Abstract
Gamma interferon (IFN-γ) regulates immune defenses against viruses, intracellular pathogens, and tumors by modulating cell proliferation, migration, invasion, and vesicle trafficking processes. The large GTPase guanylate binding protein 1 (GBP-1) is among the cellular proteins that is the most abundantly induced by IFN-γ and mediates its cell biologic effects. As yet, the molecular mechanisms of action of GBP-1 remain unknown. Applying an interaction proteomics approach, we identified actin as a strong and specific binding partner of GBP-1. Furthermore, GBP-1 colocalized with actin at the subcellular level and was both necessary and sufficient for the extensive remodeling of the fibrous actin structure observed in IFN-γ-exposed cells. These effects were dependent on the oligomerization and the GTPase activity of GBP-1. Purified GBP-1 and actin bound to each other, and this interaction was sufficient to impair the formation of actin filaments in vitro, as demonstrated by atomic force microscopy, dynamic light scattering, and fluorescence-monitored polymerization. Cosedimentation and band shift analyses demonstrated that GBP-1 binds robustly to globular actin and slightly to filamentous actin. This indicated that GBP-1 may induce actin remodeling via globular actin sequestering and/or filament capping. These results establish GBP-1 as a novel member within the family of actin-remodeling proteins specifically mediating IFN-γ-dependent defense strategies.
INTRODUCTION
Gamma interferon (IFN-γ) exerts regulatory functions in a variety of physiological and pathophysiological processes (1, 2). It is best known for its potent immunomodulatory activity (3–5). Additionally, IFN-γ plays an important role in host defenses against infection with viral and microbial pathogens (6–8). For example, IFN-γ can inhibit pathogen replication and intracellular trafficking and pathogen-induced vesicle formation (7, 9). In addition, IFN-γ is an important trigger of antitumoral immune responses (2, 10–14). The direct antitumorigenic activity of IFN-γ on tumor cells includes the induction of apoptosis (15, 16) and the inhibition of cell proliferation, migration, and invasion (3, 17–20).
Guanylate binding protein 1 (GBP-1) is among the most significantly induced proteins in cells exposed to IFN-γ (21, 22). GBP-1 belongs to the dynamin superfamily of large GTPases (23, 24), which is characterized by an oligomerization-dependent GTPase activity (25–28). GBP-1 has been shown to mediate the antibacterial and antiviral activities of IFN-γ (29, 30). Similar antipathogen effects have been observed with mouse GBPs (mGBPs), indicating that GBPs exert conserved functions in humans and rodents (9, 31–33). In addition, GBP-1 is both necessary and sufficient for the inhibitory effects of IFN-γ on cell proliferation, migration, and invasion, as shown in endothelial cells and epithelial tumor cells (17, 19, 34–37). Of note, the putative murine homologue of human GBP-1, mGBP-2, has also been shown to inhibit cell motility (38). In accordance with its cell biological activities, the expression of GBP-1 in colorectal carcinoma (CRC) has been associated with a significantly improved prognosis (39). A recent comprehensive study from the Cancer Genome Atlas Network confirmed that GBP-1 expression in CRC is associated with lower tumor aggressiveness (40).
Despite the significant clinical relevance of GBP-1 expression and its well-documented functions, the molecular mechanisms of action of the protein have not been resolved. It has been proposed that GBP-1 might inhibit cell proliferation through the suppression of β-catenin/T cell factor (TCF) signaling (19, 34). In addition, the GTPase activity of GBP-1 is required for the upregulation of integrin α4 and the inhibition of MMP-1 expression, leading to the inhibition of cell migration and invasion, respectively (35, 41). However, no direct molecular target of GBP-1 has been identified as yet. Accordingly, the goal of this study was to identify cellular binding factors of GBP-1 and to determine the function of these molecules in the regulation of the cellular response to IFN-γ.
(Parts of this work were conducted by N. Ostler in partial fulfillment of the requirements for a doctoral thesis.)
MATERIALS AND METHODS
Plasmids.
The plasmids pMCV1.4(−) and pMCV2.2(−), the latter of which contains a gentamicin resistance cassette, were obtained from Mologen (Berlin, Germany). The Flag (F) tag sequence was cloned into both vectors using the EcoRV/EcoRI restriction sites. Green fluorescent protein (GFP), GBP-1 (NCBI accession number NM_002053), and various GBP-1 mutants were inserted in frame into the pMCV1.4-Flag construct as previously described (42). The following expression vectors were created: F–GBP-1, F–GBP-1(R227E/K228) (28), F–GBP-1(R240A) (43), F–GBP-1(ΔCaaX), and F–GBP-1(K51A) (26, 44). Furthermore, an expression vector for the GFP–GBP-1 fusion protein (F–GFP–GBP-1) was generated with the pMCV1.4 construct as previously described (42). Additionally, the sequence of GBP-1 was inserted in frame into the pMCV2.2-Flag vector using the EcoRI restriction site (17).
The human β-actin gene (NCBI accession number NM_001101.3) was amplified from cDNA derived from mRNA isolated from HeLa cells. The forward primer harbors a restriction site for BamHI (underlined) (5′-CCGGGATCCAGGATGGATGATGATATCGCC-3′), and the reverse primer contains a restriction site for EcoRI (underlined) (5′-GCGGAATTCTTGAAGCATTTGCGGTGG-3′). The PCR product was subjected to restriction digestion with BamHI and EcoRI and then inserted in frame into the BamHI and EcoRI sites within the pcDNA4-Myc/His B vector (Life Technologies/Invitrogen, Karlsruhe, Germany). The final construct, pcDNA4 β-actin-Myc, was assessed via full-length sequencing of the inserted gene.
Cell culture.
HeLa cells were purchased from ATCC (CCL-2; Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM l-glutamine (both obtained from PAA Laboratories, Pasching, Austria) and 10% fetal calf serum (FCS; Biochrom, Berlin, Germany) at 37°C in a humidified atmosphere with 8.5% CO2. The cells were authenticated by the German Collection of Microorganisms and Cell Cultures (Leibniz Institute DSMZ, Braunschweig, Germany) using nonaplex PCR DNA profiling of 8 highly polymorphic sites of short tandem repeats (STRs). Primary human umbilical vein endothelial cells (HUVECs) were purchased from PromoCell (Heidelberg, Germany) and were maintained in endothelial cell growth medium (ECGM; PromoCell) supplemented with 2% FCS at 37°C in a humidified atmosphere with 5% CO2. For routine cultivation, confluent cells were washed once with phosphate-buffered saline (PBS), detached using 0.5 g/liter trypsin and 0.2 g/liter EDTA in Hanks balanced salt solution (trypsin-EDTA; PAA Laboratories) for 2 to 3 min, and passaged in a 1:4 ratio (one passage) in uncoated culture flasks (Nunc, Wiesbaden, Germany). All experiments were performed between passages 4 and 10. Human peripheral blood mononuclear cells were isolated from the blood of healthy donors by standard density gradient centrifugation using Lymphoprep (Nycomed, Zurich, Switzerland). Cells were obtained according to Good Scientific Practice Guidelines and in accordance with the requests of the Ethics Committee of the Medical University of Vienna. Cells were maintained in RPMI 1640 medium supplemented with 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (all from Life Technologies/Invitrogen) and with 10% heat-inactivated FCS (Sigma-Aldrich). T cells were either left untreated or stimulated with a combination of CD3 monoclonal antibody (MAb) OKT3 (5 μg; Ortho Pharmaceuticals, Raritan, NJ) and MAb Leu-28 (5 μg/ml; BD Biosciences, Franklin Lakes, NJ). The stimulation was carried out using 108 antibody-coated goat anti-mouse IgG Dynabeads (Life Technologies/Invitrogen) for 30 min at room temperature.
Cells of the DLD-1 colorectal carcinoma cell line (CCL-221; ATCC) were cultivated in RPMI 1640 medium supplemented with 10% FCS (Biochrom) and 2 mM l-glutamine (PAA Laboratories) at 37°C in a humidified atmosphere with 5% CO2. For the cultivation of DLD-1 cells stably expressing F–GBP-1 and pMCV2.2, 500 μg/ml G418 (PAA Laboratories) was added to the culture medium. All cells were tested monthly for mycoplasma contamination using a MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland) and tested negative each time.
For stimulation with recombinant proteins, HUVECs were seeded on a surface coated with 1.5% gelatin and incubated overnight in ECGM supplemented with 0.5% FCS (ECGM–0.5% FCS), while HeLa cells were seeded on uncoated surfaces and incubated overnight in DMEM–0.5% FCS. The cells were subsequently treated with IFN-γ (100 U/ml or 50 U/ml for cells transfected with small interfering RNA [siRNA]; Roche, Grenzach-Wyhlen, Germany) in the same medium for 24 h.
Transfection.
The transfection of HeLa cells with expression vectors was performed using the calcium phosphate method (45). For the RNA interference (RNAi) experiments, the transfection was performed with RNAiMax in Opti-MEM medium (both obtained from Life Technologies/Invitrogen, Darmstadt, Germany) according to the manufacturer's instructions. The transfection was performed in either 4-well Lab-Tek chamber slides or 6-well cell culture plates (both purchased from Nunc, Thermo Fisher Scientific, Bonn, Germany) at a cell density of 2.8 × 104 and 1.3 × 105 cells, respectively. GBP-1 Stealth Select RNAi siRNA (HSS104020) oligonucleotides specific for human GBP-1 and stealth nontargeting negative-control RNAi duplexes (medium GC content) were used at a final concentration of 16.6 nM (both were purchased from Molecular Probes/Invitrogen). HUVECs were transfected with the PromoFectin transfection reagent (PromoCell) as described by the manufacturer. To establish a stable DLD-1 cell line, cells were transfected with the expression plasmid pMCV2.2 F–GBP-1 (encoding an N-terminal F–GBP-1) using Lipofectamine 2000 (Life Technologies/Invitrogen). Selection was carried out by adding 500 μg/ml G418 (PAA Laboratories) to the culture medium, and three independent cell clones were expanded. DLD-1 cells were transfected with a pMCV2.2-Flag vector and selected as a population of cells for the negative control (17).
IP.
HeLa cells were seeded at a density of 7 × 105 cells in 10-cm culture dishes (Nunc) and transfected 24 h later with 14 μg DNA, as described above. The cells were harvested 30 h after transfection with a sterile cell scraper (Nunc) in 800 μl of ice-cold immunoprecipitation (IP) lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% Igepal, supplemented with one tablet of Complete Mini EDTA-free protease inhibitor cocktail [Roche] per 10 ml). DLD-1 cells transfected with F–GBP-1 were seeded in 10-cm culture dishes at a cell density of 8 × 105 cells and harvested after 24 h, as described for HeLa cells. The protein concentration was determined using a DC assay (Bio-Rad Laboratories, Munich, Germany) based on the Lowry method, as described by the manufacturer. One milligram of lysate was precleared via incubation with 25 μl Sepharose CL-6B (Sigma-Aldrich) for 2 h in an overhead shaker at 4°C. The immunoprecipitation reactions were performed overnight at 4°C at low-speed rotation.
