates the phosphorylation of Mint3 at T5 and S7 in an MT1-MMP-independent manner.
A phosphorylation-defective mutant form of Mint3 exhibits reduced FIH-1 binding.
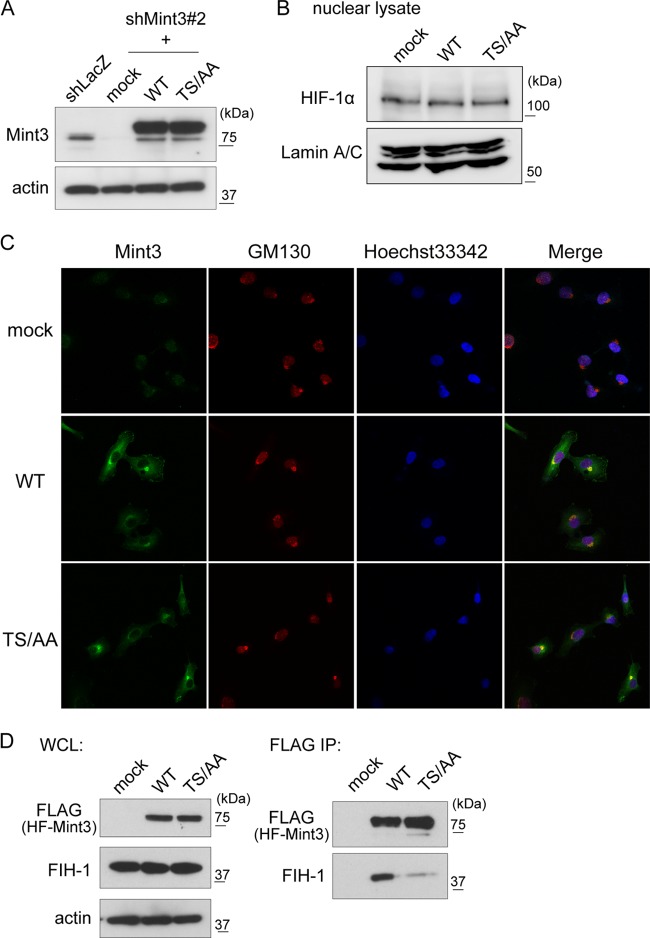
To examine the role of Mint3 phosphorylation in the MT1-MMP/Mint3 axis in HT1080 cells, we generated Mint3-depleted HT1080 (HT-KD) cells and expressed either recombinant WT or TS/AA mutant HF-Mint3 in the depleted cells (Fig. 5A). Neither WT nor TS/AA mutant HF-Mint3 expression affected HIF-1α protein levels in HT1080 cells during normoxia (Fig. 5B). Because Mint3 localizes to the Golgi compartment by interacting with APP and furin, we confirmed the localization of WT and mutant HF-Mint3 in the cells by immunostaining. Both WT and TS/AA mutant HF-Mint3 colocalized with the Golgi compartment marker protein GM130 (Fig. 5C). This localization is similar to that observed for endogenous Mint3 (14, 16). Next, we examined whether HF-Mint3 could bind FIH-1 in an immunoprecipitation assay. WT HF-Mint3 coprecipitated with FIH-1, as previously observed (14, 16), whereas TS/AA mutant HF-Mint3 coprecipitated with FIH-1 substantially less efficiently (Fig. 5D). This result is consistent with the reduced FIH-1-binding activity of Mint3 in cells exposed to rapamycin (Fig. 4A and B). Thus, Mint3 phosphorylation appears to be a prerequisite for its efficient binding to FIH-1, even in cells expressing MT1-MMP.
FIG 5.
Phosphorylation of Mint3 by mTOR promotes the binding of Mint3 to FIH-1. (A) Expression of WT or TS/AA mutant HF-Mint3 in Mint3-depleted HT-KD cells. Mint3 expression was detected by immunoblot analysis. (B) Expression of HIF-1α protein in mock-treated HT-KD cells and those expressing WT or TS/AA mutant HF-Mint3. (C) The localization of WT or TS/AA mutant Mint3 was determined in HT-KD cells. Both WT and TS/AA mutant HF-Mint3 colocalized with the Golgi compartment marker GM130. (D) TS/AA mutant Mint3 bound less efficiently to FIH-1 than did WT Mint3. Whole-cell lysates (WCL) of mock-treated HT-KD cells and those expressing WT or TS/AA mutant HF-Mint3 were subjected to immunoprecipitation (IP) with anti-FLAG antibody beads.
