Abstract
Improved methods for the detection of Histoplasma capsulatum are needed in regions with limited resources in which the organism is endemic, where delayed diagnosis of progressive disseminated histoplasmosis (PDH) results in high mortality rates. We have investigated the use of a loop-mediated isothermal amplification (LAMP) assay to facilitate rapid inexpensive molecular diagnosis of this disease. Primers for LAMP were designed to amplify the Hcp100 locus of H. capsulatum. The sensitivity and limit of detection were evaluated using DNA extracted from 91 clinical isolates of known geographic subspecies, while the assay specificity was determined using DNA extracted from 50 other fungi and Mycobacterium tuberculosis. Urine specimens (n = 6) collected from HIV-positive individuals with culture- and antigen-proven histoplasmosis were evaluated using the LAMP assay. Specimens from healthy persons (n = 10) without evidence of histoplasmosis were used as assay controls. The Hcp100 LAMP assay was 100% sensitive and specific when tested with DNA extracted from culture isolates. The median limit of detection was ≤6 genomes (range, 1 to 300 genomes) for all except one geographic subspecies. The LAMP assay detected Hcp100 in 67% of antigen-positive urine specimens (4/6 specimens), and results were negative for Hcp100 in all healthy control urine specimens. We have shown that the Hcp100 LAMP assay is a rapid affordable assay that can be used to expedite culture confirmation of H. capsulatum in regions in which PDH is endemic. Further, our results indicate proof of the concept that the assay can be used to detect Histoplasma DNA in urine. Further evaluation of this assay using body fluid samples from a larger patient population is warranted.
INTRODUCTION
Histoplasma capsulatum is a dimorphic fungus that causes histoplasmosis. In immunocompromised persons, H. capsulatum can disseminate throughout the body, causing progressive disseminated histoplasmosis (PDH), which is characterized by fever, weight loss, and hepatosplenomegaly. Without early diagnosis and antifungal intervention, PDH can cause death.
Timely detection of PDH is problematic in resource-challenged countries, since few rapid assays exist for this disease (1) and its symptoms are vague and often confused with those of mycobacterial or leishmanial infections (2, 3). Many laboratories in resource-limited areas in which the disease is endemic rely on sterile-site cultures for diagnosis of PDH; however, H. capsulatum grows slowly and may take several weeks for identification in cultures. The AccuProbe H. capsulatum culture identification test (Gen-Probe) can be used for rapid molecular identification of H. capsulatum in cultures, but this test is expensive and is not readily available in developing countries. Several additional molecular assays for detection of H. capsulatum have been developed (1, 4, 5), but none has been subjected to large-scale interlaboratory evaluation. These assays rely on PCR methodology and require expensive reagents and equipment, which may be unsustainable in laboratories with limited funding.
Here we describe the development of a loop-mediated isothermal amplification (LAMP) assay for histoplasmosis, which provides an affordable method of molecular identification that can be performed and interpreted without costly equipment in resource-challenged regions. Briefly, LAMP is a nucleic acid amplification technique that utilizes a polymerase with helicase activity, Bst from Bacillus stearothermophilus. The helicase activity allows for amplification of DNA at a constant temperature and is facilitated by four primers, 2 with cDNA, that form stem-loop DNA structures. Once formed, the stem-loop structures become the template DNA for further amplification, which occurs very rapidly (6).
Nucleic acid amplification via LAMP has several cost advantages over PCR. First, Bst polymerase is less expensive and more robust than Taq (7–9). Further, LAMP requires no thermal cycling equipment, since the assay is performed at a single temperature, allowing the use of either a heat block or a water bath to achieve nucleic acid amplification. Reactions are carried out in single tubes, and results can be visualized under UV light. In order to facilitate rapid inexpensive molecular diagnosis of PDH, we developed a LAMP assay targeting the single-copy gene Hcp100. Hcp100 is a member of the p100 gene family and is overexpressed in H. capsulatum during macrophage invasion (10). Unlike many multicopy housekeeping genes, Hcp100 shows little sequence identity with the DNA of related organisms and is not prone to false hybridization that may lead to cross-reactivity.
MATERIALS AND METHODS
H. capsulatum isolates.
H. capsulatum isolates (n = 91) used in this study were cultured from frozen mycelial stocks provided by Roche Molecular Systems (Pleasanton, CA). Mycelia were grown on brain heart infusion (BHI) agar slants and subcultured three times to ensure optimal growth and purity prior to DNA extraction. All isolates were previously identified with respect to their geographic subspecies by multilocus sequence typing (MLST) and phylogenetic analysis (11), as described by Theodoro et al. (12). The geographic subspecies of study isolates were as follows: four North American 1 (NAm 1), 65 North American 2 (NAm 2), 11 Latin American A (LAm A), five Latin American B (LAm B), two lineage H81, and one each African, Netherlands, lineage H66, and lineage H68.
Fungal DNA extraction.