The Flag IP/Myc IP lysates were incubated with either 15 μl of anti-Flag affinity gel M2 or 20 μl of anti-c-Myc agarose affinity gel (both from Sigma-Aldrich). For the actin IP, preclearing was performed with 25 μl of a protein G Plus-protein A agarose suspension (Calbiochem/Merck Millipore, Darmstadt, Germany). For immunoprecipitation, 25 μl of protein G Plus-protein A agarose was incubated with 5 μl polyclonal actin antibody (0.4 to 0.8 mg/ml; Sigma-Aldrich) and the precleared lysates. For the GBP-1 IP, beads were prepared by covalently cross-linking the GBP-1 antibody (1B1; hybridoma supernatant [22]) with the BS3 cross-linker (Thermo Fisher Scientific). Briefly, 400 μl of protein G-Sepharose 4 Fast Flow (Amersham Biosciences/GE Healthcare, Solingen, Germany) was resuspended in 650 μl 20 mM HEPES buffer. After the addition of 400 μl of 1B1 hybridoma supernatant, the suspension was incubated for 1 h at room temperature at low-speed rotation. For the cross-linking reaction, 2 mg BS3 was added and the mixture was incubated for 1 h. The reaction was stopped by the addition of Tris-HCl at a final concentration of 50 mM for 15 min. After subsequent washing, GBP-1 beads were resuspended in 650 μl PBS and 30 μl was used for each sample. As a negative control for T cell IPs, 3 μg of a rat IgG isotype control (R&D Systems, Abingdon, United Kingdom) was added.
For the IPs with purified, recombinant proteins, a preclearing step was not performed and 10 μg of protein was used. The bead-coupled protein complexes were washed four times with 1 ml IP lysis buffer and four times with IP wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% Igepal). The immunoprecipitated proteins were eluted from the beads by adding 15 μl IP wash buffer and 15 μl 2× Laemmli buffer (Bio-Rad Laboratories) and subsequent boiling of the solution for 5 min. The immunoprecipitates were analyzed via sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), followed by either silver staining or Western blotting.
Coomassie staining.
The gels were stained for at least 3 h at room temperature with a Coomassie solution (40% [vol/vol] methanol, 7% [vol/vol] acetic acid, 0.25% Coomassie brilliant blue G-250). The gels were destained using a destaining solution (40% [vol/vol] methanol, 7% [vol/vol] acetic acid).
Silver staining.
SDS-polyacrylamide gels were assessed via silver staining using a ProteoSilver Plus silver stain kit purchased from Sigma-Aldrich, and staining was carried out according to the manufacturer's instructions. The gels were fixed overnight at 4°C. After developing for 3 to 5 min, the gels were then reequilibrated in water until the bands were excised for mass spectrometry (MS).
Mass spectrometry.
Bands from the silver-stained gels were excised using a sterile scalpel and destained with the destaining solutions provided in the ProteoSilver Plus stain kit (Sigma-Aldrich), as described by the manufacturer. The proteins were identified following trypsin in-gel digestion by matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) MS via peptide mass fingerprinting using a Bruker Ultraflex TOF/TOF MALDI instrument (Bruker Daltonics, Bremen, Germany). The instrument was operated in the positive-ion reflectron mode using 2,5-dihydroxybenzoic acid and methylenediphosphonic acid as a matrix. Sum spectra consisting of 200 to 400 single spectra were acquired. For data processing and instrument control, the Compass (version 1.1) software package, consisting of FlexControl (version 2.4), FlexAnalysis (version 3.0), and BioTools (version 3.0), was used. The proteins were identified via a MASCOT peptide mass fingerprint search (Matrix Science) using the NCBInr database. For the search, a mass tolerance of 75 ppm was allowed, and the carbamidomethylation of cysteine as a global modification and the oxidation of methionine as a variable modification were used.
Western blotting.
The cells were harvested, and proteins were extracted using radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% sodium desoxycholate, 1% Igepal, and one tablet of protease inhibitor [Complete Mini; Roche] per 10 ml of buffer). The following quantities of protein were separated by SDS-PAGE and analyzed via Western blotting as previously described (46): 10 μg of protein lysate, 1/5 of the IP eluate for the pulldown control, and 4/5 of the IP eluate for binding partner codetection. The following primary antibodies were used: monoclonal mouse anti-Flag tag (M2; 1:5,000), polyclonal rabbit anti-Flag tag (1:1,000), and polyclonal rabbit antiactin (1:1,000; each from Sigma-Aldrich); monoclonal mouse anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; 1:40,000; Chemicon/Merck Millipore, Darmstadt, Germany); monoclonal mouse antigelsolin (1:1,000; Abcam, Cambridge, United Kingdom); monoclonal mouse anti-BSA (1:200; Santa Cruz, Santa Cruz, CA); monoclonal rat anti-human GBP-1 (1:500; clone 1B1 [22]); monoclonal rat anti-human GBP-2 (1:500; clone 1H2; obtained from E. Kremmer, Helmholtz Zentrum, Munich, Germany); and polyclonal rabbit anti-Myc tag (1:1,000) and monoclonal mouse anti-Myc tag (9B11; 1:3,000) (both from Cell Signaling). Rabbit anti-rat, goat anti-mouse, and goat anti-rabbit immunoglobulin G antibodies coupled to horseradish peroxidase (each from Dako) were used as secondary antibodies at a 1:5,000 dilution. Protein detection was performed using an enhanced chemiluminescence Western blot detection system (ECL Pierce Biotechnology, Rockford, IL) and Rx films (Fuji, Tokyo, Japan).
Immunofluorescence analysis.
For immunofluorescence analysis, HeLa cells and HUVECs were seeded in 4-well Lab-Tek chamber slides (Nunc) at a cell density of 2.5 × 104 cells. One microgram of plasmid was transfected per well. When indicated, the cells were treated with actin polymerization inhibitors (HeLa cells, 0.2 μM cytochalasin D-latrunculin B; HUVECs, 0.1 μM cytochalasin D-latrunculin B [both obtained from Sigma-Aldrich, Seelze, Germany]) for 30 min at 37°C. The cells were fixed for 10 min in 4% buffered paraformaldehyde and permeabilized in 0.1% Triton X-100 (both obtained from Sigma-Aldrich) at 24 h posttransfection. For anti-GBP-1 staining, an antigen target retrieval was carried out after cell fixation (10 min at 95°C using a retrieval solution buffered at pH 9 [Dako, Hamburg, Germany]). Immunofluorescence analyses were carried out as previously described (42). The following antibodies were used for immunofluorescence staining: a polyclonal rabbit anti-Flag tag antibody (diluted 1:500; ABR; Thermo Fisher Scientific, Rockford, IL) and a monoclonal rat anti-human GBP-1 (clone 1B1) antibody (diluted 1:25) (22). After washing, the cells were incubated for 1 h at room temperature with the following secondary antibodies (diluted 1:500): Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 488-conjugated goat anti-rat IgG (both from Molecular Probes/Invitrogen). Actin was stained using a fluorescently labeled phalloidin (1:70; Alexa Fluor 546-phalloidin; Molecular Probes/Invitrogen) in combination with the secondary antibody. Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole; 1:5,000; Invitrogen/Molecular Probes) or Draq5 (1:800; Cell Signaling, Danvers, MA). Fluorescence was visualized at room temperature using a confocal microscope (TCS SP5 or TCS SPE; both obtained from Leica Microsystems, Wetzlar, Germany) equipped with LAS-LAF software at a ×63 magnification. All images presented are single sections in the z plane (1 Airy unit) and are representative of the images for at least 80% of the transfected cells. For quantitative assessment of the severity of the actin disruption by GBP-1 or GBP-1 mutant forms, at least 80 images per condition were analyzed, and the status of fibrous actin was evaluated.
Computer-assisted determination of protein colocalization at the single-cell level.
To quantify the colocalization of fluorescence signals in images, ImageJ Colocalization Colormap software was used (47, 48). The software calculates the fraction of positively correlated pixels of the image on a pixel-per-pixel basis. The output is provided either as an index of correlation (Icorr) score or as an image. The correlated pixels are represented in hot colors (red and yellow). For Icorr score determination, five images per type were quantified.
Protein purification.
Actin (>99% pure) and gelsolin were purchased from Cytoskeleton (Denver, CO). Bovine serum albumin (BSA) was purchased from New England BioLabs (Ipswich, MA). His-tagged GBP-1 (with the 6× His tag being at the C terminus), His–GBP-3, and His-GFP were expressed from a pQE9 vector in Escherichia coli strain M15 (Qiagen). The induction of His–GBP-1, His–GBP-3, and His-enhanced GFP expression was carried out at an optical density at 600 nm of ∼0.6 using 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG; Peqlab, Erlangen, Germany). The proteins were purified under native conditions using standard Ni-nitrilotriacetic acid affinity Sepharose column chromatography as previously described (22, 44). Eluted proteins were dialyzed against PBS, and dithiothreitol (DTT; Sigma-Aldrich) was added at a final concentration of 2 mM. The purity of the protein extracts was assessed via SDS-PAGE, subsequent Coomassie staining, and Western blotting.
AFM.
Freshly cleaved mica (V1 quality, round, 9.5 mm; Electron Microscopy Sciences, Hatfield, PA) was completely covered by 100 mM MgCl2 (100 μl) and incubated for 5 min at room temperature (RT). The fluid was removed and washed once with water (100 μl), and afterwards the mica was air dried. Precoating with MgCl2 was previously used to increase the absorption of biomolecules to mica surfaces (49, 50). Actin (Cytoskeleton) was solved in PBS with 2 mM DTT to a final concentration of 2 mg/ml and polymerized by actin polymerization buffer (50 mM KCl, 2 mM MgCl2, 1 mM ATP; Cytoskeleton) in the presence or absence of GBP-1 (2 mg/ml) for 1 h. GBP-1 and buffer control (PBS plus 2 mM DTT) samples were treated equally. Five microliters of protein sample was deposited on MgCl2-pretreated mica, and the combination was incubated for 5 min at RT and dried in a stream of nitrogen for 3 s. Next, the mica was washed twice with 10 μl of distilled water and dried by a stream of nitrogen as described above. Atomic force microscopy (AFM) data were obtained using a non-contact-mode AFM (XE-100; Park Systems, Santa Clara, CA) with a silicon tip (ACTA; Park Systems) under ambient conditions and a scan rate of 1 Hz. Images were flattened and equalized with WSxM Develop (version 6.2) software (51). Relative actin filament length was determined by calculating the quotient of (i) the number of fibers and (ii) the number of open filament ends counted in five defined optical fields.
DLS.
Actin was polymerized in the presence and absence of GBP-1. Protein solutions were prepared as described above for AFM analysis, and the final concentration was adjusted to 0.33 mg/ml. All samples were applied to a dynamic light scattering (DLS) particle size analyzer (HORIBA LB-550; Retsch Technology GmbH, Haan, Germany) at 650 nm for 2 min. The median (50%) hydrodynamic radius (D50), resembling the polymer size, including the hydration shell, was averaged over 6 independent measurements. The standard deviation was calculated from the D50 values.
Actin polymerization assay.
The actin polymerization assay was performed in the presence and absence of GBP-1 using an actin polymerization Biochem kit (Cytoskeleton) according to the manufacturer's instructions. GBP-1 was replaced by BSA and GFP as nonrelevant proteins serving as a negative control. In addition, GBP-3 was applied as the closest homologue of GBP-1. Actin was dissolved in monomeric globular actin (G-actin) buffer (5 mM Tris-HCl, pH 8.0, 233 μM ATP, 0.2 mM CaCl2) to a final concentration of 0.5 mg/ml. For all experiments, proteins were applied in a 3 μM concentration. The baseline fluorescence of pyrene actin was measured once every minute for 4 min. Next, the above-mentioned proteins, the corresponding volume of the same buffer, and actin polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP) were mixed and added as a master mix. The total volumes and actin concentration were kept constant for all samples. The samples were mixed for 5 s, and the fluorescence was measured every 30 s for 1 h in duplicate in a black 96-well plate with a Tecan Infinite m200 instrument (Tecan, Männedorf, Switzerland) using a 350-nm excitation filter and a 410-nm detection filter.
Actin cosedimentation assay.