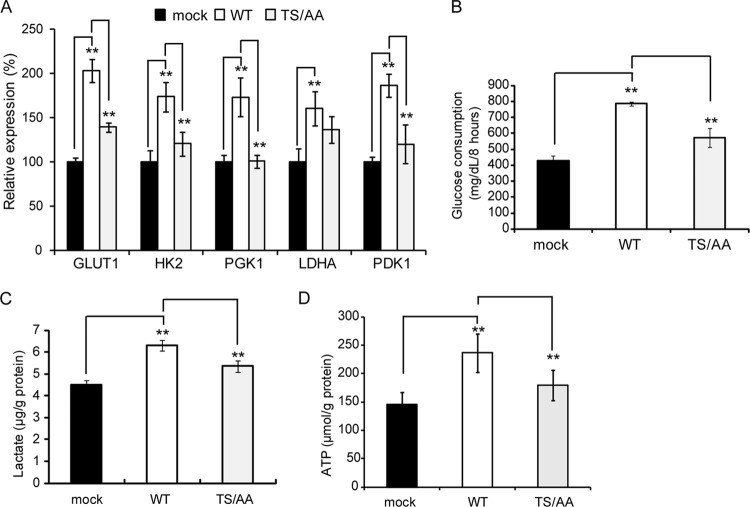
Activation of HIF-1α promotes HIF-1 target gene expression and glycolytic activity (14, 16, 22). Thus, we examined whether Mint3 phosphorylation is indeed associated with increased expression of HIF-1 target genes. The expression of WT HF-Mint3 in Mint3-depleted HT-KD cells promoted the expression of GLUT1, HK2, PGK1, LDHA, and PDK1, whereas the effect of TS/AA mutant HF-Mint3 on gene expression was substantially reduced even when it is expressed at a level higher than that of endogenous Mint3 (Fig. 5A and 6A). Glycolytic activity was also evaluated by measuring glucose consumption (Fig. 6B), lactate production (Fig. 6C), and ATP production (Fig. 6D). Cells expressing TS/AA mutant HF-Mint3 exhibited lower activity than those expressing WT HF-Mint3. Thus, mTOR-mediated phosphorylation of Mint3 regulates the expression of HIF-1 target genes and glycolysis in cells expressing MT1-MMP.
FIG 6.
A nonphosphorylatable mutant form of Mint3 leads to defects in HIF-1 activation and glycolysis. (A) Expression of HIF-1 target genes in Mint3-depleted, mock-treated HT-KD cells and those expressing WT or TS/AA Mint3. (B to D) Mock-treated Mint3-depleted HT-KD cells and those expressing WT or TS/AA mutant HF-Mint3 were analyzed for glucose consumption (B, n = 3), lactate production (C, n = 3), and ATP (D, n = 6). The data in panels B to D were analyzed with the Student t test. **, P < 0.01.
Effect of Mint3 phosphorylation on tumor growth.
