Abstract
The United States has experienced a substantial increase in the reported incidence of cryptosporidiosis since 2005. Accompanying this is the emergence of a new subtype of Cryptosporidium hominis based on variation at the 60-kDa glycoprotein (gp60) locus, IaA28R4, which has become a frequently identified subtype in both sporadic and outbreak-related cases. In this study, using multilocus sequence typing (MLST) at eight genetic loci, we characterized 62 specimens of IaA28R4 and 33 specimens of three other gp60 subtypes of C. hominis from four U.S. states with increased cryptosporidiosis incidences during the summer of 2008. Extensive genetic heterogeneity was seen within the gp60 subtype IaA28R4, but specimens from Ohio and southwestern states formed two distinct subpopulations, suggesting that there were at least two origins of IaA28R4 within the United States. Discordance in typing results was observed between gp60 and other genetic markers, especially DZ-HRGP, and this discordance was largely the result of genetic recombination within the gp60 subtype IaA28R4. The results of population genetic analyses supported the presence of two subpopulations of IaA28R4 and the occurrence of genetic recombination within this gp60 subtype. Thus, the IaA28R4 subtype at gp60 is likely a fitness marker for C. hominis, and genetic recombination is potentially a driving force in the emergence of the virulent IaA28R4 subtype in the United States. A rapid evolution of IaA28R4 was indicated by the observation of multiple MLST subtypes of IaA28R4 within two large outbreaks that lasted for extended periods and involved multiple swimming pools.
INTRODUCTION
Cryptosporidiosis is one of the most important causes of moderate to severe diarrhea and diarrhea-associated deaths among children in developing countries (1) and a major cause of waterborne outbreaks of human illness in industrialized nations (2, 3). In addition, it was a major cause of chronic diarrhea and deaths among immunocompromised persons such as HIV/AIDS patients prior to the introduction of highly active antiretroviral therapy (4). In the United States, the number of annually reported cases of cryptosporidiosis has more than doubled since 2005 (5–8). Cryptosporidiosis incidence has a very seasonal distribution, with peak transmission occurring during summer months, largely as a result of outbreaks associated with recreational water use (9). Currently, it is estimated that ∼750,000 cases of cryptosporidiosis occur in the United States each year (3).
Genotyping and subtyping tools have been used widely to investigate Cryptosporidium transmission patterns. The results of these studies have shown that, among the many established Cryptosporidium species and genotypes, Cryptosporidium hominis is responsible for most human cryptosporidiosis cases in most countries. This species is largely human specific and thus is transmitted anthroponotically (10). Within C. hominis, nucleotide sequence analysis of the 60-kDa glycoprotein (gp60) gene has identified many subtype families, such as Ia, Ib, Id, and Ie. Among them, subtype IbA10G2 is a dominant subtype responsible for C. hominis-associated outbreaks of cryptosporidiosis in the United States, Europe, and Australia (10–15). Since 2005, a new subtype within the Ia subtype family, IaA28R4, has become a frequently identified subtype for waterborne outbreaks of cryptosporidiosis in the United States. It was first detected during an investigation of a swimming pool-associated outbreak in Ohio in 2005 but has since been detected in Pennsylvania, Missouri, South Carolina, Texas, New Mexico, Colorado, Utah, and Idaho (16–20). It is unclear whether the increased cryptosporidiosis incidence in the United States is partially attributable to the emergence of this new subtype of C. hominis. In this study, we conducted multilocus sequence typing (MLST) of IaA28R4 and multiple other C. hominis subtypes circulating in four states in the United States, where cryptosporidiosis incidence increased during the summer of 2008.
MATERIALS AND METHODS
Specimens and subtype determination.
Ninety-five C. hominis specimens from humans were used in this study. They were from outbreak-related and sporadic cases that occurred in three southwestern states (Texas, New Mexico, and Arizona) and Ohio in 2008, except for two specimens from an outbreak in Ohio in 2005 (Table 1). The Texas specimens included 16 specimens from sporadic cases and 36 from a prolonged cryptosporidiosis outbreak in two neighboring counties. The outbreak affected >2,000 persons, was associated with a man-made chlorinated lake (17) and multiple other recreational water venues (e.g., swimming pools and interactive fountains), and probably spread via poor swimmer hygiene (e.g., swimming when ill with diarrhea and swallowing the water) (7). The 13 New Mexico specimens were from 11 sporadic cases and 1 day care-associated outbreak. The three Arizona specimens were from a swimming pool-associated outbreak. The 27 Ohio specimens were from two outbreaks associated with swimming pools in 2005 (2 specimens) and 2008 (25 specimens).
TABLE 1.
Sources and gp60 subtype identities of 95 Cryptosporidium hominis specimens used in this study
| Specimen | Yr | Sample size | Sample nature | No. with gp60 subtype of: |
|||
|---|---|---|---|---|---|---|---|
| IaA28R4 | IaA15R3 | IdA15G1 | IgA27 | ||||
| Texas 1a | 2008 | 18 | Outbreak | 9 | 5 | 2 | 2 |
| Texas 2a | 2008 | 18 | Outbreak | 11 | 3 | 4 | 0 |
| Texas 3 | 2008 | 16 | Sporadic | 8 | 3 | 4 | 1 |
| New Mexico | 2008 | 11 | Sporadic | 4 | 3 | 4 | 0 |
| 2008 | 2 | Outbreak | 0 | 2 | 0 | 0 | |
| Arizona | 2008 | 3 | Outbreak | 3 | 0 | 0 | 0 |
| Ohio | 2005 | 2 | Outbreak | 2 | 0 | 0 | 0 |
| 2008 | 25 | Outbreak | 25 | 0 | 0 | 0 | |
| Total | 62 | 16 | 14 | 3 | |||
Texas 1 and Texas 2 are specimens from the same outbreak in two neighboring counties.
