Abstract
Early detection of Mycobacterium tuberculosis complex (MTBC) and markers conveying drug resistance can have a beneficial impact on preventive public health actions. We describe here a new molecular point-of-care (POC) system, the Genedrive, which is based on simple sample preparation combined with PCR to detect MTBC and simultaneously detect mutation markers in the rpoB gene directly from raw sputum sample. Hybridization probes were used to detect the presence of the key mutations in codons 516, 526, and 531 of the rpoB gene. The sensitivities for MTBC and rpoB detection from sputum samples were assessed using model samples spiked with known numbers of bacteria prepared from liquid cultures of M. tuberculosis. The overall sensitivities were 90.8% (95% confidence interval [CI], 81, 96.5) for MTBC detection and 72.3% (95% CI, 59.8, 82.7) for rpoB detection. For samples containing ≥1,000 CFU/ml, the sensitivities were 100% for MTBC and 85.7% for rpoB detection, while for samples containing ≤100 CFU/ml, the sensitivities were 86.4% and 65.9% for MTBC and rpoB detection, respectively. The specificity was shown to be 100% (95% CI, 83.2, 100) for MTBC and rpoB. The clinical sputum samples were processed using the same protocol and showed good concordance with the data generated from the model. Tuberculosis-infected subjects with smear samples assessed as scanty or negative were detectable by the Genedrive system. In these paucibacillary patients, the performance of the Genedrive system was comparable to that of the GeneXpert assay. The characteristics of the Genedrive platform make it particularly useful for detecting MTBC and rifampin resistance in low-resource settings and for reducing the burden of tuberculosis disease.
INTRODUCTION
Nearly one-third of the global population is believed to be infected with a member of the Mycobacterium tuberculosis complex (MTBC). It is estimated that as many as 8 million new cases of tuberculosis (TB) occur annually, leading to up to 1.5 million deaths (1). Health actions that are critically important for treating and preventing the spread of TB have focused on downstream factors, such as reducing the underlying causes of poverty or lowering the incidence of the progenitors of coinfection, such as infection by human immunodeficiency virus (HIV) (2), and on upstream factors, such as the early identification of infected patients (3) and the availability of effective drug therapies (4). It has been reported in studies from HIV-positive cohorts that the mortality rates are particularly high for patients presenting with smear-negative sputum (5). This has also been shown in samples containing MTBC strains that are resistant to rifampin (RIF), one of the most widely used front-line antibiotics, along with isoniazid.
Diagnosing infected individuals represents a vital part of TB control. Before infection can be treated efficiently, an accurate and timely diagnosis is required, preferably at the point of care (POC) using tools that are field friendly. The World Health Organization (WHO) has set guidelines for what a POC TB test should provide (1). The WHO reports that for any TB program, there is a significant proportion of patients in high-burden settings who fail to return to collect their smear microscopy result. There is a clear and present need to provide rapid screening tools that enable the clinician to place patients on the appropriate therapies. Although the definition of POC has become contentious, it should at least encompass the concept of diagnosis at the point where a patient presents himself or herself to the clinician, thus enabling the result to be translated into an appropriate and timely treatment (6).
The Genedrive system (Epistem, United Kingdom) provides POC detection of MTBC directly from sputum samples, and it simultaneously identifies mutations involved in conferring RIF resistance. The system uses a simple paper-based extraction method coupled with an asymmetric PCR and proprietary hybridization probe technology (Highlighter Probes) that provides genotypic information. The composite paper is chemically treated to enable the extraction of nucleic acids from bacteria and simultaneously decontaminate the sample. Lyophilized PCR reagents are provided that are stable at ambient temperatures, removing the need for cold chain storage.