The actin cosedimentation assay was performed using an actin binding protein Biochem kit (Cytoskeleton) following the manufacturer's protocol. Briefly, 16 μM actin was incubated with 2 μM BSA, 2 μM α-actinin, or 2.8 μM GBP-1 in G-actin buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2) supplemented with 0.2 mM ATP for 30 min at 4°C. Actin polymerization was induced by addition of polymerization buffer (50 mM KCl, 2 mM MgCl2, 1 mM ATP [final concentrations]) and incubation for 30 min at 24°C. Following centrifugation at 125,000 × g for 90 min at 24°C in a Hitachi Himac CS-FNX ultracentrifuge (Hitachi Koki, Willich, Germany), equal volumes of supernatants and pellets were analyzed by SDS-PAGE and Coomassie blue staining.
G-actin binding assay.
Binding of GBP-1 to G actin was monitored by incubating up to 10 μM recombinant GBP-1 and actin in G-actin buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2) supplemented with 0.2 mM ATP and 0.2 mM GTP for 30 min at 25°C. Proteins were then separated by nondenaturing 10% polyacrylamide gel electrophoresis in the presence of 0.2 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT, and 0.2 mM GTP at 200 V for 2 h. Gels were stained with Coomassie blue, destained, and subjected to Western blotting for the immunodetection of actin and GBP-1. Densitometric analysis was performed using ImageJ software (48).
Statistical analyses.
Two groups were compared by the appropriate Student's t test, and multiple groups were compared by analysis of variance (ANOVA) extended with an honestly significant difference (HSD) post hoc test. Statistical analysis was performed using SPSS (version 16) software (SPSS Inc., Chicago, IL) or Statistica (version 8) software (StatSoft, Tulsa, OK). A P value of <0.05 was considered significant.
RESULTS
Actin is a cellular binding factor of GBP-1.
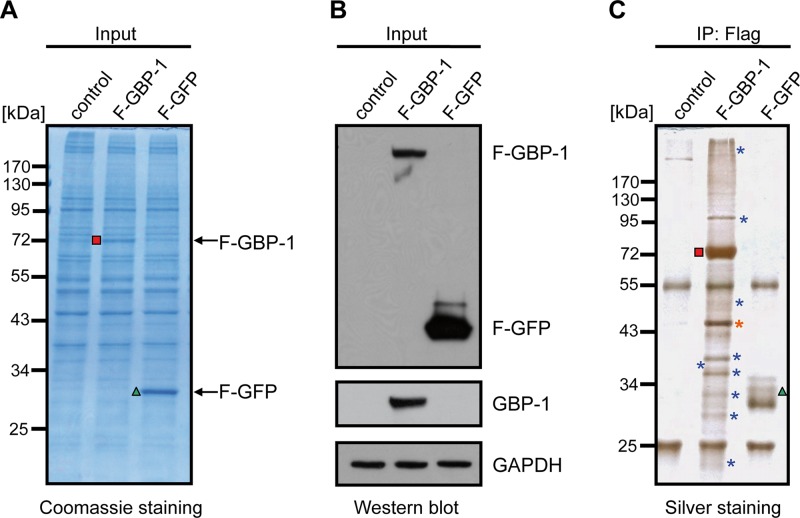
In order to identify putative interaction partners of GBP-1, an immunoprecipitation approach based on the transient expression of Flag-tagged GBP-1 (F–GBP-1) in HeLa cells was chosen. Cells transfected with an empty vector (control) or Flag-tagged GFP (F-GFP) served as negative controls. The expression of F–GBP-1 and F-GFP was confirmed by Coomassie staining (Fig. 1A, red square and green triangle, respectively) and anti-Flag tag Western blot analysis (Fig. 1B). Silver staining of immunoprecipitates obtained with an anti-Flag tag antibody yielded different band patterns with F–GBP-1, F-GFP, and control transfected cells (Fig. 1C). Bands appearing specifically in the F–GBP-1 precipitate were considered potential GBP-1 binding factors (Fig. 1C, asterisks) and were identified via MALDI-TOF mass spectrometry and peptide mass fingerprinting (Table 1). Beta-actin (subsequently termed actin) displayed the most prominent signal in the silver stain (Fig. 1C, orange asterisk) and was further investigated.
FIG 1.
Identification of cellular interaction partners of human GBP-1 in HeLa cells. (A) HeLa cells were transfected with Flag-tagged GBP-1 (F–GBP-1) or with Flag-tagged GFP (F-GFP) or the empty parental vector (control) as controls. The protein lysates were separated via 10% SDS-PAGE and stained using Coomassie. Red square, GBP-1; green triangle, GFP. (B) Western blot analysis using antibodies against GBP-1 and the Flag tag. The detection of the GAPDH protein demonstrates equal protein loading. (C) Silver staining analysis of precipitates obtained via immunoprecipitation (IP) using an anti-Flag affinity gel with the various transfected cells, as described in the legend to panel A. Red square, F–GBP-1; green triangle, F-GFP; blue asterisks, potential interaction partners, which varied between the F–GBP-1 precipitate and the control precipitates; orange asterisk, actin.
TABLE 1.
Potential binding factors of GBP-1 analyzed by MALDI-TOF mass spectrometry
| Gene name | Protein name | Molecular mass (kDa) | Accession no. |
|
|---|---|---|---|---|
| Uniprot | RefSeqa | |||
| HSP90AB1 | Heat shock protein HSP 90-beta | 83.3 | P08238 | NP_031381.2 |
| GBP-1 | Guanylate-binding protein 1 | 67.0 | P32455 | NP_002044.2 |
| TUBB | Tubulin, beta polypeptide | 49.7 | P07437 | NP_821133.1 |
| ACTB | Beta-actin | 42.7 | P60709 | NP_001092.1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 36.0 | P04406 | NP_002037.2 |
| LDHA | Lactate dehydrogenase A | 36.4 | P00338 | NP_005557.1 |
| RPL5 | 60S ribosomal protein L5 | 34.4 | P46777 | NP_000960.2 |
| SLC25A6 | ADP, ATP carrier protein, liver isoform T2 | 35.0 | P12236 | NP_001627.2 |
| YWHAE | 14-3-3 protein epsilon | 29.3 | P62258 | NP_006752.1 |
| PRDX1 | Peroxiredoxin 1 | 22.3 | Q06830 | NP_002565.1 |
RefSeq, reference sequence.
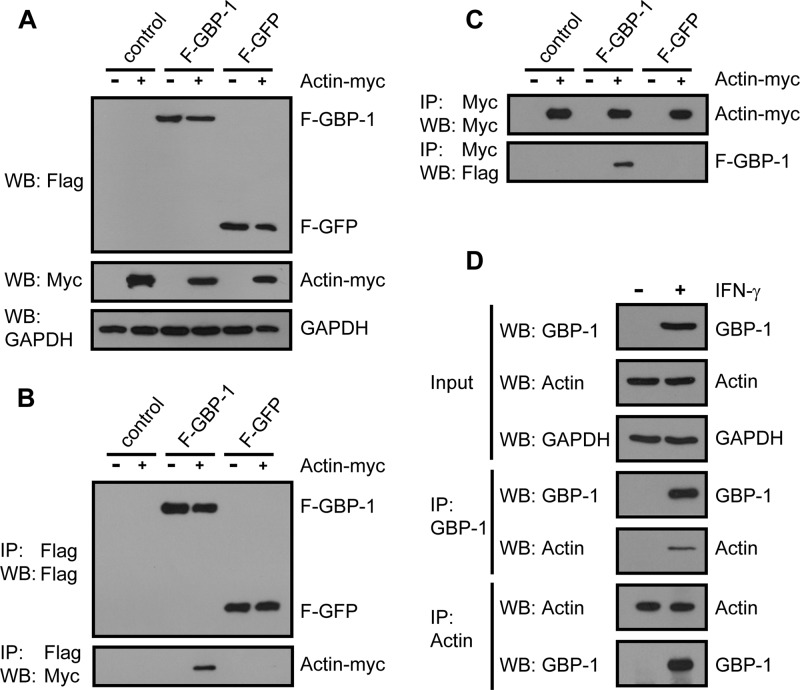
The interaction between GBP-1 and actin was confirmed by different approaches, using overexpressed GBP-1 and actin as well as the endogenous proteins. First, HeLa cells were cotransfected with F–GBP-1, F-GFP, or the empty vector together with an expression plasmid encoding Myc-tagged actin (actin-Myc) or the corresponding empty control vector. The expression of the recombinant proteins was confirmed by Western blotting analyses (Fig. 2A). F–GBP-1 and actin-Myc were coimmunoprecipitated in a reciprocal manner via either a Flag pulldown (Fig. 2B, bottom) or a Myc pulldown (Fig. 2C, bottom). No signal was obtained with either the control vector or F-GFP, thereby demonstrating the specificity of the interaction.
FIG 2.
Interaction of tagged and endogenous GBP-1 and actin proteins shown by reciprocal coimmunoprecipitation. (A) HeLa cells were cotransfected with F–GBP-1, F-GFP, or the empty parental vector each together with an expression plasmid encoding Myc-tagged actin (actin-Myc) or the corresponding control vector. Subsequently, 10 μg of lysate was analyzed via Western blotting (WB) using antibodies against the Flag and Myc tags. GAPDH was used as a loading control. (B) Immunoprecipitation analysis of the extracts from panel A with anti-Flag antibody and subsequent Western blot analysis with antibodies against either the Flag or Myc tag. (C) Lysates, transfected and analyzed as described in the legend to panel A, were immunoprecipitated using an antibody against the Myc tag. The precipitated proteins were subsequently analyzed via Western blotting using the indicated antibodies. (D) HeLa cells were either left untreated or treated with IFN-γ (100 U/ml, 24 h). The protein extracts were immunoprecipitated using an antibody against GBP-1 or actin and analyzed via Western blotting. GBP-1 and actin were detected using specific antibodies.
Second, the interaction between recombinant F–GBP-1 and endogenous actin was confirmed using a reciprocal precipitation approach with the colorectal carcinoma cells (DLD-1), which stably expressed F–GBP-1 (data not shown).
Third, the interaction between endogenous GBP-1 and actin was investigated using HeLa cells treated with recombinant IFN-γ (100 U/ml for 24 h). IFN-γ treatment resulted in robust GBP-1 expression, while actin expression remained constant (Fig. 2D, input). The reciprocal coprecipitations demonstrated that endogenous IFN-γ-induced GBP-1 also efficiently interacted with endogenous actin (Fig. 2D, IP: GBP-1 and IP: Actin, bottom). The interaction of the two endogenous proteins was also confirmed in human primary T cells, which constitutively express GBP-1 (data not shown). Of note, Western blot analysis indicated similar levels of expression of ectopically expressed (Fig. 2A) and IFN-γ-induced GBP-1 (Fig. 2D).
GBP-1 colocalizes with actin and disrupts stress fibers.