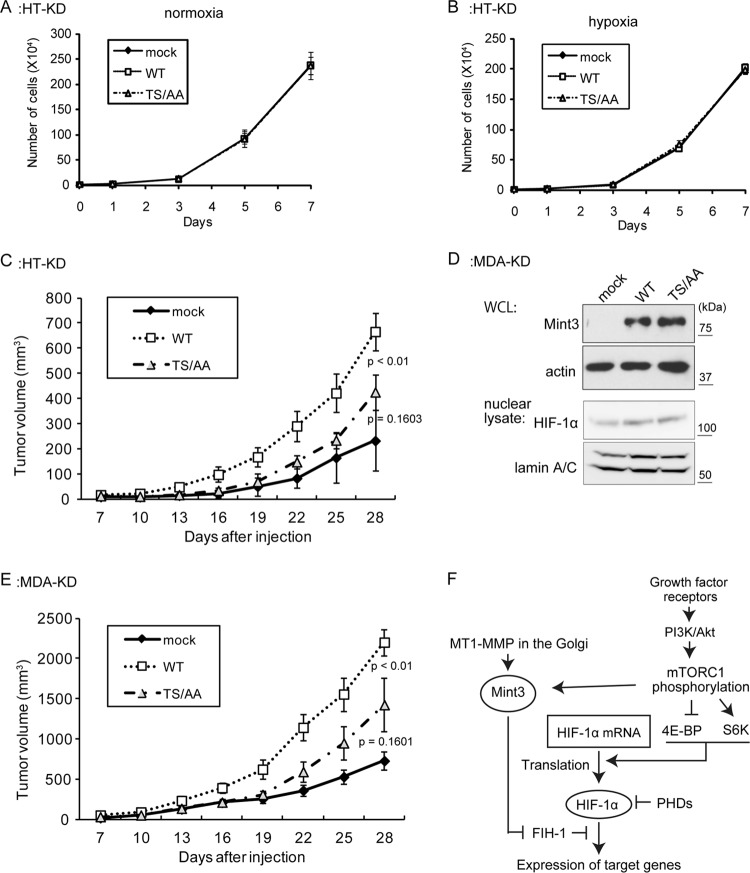
In a previous study, we reported that human tumor cells expressing MT1-MMP can form tumors in immunodeficient mice. Depletion of Mint3 in these cells significantly reduces tumorigenicity but does not affect cell growth under conventional culture conditions in vitro (14). Therefore, we examined whether Mint3 phosphorylation plays a role in the tumorigenicity of cells expressing MT1-MMP by using mutant Mint3. Expression of WT or TS/AA mutant HF-Mint3 or the vector alone in HT-KD cells did not affect cell growth in culture under normoxic or hypoxic conditions (Fig. 7A and B). We next implanted these cells subcutaneously into immunodeficient mice and monitored tumor growth for 28 days. WT HF-Mint3-expressing cells formed tumors that were approximately three times larger than those from control cells (Fig. 7C). In contrast, TS/AA mutant HF-Mint3-expressing cells produced tumors that were only 1.8-fold larger (Fig. 7C). Similar results were obtained with another invasive breast cancer cell line, MDA-MB-231, which also expresses MT1-MMP (14). For this assay, we also generated Mint3-depleted MDA-MB-231 cells (MDA-KD) and expressed either WT or TS/AA mutant HF-Mint3 in these cells (Fig. 7D). Similar to the effects in HT1080 cells, expression of WT HF-Mint3 promoted MDA-KD tumorigenicity, whereas expression of TS/AA mutant HF-Mint3 was less effective (Fig. 7E). Taken together, we have shown that mTOR-mediated phosphorylation of Mint3 is important for HIF-1 activation by the MT1-MMP/Mint3 axis and that this modification contributes to tumor growth in vivo.
FIG 7.
The nonphosphorylatable mutant form of Mint3 reduces tumorigenicity. (A and B) In vitro cell proliferation was analyzed during normoxia (A) and hypoxia (B). (C) Tumor growth of Mint3 KD HT1080 cells was evaluated upon subcutaneous implantation into immunodeficient mice (n = 8). (D) Preparation of mock-treated Mint3 KD MDA-MB-231 cells and those expressing WT or TS/AA Mint3. Mint3 and HIF-1α expression was detected by immunoblot analysis. WCL, whole-cell lysate. (E) Tumor growth of Mint3 KD MDA-MB-231 cells was evaluated upon subcutaneous implantation into immunodeficient mice (n = 8). (F) Schematic summary of HIF-1α regulation by MT1-MMP and mTOR.