Genomic DNA was extracted using a Qiagen DNeasy tissue kit (Qiagen, Valencia, CA), with several modifications to the manufacturer's instructions. Briefly, a portion of the fungal mat was transferred to 5-ml polypropylene tubes containing 800 μl Qiagen ATL buffer and 60 U of proteinase K and was homogenized inside a biological safety cabinet using an Omni tissue homogenizer (Omni International, Kennesaw, GA) at slow speed for 30 s and then at high speed for 30 s, using a clean probe for each isolate. Homogenates were capped, incubated at 55°C for 1 h with frequent vortex mixing, and then cooled to room temperature (RT). For each homogenate, RNase A (Sigma-Aldrich Corp., St. Louis, MO) was added to a final concentration of 1 mg/ml and the mixture was incubated for 5 min at RT, followed by the addition of 900 μl Qiagen buffer AL and vortex mixing. Homogenates were incubated at 70°C for 10 min, transferred to 1.7-ml microcentrifuge tubes, and centrifuged at 15,000 × g for 10 min. Clear supernatants (1 ml each) were transferred to clean microcentrifuge tubes, and 500 μl of 200-proof genomic-grade ethanol (Sigma-Aldrich Corp.) was added to each tube. The suspensions were vortex mixed and transferred to Qiagen DNeasy columns; the manufacturer's instructions were followed throughout the remainder of the procedure, except that DNA was eluted with 0.01 M Tris. DNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Archived fungal DNA samples used as controls were buffer exchanged into 0.01 M Tris using the Qiagen protocol for cleanup of genomic DNA (13), in order to eliminate EDTA and other additives known to interfere with subsequent applications.
PCR and sequencing of the Hcp100 genetic locus.
Portions of the H. capsulatum Hcp100 gene were PCR amplified using Hc I (5′-GCGTTCCGAGCCTTCCACCTCAAC-3′) and Hc II (5′-ATGTCCCATCGGGCGCCGTGTAGT-3′) primers, as described previously (14). Each 25-μl reaction mixture contained 0.2 μM each primer, 0.25 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), and 0.625 U Taq DNA polymerase in a buffer of 10 mM Tris (pH 8.3), 50 mM KCl (Roche Carolina Inc., Florence, SC). Thermal cycling was performed in MicroAmp 96-well optical reaction plates, using a GeneAmp PCR system 9700 (Applied Biosystems, Inc., Foster City, CA), as follows: 1 cycle of 94°C for 5 min; 40 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min; and a final cycle of 72°C for 5 min. The resulting amplicons were sized on 1.75% agarose gels, visualized with ethidium bromide under UV light, and cleaned using Exo-SAP-IT (USB Corp., Cleveland, OH), according to the manufacturer's instructions. Amplicons were sequenced using the amplification primers Hc I and Hc IV (5′-AGGAGAGAACTGTATCGGTGGCTT-3′) (14) and a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems Inc.), and reactions were analyzed with a 3730 DNA analyzer (Applied Biosystems, Inc.).
Comparative sequence analysis and LAMP primer design.
Consensus sequences among 14 isolates from geographically and genetically diverse clades were generated from raw data using Sequencher 4.9 (Gene Codes Corp., Ann Arbor, MI) and were aligned using MUSCLE (15) in the MEGA 5.05 software package (16). Primers for LAMP were designed on the basis of conserved regions of the reverse complement of Hcp100 using Primer Explorer 4, a Web-based, open-use, primer design program (Eiken Chemical Co., Ltd., Tokyo, Japan) (http://primerexplorer.jp/elamp4.0.0/index.html). Primers used for LAMP were forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3), and backward outer primer (B3) (Table 1).
TABLE 1.
Primers used for LAMP of the Hcp100 genetic locus of H. capsulatum
| Primer | Sequence (5′ to 3′) |
|---|---|
| FIP | TCCCCGCGTCTCCCGAATACCGATCCAATGTCCGTTCACC |
| BIP | TCTGCACGGAAAACTGCGGCCTACGGCAACTCCGAAACC |
| F3 | GTAGTCGACGTTCGCAACT |
| B3 | GCCGACGTCGTTTACATCG |
Clinical specimens.
Six urine specimens (one specimen per person) from HIV-infected persons with symptoms consistent with PDH, positive Histoplasma urine antigen test results, and culture-confirmed H. capsulatum infections were collected between 2004 and 2009 at the Clinica Familiar Luis Angel Garcia (Guatemala City, Guatemala) (17), under conditions reviewed and approved by the internal review boards of the Centers for Disease Control and Prevention and the Universidad del Valle de Guatemala. Ten urine specimens were collected from healthy persons who had no indication of histoplasmosis. Two “mock-positive” samples were prepared from different pools of histoplasmosis-negative urine samples, collected from healthy persons, by spiking with 1 ng H. capsulatum DNA prior to extraction.
Urine DNA extraction and PCR.
A QIAmp circulating nucleic acid kit (Qiagen) was used to extract DNA from urine specimens, according to the manufacturer's instructions. Briefly, 4 ml urine was centrifuged at 16,000 × g for 10 min, and the pellet was resuspended in 0.01 M phosphate-buffered saline (PBS) (pH 7.2). Both the supernatant and the resuspended pellets were subjected to DNA extraction using lysis buffers ATL and ACB, according to the manufacturer's instructions. A vacuum manifold designed for DNA extraction (Promega, Madison, WI) was attached to a rotary vane vacuum pump (Gast, Benton Harbor, MI) with a filter trap, and Qiagen Mini columns were attached to the vacuum using adapters provided in the kit. After lysis, the urine specimens were pulled through the columns with the vacuum pump at a pressure of −85,000 pascals, bound to the resin columns, and washed. Nucleic acids were eluted in 30 μl of 0.01 M Tris-HCl by centrifugation and were quantified using a NanoDrop ND-1000 spectrophotometer.