The assay targets specific sequences in the MTBC genomes and has a broad detection profile covering clinically relevant strains and subtypes; these include M. tuberculosis, Mycobacterium bovis, and Mycobacterium africanum, as all three species are clinically significant human pathogens (7, 8). Two regions of the MTBC genomes are targeted: a short repetitive region, REP13E12 (9, 10), and an 81-bp core region of rpoB, which encodes the β-subunit of RNA polymerase. Ninety-six percent of rifampin-resistant clinical isolates of M. tuberculosis have mutations in the rpoB gene within this 81-bp core region. Hybridization probes based on a proprietary chemistry were shown to report the presence of the key mutations in codons 516, 526, and 531, which were previously shown to occur at a high frequency (11).
The data obtained from spiked sputum models and clinical sputum samples demonstrate the effectiveness of a simple workflow in processing and diagnosing the presence of MTBC. Further, the same process provided a sensitive analysis of complex genotyping information around the RIF status of MTBC from both smear-positive and smear-negative patients. The limit of detection (LOD), accuracy, and reproducibility of the Genedrive system for both MTBC and RIF resistance were determined. Comparisons were made with the GeneXpert assay (Cepheid, Inc., USA), a laboratory-based instrument targeting the same organisms and resistance markers (12).
MATERIALS AND METHODS
Sputum collection and characterization.
Primary isolation and culturing of M. tuberculosis isolates from sputum specimens was done using standardized culture procedures (13). All isolates used in this study were collected from patients infected with M. tuberculosis and referred to the Mycobacteriology Laboratory of Hospital Carlos III and/or the Microbiology Service of Hospital La Paz (Madrid, Spain). MTBC was detected using conventional acid-fast stain microscopy and confirmed by the GenoType Mycobacterium CM system (Hain Diagnostics, Germany). Drug susceptibility tests for RIF were performed using the MTBDRplus system (Hain Diagnostics), and standard antibiogram methods in liquid culture were performed using MGIT 960 (Becton, Dickinson, Sparks, MD, USA). The critical concentration of RIF used in this study was 1 μg/ml, and the instrument was set to perform the final interpretation of data, automatically reporting the susceptibility results (14).
Preparation of sputum samples spiked with known loads of mycobacteria.
Liquid cultures were grown on the MGIT 960 (Becton, Dickinson) according to the manufacturer's standard protocol. The cultures were incubated until the instrument automatically called a positive result; this has previously been characterized to approximate to an exponential growth phase equivalent to around 106 CFU/ml (Becton, Dickinson, personal communication). The concentration of M. tuberculosis was further confirmed to be 106 CFU/ml using quantitative PCR (qPCR). Primers targeting 16S ribosomal DNA (rDNA) were used to quantitate the number of bacteria according to previously published methods (15). Samples that indicated discordant quantities by the cycle threshold (CT) were discarded. Prior to dilution, the liquid cultures were processed in order to reduce the amount of bacterial clumping, which skews the number of CFU in the dilution series. Three successive rounds of vigorous vortexing was performed with 1.8-ml screw-top cryotubes containing 1 ml of the freshly positive culture. Each round of vortexing was performed for a minimum of 20 min using a benchtop vortex. After each round of vortexing, a Ziehl-Neelsen stain was performed in order to confirm the reduction in the size of the bacterial cords. The cords were observed at 40× and 100× magnification. After the three successive rounds of vortexing and final microscopy confirmation, a 10-fold serial dilution was prepared from 106 to 102 CFU/ml using ultrapure water as the diluent. Each of the samples in the dilution series was confirmed by 16S rDNA qPCR (15) using a StepOne real-time PCR instrument (Life Technologies, USA). Further confirmation was obtained using Lowenstein-Jensen solid culture medium. Each dilution from 100 to 10−6 was plated and incubated for 2 weeks. Slant counts were used to correlate back to the original concentrations and fine-tune the final concentrations when required. Each of the dilution series was added into fresh smear-negative sputum to provide a model sample containing a known number of CFU.