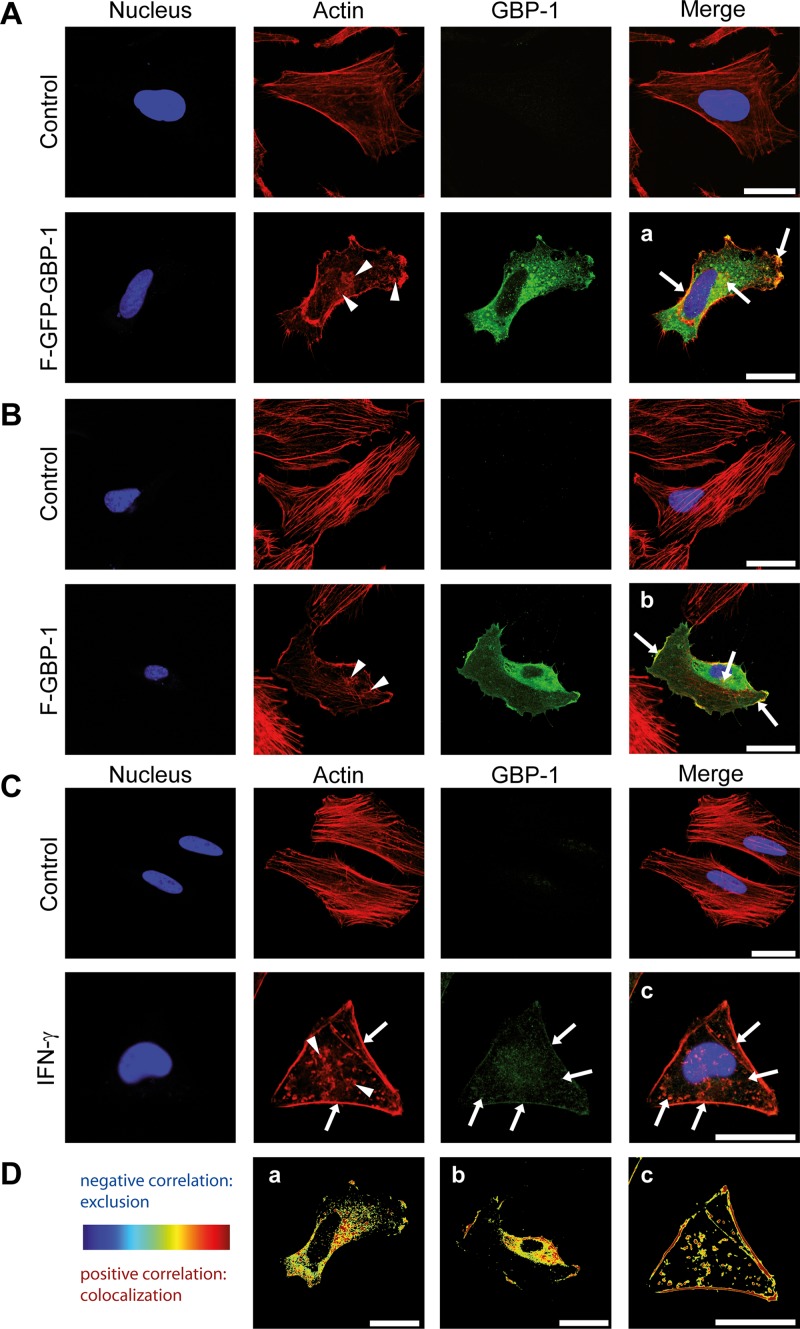
The intracellular colocalization of GBP-1 and actin was assessed via immunofluorescence analysis and confocal microscopy. To achieve this goal, HeLa cells were transfected with either an expression plasmid encoding GFP-tagged GBP-1 (F–GFP–GBP-1) or an empty parental vector (control). We have previously demonstrated that the GFP tag does not affect the intracellular localization of GBP-1 compared with that of the endogenously expressed protein (42, 52). In agreement with previous findings, F–GFP–GBP-1 appeared in granular structures within the cytoplasm and at the plasma membrane (Fig. 3A, GBP-1, green) (42). Filamentous actin (F-actin) was stained with fluorescently labeled phalloidin. In the cells transfected with the control vector, F-actin localized as membrane-associated cortical actin and in parallel-oriented, cytoplasmic stress fibers (Fig. 3A, control, red staining). Interestingly, the stress fibers were considerably impaired in the cells transfected with F–GFP–GBP-1 and F–GBP-1 (Fig. 3A, arrowheads). The colocalization of F–GFP–GBP-1 and actin was observed in granular structures within the cytoplasm and most prominently at the cell membrane (Fig. 3A, arrows).
FIG 3.
GBP-1 colocalizes with cellular actin and disrupts the stress fibers of the actin cytoskeleton. Images were taken using a Leica TCS SPE confocal microscope with z-plane focusing on the attachment site of the cell culture slide. Nuclei were counterstained with Draq5. (A) HeLa cells were transfected with an expression plasmid for Flag- and GFP-tagged GBP-1 (F–GFP–GBP-1) or the empty parental vector (control). F–GFP–GBP-1 was visualized by the fluorescence of the GFP tag, and actin was visualized using Alexa Fluor 546-labeled phalloidin. Arrowheads, the disrupted actin structure, observed as a granular distribution of actin proteins in the cytoplasm of F–GFP–GBP-1-transfected cells; arrows, colocalization of actin and F–GFP–GBP-1 observed at the plasma membrane and actin aggregates. (B) HUVECs were transfected with either F–GBP-1 or the empty parental vector (control). GBP-1 was detected in a subsequent immunofluorescence staining using an antibody against the Flag tag. Actin was detected by a fluorescently labeled phalloidin. The staining pattern was similar to that described in the legend to panel A. Arrows, colocalization. (C) HeLa cells were either left untreated (control) or treated with IFN-γ (100 U/ml, 24 h). Endogenous GBP-1 was stained using a GBP-1 antibody and immunofluorescence analysis. Arrowheads, impaired actin cytoskeleton shown as actin aggregates throughout the cytoplasm; arrows, colocalization of GBP-1 and actin. (D) The images from panels A to C (a, b, and c, respectively) were quantified using the ImageJ Colocalization Colormap software. The results are presented as images using hot colors (red and yellow), which display a positive correlation, and cold colors (blue), which display a negative correlation. A clear colocalization of GBP-1 and actin at the cell membrane, granular structures, and actin aggregates is shown. All results were independently reproduced by a second researcher. Bars = 25 μm.
These results were confirmed in primary human umbilical vein endothelial cells (HUVECs) using F–GBP-1. Also in these experiments, the fibrous actin cytoskeleton was disrupted in the presence of F–GBP-1 (Fig. 3B, arrowheads) but not in the control transfected cells (Fig. 3B). The colocalization of GBP-1 and actin was again observed at the cell membrane and in cytoplasmic granules (Fig. 3B, arrows).
Finally, we investigated the colocalization of endogenous IFN-γ-induced GBP-1 with actin. In agreement with previous reports, IFN-γ treatment resulted in significant remodeling of the cytoskeleton (53–56). IFN-γ exposure led to a severe disruption of the actin stress fibers (Fig. 3C). The observed changes were similar to those observed with the overexpression of recombinant GBP-1. Endogenous GBP-1 and actin colocalized at the plasma membrane and in cytoplasmic granules (Fig. 3C, arrows). The low signal intensity of endogenous GBP-1 (Fig. 3C) was due to the limited compatibility of the staining protocol for actin and GBP-1. However, the colocalization of both proteins was clearly demonstrated and could be quantified using the ImageJ Colocalization Colormap software. The correlated pixels from the stainings described above are represented in hot colors (red, yellow) (Fig. 3Da, b, and c). The Icorr scores obtained with HeLa cells (Icorr = 0.37) and HUVECs (Icorr = 0.36) demonstrated that the signals of GBP-1 and actin significantly correlated. In contrast, the signals obtained with the background detection (control, Icorr = 0) and F-GFP (Icorr = 0.15), a highly overexpressed control protein, did not correlate with the actin signal. These data confirmed the intracellular colocalization of GBP-1 and actin and demonstrated that ectopic GBP-1 expression and IFN-γ treatment of cells induce severe changes in the actin cytoskeleton structure.
GBP-1 is necessary for IFN-γ-induced remodeling of the actin cytoskeleton.
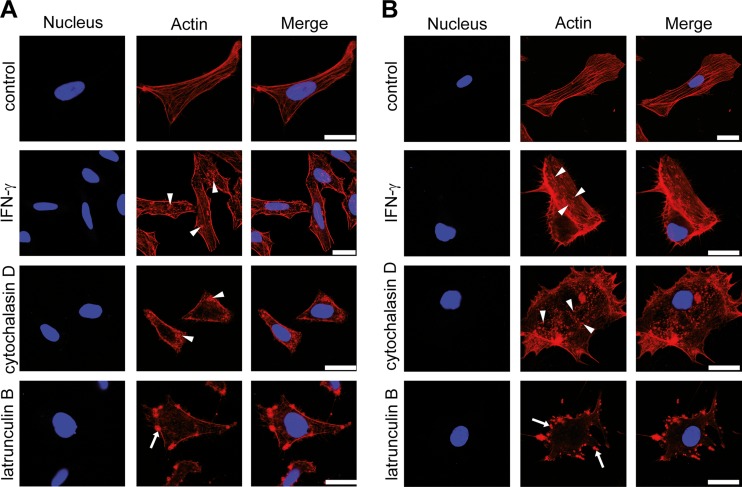
The effects of IFN-γ on the actin cytoskeleton were compared with those of established polymerization inhibitors, such as cytochalasin D and latrunculin B (57, 58), in HeLa cells and HUVECs. To achieve this goal, cells were treated with IFN-γ or actin polymerization inhibitors or were left untreated. Subsequent actin staining revealed that IFN-γ induced in the cytoskeleton a disruption pattern similar to that observed in the presence of cytochalasin D (Fig. 4A and B). This pattern was characterized by the absence of stress fibers and the appearance of actin granules distributed throughout the cell cytoplasm (Fig. 4A and B, arrowheads). In contrast, latrunculin B induced the formation of bleb-like actin aggregates, which were preferentially localized at the cell membrane (Fig. 4A and B, arrows).
FIG 4.
The IFN-γ-induced disruption of the actin cytoskeleton mimics the effect of cytochalasin D. (A) HeLa cells were left untreated, treated with IFN-γ (100 U/ml, 24 h), or treated with different actin polymerization inhibitors (cytochalasin D and latrunculin B) (both at 0.2 μM, 30 min). Subsequently, the actin cytoskeleton was stained using fluorescently labeled phalloidin. Arrowheads, disrupted actin cytoskeleton, which was visualized as small, granular structures in the cytoplasm; arrows, bleb-like aggregates at the cell membrane in the latrunculin B-treated cells. The nuclei were counterstained with DAPI. (B) HUVECs were treated as described in the legend to panel A, with the exception that the polymerization inhibitor was utilized at a concentration of 0.1 μM. The actin pattern was observed as described in the legend to panel A and is indicated in the same manner described in the legend to panel A. The nuclei were counterstained with DAPI. Bars = 25 μm.