All urine specimen DNA was subjected to PCR using positive-control primers Beta 2 and Beta 3, which amplify portions of the human β-globin (BG) gene (Beta 2 [G1], 5′-GAAGAGCCAAGGACAGGTAC; Beta 3 [G2], 5′-CAACTTCATCCACGTTCACC) (18), and H. capsulatum Hc I/Hc IV primers specific for the Hcp100 genetic locus (14). The resulting amplicons were visualized on agarose gels as described above.
LAMP method.
Reaction mixtures were prepared according to the manufacturer's specifications, using a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd). Each 25-μl reaction mixture contained 12.5 μl 2× reaction buffer [40 mM Tris-HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)SO4, 1.6 M betaine, 0.2% Tween 20, 2.8 mM each DNTP], 40 pmol FIP, 40 pmol BIP, 10 pmol F3, 10 pmol B3, 1 μl Bst polymerase (Loopamp fluorescent detection reagent; Eiken Chemical Co., Ltd.), 2 μl of DNA, and 1 μl calcein-MnCl2 dye. Reaction mixtures were incubated at 63°C for 1.5 h, and the reactions were stopped at 95°C for 2 min. Amplification of H. capsulatum DNA was detected as calcein fluorescence directly visualized over a UV light box, in comparison with a lack of fluorescence in control reactions to which either non-H. capsulatum DNA or no DNA template was added (19).
LAMP assay validation.
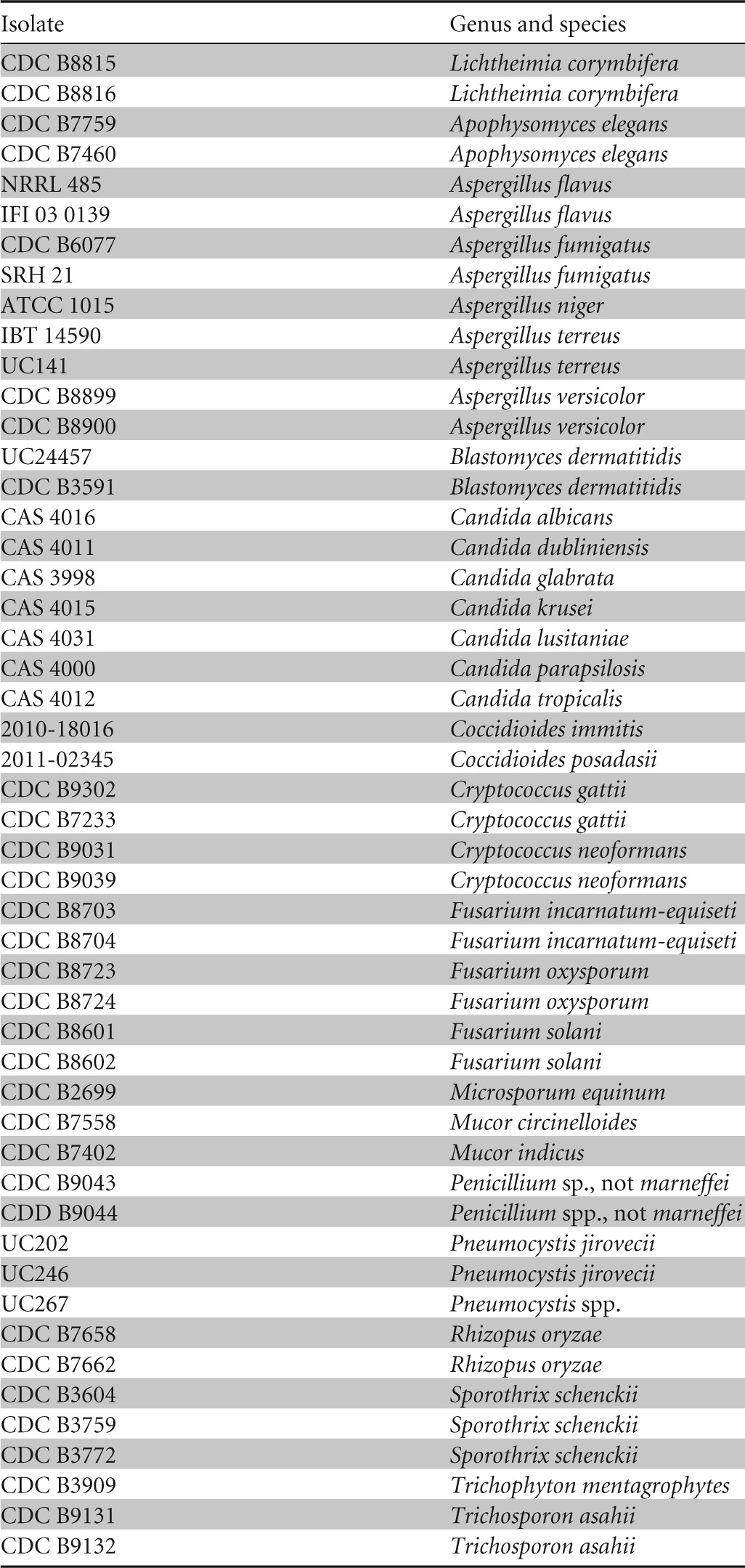
All H. capsulatum clinical isolates (n = 91) were assayed using the Hcp100 LAMP assay to determine its sensitivity. To define the limit of detection (LOD), DNA from 39 H. capsulatum isolates, including at least one representative of each geographic subspecies, was diluted 10-fold (from 1 ng/ml to 10 fg/ml) and assayed using LAMP. To test for potential cross-reactivity, archived DNA extracted from clinical fungal isolates maintained by the Mycotic Diseases Branch (n = 50) (Table 2) was assayed at concentrations of ≥2 ng/μl. Tenfold dilutions (50 ng/μl to 5 fg/μl) of Mycobacterium tuberculosis (ATCC 25177D-5) and human (catalog no. G304A; Promega, Madison, WI) genomic DNA were also assayed. All DNA and clinical specimen extracts were tested in duplicate. Two negative controls (human DNA and no DNA template) and at least one positive control were included in each LAMP procedure, and assays were repeated twice, to ensure reproducibility.