To analyze specificity, spiked sputum samples were made from cultures of Staphylococcus aureus, Citrobacter freundii, Pseudomonas aeruginosa, nontuberculous mycobacteria (Mycobacterium abscessus, M. avium, Mycobacterium celatum, Mycobacterium chelonae, Mycobacterium fortuitum, Mycobacterium gordonae, Mycobacterium intracellulare, Mycobacterium kansasii, Mycobacterium lentiflavum, Mycobacterium marinum, Mycobacterium mucogenicum, Mycobacterium peregrinum, and Mycobacterium xenopi), and other mycobacteria from MTBC (M. africanum, M. bovis, and M. bovis BCG). Known amounts of these samples were spiked into smear-negative sputum samples using sterile procedures.
Semiquantification of clinical sputum samples using microscopy.
Sputum samples were obtained from patients infected with M. tuberculosis and were used immediately or refrigerated (4°C) for ≤72 h. A standard microscopic Ziehl-Neelsen test was used to assess the smear status of the samples. Smear-negative samples were further assessed using 300 fields of view. Smear-positive samples for acid-fast bacilli were graded according to the WHO/International Union against Tuberculosis and Lung Disease Method and scored as scanty, 1+, 2+, or 3+ (16).
Processing of samples for the Genedrive system.
The sample processing cassette from Epistem provides simultaneous sample decontamination and nucleic acid extraction directly from raw sputum samples. The extraction process consists of a filter paper that is functionalized with a proprietary organic moiety capable of penetrating and rupturing bacterial cell membranes. The cellular exudate is released and dries in a stable state on the paper. This is compatible with PCR analysis. Twenty microliters of the most purulent portion of a sputum sample was pipetted onto the paper and encapsulated within the plastic housing. The paper was allowed to air dry for 10 min at room temperature. A 1-mm sterile disposable biopsy punch (Miltex, NY, USA) was used to remove a disc from the cassette from each of the three marked locations, and each disc was added individually to each of the three sample positions of a Genedrive cartridge. The dynamics of decontamination and parallel nucleic acid isolation are currently being compiled for peer review publication and are not shown here.
Analysis using the Genedrive system.
Cartridges were run on the Genedrive system using the following M. tuberculosis program: a 95°C/5 min isothermal step, followed by 45 cycles each of 95°C/9 s, 53°C/13 s, and 72°C/7 s. A melt curve analysis was performed from 40°C to 80°C (entire analysis time, 60 min). The melt curve analysis identifies the temperature at which each of the Highlighter Probes dissociates. The presence of mutations in the rpoB gene destabilizes the probe amplicon hybridization and results in a lower melting temperature. The results are automatically called on the device's display, indicating that the sample has no detectable M. tuberculosis (undetected) or is either a RIF-sensitive wild-type (detected sensitive) or a RIF-resistant (detected resistant) strain.
The performance was compared against laboratory-based StepOne qPCR equipment. Lyophilized reagents from the Genedrive cartridges were transferred into microtiter plates, and PCR with melt curve data using several sets of titrated M. tuberculosis cultures of known RIF status were obtained. The data provided 100% concordance with the same samples analyzed on the Genedrive system.
Assessment of the Genedrive analytical limit of detection.
The analytical limit of detection (LOD) for the Genedrive system was determined using an M. tuberculosis strain H37Rv genomic DNA standard (ATCC 25618). The standards were resuspended in ultrapure sterile Milli-Q water and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). A serial dilution was prepared from 106 to 10 copies in ultrapure sterile Milli-Q water.
GeneXpert assay performance benchmarking in clinical sputum samples.
The GeneXpert assay provides an automated laboratory-based diagnosis of MTBC from clinical sputum samples (17) and was used in this work to provide a standardized molecular diagnostic test with which to benchmark the performance of the Genedrive workflow. After removing 20 μl of a clinical sputum sample for the Genedrive test, 1.5 ml of the remaining sputum sample was digested and decontaminated by the N-acetyl-l-cysteine/NaOH method according to the standard GeneXpert protocol (13). The decontaminated sample was then placed into the GeneXpert cartridge and processed according to the GeneXpert instructions for use. The cartridges were run and the results were collected.