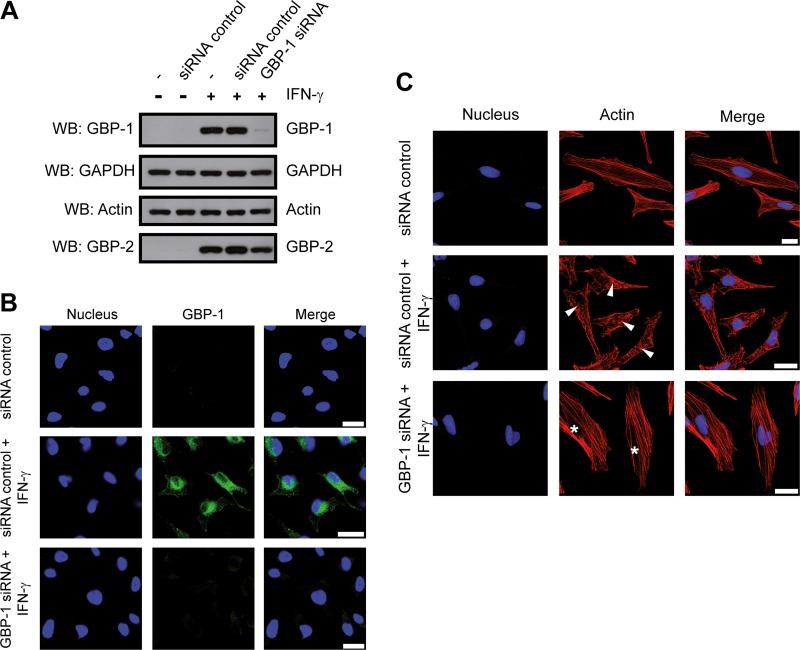
The binding of GBP-1 to actin suggested that GBP-1 might be required for the IFN-γ-induced rearrangements of the cytoskeleton. To analyze whether GBP-1 is necessary for the effects of IFN-γ on fibrous actin, we employed a GBP-1 gene-silencing approach. HeLa cells were transfected with either an siRNA directed against GBP-1 or a control siRNA with a similar GC content. The cells were then either left untreated or treated with IFN-γ (100 U/ml, 24 h). Western blot analysis revealed that GBP-1 expression was highly induced by IFN-γ (Fig. 5A). GBP-1 expression was not affected by the control siRNA, while it was efficiently inhibited by the GBP-1 siRNA (Fig. 5A). Of note, neither the expression of actin nor the expression of GBP-2 (another member of the GBP family) was altered, underscoring the specificity of the GBP-1 knockdown (Fig. 5A). Efficient GBP-1 silencing was also confirmed at the single-cell level via immunofluorescence staining (Fig. 5B). In IFN-γ-treated HeLa cells, the actin cytoskeleton exhibited a distorted structure in the presence of the control siRNA (Fig. 5C, middle, arrowheads), but this effect was fully abrogated with the GBP-1 siRNA (Fig. 5C, asterisks). These results demonstrate that GBP-1 is necessary for mediating the actin cytoskeleton rearrangements induced by IFN-γ.
FIG 5.
GBP-1 is necessary to mediate remodeling of the actin cytoskeleton induced by IFN-γ. (A) HeLa cells were transfected with either control siRNA or GBP-1 siRNA, as indicated. After 6 h, the cells were either left untreated or treated with IFN-γ (100 U/ml, 24 h). Next, 10 μg of protein lysate was separated via 10% SDS-PAGE and analyzed via Western blotting. GBP-1, actin, and GBP-2 (used as an siRNA specificity control) were detected using specific antibodies. (B) HeLa cells were transfected and treated with IFN-γ as described in the legend to panel A. The cells were stained for GBP-1 following antigen target retrieval (pH 9). The nuclei were counterstained with Draq5. (C) HeLa cells were treated as described in the legend to panel A and subsequently stained for actin. Asterisks, organized, filamentous actin cytoskeletons; arrowheads, disruption of the filamentous actin organization following IFN-γ treatment. The nuclei were counterstained with Draq5. All results were independently reproduced by a second researcher. Bars = 25 μm.
The GTPase activity of GBP-1 is required for actin cytoskeleton remodeling.
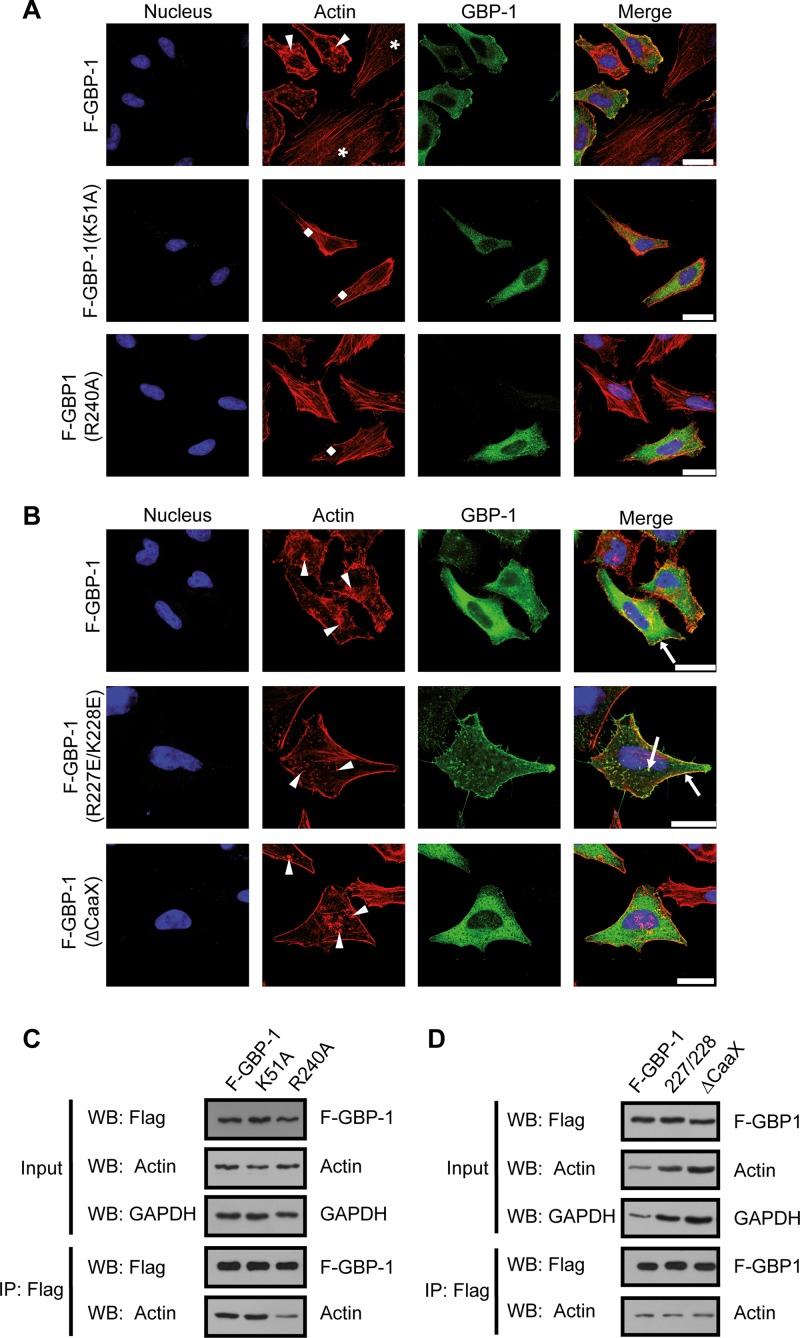
We performed a mutational analysis to determine which functional parts of GBP-1 participate in actin remodeling. First, the involvement of the GTPase function was investigated. F–GBP-1 and two Flag-tagged mutant GTPase forms of GBP-1 [F–GBP-1(K51A) and F–GBP-1(R240A)] (26, 43) were expressed in HeLa cells. Both mutants exhibited significantly reduced GTPase activity [for F–GBP-1(K51A), kcat < 0.05 min−1; for F–GBP-1(R240A), kcat = 13.2 min−1] compared to that of wild-type GBP-1 (kcat = 22.8 min−1) and were predominantly present in a monomeric form (43, 44). The actin stress fibers were selectively impaired in HeLa cells expressing recombinant F–GBP-1 (Fig. 6A, arrowheads) but were not significantly affected by the two mutated proteins [Fig. 6A, F–GBP-1(K51A) and F–GBP-1(R240A), lozenges]. Nontransfected cells plated in the same culture dish showed intact stress fibers (Fig. 6A, asterisks). Of note, F–GBP-1(K51A) and F–GBP-1(R240A) were expressed at levels similar to those at which F–GBP-1 was expressed (Fig. 6A, green), as demonstrated by immunofluorescence staining.
FIG 6.
GBP-1 GTPase activity is required to induce remodeling of the actin cytoskeleton. (A) HeLa cells were transfected with expression constructs encoding F–GBP-1 and two GTPase-deficient mutant forms, F–GBP-1(R240A) and F–GBP-1(K51A). The immunocytochemical detection of GBP-1 was performed using an antibody against the Flag tag. Actin was visualized with fluorescently labeled phalloidin. Arrowheads, disrupted actin structures; lozenges, organized actin filaments. The nuclei are counterstained with Draq5. Note the different organizations of the cytoskeleton in F–GBP-1-expressing cells (arrowheads) and the nonexpressing cells (asterisks) in the upper panel. (B) F–GBP-1, a constitutively dimeric mutant [F–GBP-1(R227E/K228E)], and a mutant lacking the prenylation motif [F–GBP-1(ΔCaaX)] were overexpressed in HeLa cells. Immunofluorescence staining was performed as described in the legend to panel A. Arrowheads, a disrupted actin cytoskeleton structure; arrows, colocalization of GBP-1 and actin. The results shown in panels A and B were independently reproduced by a second researcher. (C) Flag immunoprecipitation and subsequent Western blot analysis of HeLa cells overexpressing proteins (as described in the legend to panel A). R240A, F–GBP-1(R240A); K51A, F–GBP-1(K51A). (D) Flag immunoprecipitation and Western blot analysis with HeLa cells overexpressing proteins (as described in the legend to panel B). 227/228, F–GBP-1(R227E/K228E); ΔCaaX, F–GBP-1(ΔCaaX). Bars = 25 μm.
To investigate the potential further influence of the dimerization and membrane-targeting properties of GBP-1, we applied the following two additional mutants: F–GBP-1(R227E/K228E) and F–GBP-1(ΔCaaX). The R227E/K228E double mutation induced constitutive dimer formation of GBP-1, and the mutant had a greater tendency to form tetramers and a slightly increased GTPase activity (kcat = 36.2 min−1) (28). F–GBP-1(R227E/K228E) localized to the plasma membrane and in granular structures distributed throughout the cytoplasm (Fig. 6B, green), which was in agreement with previous reports (42). This mutant severely disrupted actin stress fibers (Fig. 6B, arrowheads) and colocalized with actin at the plasma membrane and in granular cytoplasmic structures (Fig. 6B, arrows).
A mutant with a deleted CaaX motif [F–GBP-1(ΔCaaX)] was used to investigate the role of the membrane association of GBP-1 in the remodeling of the cytoskeleton. It has been shown that this mutant is not farnesylated and does not associate with the plasma membrane (42), which is in agreement with the diffuse cytoplasmic staining pattern observed in the immunofluorescence analysis (Fig. 6B, green). The staining of the actin cytoskeleton in F–GBP-1(ΔCaaX)-expressing cells showed that the lack of GBP-1 membrane association does not abrogate its effect on the remodeling of the actin cytoskeleton (Fig. 6B, arrowheads).
To determine quantitatively the disruptive effect of wild-type and mutant GBP-1 on the actin cytoskeleton, the percentage of cells with a disrupted cytoskeleton was determined and defined as the stress fiber disruption coefficient (SFD). A low level of disruption of the fibrous actin cytoskeleton was observed for control transfected cells (SFD = 13.9%) and cells expressing the GTPase-deficient mutant GBP-1 [for GBP-1(K51A), SFD = 7.7%; for GBP-1(R240A), SFD = 18.2%]. Severe disruption was observed in cells expressing wild-type GBP-1 (SFD = 83.4%), the constitutive dimeric mutant GBP-1 [GBP-1(R227E/K228E); SFD = 91.3%], and GBP-1-ΔCaaX (SFD = 70%).
Hence, the GTPase activity and, likely, the dimerization of GBP-1 were required for the remodeling of the cytoskeleton, whereas the membrane association of the protein was dispensable for this effect. None of the mutations described above abrogated the binding of the respective proteins to actin, as shown by coimmunoprecipitation analyses (Fig. 6C and D, bottom).
Purified GBP-1 directly interacts with actin and is sufficient to control actin remodeling in vitro.