TABLE 2.
Fungal isolates (n = 50) tested in the LAMP assay for detection of H. capsulatum
RESULTS
LAMP assay validity with Histoplasma isolates.
The Hcp100 LAMP assay was able to detect DNA of all geographically diverse H. capsulatum isolates. The LOD was strain dependent, and values fell between 10 fg/μl and 1 pg/μl (1 to 30 genomes per reaction) of H. capsulatum (Table 3), with a median of 6 genomes. Sixteen strains were detected at concentrations of ≤100 fg/μl (1 to 6 genomes), and 19 were reactive at ≤10 fg/μl (one genome). When Hcp100 LAMP was compared with traditional PCR of Hcp100, the LAMP assay showed a 10-fold lower LOD (data not shown).
TABLE 3.
LOD of the LAMP assay for detection of isolates of different geographic subspecies (n = 39)
| Geographic subspecies | LOD (no. of genomes)a |
|
|---|---|---|
| Median | Range | |
| NAm 1 (4 isolates) | ≤6 | 1–30 |
| NAm 2 (13 isolates) | ≤1 | 1–6 |
| LAm A (11 isolates) | ≤6 | 1–30 |
| LAm B (5 isolates) | ≤6 | 1–30 |
| Other (6 isolates) | ≤6 | 1–300 |
| Avg | ≤6 | 1–30 |
The number of genomes was calculated by titration of DNA, based on the estimated genome size of H. capsulatum of 33 Mb.
The specificity of the LAMP assay was determined by testing the reactivity of LAMP primers against DNA of other clinically relevant yeasts and molds (Table 2), as well as human and mycobacterial DNA. No cross-reactivity occurred with any other organism tested, and the assay was 100% specific. When the assay incubation time was increased from 1.5 to 2 h, however, cross-reactivity did occur with some negative-control DNA samples.
LAMP assay validity with human urine specimens.
The two mock-positive samples that were prepared by spiking control urine samples with known concentrations of H. capsulatum DNA were assayed using LAMP and conventional PCR. These samples showed strong signals in the LAMP assay, and Hcp100 bands were present with PCR amplification (data not shown).
In addition, we tested six urine samples from persons with HIV infection and proven histoplasmosis (Table 4). Four of these samples (67%) showed strong signals in the Hcp100 LAMP assay, three with fluorescence in both the pellet and supernatant fractions and one with fluorescence in the pellet fraction only (Table 4). None of the 6 samples showed Hcp100 bands when DNA was amplified using traditional PCR and visualized using ethidium bromide (data not shown). Two samples failed to show amplification with the human β-globin (BG) primers; one of these samples was negative by both the HCP LAMP and BG PCR assays, while one sample was positive by BG PCR but negative by HCP LAMP. Furthermore, one sample that was positive by the LAMP assay failed to show amplification with BG primers. Conversely, no fluorescence was observed in the pellet or supernatant fractions of urine samples from healthy individuals. LAMP amplification products from positive urine samples and isolate DNA were visualized on agarose gels and produced similar characteristic patterns (Fig. 1).
TABLE 4.
Detection of H. capsulatum in human urine specimens using the LAMP assay
| Urine specimen no. | Health statusa | ELISAb antigenuria (ng/μl) | LAMP resultc | Human β-globin (PCR) |
|---|---|---|---|---|
| 208 | HIV infection, culture-proven histoplasmosis | 25.3 | + S, P | + |
| 209 | HIV infection, culture-proven histoplasmosis | 12.4 | − | − |
| 260 | HIV infection, culture-proven histoplasmosis | 13.8 | + P | + |
| 258 | HIV infection, culture-proven histoplasmosis | 12.9 | − | + |
| 256 | HIV infection, culture-proven histoplasmosis | 13.1 | + S, P | + |
| 548 | HIV infection, culture-proven histoplasmosis | 12.6 | + S, P | − |
| C5 | Healthy | 0.0 | − | + |
| C6 | Healthy | 0.0 | − | + |
| C7 | Healthy | 0.0 | − | + |
| C12 | Healthy | 0.0 | − | + |
| C13 | Healthy | 0.0 | − | + |
| C15 | Healthy | 0.0 | − | + |
| C16 | Healthy | 0.0 | − | + |
| C17 | Healthy | 0.0 | − | + |
| UP1 | Healthy | 0.0 | − | + |
| UP2 | Healthy | 0.0 | − | + |
Urine specimens were collected from persons with HIV infection and histoplasmosis (n = 6) and from healthy control subjects (n = 10).