Statistical analysis.
The standard deviation and coefficient of variation (CV) were calculated using Excel software (Microsoft Corp., Redmond, WA, USA). A chi-square test for linear trend was performed to analyze the smear-positive grade by microscopy against bacterial load by the GeneXpert assay using SPSS version 19 (Chicago, IL, USA). Significance was assigned at a P value of <0.05, and a 95% confidence interval (CI) was used.
Ethical approval for the study.
This study was approved by the ethics committees of Hospital Carlos III and Hospital La Paz.
RESULTS
LOD of MTBC and rpoB mutation markers.
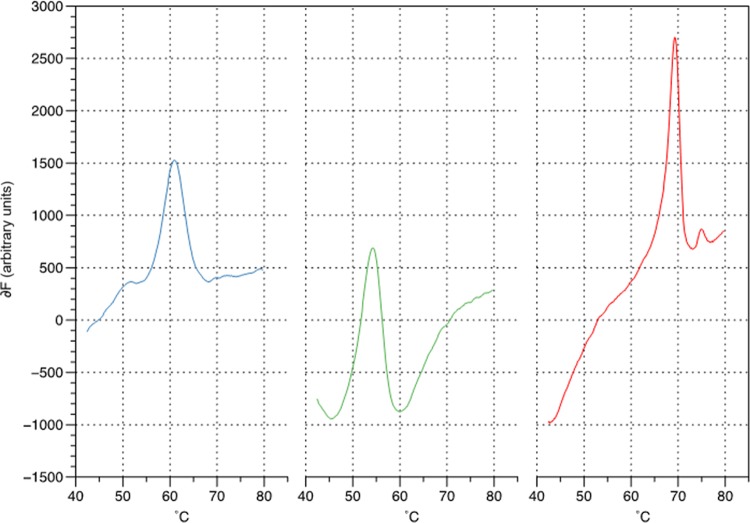
The Genedrive system amplifies MTBC and the core 81-bp region of rpoB. Both reactions were shown to have an analytical LOD (CI > 95%) that was capable of detecting 5 genomic DNA copies (Fig. 1). The Highlighter Probe for each target was capable of generating a simple melt curve profile providing clear peaks for each of the amplicons at the LOD. No-template controls (NTCs) yielded a single peak in the internal inhibition control reaction, and no peaks were visible in the two test reactions (data not shown).
FIG 1.
Representative Highlighter Probe melt curve data from 5 copies of genomic DNA from cultured strain H37Rv (ATCC 25618, USA) amplified using the Genedrive system. The data show first-order derivative melt peaks for M. tuberculosis at 62°C (blue), internal positive control at 54°C (green), and RIF wild type (WT) at 69°C (red).
Sensitivity and specificity for MTBC and rpoB mutation marker detection using spiked sputum samples.
A total of 85 sputum samples were spiked using known amounts of CFU obtained from liquid cultures. The overall sensitivity of the Genedrive system compared with culture was 90.8% (95% CI, 81, 96.5) for MTBC detection and 72.3% (95% CI, 59.8, 82.7) for rpoB detection. For samples containing ≥1,000 CFU/ml, the sensitivities were 100% for MTBC and 85.7% for rpoB detection, while for samples containing ≤100 CFU/ml, the sensitivities were 86.4% and 65.9% for MTBC and rpoB detection, respectively (Table 1). In all cases, RIF susceptibility detection was concordant with the detection using genotypic and phenotypic methods. Eight samples were known RIF-resistant strains (six within the ≥1,000 CFU/ml group and two within the ≤100 CFU/ml group) which were correctly identified by the Genedrive system as RIF resistant. All 20 spiked sputum samples from bacteria and nontuberculous mycobacteria, as well as three samples corresponding to other mycobacteria from MTBC, were classified correctly by the Genedrive system (specificity, 100%; 95% CI, 83.2%, 100%).