DISCUSSION
Rapamycin and Akt inhibitor VIII were shown to inhibit HIF-1α activation via the MT1-MMP/Mint3 axis after the screening of 252 inhibitors of various signaling pathways. The results indicate that the phosphatidylinositol 3-kinase (PI3K)/Akt/mTOR pathway regulates the MT1-MMP/Mint3 axis. The phosphorylation of Mint3 at N-terminal amino acid residues was catalyzed by mTOR, which led to increased binding of Mint3 to FIH-1 and inhibition of FIH-1 in the presence of MT1-MMP. The regulation of Mint3 phosphorylation by mTOR is independent of MT1-MMP, as indicated by the results observed in HEK293FT cells lacking MT1-MMP (Fig. 4E to G) and in HT1080 cells in which MT1-MMP expression was knocked down (Fig. 3D). The T5 threonine and S7 serine residues of Mint3 are the most likely phosphorylation sites, and the replacement of both residues with alanines completely abolished mTOR-dependent modification of Mint3 (Fig. 4D and F). Mint3 may be directly phosphorylated by the serine/threonine kinase activity of mTOR, although other serine/threonine kinases could also mediate Mint3 phosphorylation upon activation by mTOR. The mTOR-mediated phosphorylation of Mint3 itself is not sufficient to induce HIF-1α activation in the absence of MT1-MMP, as shown in HEK293 cells (Fig. 2B). However, Mint3 phosphorylation promoted HIF-1α activation upon the expression of recombinant MT1-MMP in the cells. Thus, Mint3 integrates the actions of MT1-MMP and mTOR to inhibit FIH-1 and thereby activate HIF-1. In other words, activation of HIF-1 by the MT1-MMP/Mint3 axis is coordinated with the activity of mTOR, which monitors cellular growth conditions. Such coordination of different signals via Mint3 also indicates that the Mint3-mediated pathway is an important contributor to HIF-1 regulation during cell growth control. The results of this study are schematically summarized in Fig. 7F.
MT1-MMP, HIF-1, and mTOR are frequently expressed and activated in many types of tumor cells, and each protein is an important molecular hub that regulates different but overlapping phenotypes of malignant tumor cells. mTOR plays a central role in unifying multiple aspects of cell metabolism to maintain the integrity of cell responses by monitoring cellular levels of energy, nutrients, growth signals, and environmental conditions (32, 33, 43). Growth factors stimulate Ras/mitogen-activated protein kinase signals to drive cell growth, and PI3K/Akt signals to activate mTOR to promote survival and metabolic activity. mTOR is a serine/threonine kinase that forms the large multiprotein complexes mTORC1 and mTORC2. Through these complexes, mTOR interacts with multiple signaling molecules, some of which are directly phosphorylated by mTOR. Activation of mTORC1 is known to shift cellular metabolism from a catabolic to an anabolic state to support cell division (32, 33). mTOR phosphorylates S6 kinase and 4E-BP, which are known to promote mRNA translation and increase protein levels during cell growth. HIF-1α is a downstream target of these factors; therefore, mTOR increases the amount of HIF-1 protein (44). However, even when mTOR enhances the translation of HIF-1α during normoxia, the posttranslational regulators PHDs and FIH-1 can suppress HIF-1 activity. We show here that suppression of HIF-1α by FIH-1 can also be released by mTOR in cells expressing MT1-MMP. Signals that activate mTOR are also aberrantly activated in many types of tumor cells; the PI3K pathway is well known to be constitutively activated by gain-of-function mutations in upstream pathway genes or by loss-of-function mutations in the gene encoding PTEN (32, 33, 45). mTOR activity is believed to contribute to the Warburg effect and glucose addiction via HIF-1 activation (32).
Because mTOR can be activated by cell growth signals, we must remember that MT1-MMP promotes cell growth, particularly under three-dimensional culture conditions, such as in collagen gel or in mice (24, 46). Without the collagenase activity provided by MT1-MMP, tumor cells struggle to grow under 3D conditions. MT1-MMP can also modulate the growth factor activity of heparin-binding epidermal growth factor-like growth factor (HB-EGF) by cleaving the N-terminal heparin-binding domain, thereby converting the protein into a potent heparin-independent growth factor (47, 48). HB-EGF is frequently overexpressed in many types of tumors, including ovarian, bladder, pancreatic, and colon tumors, and HB-EGF is a major ligand for ErbB receptors (49–51). MT1-MMP and HB-EGF are coexpressed in ovarian tumor cells, and the processed form of HB-EGF can be detected in ovarian cancer tissue (47, 48). Although the complete mechanism by which MT1-MMP promotes tumor growth in vivo remains to be established, MT1-MMP-mediated proteolytic events that promote tumor growth may also stimulate the signals that activate mTOR. Thus, the protease activity of MT1-MMP, which activates mTOR, may cooperate with the nonproteolytic activation of HIF-1 by the MT1-MMP/Mint3 axis. Thus, the integrated cooperation of the proteolytic and nonproteolytic functions of MT1-MMP appears to effectively promote tumor growth.