ELISA, enzyme-linked immunosorbent assay.
S, supernatant fraction; P, pellet fraction.
FIG 1.
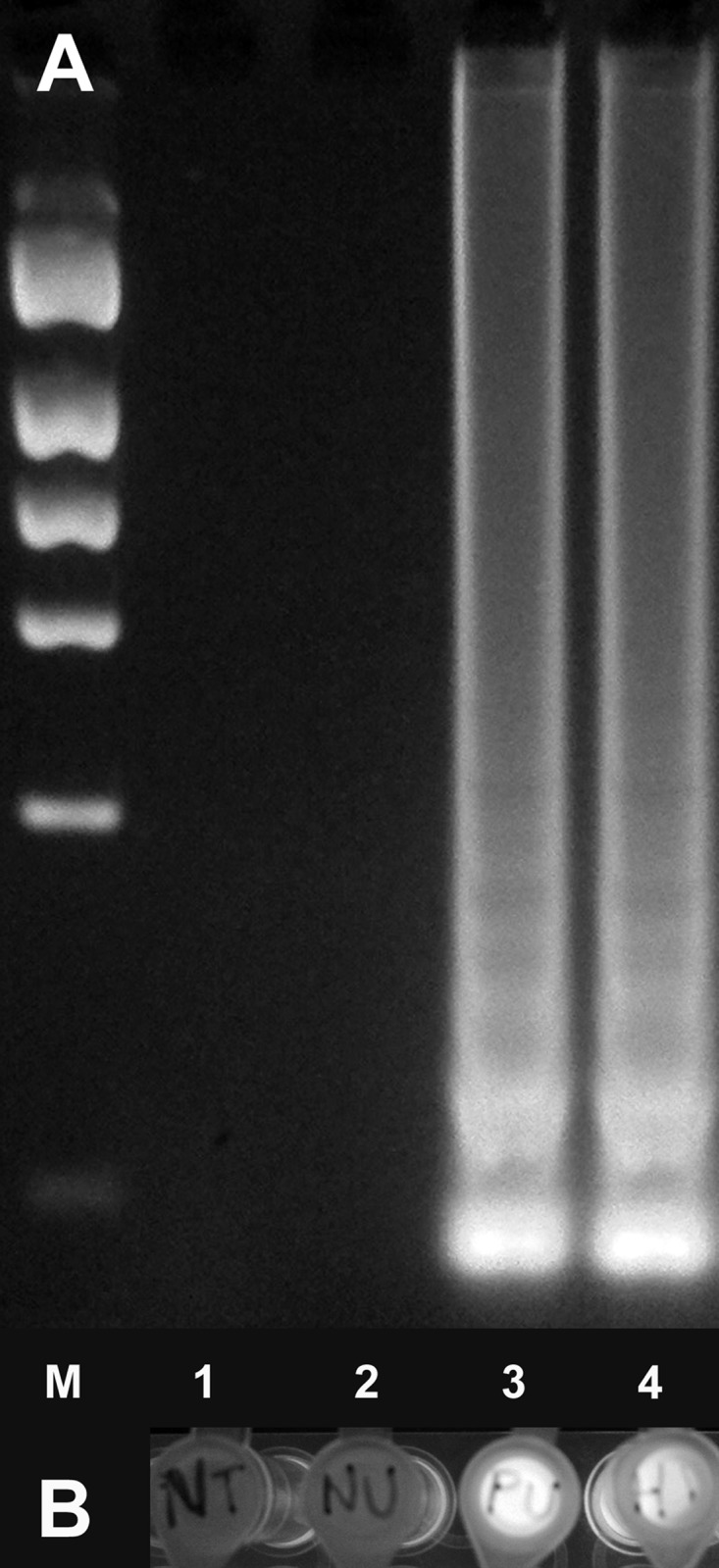
Hcp100 LAMP assay of DNA from human urine specimens and cultured H. capsulatum. (A) LAMP products were visualized on a 1.75% agarose gel. Characteristic “ladder-type” banding is shown with H. capsulatum antigen-positive urine and cultured H. capsulatum DNA (lanes 3 and 4, respectively), while no DNA amplification was seen after LAMP of healthy control urine (lane 2). Lane M, molecular size marker; lane 1, no-template control. (B) Corresponding tubes were visualized under UV light; fluorescent signals in tubes with antigen-positive urine and cultured H. capsulatum DNA are shown (tubes 3 and 4, respectively). Tube 1, no-template control; tube 2, healthy control urine.