TABLE 1.
Estimation of sensitivity of Genedrive assay according to bacterial load in sputum samples spiked with mycobacterial cultures
| Bacterial load and RIF susceptibility for MTBC | MTBC identifications by Genedrive (no. [%]) |
Total no. (%) | ||
|---|---|---|---|---|
| Positive | Positive plus rpoB detectiona | Negative | ||
| ≥1,000 CFU/ml | ||||
| RIF susceptible | 15 (100) | 12 (81) | 15 (100) | |
| RIF resistant | 6 (100) | 6 (100) | 6 (100) | |
| ≤100 CFU/ml | ||||
| RIF susceptible | 36 (85.7) | 27 (64.3) | 6 (14.3) | 42 (100) |
| RIF resistant | 2 (100) | 2 (100) | 2 (100) | |
| Total | 59 (90.8) | 47 (72.3) | 6 (9.2) | 65 (100) |
RIF-susceptible and RIF-resistant samples were identified correctly in all cases.
The accuracy of the melting temperature for peaks reported by the Genedrive system for each of the targets is shown in Table 2; the data show the interval analysis for peaks corresponding to the internal inhibition control, MTBC, and rpoB. There was no overlapping of the peak intervals, including for the RIF-sensitive and RIF-resistant strains (Fig. 2). A much larger study of rpoB mutations is being undertaken by the authors and will be presented in a future publication.
TABLE 2.
Statistical analysis for detection peaks corresponding to melting temperatures of internal control, MTBC detection, and RIF susceptibility
| Tm typea | No. of detection peaks | Melting temp (°C) |
CV (%)b | |||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | |||
| Internal control | 71 | 53.6 | 57.1 | 55.7 | 0.9 | 1.6 |
| MTBC detection | 56 | 59.4 | 63.5 | 62.3 | 0.9 | 1.4 |
| RIF susceptible | 37 | 70 | 71.9 | 70.9 | 0.4 | 0.6 |
| RIF resistant | 8 | 66.2 | 69.6 | 68.1 | 1.1 | 1.6 |
Tm, melting temperature.
CV, coefficient of variation.
FIG 2.
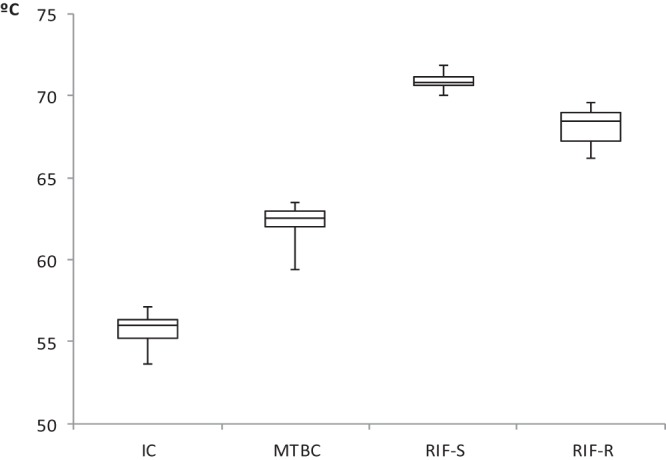
Box-and-whisker graph of melting temperatures corresponding to internal control (IC), M. tuberculosis complex (MTBC), and rifampin-susceptible (RIF-S) or RIF-resistant (RIF-R) samples.
GeneXpert assay performance benchmarking in clinical sputum samples.