We further showed that rapamycin, Akt inhibitor VIII, and KT5823 suppressed HIF-1α activation by the MT1-MMP/Mint3 axis. Unlike rapamycin and Akt inhibitor VIII, the PKG inhibitor KT5823 did not affect the isoelectric points of Mint3 or FIH-1 (Fig. 3B). PKG is a cyclic-GMP-dependent kinase involved in the relaxation of vascular muscle cells by nitric oxide, although its role in tumor progression remains unclear (52). However, inhibition of the MT1-MMP/Mint3 pathway by KT5823 indicates the existence of as-yet-unknown mechanisms that regulate this pathway.
Although Mint3 can be phosphorylated by mTOR (see Fig. S4 in the supplemental material) in HEK293 cells that do not express endogenous MT1-MMP, phosphorylated Mint3 cannot bind and inhibit FIH-1 without the forced expression of MT1-MMP (21). Thus, phosphorylation of Mint3 by mTOR is necessary but not sufficient for inhibition of FIH-1 by Mint3. We also observed that Mint3 isolated from rapamycin-treated HT1080 cells, as well as TS/AA mutant Mint3, retained the ability to bind GST-FIH-1 in vitro (see Fig. S5 in the supplemental material). Nevertheless, phosphorylated Mint3 binds FIH-1 preferentially in MT1-MMP expressing cells. Phosphorylation of Mint3 may increase its recruitment to the Golgi compartment or facilitate its correct localization within the membrane compartment so as to interact with FIH-1 and MT1-MMP. It is also possible that phosphorylation of Mint3 increases its affinity for FIH-1 specifically in cells through interaction with other factors.
Glycolysis is mainly regulated by HIF-1, but another HIF family member, HIF-2, also contributes to tumor malignancy (4). HIF-2 activity is also suppressed by PHDs and FIH-1 during normoxia (4). Because the MT1-MMP/Mint3 axis activates HIF-1 by suppressing FIH-1, we examined whether HIF-2 is regulated similarly to HIF-1 by using a HIF-2α specific reporter assay. The activity of the HIF-2α reporter was enhanced by the MT1-MMP/Mint3 axis in HEK293 and HT1080 cells in a rapamycin-sensitive manner, although the intensity of HIF-2α activation was less than that of HIF-1α (see Fig. S6A and B in the supplemental material). These results might reflect the fact that FIH-1 inactivates HIF-1α more efficiently than HIF-2α (53, 54). The impact of HIF-2 activation by the MT1-MMP/Mint3 axis upon tumor malignancy and other biological events needs to be addressed in future studies.
In summary, this study revealed a novel cross talk between two important hub proteins that regulate tumor malignancy, MT1-MMP and mTOR. Phosphorylation of Mint3 by mTOR cooperates with MT1-MMP to inhibit FIH-1 and thereby activates HIF-1. The integrated influence of MT1-MMP and mTOR on Mint3 may suggest interesting targets for tumor therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Naohiko Koshikawa and Daisuke Hoshino for helpful discussions. We also thank Kouhei Mimura and Yuka Takahashi for technical support.
SCADS inhibitor kits I to III were kindly provided by Screening Committee of Anticancer Drugs supported by a Grant-in-Aid for Scientific Research on Innovative Areas, Scientific Support Programs for Cancer Research, from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT). This work was supported by a Grant-in-Aid for Scientific Research (C), a Grant-in-Aid for Scientific Research on Innovative Areas, and the P-DIRECT (Project for Development of Innovative Research on Cancer Therapeutics) from MEXT to T.S., by a Grant-in-Aid for Scientific Research (S) and on Priority Areas, i.e., the Integrative Research toward the Conquest of Cancer, from MEXT to M.S., and by a Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science to T.H.
Footnotes
Published ahead of print 28 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01169-13.