DISCUSSION
The ability to provide laboratory diagnoses in resource-poor regions remains challenging. Many institutions in the developing world do not have the resources to detect and to identify infectious agents rapidly and precisely (20). In the case of PDH associated with HIV, the need for straightforward and inexpensive laboratory diagnostic tools remains important, because PDH can cause death in 95% of cases (21) within months if it is undiagnosed or misdiagnosed. Delayed antifungal treatment of PDH results in mortality rates between 30 and 42% (22–26) in regions where the disease is endemic and where there are underserved populations or limited diagnostic resources. Molecular diagnostic tests have the potential to detect very small amounts of DNA with high specificity, and the LAMP method is both rapid and inexpensive. We developed a LAMP assay to assist in rapid molecular identification of cultured H. capsulatum isolates, as well as to detect H. capsulatum DNA in urine from patients with disseminated disease. Using DNA from cultured isolates, we showed that the Hcp100 LAMP assay could detect less than 30 Histoplasma genomes, a sensitivity 10-fold greater than that of traditional PCR assays. Further, the LAMP assay did not show cross-reactions with DNA of other fungal organisms or with mycobacteria; the latter cause disseminated infections with symptoms nearly identical to those of PDH in persons with HIV/AIDS.
This novel LAMP assay may have two potential applications for the diagnosis of histoplasmosis in limited-resource settings. First, in a pilot validation study, we found that the LAMP assay was more sensitive than traditional PCR assays in detecting Hcp100 DNA in urine, and the LAMP assay showed no cross-reactions with DNA isolated from urine specimens from healthy persons, suggesting that this assay can be useful for direct detection of H. capsulatum DNA in clinical samples. Second, our data demonstrate that the LAMP assay can be used for rapid confirmation of H. capsulatum in culture when the AccuProbe test is not available. The advantages of and caveats for each application are discussed below.
The LAMP method has proven successful in detecting fungal DNA in specimen types in which whole intact organisms are localized. Pneumocystis species DNA has been detected in sputum and bronchiolar lavage (BAL) fluid specimens from patients with pneumonia (27), Paracoccidioides brasiliensis DNA has been detected in sputum specimens (28), and Penicillium marneffei DNA has been detected in formalin-fixed, paraffin-embedded (FFPE) tissue specimens (29). In our study, we targeted residual intracellular (in leukocyte debris) and free circulating fungal DNA in urine. Obvious advantages of urine are that sample collection is noninvasive and large quantities can easily be obtained from individual patients. Examination of urine sediment reveals a variety of cells including phagocytes (30), in which H. capsulatum survives by avoidance of lytic digestion (31). Additionally, DNA released from dying fungal cells is known to cross the renal barrier and is subsequently excreted in urine as cell-free DNA in lengths suitable for detection using PCR (32). In fact, urine is now commonly utilized in molecular diagnostic testing, and many organisms that cause systemic infections are detected in this bodily fluid (33–37).
We tested urine specimens collected from persons with HIV infection who had culture-proven PDH and positive antigenuria to determine LAMP assay sensitivity. Although an obvious caveat of our study is that only a small number of culture-positive urine specimens were available for testing, these pilot data demonstrated that 67% of samples (4/6 samples) were positive in the Hcp100 LAMP assay. One of the two urine samples for which negative LAMP results were obtained was also negative with PCR amplification with human β-globin primers, suggesting that no PCR-amplifiable human DNA was present in that sample (Table 4). In our prior experience using FFPE tissue biopsy specimens, we were seldom able to amplify fungal DNA with PCR when the human β-globin locus did not show amplification (38, 39). Human globin DNA could be amplified from a second urine sample that did not react in the LAMP assay. We assume that this sample contained insufficient fungal DNA to be detected even with the sensitive LAMP assay.
We have ruled out the presence of DNA polymerase inhibitors as a cause of insensitivity, since mock-positive urine specimens amplified Hcp100 strongly in both the LAMP and PCR assays. These samples were spiked with H. capsulatum DNA and immediately processed for DNA extraction. All were positive, further suggesting that DNA degradation contributed to decreased sensitivity of LAMP detection in urine. Overall, our data suggest that LAMP can be used to detect H. capsulatum DNA in urine samples; however, a large number of urine samples will need to be tested to determine the sensitivity of this method. In addition, the difficulty of extracting high-quality fungal DNA from clinical specimens using a rapid inexpensive method poses the greatest challenge in making LAMP available as a sustainable diagnostic method for fungal infections in resource-challenged laboratories.
The Hcp100 LAMP assay was highly sensitive in confirming the identification of H. capsulatum from DNA prepared from cultured isolates, a feature that can be helpful in countries with limited resources, where culture of blood and/or bone marrow samples is frequently the primary method for diagnosis of PDH. In these countries, diagnostic confirmation of cultured isolates is frequently made using morphological observations alone, and Histoplasma can be confused with other yeasts of similar size, such as Candida glabrata. Using the Hcp100 LAMP assay, only a small amount of yeast growth is necessary for DNA extraction and confirmation of H. capsulatum in culture.
The purpose of our study was to develop a DNA-based method for detection of disseminated histoplasmosis that could be performed in resource-challenged laboratories. We have shown proof of the concept that LAMP may be a valuable tool for detecting disseminated histoplasmosis. Further evaluation of LAMP using fresh-frozen urine, serum, or whole-blood samples is required, and a simpler and less expensive DNA extraction method should be evaluated for use in resource-limited countries.