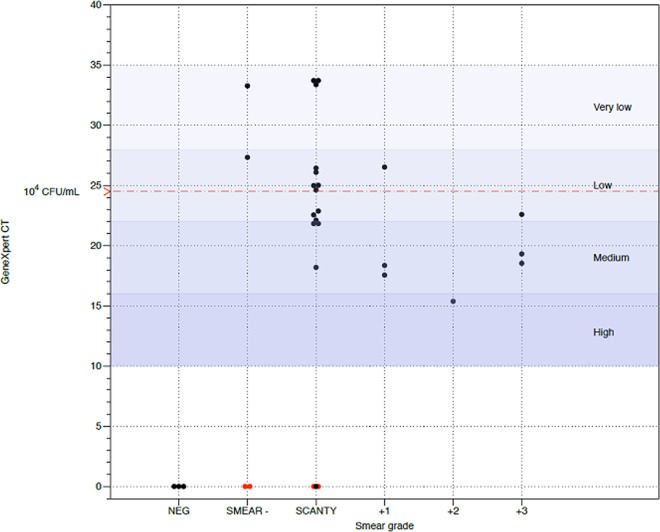
The performance of the Genedrive system was compared against that of the GeneXpert assay using clinical sputum samples. In order to know the performance of both assays for M. tuberculosis-infected subjects with a negative or scanty smear result, a group of patients who had recently initiated anti-TB treatment was included. The patients had been undergoing treatment for ≤2 weeks and as a result, the cultures were still positive. All patients were susceptible to RIF. The samples were assessed according to standard microscopy methods in order to obtain a smear status. A total of 31 fresh clinical sputum samples were processed, providing 24 smear-positive and 7 smear-negative samples, of which 3 were true negative samples and 4 were shown to have ≤2 bacilli per 300 optical fields. Figure 3 shows that 100% of the samples containing MTBC were correctly identified with the Genedrive system compared to 93.5% with the GeneXpert assay. The Genedrive system was able to correctly identify all samples with ≤2 bacilli per 300 fields of view. All assays correctly identified RIF susceptibility, with a sensitivity of 89.3% for the Genedrive system. As expected, a linear trend was observed between the results of bacterial load by the GeneXpert assay and the smear grade by microscopy (P = 0.001).
FIG 3.
Smear grade and GeneXpert results of clinical sputum samples. Black dots represent samples in which the Genedrive and GeneXpert results were concordant; red dots represent samples that were Genedrive positive and GeneXpert negative. NEG, no MTBC samples.
DISCUSSION
Early and effective TB diagnosis is pivotal as a global strategy for controlling the spread of the disease. In recent years, the wider availability of molecular tests as in the laboratory has boosted the diagnosis rate of TB and will help limit the spread and the potential for this disease to become epidemic (1). We have described a new POC platform (Genedrive) and demonstrated its capability to process raw sputum samples, detect the presence of MTBC, and simultaneously genotype rpoB, a gene with mutations conferring resistance to rifampin.
The Genedrive workflow was shown to be effective, providing a simple and easy-to-use process comparable to those of standard staining and microscopy methods. Sputum is known to offer particular challenges to PCR in terms of the amount of inhibitors in the matrix. The paper-based processing of raw sputum offers a compelling workflow for diagnostics in the field, and this might be particularly compelling in low-resource settings (18, 19). We have shown that the sample extraction process workflow is capable of processing the most purulent portion of sputum (the “core”), and this tended to generate the best PCR amplification profiles in comparison to samples taken from the more fluid peripheral parts of the sputum samples. However, other factors, such as the condition of the specimen (fresh versus frozen) or the specimen preparation (decontamination), were found to alter the performance of the Genedrive system to some degree (data not shown), as has been reported for the GeneXpert system (20). An advantage of the sample processing workflow in the Genedrive instructions for use is the ability to use a small sample volume. This eliminates the requirement for pipetting a specific minimal volume of sputum in order to obtain an accurate measure; for example, the GeneXpert assay requires a minimum of 1 ml of sputum. This can be particularly advantageous in the field, where collecting sufficient sample volume from patients may represent an issue.