Using a defined in vitro approach with purified recombinant proteins, we investigated whether further cofactors may be required for the binding of GBP-1 to actin as well as for the regulation of actin remodeling. To achieve this goal, purified recombinant GBP-1, bovine serum albumin (BSA), gelsolin, and actin proteins were applied (Fig. 7A). Reciprocal coimmunoprecipitation experiments showed that actin and GBP-1 coprecipitated in the absence of additional factors (Fig. 7B). In contrast, the negative-control protein BSA failed to coprecipitate (Fig. 7B). Gelsolin, which is known to interact with actin (59), served as a positive control and was also coprecipitated with actin (Fig. 7B, bottom). In experiments where GBP-1, gelsolin, and actin were simultaneously added, GBP-1 and gelsolin were pulled down with actin, but gelsolin did not precipitate with GBP-1 (Fig. 7B). These results demonstrate that GBP-1, similarly to gelsolin, binds directly to actin and that both GBP-1 and gelsolin are not present in the same complex.
FIG 7.
GBP-1 directly interacts with actin in vitro. (A) Recombinant GBP-1, actin, gelsolin, and BSA proteins were detected via Western blotting using the indicated antibodies. (B) The recombinant proteins were mixed as indicated, and immunoprecipitations with either GBP-1 or actin were performed. The precipitates were analyzed via Western blotting using specific antibodies against GBP-1, BSA, gelsolin, or actin.
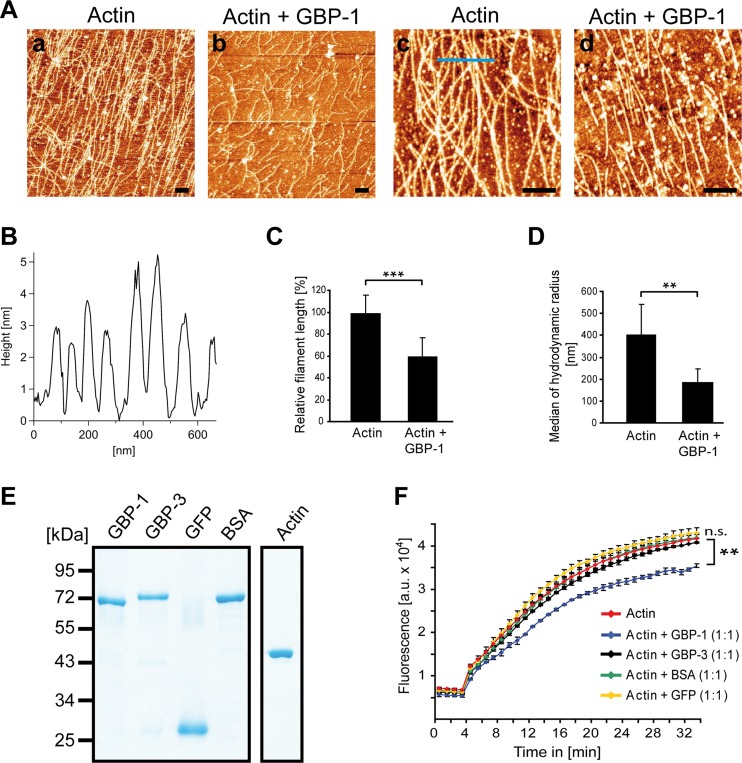
Then, we investigated whether purified GBP-1 alone was sufficient to induce remodeling of actin filaments. To achieve this goal, actin polymerization was initiated in vitro in the presence and absence of GBP-1, and actin filaments were analyzed using atomic force microscopy (AFM). Under the established in vitro conditions, actin polymerized to long filaments (Fig. 8Aa and c). In the presence of GBP-1, the filaments were clearly shorter (Fig. 8Ab and d). The filament diameter was 4 nm, on average (Fig. 8B), which was well in agreement with the results of previous AFM studies of F-actin (60–62). Calculating the relative filament length (RFL) in identical optical fields confirmed that the filament length was significantly reduced in the presence of GBP-1 (Fig. 8C).
FIG 8.
GBP-1 controls actin remodeling by reduction of filament length in vitro. (A) Actin (2 mg/ml) was polymerized alone or in the presence of GBP-1 (2 mg/ml). Polymerized actin filaments were deposited onto freshly cleaved, MgCl2-coated mica carriers and analyzed by AFM. Actin filaments were long in the absence of GBP-1 (a and c) and short in the presence of GBP-1 (b and d). White granular structures are due to salt precipitates. Bars = 400 nm. (B) Representative topography of actin filaments along the blue line (panel Ac) is given. Height measurements indicate an average diameter of actin filaments of 4 nm. (C) The relative actin filament length was quantitatively assessed by calculating the quotient of the total numbers of fibers and of the free polymer ends in five different optical fields. Actin fibers were significantly shorter in the presence of GBP-1 (***, P ≤ 0.001). (D) Actin (0.33 mg/ml) was polymerized in the presence or absence of GBP-1 (0.33 mg/ml), as described in the legend to panel A. Solutions were directly analyzed by DLS. For statistical analyses, the median of the hydrodynamic radius (D50) was averaged over six replicates per type. The standard deviation was calculated from the D50 values. **, P ≤ 0.01. (E) All purified proteins applied to the in vitro polymerization assays were separated by SDS-PAGE and visualized by Coomassie staining. (F) Actin polymerization using pyrene-labeled actin in combination with the indicated proteins was monitored over time by fluorescence intensity analysis. The polymerization assay was performed with pyrene-labeled actin alone or pyrene-labeled actin in combination with either GBP-1, GBP-3, BSA, or GFP. All proteins were applied at a 3 μM concentration. The results of one representative assay of three are shown. Significant differences between the experiments with GBP-1 and all other experimental conditions were observed after 5 min [ANOVA; F(7, 8) = 261; P ≤ 0.001]. GBP-1 showed a lower fluorescence than all other groups (P ≤ 0.008 each, HSD post hoc tests). a.u., arbitrary units. **, P < 0.01; n.s., not significant.
In order to confirm the significant reduction of the actin filament length by GBP-1 in vitro using an alternative method, dynamic light scattering (DLS) analysis was applied. Hydrodynamic radii derived from DLS measurements have previously been used to determine the size and shape of proteins (63, 64). Actin polymerization was initiated again in the presence or absence of GBP-1, but in contrast to AFM, the DLS method determined fragment length directly in solution. In close agreement with the AFM results, the median of the hydrodynamic radius was significantly reduced in samples containing GBP-1 compared to that in samples containing actin alone (Fig. 8D).
In order to determine the specificity of the effects of GBP-1 on actin filament remodeling, a fluorescence-based polymerization assay using pyrene-labeled actin was used (65). This test provides an easy approach for comparison of the effects of several different proteins on actin filament remodeling. GBP-3 is the closest human homologue of GBP-1 and was used as a stringent specificity control (52). GFP and BSA were used as additional unrelated control proteins. All proteins showed comparable integrity and purity (Fig. 8E). Only GBP-1 (Fig. 8F, blue) significantly inhibited actin polymerization, whereas none of the control proteins had an effect (Fig. 8F). These results demonstrate that actin remodeling is specifically caused by GBP-1 and does not require additional factors.
GBP-1 interacts with both globular and filamentous actin.
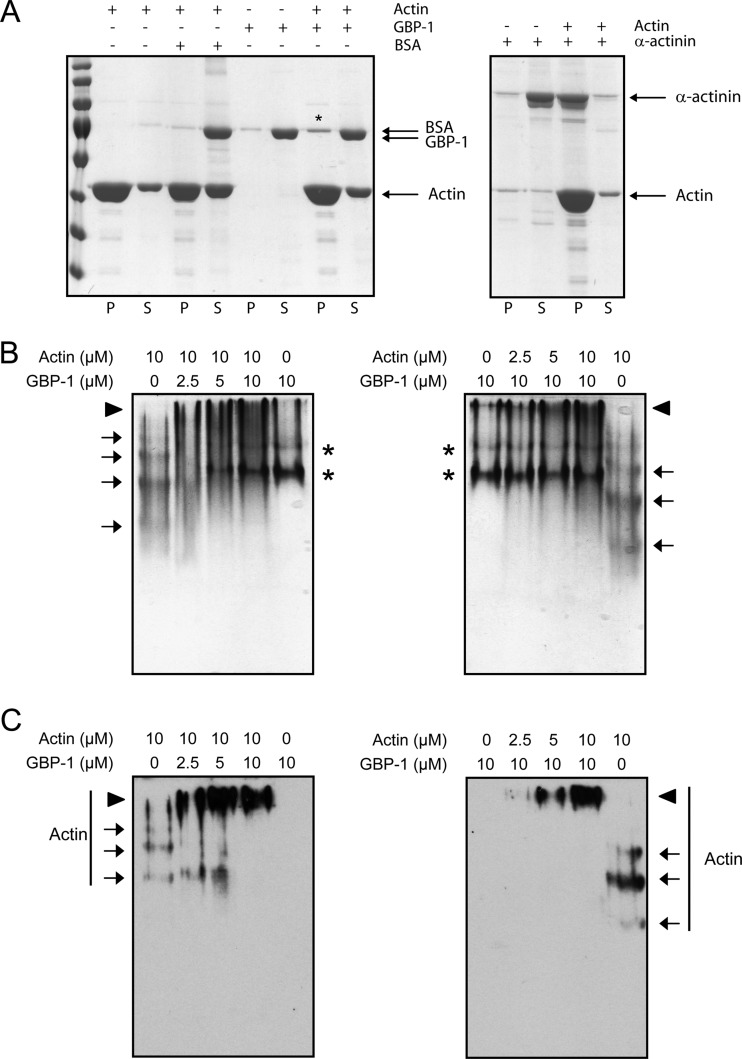
Finally, we investigated whether GBP-1 interacts with G-actin, F-actin, or both. First, we performed an F-actin cosedimentation assay. Actin was polymerized in the presence of GBP-1, BSA, or α-actinin and sedimented by ultracentrifugation. Cosedimented proteins were analyzed by denaturing polyacrylamide gel electrophoresis (Fig. 9A). The amount of GBP-1 cosedimented in the pellet (20%) was four times higher than that of BSA (5%) but clearly lower than that of α-actinin (87%), a known F-actin bundling factor (Fig. 9A).
FIG 9.
GBP-1 binds to both G-actin and F-actin. (A) F-actin was cosedimented by ultracentrifugation with GBP-1, BSA as a negative control, or α-actinin as a positive control. Equal amounts of pellets (lanes P) and supernatants (lanes S) were separated by SDS-PAGE. Gels were stained with Coomassie blue. An increase of GBP-1 in the pellet fraction was observed in the presence of F-actin, as indicated by an asterisk. (B) G-actin was incubated with GBP-1, and proteins were separated by native polyacrylamide gel electrophoresis. Gels were stained with Coomassie blue. Arrows, actin, which formed a ladder when incubated alone, indicating partial polymerization; asterisks, GBP-1 in its monomeric or dimeric form; arrowheads, actin–GBP-1 complexes. (C) Actin was detected using a specific antibody in a subsequent Western blot. Arrows, corresponding actin bands; arrowheads, larger protein complexes.
Second, we performed a G-actin binding assay. G-actin was incubated with increasing amounts of GBP-1 in G-actin buffer (Fig. 9B and C, left). Alternatively, the concentration of GBP-1 was kept constant and increasing amounts of G-actin were used (Fig. 9B and C, right). Proteins were subsequently separated by native polyacrylamide gel electrophoresis. A shift of G-actin was already observed in the polyacrylamide gel in the presence of GBP-1 at a GBP-1 concentration four times lower than the G-actin concentration (Fig. 9B). Western blot analyses confirmed that G actin formed a complex with GBP-1 (Fig. 9C). In conclusion, we found that GBP-1 binds strongly to G-actin and, to a lower extent, to F-actin.