ACKNOWLEDGMENTS
We thank the Bevier Public Health Summer Internship at Agnes Scott College for providing financial support to Yitian Zhou during her tenure at the Mycotic Diseases Branch of the CDC.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Munoz C, Gomez BL, Tobon A, Arango K, Restrepo A, Correa MM, Muskus C, Cano LE, Gonzalez A. 2010. Validation and clinical application of a molecular method for identification of Histoplasma capsulatum in human specimens in Colombia, South America. Clin. Vaccine Immunol. 17:62–67. 10.1128/CVI.00332-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couppie P, Aznar C, Carme B, Nacher M. 2006. American histoplasmosis in developing countries with a special focus on patients with HIV: diagnosis, treatment, and prognosis. Curr. Opin. Infect. Dis. 19:443–449. 10.1097/01.qco.0000244049.15888.b9 [DOI] [PubMed] [Google Scholar]
- 3.Zollner MS, Rezende KM, Birman S, Elias CP, Arisawa EA, Santos MA. 2010. Clinical and evolutionary characteristics of four patients with pulmonary histoplasmosis reported in the Paraiba Paulista Valley region. Rev. Soc. Bras. Med. Trop. 43:599–601. 10.1590/S0037-86822010000500028 [DOI] [PubMed] [Google Scholar]
- 4.Babady NE, Buckwalter SP, Hall L, Le Febre KM, Binnicker MJ, Wengenack NL. 2011. Detection of Blastomyces dermatitidis and Histoplasma capsulatum from culture isolates and clinical specimens by use of real-time PCR. J. Clin. Microbiol. 49:3204–3208. 10.1128/JCM.00673-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon S, Veron V, Boukhari R, Blanchet D, Aznar C. 2010. Detection of Histoplasma capsulatum DNA in human samples by real-time polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 66:268–273. 10.1016/j.diagmicrobio.2009.10.010 [DOI] [PubMed] [Google Scholar]
- 6.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, Notomi T, Chiodini PL, Sutherland CJ. 2010. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J. Clin. Microbiol. 48:2866–2871. 10.1128/JCM.00355-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiddle G, Hardinge P, Buttigieg N, Gandelman O, Pereira C, McElgunn CJ, Rizzoli M, Jackson R, Appleton N, Moore C, Tisi LC, Murray JA. 2012. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 12:15. 10.1186/1472-6750-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earley JJ, Kuivaniemi H, Prockop DJ, Tromp G. 1993. Efficient DNA sequencing on microtiter plates using dried reagents and Bst DNA polymerase. DNA Seq. 4:79–85 [DOI] [PubMed] [Google Scholar]
- 10.Porta A, Colonna-Romano S, Callebaut I, Franco A, Marzullo L, Kobayashi GS, Maresca B. 1999. An homologue of the human 100-kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem. Biophys. Res. Commun. 254:605–613. 10.1006/bbrc.1998.9894 [DOI] [PubMed] [Google Scholar]
- 11.Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castaneda E, Da Silva Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancope-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 12:3383–3401. 10.1046/j.1365-294X.2003.01995.x [DOI] [PubMed] [Google Scholar]
- 12.Theodoro RC, Scheel CM, Brandt ME, Kasuga T, Bagagli E. 2013. PRP8 intein in cryptic species of Histoplasma capsulatum: evolution and phylogeny. Infect. Genet. Evol. 18:174–182. 10.1016/j.meegid.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 13.Qiagen 2010. QIAmp DNA Micro handbook, 2nd ed, p 31–32 Qiagen, Valencia, CA [Google Scholar]
- 14.Bialek R, Feucht A, Aepinus C, Just-Nubling G, Robertson VJ, Knobloch J, Hohle R. 2002. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 40:1644–1647. 10.1128/JCM.40.5.1644-1647.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheel CM, Samayoa B, Herrera A, Lindsley MD, Benjamin L, Reed Y, Hart J, Lima S, Rivera BE, Raxcaco G, Chiller T, Arathoon E, Gomez BL. 2009. Development and evaluation of an enzyme-linked immunosorbent assay to detect Histoplasma capsulatum antigenuria in immunocompromised patients. Clin. Vaccine Immunol. 16:852–858. 10.1128/CVI.00066-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bialek R, Konrad F, Kern J, Aepinus C, Cecenas L, Gonzalez GM, Just-Nubling G, Willinger B, Presterl E, Lass-Florl C, Rickerts V. 2005. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 58:1180–1184. 10.1136/jcp.2004.024703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3:877–882. 10.1038/nprot.2008.57 [DOI] [PubMed] [Google Scholar]
- 20.Petti CA, Polage CR, Quinn TC, Ronald AR, Sande MA. 2006. Laboratory medicine in Africa: a barrier to effective health care. Clin. Infect. Dis. 42:377–382. 10.1086/499363 [DOI] [PubMed] [Google Scholar]
- 21.Ramos-e-Silva M, Lima CM, Schechtman RC, Trope BM, Carneiro S. 2012. Systemic mycoses in immunodepressed patients (AIDS). Clin. Dermatol. 30:616–627. 10.1016/j.clindermatol.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 22.Pontes LB, Leitao Tdo M, Lima GG, Gerhard ES, Fernandes TA. 2010. Clinical and evolutionary characteristics of 134 patients with disseminated histoplasmosis associated with AIDS in the State of Ceara. Rev. Soc. Bras. Med. Trop. 43:27–31. 10.1590/S0037-86822010000100007 [DOI] [PubMed] [Google Scholar]