Spiked sputum samples were used as a model to predict the performance of the test using samples derived from liquid culture and titrated against known numbers of CFU spiked into sputum. A number of independent methods were used to assess the quantity of cells in the titration, and good concordance was obtained with each of the methods used. These samples provided a good assessment of sensitivity, showing that the best performance for the system was with samples bearing ≥1,000 CFU/ml. However, the system still showed a value of 86% in samples with ≤100 CFU/ml. Several studies have reported that the threshold for detecting bacilli using light microscopy is between 5,000 and 10,000 CFU/ml (21, 22). Sputum samples that are smear positive and culture positive for TB correspond to patients that are highly contagious as a result of a high bacterial burden. In contrast, smear-negative/culture-positive sputum samples are found in paucibacillary patients, such as HIV-infected subjects, for whom the risk of contracting TB is higher than for other groups of individuals (1, 5). According to our data, the Genedrive system is able to diagnose TB even in the group of smear-negative/culture-positive patients. These in vitro results were corroborated to some extent using clinical sputum samples from paucibacillary patients and demonstrates the capability of the Genedrive system to detect TB in these challenge samples.
Finally, we have found that the Genedrive system provides comparable performance to the GeneXpert assay, correctly identifying the presence of MTBC and rpoB mutations in the pool of clinical samples tested. In some instances, the Genedrive system identified samples that were missed by the GeneXpert. These are ultralow titer samples that were smear negative or contained ≤2 bacilli per 300 fields of view. Even though the Genedrive system failed to detect rpoB in a small number of samples, it correctly identified the MTBC target. This is because the RIF assay targets a single copy gene compared to the multicopy MTBC target (23). The overall sensitivity for rpoB detection was comparable to that of the GeneXpert assay. However, all these data should be interpreted cautiously, for two reasons: first, we used raw sputum in the Genedrive assay, while in the GeneXpert assay, processed samples were employed, which can diminish its performance (20); second, the sample size was limited and it was a nonblinded study. A large field study will be conducted to confirm these data in the near future.
There is a need for better solutions to diagnose disease in the field, and this is particularly acute for the diagnosis of TB in low-resource settings. The data presented here demonstrate a workflow that is compatible with the POC and the specific challenges around the ease of use and the requirement for a simplified workflow that is compatible with low-resource settings. The effectiveness of these tools at the POC will not only facilitate early and appropriate therapy but also limit the impact of patient dropout, which is one of the most significant factors responsible for the spread of TB.
ACKNOWLEDGMENTS
We are indebted to the staff of the Bioincubator in Manchester, United Kingdom, for their help with the revision of the manuscript and their excellent technical assistance.
This work was partially supported by Epistem, Ltd. (Manchester, United Kingdom).
Pablo Castan and Benjamin D. Cobb are staff members of Epistem, Ltd. José Miguel Rubio, Jesús Mingorance, and Carlos Toro receive research funding from Epistem, Ltd. All other authors declare no conflicts of interest.
Footnotes
Published ahead of print 4 December 2013
REFERENCES
- 1.WHO 2012. Global tuberculosis report 2012. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/75938/1/9789241564502_eng.pdf [Google Scholar]
- 2.Marais BJ, Lönnroth K, Lawn SD, Migliori GB, Mwaba P, Glaziou P, Bates M, Colagiuri R, Zijenah L, Swaminathan S, Memish ZA, Pletschette M, Hoelscher M, Abubakar I, Hasan R, Zafar A, Pantaleo G, Craig G, Kim P, Maeurer M, Schito M, Zumla A. 2013. Tuberculosis comorbidity with communicable and non-communicable diseases: integrating health services and control efforts. Lancet Infect. Dis. 13:436–448. 10.1016/S1473-3099(13)70015-X [DOI] [PubMed] [Google Scholar]