DISCUSSION
IFN-γ regulates adaptive and innate immune defenses against viruses, intracellular pathogens, and tumors. GBP-1 is a major IFN-γ-induced protein in eukaryotic cells (22, 66). In this study, GBP-1 was found to bind strongly to actin. The interaction between GBP-1 and actin was systematically and consistently confirmed under various experimental conditions using ectopically and endogenously expressed proteins. In addition, these results were reproduced in different cell types, highlighting the general relevance of our findings. Importantly, we showed that purified GBP-1 and actin can be coimmunoprecipitated in vitro. Thus, both proteins are interacting in the absence of additional factors. The interaction of GBP-1 with the cytoskeleton is in line with independent studies showing that GBP-1 can associate with the cytoskeletal proteins spectrin, plastin (F. Forster, personal communication), and βIII-tubulin (67).
GBP-1 was demonstrated to be both necessary and sufficient for actin cytoskeleton remodeling induced by IFN-γ, as shown by ectopic overexpression and inhibition of GBP-1 expression. In this framework, it is noteworthy that GBP-1 can heterodimerize with various members of the GBP family (42). However, since other GBPs failed to compensate for the remodeling effects on the cytoskeleton when GBP-1 was silenced in IFN-γ-treated cells, GBP-1 is likely to play a dominant role in actin remodeling. Mutant analyses demonstrated that the GTPase function of GBP-1 is required for actin fiber disruption, whereas membrane association is not. Interestingly, a mutant with an increased oligomerization capability [GBP-1(K227E/R228E)] (28) also exhibited increased remodeling activity. Oligomerization requires both the globular domain of the protein, which encloses the GTPase function, and the C-terminal helical domain, which has been shown to inhibit cell proliferation (27, 28). GBP-3, the closest relative of GBP-1 (88% sequence homology) (52, 68), did not significantly affect the total F-actin content in vitro. The sequences of both proteins are almost identical in the globular domain and vary predominantly at the C terminus of the helical domain (69). Interestingly, this region has been found to be involved in tetramerization (27). This suggests that differential oligomerization capabilities of GBP-1 and GBP-3 may be responsible for the different activities of the two proteins in actin remodeling. Hence, the actin remodeling activity of GBP-1 may depend on two important features of the molecule: (i) the GTPase function residing in the globular domain and (ii) oligomerization, which is mediated by two areas of the protein, one resident in the globular domain and the other resident in the helical domain.
Using purified proteins, we demonstrated that in vitro binding of GBP-1 to actin is sufficient for the actin remodeling process. Analyses of actin filament formation in the presence and absence of GBP-1 using AFM, DLS, and a fluorescence-based actin polymerization assay consistently showed that GBP-1 significantly inhibits actin filament formation. These effects observed under defined in vitro conditions convincingly supported the suggestion that the actin remodeling activity of GBP-1 observed in vivo does not require additional cofactors. Accordingly, GBP-1 may be regarded as a novel member of the family of actin polymerization regulatory factors, which, among others, includes profilin, thymosin-β4, gelsolin, villin, capping protein, and CapZ. These factors regulate actin filaments either by sequestering of actin monomers, by severing of preexisting filaments, or by capping of filament ends (70–73).
We showed that GBP-1 binds efficiently to G-actin and, to a lower extent, to F-actin. Efficient binding of GBP-1 to G-actin suggested that GBP-1 may act predominantly as a sequestering factor for actin monomers, thereby reducing the G-actin pool available for polymerization. This mechanism has been described for the proteins profilin and thymosin-β4 and for the latrunculin inhibitors (57, 71, 74) and is in line with the fact that GBP-1 and actin colocalize in large aggregates.
By binding to F-actin, GBP-1 might, in addition, be able to shrink actin filaments by severing or by capping of actin filament barbed ends. However, a severing activity of GBP-1 would by associated with increased numbers of short filaments, which was not observed either in vitro or in vivo (Fig. 3 and 8A and B). In contrast, the presently available results do not exclude the possibility that a capping activity contributes to the actin remodeling activity of GBP-1. Altogether, our data indicate that GBP-1 is a novel member within the family of actin remodeling proteins, acting predominantly as a sequestering factor.
The regulation of the cytoskeleton by IFN-γ has been shown to be of great relevance in manifold physiological and pathophysiological processes, including cell migration, invasion, proliferation, defense against pathogens, and barrier function (8, 17, 18, 20, 75, 76). Importantly, GBP-1 has been shown to regulate all of these processes in response to IFN-γ (17, 19, 30, 34, 77). By identifying GBP-1 as a new actin remodeling protein we provide for the first time a molecular mechanism explaining the effects of IFN-γ on the cytoskeleton. As yet, regulation of the actin cytoskeleton has been predominantly attributed to the activity of the Rho protein family, including Rho, Rac, and Cdc42 (78, 79). Our data demonstrate that the large GTPase GBP-1 is both necessary and sufficient to remodel the actin cytoskeleton in the absence of additional cofactors. In this framework, GBP-1-mediated actin remodeling may contribute to the regulation of innate and adaptive immune defense.
ACKNOWLEDGMENTS
We thank Gertrud Hoffmann, Katja Petter and Mahimaidos Manoharan (Division of Molecular and Experimental Surgery, University of Erlangen-Nuremberg) for excellent technical assistance. We are grateful to Thomas Seyller (Institute of Physics, Chemnitz University of Technology) for providing the atomic force microscope and to Wei Xiang (University of Erlangen-Nuremberg) for the access to the ultracentrifuge.
This work was supported by grants awarded to M.S. from the German Research Foundation (DFG; KFO257 [subproject 4], SFB 796 [subproject B9], GK1071 [subproject A2]), the German Cancer Aid (109510), the German Federal Ministry of Education and Research (BMBF; 01ES0807, 01ES1001), the emerging fields initiative of the FAU Erlangen, and the Interdisciplinary Center for Clinical Research (IZKF) of the Clinical Center Erlangen. N.O. was supported by a Bavarian equal opportunities sponsorship, which promotes equal opportunities for women in research and teaching.
The sponsors played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 4 November 2013
REFERENCES
- 1.Damsker JM, Hansen AM, Caspi RR. 2010. Th1 and Th17 cells: adversaries and collaborators. Ann. N. Y. Acad. Sci. 1183:211–221. 10.1111/j.1749-6632.2009.05133.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeda H, Old LJ, Schreiber RD. 2002. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 13:95–109. 10.1016/S1359-6101(01)00038-7 [DOI] [PubMed] [Google Scholar]
- 3.Bach EA, Aguet M, Schreiber RD. 1997. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15:563–591. 10.1146/annurev.immunol.15.1.563 [DOI] [PubMed] [Google Scholar]
- 4.Farrar MA, Schreiber RD. 1993. The molecular cell biology of interferon-gamma and its receptor. Annu. Rev. Immunol. 11:571–611. 10.1146/annurev.iy.11.040193.003035 [DOI] [PubMed] [Google Scholar]
- 5.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264. 10.1146/annurev.biochem.67.1.227 [DOI] [PubMed] [Google Scholar]
- 6.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267. 10.1098/rspb.1957.0048 [DOI] [PubMed] [Google Scholar]
- 7.Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809. 10.1128/CMR.14.4.778-809.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shtrichman R, Samuel CE. 2001. The role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 4:251–259. 10.1016/S1369-5274(00)00199-5 [DOI] [PubMed] [Google Scholar]
- 9.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. 2011. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332:717–721. 10.1126/science.1201711 [DOI] [PubMed] [Google Scholar]
- 10.Dunn GP, Koebel CM, Schreiber RD. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6:836–848. 10.1038/nri1961 [DOI] [PubMed] [Google Scholar]
- 11.Dunn GP, Old LJ, Schreiber RD. 2004. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22:329–360. 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- 12.Fridman WH, Dieu-Nosjean MC, Pages F, Cremer I, Damotte D, Sautes-Fridman C, Galon J. 2013. The immune microenvironment of human tumors: general significance and clinical impact. Cancer Microenviron. 6:117–122. 10.1007/s12307-012-0124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. 1998. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 95:7556–7561. 10.1073/pnas.95.13.7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410:1107–1111. 10.1038/35074122 [DOI] [PubMed] [Google Scholar]
- 15.Chin YE, Kitagawa M, Kuida K, Flavell RA, Fu XY. 1997. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 17:5328–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S. 2001. Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut 49:251–262. 10.1136/gut.49.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britzen-Laurent N, Lipnik K, Ocker M, Naschberger E, Schellerer VS, Croner RS, Vieth M, Waldner M, Steinberg P, Hohenadl C, Stürzl M. 2013. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis 34:153–162. 10.1093/carcin/bgs310 [DOI] [PubMed] [Google Scholar]
- 18.Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. U. S. A. 93:7673–7678. 10.1073/pnas.93.15.7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capaldo CT, Beeman N, Hilgarth RS, Nava P, Louis NA, Naschberger E, Stürzl M, Parkos CA, Nusrat A. 2012. IFN-γ and TNF-α-induced GBP-1 inhibits epithelial cell proliferation through suppression of β-catenin/TCF signaling. Mucosal Immunol. 5:681–690. 10.1038/mi.2012.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong Q, Vassilieva EV, Ivanov AI, Wang Z, Brown GT, Parkos CA, Nusrat A. 2005. Interferon-gamma inhibits T84 epithelial cell migration by redirecting transcytosis of beta1 integrin from the migrating leading edge. J. Immunol. 175:4030–4038 [DOI] [PubMed] [Google Scholar]
- 21.Cheng YS, Colonno RJ, Yin FH. 1983. Interferon induction of fibroblast proteins with guanylate binding activity. J. Biol. Chem. 258:7746–7750 [PubMed] [Google Scholar]
- 22.Lubeseder-Martellato C, Guenzi E, Jörg Töpolt AK, Naschberger E, Kremmer E, Zietz C, Tschachler E, Hutzler P, Schwemmle M, Matzen K, Grimm T, Ensoli B, Stürzl M. 2002. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 161:1749–1759. 10.1016/S0002-9440(10)64452-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Praefcke GJ, McMahon HT. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147. 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- 24.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. 2000. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403:567–571. 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- 25.Kunzelmann S, Praefcke GJ, Herrmann C. 2006. Transient kinetic investigation of GTP hydrolysis catalyzed by interferon-gamma-induced hGBP1 (human guanylate binding protein 1). J. Biol. Chem. 281:28627–28635. 10.1074/jbc.M604911200 [DOI] [PubMed] [Google Scholar]
- 26.Praefcke GJ, Geyer M, Schwemmle M, Robert Kalbitzer H, Herrmann C. 1999. Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J. Mol. Biol. 292:321–332. 10.1006/jmbi.1999.3062 [DOI] [PubMed] [Google Scholar]