REFERENCES
- 1.Denko NC. 2008. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 8:705–713. 10.1038/nrc2468 [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, Ratcliffe PJ. 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30:393–402. 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL. 2010. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20:51–56. 10.1016/j.gde.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keith B, Johnson RS, Simon MC. 2012. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12:9–22. 10.1038/nrc3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. 2004. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 36:1–12. 10.1038/emm.2004.1 [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL, Roth PH, Fang HM, Wang GL. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269:23757–23763 [PubMed] [Google Scholar]
- 7.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 22:4082–4090. 10.1093/emboj/cdg392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaelin WG., Jr 2008. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8:865–873. 10.1038/nrc2502 [DOI] [PubMed] [Google Scholar]
- 9.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466–1471. 10.1101/gad.991402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahon PC, Hirota K, Semenza GL. 2001. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675–2686. 10.1101/gad.924501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatenby RA, Gillies RJ. 2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4:891–899. 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 12.Warburg O. 1956. On the origin of cancer cells. Science 123:309–314. 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 13.Ward PS, Thompson CB. 2012. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21:297–308. 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto T, Niiya D, Seiki M. 2011. Targeting the Warburg effect that arises in tumor cells expressing membrane type-1 matrix metalloproteinase. J. Biol. Chem. 286:14691–14704. 10.1074/jbc.M110.188714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. 2003. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 112:645–657. 10.1016/S0092-8674(03)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto T, Seiki M. 2009. Mint3 enhances the activity of hypoxia-inducible factor-1 (HIF-1) in macrophages by suppressing the activity of factor inhibiting HIF-1. J. Biol. Chem. 284:30350–30359. 10.1074/jbc.M109.019216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto M, Sudhof TC. 1998. Mint 3: a ubiquitous mint isoform that does not bind to munc18-1 or -2. Eur. J. Cell Biol. 77:161–165. 10.1016/S0171-9335(98)80103-9 [DOI] [PubMed] [Google Scholar]
- 18.Tanahashi H, Tabira T. 1999. X11L2, a new member of the X11 protein family, interacts with Alzheimer's beta-amyloid precursor protein. Biochem. Biophys. Res. Commun. 255:663–667. 10.1006/bbrc.1999.0265 [DOI] [PubMed] [Google Scholar]
- 19.Han J, Wang Y, Wang S, Chi C. 2008. Interaction of Mint3 with furin regulates the localization of furin in the trans-Golgi network. J. Cell Sci. 121:2217–2223. 10.1242/jcs.019745 [DOI] [PubMed] [Google Scholar]
- 20.Okamoto M, Nakajima Y, Matsuyama T, Sugita M. 2001. Amyloid precursor protein associates independently and collaboratively with PTB and PDZ domains of mint on vesicles and at cell membrane. Neuroscience 104:653–665. 10.1016/S0306-4522(01)00124-5 [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto T, Seiki M. 2010. A membrane protease regulates energy production in macrophages by activating hypoxia-inducible factor-1 via a non-proteolytic mechanism. J. Biol. Chem. 285:29951–29964. 10.1074/jbc.M110.132704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, Mimura K, Abe T, Shioi G, Seiki M, Sakamoto T. 2011. Deletion of the Mint3/Apba3 gene in mice abrogates macrophage functions and increases resistance to lipopolysaccharide-induced septic shock. J. Biol. Chem. 286:32542–32551. 10.1074/jbc.M111.271726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, Mimura K, Seiki M, Sakamoto T. 2011. Genetic dissection of proteolytic and non-proteolytic contributions of MT1-MMP to macrophage invasion. Biochem. Biophys. Res. Commun. 413:277–281. 10.1016/j.bbrc.2011.08.085 [DOI] [PubMed] [Google Scholar]
- 24.Seiki M, Yana I. 2003. Roles of pericellular proteolysis by membrane type-1 matrix metalloproteinase in cancer invasion and angiogenesis. Cancer Sci. 94:569–574. 10.1111/j.1349-7006.2003.tb01484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakamoto T, Seiki M. 2009. Cytoplasmic tail of MT1-MMP regulates macrophage motility independently from its protease activity. Genes Cells 14:617–626. 10.1111/j.1365-2443.2009.01293.x [DOI] [PubMed] [Google Scholar]
- 26.Yoshino S, Hara T, Weng JS, Takahashi Y, Seiki M, Sakamoto T. 2012. Genetic screening of new genes responsible for cellular adaptation to hypoxia using a genome-wide shRNA library. PLoS One 7:e35590. 10.1371/journal.pone.0035590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, Tamanoi F. 2007. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 104:3514–3519. 10.1073/pnas.0608510104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawada M, Inoue H, Masuda T, Ikeda D. 2006. Insulin-like growth factor I secreted from prostate stromal cells mediates tumor-stromal cell interactions of prostate cancer. Cancer Res. 66:4419–4425. 10.1158/0008-5472.CAN-05-4239 [DOI] [PubMed] [Google Scholar]