- 3.WHO 2011. Same-day diagnosis of tuberculosis by microscopy: WHO policy statement. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 4.Böttger EC, Springer B. 2008. Tuberculosis: drug resistance, fitness, and strategies for global control. Eur. J. Pediatr. 167:141–148. 10.1007/s00431-007-0606-9 [DOI] [PubMed] [Google Scholar]
- 5.Colebunders R, Bastian I. 2000. A review of the diagnosis and treatment of smear-negative pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 4:97–107 [PubMed] [Google Scholar]
- 6.Parsons LM, Somoskövi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, Spector S, Roscigno G, Nkengasong J. 2011. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin. Microbiol. Rev. 24:314–350. 10.1128/CMR.00059-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer MV. 2007. Tuberculosis: a reemerging disease at the interface of domestic animals and wildlife. Curr. Top. Microbiol. Immunol. 315:195–215. 10.1007/978-3-540-70962-6_9 [DOI] [PubMed] [Google Scholar]
- 8.de Jong BC, Antonio M, Gagneux S. 2010. Mycobacterium africanum–review of an important cause of human tuberculosis in West Africa. PLoS Negl. Trop. Dis. 4:e744. 10.1371/journal.pntd.0000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TY, Lee TJ, Belisle JT, Brennan PJ, Kim SK. 1997. A novel repeat sequence specific to Mycobacterium tuberculosis complex and its implications. Tuber. Lung Dis. 78:13–19. 10.1016/S0962-8479(97)90011-3 [DOI] [PubMed] [Google Scholar]
- 10.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881–892. 10.1099/13500872-145-4-881 [DOI] [PubMed] [Google Scholar]
- 11.Musser JM. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raja S, Ching J, Xi L, Hughes SJ, Chang R, Wong W, McMillan W, Gooding WE, McCarty KS, Jr, Chestney M, Luketich JD, Godfrey TE. 2005. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin. Chem. 51:882–890. 10.1373/clinchem.2004.046474 [DOI] [PubMed] [Google Scholar]
- 13.Pfyffer GE. 2007. Mycobacterium: general characteristics, laboratory detection, and staining procedures, p 543–572 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 14.Woods GL, Warren NG, Inderlied CB. 2007. Susceptibility test methods: mycobacteria, Nocardia, and other actinomycetes, p 1223–1247 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA.(ed), Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 15.Romero Gómez MP, García-Perea A, Ruiz Carrascoso G, Bajo MA, Mingorance J. 2010. Campylobacter fetus peritonitis and bacteremia in a patient undergoing continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 48:336–337. 10.1128/JCM.01625-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO 1998. Laboratory services in tuberculosis control. Part 2: microscopy. Publication number WHO_TB_98.258. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/hq/1998/WHO_TB_98.258_(part2).pdf [Google Scholar]
- 17.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J. Acquir. Immune Defic. Syndr. 38:615–617. 10.1097/01.qai.0000143604.71857.5d [DOI] [PubMed] [Google Scholar]
- 19.Chakravarti A, Rawat D, Yadav S. 2003. Whole blood samples as an alternative to serum for detection of immunity to measles virus by ELISA. Diagn. Microbiol. Infect. Dis. 47:563–567. 10.1016/S0732-8893(03)00166-4 [DOI] [PubMed] [Google Scholar]
- 20.Steingart KR, Sohn H, Schiller I, Kloda LA, Boehme CC, Pai M, Dendukuri N. 2013. Xpert MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst. Rev. 2013:CD009593. 10.1002/14651858.CD009593.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobby GL, Holman AP, Iseman MD, Jones JM. 1973. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob. Agents Chemother. 4:94–104. 10.1128/AAC.4.2.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behr MA, Warren SA, Salamon H, Hopewell PC, Ponce de Leon A, Daley CL, Small PM. 1999. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet 353:444–449. 10.1016/S0140-6736(98)03406-0 [DOI] [PubMed] [Google Scholar]
- 23.Donnabella V, Martiniuk F, Kinney D, Bacerdo M, Bonk S, Hanna B, Rom WN. 1994. Isolation of the gene for the beta subunit of RNA polymerase from rifampicin-resistant Mycobacterium tuberculosis and identification of new mutations. Am. J. Respir. Cell Mol. Biol. 11:639–643. 10.1165/ajrcmb.11.6.7946393 [DOI] [PubMed] [Google Scholar]