- 27.Syguda A, Bauer M, Benscheid U, Ostler N, Naschberger E, Ince S, Stürzl M, Herrmann C. 2012. Tetramerization of human guanylate-binding protein 1 is mediated by coiled-coil formation of the C-terminal alpha-helices. FEBS J. 279:2544–2554. 10.1111/j.1742-4658.2012.08637.x [DOI] [PubMed] [Google Scholar]
- 28.Vöpel T, Syguda A, Britzen-Laurent N, Kunzelmann S, Lüdemann MB, Dovengerds C, Stürzl M, Herrmann C. 2010. Mechanism of GTPase-activity-induced self-assembly of human guanylate binding protein 1. J. Mol. Biol. 400:63–70. 10.1016/j.jmb.2010.04.053 [DOI] [PubMed] [Google Scholar]
- 29.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50:1727–1737. 10.1002/hep.23195 [DOI] [PubMed] [Google Scholar]
- 30.Tietzel I, El-Haibi C, Carabeo RA. 2009. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One 4:e6499. 10.1371/journal.pone.0006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degrandi D, Konermann C, Beuter-Gunia C, Kresse A, Wurthner J, Kurig S, Beer S, Pfeffer K. 2007. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179:7729–7740 [DOI] [PubMed] [Google Scholar]
- 32.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HW, IV, Macmicking JD, Sibley LD. 2013. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 9:e1003320. 10.1371/journal.ppat.1003320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. 2012. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37:302–313. 10.1016/j.immuni.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 34.Guenzi E, Töpolt K, Cornali E, Lubeseder-Martellato C, Jörg Matzen AK, Zietz C, Kremmer E, Nappi F, Schwemmle M, Hohenadl C, Barillari G, Tschachler E, Monini P, Ensoli B, Stürzl M. 2001. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 20:5568–5577. 10.1093/emboj/20.20.5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guenzi E, Töpolt K, Lubeseder-Martellato C, Jörg Naschberger AE, Benelli R, Albini A, Stürzl M. 2003. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 22:3772–3782. 10.1093/emboj/cdg382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammon M, Herrmann M, Bleiziffer O, Pryymachuk G, Andreoli L, Munoz LE, Amann KU, Mondini M, Gariglio M, Airo P, Schellerer VS, Hatzopoulos AK, Horch RE, Kneser U, Stürzl M, Naschberger E. 2011. Role of guanylate binding protein-1 in vascular defects associated with chronic inflammatory diseases. J. Cell. Mol. Med. 15:1582–1592. 10.1111/j.1582-4934.2010.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipnik K, Naschberger E, Gonin-Laurent N, Kodajova P, Petznek H, Rungaldier S, Astigiano S, Ferrini S, Stürzl M, Hohenadl C. 2010. Interferon gamma-induced human guanylate binding protein 1 inhibits mammary tumor growth in mice. Mol. Med. 16:177–187. 10.2119/molmed.2009.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messmer-Blust AF, Balasubramanian S, Gorbacheva VY, Jeyaratnam JA, Vestal DJ. 2010. The interferon-gamma-induced murine guanylate-binding protein-2 inhibits rac activation during cell spreading on fibronectin and after platelet-derived growth factor treatment: role for phosphatidylinositol 3-kinase. Mol. Biol. Cell 21:2514–2528. 10.1091/mbc.E09-04-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naschberger E, Croner RS, Merkel S, Dimmler A, Tripal P, Amann KU, Kremmer E, Brueckl WM, Papadopoulos T, Hohenadl C, Hohenberger W, Stürzl M. 2008. Angiostatic immune reaction in colorectal carcinoma: impact on survival and perspectives for antiangiogenic therapy. Int. J. Cancer 123:2120–2129. 10.1002/ijc.23764 [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinländer K, Naschberger E, Lehmann MH, Tripal P, Paster W, Stockinger H, Hohenadl C, Stürzl M. 2008. Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin alpha4 expression. FASEB J. 22:4168–4178. 10.1096/fj.08-107524 [DOI] [PubMed] [Google Scholar]
- 42.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, Naschberger E, Herrmann C, Stürzl M. 2010. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5:e14246. 10.1371/journal.pone.0014246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wehner M, Kunzelmann S, Herrmann C. 2012. The guanine cap of human guanylate-binding protein 1 is responsible for dimerization and self-activation of GTP hydrolysis. FEBS J. 279:203–210. 10.1111/j.1742-4658.2011.08415.x [DOI] [PubMed] [Google Scholar]
- 44.Praefcke GJ, Kloep S, Benscheid U, Lilie H, Prakash B, Herrmann C. 2004. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J. Mol. Biol. 344:257–269. 10.1016/j.jmb.2004.09.026 [DOI] [PubMed] [Google Scholar]
- 45.Chen CA, Okayama H. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 6:632–638 [PubMed] [Google Scholar]
- 46.Naschberger E, Lubeseder-Martellato C, Meyer N, Gessner R, Kremmer E, Gessner A, Stürzl M. 2006. Human guanylate binding protein-1 is a secreted GTPase present in increased concentrations in the cerebrospinal fluid of patients with bacterial meningitis. Am. J. Pathol. 169:1088–1099. 10.2353/ajpath.2006.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaskolski F, Mulle C, Manzoni OJ. 2005. An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods 146:42–49. 10.1016/j.jneumeth.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naldi M, Vasina E, Dobroiu S, Paraoan L, Nicolau DV, Andrisano V. 2009. Self-assembly of biomolecules: AFM study of F-actin on unstructured and nanostructured surfaces. Proc. SPIE 7188:Q7–Q9 10.1117/12.822800 [DOI] [Google Scholar]
- 50.Pastre D, Pietrement O, Fusil S, Landousy F, Jeusset J, David MO, Hamon L, Le Cam E, Zozime A. 2003. Adsorption of DNA to mica mediated by divalent counterions: a theoretical and experimental study. Biophys. J. 85:2507–2518. 10.1016/S0006-3495(03)74673-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horcas I, Fernandez R, Gomez-Rodriguez JM, Colchero J, Gomez-Herrero J, Baro AM. 2007. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78:013705. 10.1063/1.2432410 [DOI] [PubMed] [Google Scholar]
- 52.Tripal P, Bauer M, Naschberger E, Mortinger T, Hohenadl C, Cornali E, Thurau M, Stürzl M. 2007. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 27:44–52. 10.1089/jir.2007.0086 [DOI] [PubMed] [Google Scholar]
- 53.Asakawa H, Miyagawa J, Hanafusa T, Katsura H, Miyazaki A, Otsuka A, Nakagawa C, Yamagata K, Tajima K, Mashita K, Kono N, Tarui S. 1990. Interferon-gamma reduces actin filaments and inhibits thyroid-stimulating hormone-induced formation of microvilli and pseudopods in mouse monolayer thyrocytes. Endocrinology 127:325–329. 10.1210/endo-127-1-325 [DOI] [PubMed] [Google Scholar]
- 54.Molony L, Armstrong L. 1991. Cytoskeletal reorganizations in human umbilical vein endothelial cells as a result of cytokine exposure. Exp. Cell Res. 196:40–48. 10.1016/0014-4827(91)90454-3 [DOI] [PubMed] [Google Scholar]
- 55.Muros MA, Aranega AE, Velez C, Melguizo C, Alvarez L, Aranega A. 1994. Modulation of contractile proteins in embryonic and fetal chick cardiac cells by phorbol ester, gamma-interferon, 5-azacytidine and diacylglycerols. Life Sci. 54:171–183. 10.1016/0024-3205(94)00586-9 [DOI] [PubMed] [Google Scholar]
- 56.Surin B, Rouillard D, Bauvois B. 2000. Loss of alpha5beta1-mediated adhesion of monocytic cells to fibronectin by interferons beta and gamma is associated with changes in actin and paxillin cytoskeleton. Hematol. J. 1:172–180. 10.1038/sj.thj.6200027 [DOI] [PubMed] [Google Scholar]
- 57.Coué M, Brenner SL, Spector I, Korn ED. 1987. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213:316–318. 10.1016/0014-5793(87)81513-2 [DOI] [PubMed] [Google Scholar]
- 58.Schliwa M. 1982. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 92:79–91. 10.1083/jcb.92.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurth MC, Wang LL, Dingus J, Bryan J. 1983. Purification and characterization of a gelsolin-actin complex from human platelets. Evidence for Ca2+-insensitive functions. J. Biol. Chem. 258:10895–10903 [PubMed] [Google Scholar]
- 60.Ikawa T, Hoshino F, Watanabe O, Li Y, Pincus P, Safinya CR. 2007. Molecular scale imaging of F-actin assemblies immobilized on a photopolymer surface. Phys. Rev. Lett. 98:018101. 10.1103/PhysRevLett.98.018101 [DOI] [PubMed] [Google Scholar]
- 61.Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. 2011. Atomic force microscopy reveals drebrin induced remodeling of F-actin with subnanometer resolution. Nano Lett. 11:825–827. 10.1021/nl104159v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Wang YL, Gu L, Pan J. 2003. Atomic force microscopy of actin. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35:489–494 [PubMed] [Google Scholar]
- 63.Czurylo EA, Eimer W, Kulikova N, Hellweg T. 2000. Size, shape and secondary structure of calponin. Acta Biochim. Pol. 47:791–806 [PubMed] [Google Scholar]
- 64.Pivovarova AV, Chebotareva NA, Chernik IS, Gusev NB, Levitsky DI. 2007. Small heat shock protein Hsp27 prevents heat-induced aggregation of F-actin by forming soluble complexes with denatured actin. FEBS J. 274:5937–5948. 10.1111/j.1742-4658.2007.06117.x [DOI] [PubMed] [Google Scholar]
- 65.Cooper JA, Walker SB, Pollard TD. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4:253–262. 10.1007/BF00712034 [DOI] [PubMed] [Google Scholar]
- 66.Naschberger E, Werner T, Vicente AB, Guenzi E, Töpolt K, Leubert R, Lubeseder-Martellato C, Nelson PJ, Stürzl M. 2004. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem. J. 379:409–420. 10.1042/BJ20031873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Donato M, Mariani M, Petrella L, Martinelli E, Zannoni GF, Vellone V, Ferrandina G, Shahabi S, Scambia G, Ferlini C. 2012. Class III beta-tubulin and the cytoskeletal gateway for drug resistance in ovarian cancer. J. Cell. Physiol. 227:1034–1041. 10.1002/jcp.22813 [DOI] [PubMed] [Google Scholar]
- 68.Naschberger E, Bauer M, Stürzl M. 2005. Human guanylate binding protein-1 (hGBP-1) characterizes and establishes a non-angiogenic endothelial cell activation phenotype in inflammatory diseases. Adv. Enzyme Regul. 45:215–227. 10.1016/j.advenzreg.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 69.Olszewski MA, Gray J, Vestal DJ. 2006. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 26:328–352. 10.1089/jir.2006.26.328 [DOI] [PubMed] [Google Scholar]
- 70.Cooper JA, Sept D. 2008. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267:183–206. 10.1016/S1937-6448(08)00604-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- 72.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. 2004. Gelsolin superfamily proteins: key regulators of cellular functions. Cell. Mol. Life Sci. 61:2614–2623. 10.1007/s00018-004-4225-6 [DOI] [PubMed] [Google Scholar]
- 73.Weeds A, Maciver S. 1993. F-actin capping proteins. Curr. Opin. Cell Biol. 5:63–69. 10.1016/S0955-0674(05)80009-2 [DOI] [PubMed] [Google Scholar]
- 74.Spector I, Shochet NR, Blasberger D, Kashman Y. 1989. Latrunculins—novel marine macrolides that disrupt microfilament organization and affect cell growth. I. Comparison with cytochalasin D. Cell Motil. Cytoskeleton 13:127–144. 10.1002/cm.970130302 [DOI] [PubMed] [Google Scholar]
- 75.Ivanov AI, Parkos CA, Nusrat A. 2010. Cytoskeletal regulation of epithelial barrier function during inflammation. Am. J. Pathol. 177:512–524. 10.2353/ajpath.2010.100168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Youakim A, Ahdieh M. 1999. Interferon-gamma decreases barrier function in T84 cells by reducing ZO-1 levels and disrupting apical actin. Am. J. Physiol. 276:G1279–G1288 [DOI] [PubMed] [Google Scholar]
- 77.Schnoor M, Betanzos A, Weber DA, Parkos CA. 2009. Guanylate-binding protein-1 is expressed at tight junctions of intestinal epithelial cells in response to interferon-gamma and regulates barrier function through effects on apoptosis. Mucosal Immunol. 2:33–42. 10.1038/mi.2008.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509–514. 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- 79.Ridley AJ. 2001. Rho GTPases and cell migration. J. Cell Sci. 114:2713–2722 [DOI] [PubMed] [Google Scholar]