- 29.Tsuchiya A, Tashiro E, Yoshida M, Imoto M. 2007. Involvement of protein phosphatase 2A nuclear accumulation and subsequent inactivation of activator protein-1 in leptomycin B-inhibited cyclin D1 expression. Oncogene 26:1522–1532. 10.1038/sj.onc.1209962 [DOI] [PubMed] [Google Scholar]
- 30.Kozuka-Hata H, Nasu-Nishimura Y, Koyama-Nasu R, Ao-Kondo H, Tsumoto K, Akiyama T, Oyama M. 2012. Phosphoproteome of human glioblastoma initiating cells reveals novel signaling regulators encoded by the transcriptome. PLoS One 7:e43398. 10.1371/journal.pone.0043398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino D, Koshikawa N, Suzuki T, Quaranta V, Weaver AM, Seiki M, Ichikawa K. 2012. Establishment and validation of computational model for MT1-MMP dependent ECM degradation and intervention strategies. PLoS Comput. Biol. 8:e1002479. 10.1371/journal.pcbi.1002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatini DM. 2006. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6:729–734. 10.1038/nrc1974 [DOI] [PubMed] [Google Scholar]
- 34.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177–189. 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- 35.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- 36.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122–1128. 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- 37.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39:171–183. 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. 2002. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22:7004–7014. 10.1128/MCB.22.20.7004-7014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huffman TA, Mothe-Satney I, Lawrence JC., Jr 2002. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. U. S. A. 99:1047–1052. 10.1073/pnas.022634399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146:408–420. 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi JH, Bertram PG, Drenan R, Carvalho J, Zhou HH, Zheng XF. 2002. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 3:988–994. 10.1093/embo-reports/kvf197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokogami K, Wakisaka S, Avruch J, Reeves SA. 2000. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10:47–50. 10.1016/S0960-9822(99)00268-7 [DOI] [PubMed] [Google Scholar]
- 43.Guertin DA, Sabatini DM. 2007. Defining the role of mTOR in cancer. Cancer Cell 12:9–22. 10.1016/j.ccr.2007.05.008 [DOI] [PubMed] [Google Scholar]
- 44.Wouters BG, Koritzinsky M. 2008. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer 8:851–864. 10.1038/nrc2501 [DOI] [PubMed] [Google Scholar]
- 45.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr 2003. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 4:147–158. 10.1016/S1535-6108(03)00187-9 [DOI] [PubMed] [Google Scholar]
- 46.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. 2003. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell 114:33–45. 10.1016/S0092-8674(03)00513-0 [DOI] [PubMed] [Google Scholar]
- 47.Koshikawa N, Mizushima H, Minegishi T, Eguchi F, Yotsumoto F, Nabeshima K, Miyamoto S, Mekada E, Seiki M. 2011. Proteolytic activation of heparin-binding EGF-like growth factor by membrane-type matrix metalloproteinase-1 in ovarian carcinoma cells. Cancer Sci. 102:111–116. 10.1111/j.1349-7006.2010.01748.x [DOI] [PubMed] [Google Scholar]
- 48.Koshikawa N, Mizushima H, Minegishi T, Iwamoto R, Mekada E, Seiki M. 2010. Membrane type 1-matrix metalloproteinase cleaves off the NH2-terminal portion of heparin-binding epidermal growth factor and converts it into a heparin-independent growth factor. Cancer Res. 70:6093–6103. 10.1158/0008-5472.CAN-10-0346 [DOI] [PubMed] [Google Scholar]
- 49.Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. 2003. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. U. S. A. 100:3221–3226. 10.1073/pnas.0537588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto S, Yagi H, Yotsumoto F, Kawarabayashi T, Mekada E. 2006. Heparin-binding epidermal growth factor-like growth factor as a novel targeting molecule for cancer therapy. Cancer Sci. 97:341–347. 10.1111/j.1349-7006.2006.00188.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raab G, Klagsbrun M. 1997. Heparin-binding EGF-like growth factor. Biochim. Biophys. Acta 1333:F179–F199 [DOI] [PubMed] [Google Scholar]
- 52.Pearce LR, Komander D, Alessi DR. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22. 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- 53.Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, Peet DJ. 2006. Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 281:22575–22585. 10.1074/jbc.M600288200 [DOI] [PubMed] [Google Scholar]
- 54.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. 2004. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J. Biol. Chem. 279:9899–9904. 10.1074/jbc.M312254200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.