- 23.Daher EF, Silva GB, Jr, Barros FA, Takeda CF, Mota RM, Ferreira MT, Oliveira SA, Martins JC, Araujo SM, Gutierrez-Adrianzen OA. 2007. Clinical and laboratory features of disseminated histoplasmosis in HIV patients from Brazil. Trop. Med. Int. Health 12:1108–1115. 10.1111/j.1365-3156.2007.01894.x [DOI] [PubMed] [Google Scholar]
- 24.Chang MR, Taira CL, Paniago AM, Taira DL, Cunha RV, Wanke B. 2007. Study of 30 cases of histoplasmosis observed in the Mato Grosso do Sul State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 49:37–39. 10.1590/S0036-46652007000100007 [DOI] [PubMed] [Google Scholar]
- 25.Huber F, Nacher M, Aznar C, Pierre-Demar M, El Guedj M, Vaz T, Vantilcke V, Mahamat A, Magnien C, Chauvet E, Carme B, Couppie P. 2008. AIDS-related Histoplasma capsulatum var. capsulatum infection: 25 years experience of French Guiana. AIDS 22:1047–1053. 10.1097/QAD.0b013e3282ffde67 [DOI] [PubMed] [Google Scholar]
- 26.Baddley JW, Sankara IR, Rodriquez JM, Pappas PG, Many WJ., Jr 2008. Histoplasmosis in HIV-infected patients in a southern regional medical center: poor prognosis in the era of highly active antiretroviral therapy. Diagn. Microbiol. Infect. Dis. 62:151–156. 10.1016/j.diagmicrobio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 27.Uemura N, Makimura K, Onozaki M, Otsuka Y, Shibuya Y, Yazaki H, Kikuchi Y, Abe S, Kudoh S. 2008. Development of a loop-mediated isothermal amplification method for diagnosing Pneumocystis pneumonia. J. Med. Microbiol. 57:50–57. 10.1099/jmm.0.47216-0 [DOI] [PubMed] [Google Scholar]
- 28.Tatibana BT, Sano A, Uno J, Kamei K, Igarashi T, Mikami Y, Miyaji M, Nishimura K, Itano EN. 2009. Detection of Paracoccidioides brasiliensis gp43 gene in sputa by loop-mediated isothermal amplification method. J. Clin. Lab. Anal. 23:139–143. 10.1002/jcla.20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Li X, Zeng H, Xie Z, Lu C, Xi L, de Hoog GS. 2010. Development and evaluation of loop-mediated isothermal amplification (LAMP) for the rapid diagnosis of Penicillium marneffei in archived tissue samples. FEMS Immunol. Med. Microbiol. 58:381–388. 10.1111/j.1574-695X.2010.00647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokota M, Tatsumi N, Tsuda I, Takubo T, Hiyoshi M. 1998. DNA extraction from human urinary sediment. J. Clin. Lab. Anal. 12:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA. 2012. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 8:e1002713. 10.1371/journal.ppat.1002713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botezatu I, Serdyuk O, Potapova G, Shelepov V, Alechina R, Molyaka Y, Ananev V, Bazin I, Garin A, Narimanov M, Knysh V, Melkonyan H, Umansky S, Lichtenstein A. 2000. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 46:1078–1084 [PubMed] [Google Scholar]
- 33.Enk MJ, Oliveira e Silva G, Rodrigues NB. 2012. Diagnostic accuracy and applicability of a PCR system for the detection of Schistosoma mansoni DNA in human urine samples from an endemic area. PLoS One 7:e38947. 10.1371/journal.pone.0038947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mharakurwa S, Simoloka C, Thuma PE, Shiff CJ, Sullivan DJ. 2006. PCR detection of Plasmodium falciparum in human urine and saliva samples. Malar. J. 5:103. 10.1186/1475-2875-5-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrea G, Van de Perre P, Ouedraogo M, Zougba A, Sawadogo A, Dingtoumda B, Diallo B, Defer MC, Sombie I, Zanetti S, Sechi LA. 2005. PCR-based detection of the Mycobacterium tuberculosis complex in urine of HIV-infected and uninfected pulmonary and extrapulmonary tuberculosis patients in Burkina Faso. J. Med. Microbiol. 54:39–44. 10.1099/jmm.0.45688-0 [DOI] [PubMed] [Google Scholar]
- 36.Exner MM, Lewinski MA. 2003. Isolation and detection of Borrelia burgdorferi DNA from cerebral spinal fluid, synovial fluid, blood, urine, and ticks using the Roche MagNA Pure system and real-time PCR. Diagn. Microbiol. Infect. Dis. 46:235–240. 10.1016/S0732-8893(03)00080-4 [DOI] [PubMed] [Google Scholar]
- 37.Tang YW, Li H, Durkin MM, Sefers SE, Meng S, Connolly PA, Stratton CW, Wheat LJ. 2006. Urine polymerase chain reaction is not as sensitive as urine antigen for the diagnosis of disseminated histoplasmosis. Diagn. Microbiol. Infect. Dis. 54:283–287. 10.1016/j.diagmicrobio.2005.10.008 [DOI] [PubMed] [Google Scholar]
- 38.Munoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, Gomez BL. 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 48:2147–2153. 10.1128/JCM.00459-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campos PF, Gilbert TM. 2012. DNA extraction from formalin-fixed material. Methods Mol. Biol. 840:81–85. 10.1007/978-1-61779-516-9_11 [DOI] [PubMed] [Google Scholar]