DNA was extracted from 200 μl of Cryptosporidium-positive specimens using the FastDNA SPIN kit for soil (MP Biomedicals, Irvine, CA). Cryptosporidium hominis in the specimens was subtyped by DNA sequence analysis of the gp60 gene (21). The 95 C. hominis specimens used belonged to four subtypes based on variation at the gp60 locus, i.e., IaA28R4 (n = 62), IaA15R3 (n = 16), IdA15G1 (n = 14), and IgA27 (n = 3) (Table 1).
MLST analysis.
In addition to gp60, each specimen was analyzed at seven other polymorphic loci, including 47-kDa protein (CP47), 56-kDa transmembrane protein (CP56), hydroxyproline-rich glycoprotein (DZ-HRGP), 70-kDa heat shock protein (HSP70), serine repeat antigen (MSC6-7), mucin protein (Mucin1), and retinitis pigmentosa GTPase regulator (RPGR), using PCR primers and conditions described previously (20, 22–24). All PCR analyses were done in duplicate, using 1 μl of extracted DNA per PCR and 400 ng/μl of nonacetylated bovine serum albumin (Sigma-Aldrich, St. Louis, MO) in the primary PCR. The secondary PCR products were sequenced in both directions using the ABI BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI3130 genetic analyzer (Applied Biosystems). The nucleotide sequences obtained were aligned with reference sequences downloaded from GenBank by using ClustalX (http://www.clustal.org).
Population genetic analysis.
Because gp60 had far more segregation (single-nucleotide polymorphism [SNP]) sites than other genetic loci, multilocus analyses were mostly conducted with allelic data. Multilocus linkage disequilibrium (LD) in allelic data with and without gp60 was assessed by calculating the standardized index of association (ISA) using LIAN 3.5 (http://pubmlst.org/cgi-bin/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=lian&referer=pubmlst.og). LD would be indicated by a positive value and linkage equilibrium (LE) by 0 or a negative value. In addition, the variance of pairwise differences (VD) and the 95% critical value (L) for VD were calculated to assess the population structure. If the VD is less than L, then the population is in LE, with a panmictic population structure; otherwise, the population is nonpanmictic and a degree of LD exists. To determine whether there was population differentiation within C. hominis and subtype IaA28R4, we used the Bayesian tool Structure v2.3 (http://pritchardlab.stanford.edu/structure.html) to analyze the allelic data. Several analyses of the data were performed using K values (likely populations) ranging from 2 to 6, with 50,000 iterations after a burn-in of 50,000 iterations. The output at a K value of 5 provided the best fit to the MLST data and thus was used in further analyses. The population substructure was further supported by eBURST analysis (http://eburst.mlst.net) of the MLST data. To assess the robustness of the substructuring, Wright's fixation index (FST) was calculated between subpopulations using Arlequin (http://cmpg.unibe.ch/software/arlequin35). LD and recombination rates also were compared across subpopulations. Multilocus LD (as indicated by |D′|[LD where y is the LD value and x is the nucleotide distance in kb]) and recombination analyses were further conducted on concatenated sequences excluding gp60 by using DnaSP 5.10.1 (http://www.ub.es/dnasp).
Nucleotide sequence accession numbers.
Unique nucleotide sequences obtained during the study were deposited in the GenBank database under accession numbers KF682371 to KF682391.
RESULTS
Sequence polymorphisms at MLST loci.
All specimens were successfully subtyped at the CP47, CP56, DZ-HRGP, HSP70, Mucin1, MSC6-7, and RPGR loci. The Mucin1 and RPGR loci were monomorphic, while most other loci had 2 or 3 subtypes, except for CP47, which had 5 subtypes (data not shown). Genetic heterogeneity was seen in each of the four gp60 subtypes at other loci. Although there was general agreement in subtyping results between g60 and CP47, DZ-HRGP and MSC6-7 showed discordant typing results. Altogether, 24 MLST types were seen among the 95 C. hominis specimens. Most of the sequence polymorphisms in these loci were variations in the numbers of insertions and deletions in the microsatellite and minisatellite regions. CP56, MSC6-7, and HSP70, however, each had 1 or 2 SNPs, far fewer than the values seen for the three subtype families (Ia, Id, and Ig) based on variation at the gp60 locus. As a result, most of the multilocus analyses were performed with allelic data, including all loci or concatenated sequences except gp60.
Overall population genetic structure.
Initially, multilocus LD was assessed by calculating ISA between alleles for all pairwise combinations of all loci including gp60, using LIAN. In the analysis of allelic data from 95 specimens, the value of ISA (0.1005) was positive and the VD (1.7013) was more than L (1.0951). A Monte Carlo analysis was further used to test the significance of LD, leading to the generation of a significant P value of <0.001 (Table 2). Thus, results of the analysis indicated an overall nonpanmictic population structure with a high level of LD. The LD was maintained when the interlocus ISA and variance analyses treated each group of specimens with the same multilocus genotype (MLG) as one individual (Table 2). As gp60 contributed more genetic heterogeneity than other loci, an interlocus LD analysis was also conducted on allelic data excluding gp60, but similar observations of LD were obtained (Table 3). Furthermore, a strong but incomplete LD (|D′| of y = 0.6618 + 0.0464x) was detected in the analysis of concatenated nucleotide sequences excluding gp60 by DnaSP (Table 4). Thus, recombination might exist because of the incomplete LD. This was confirmed by an intergenic recombination test, which identified three potential recombination events (Table 4).
TABLE 2.
Multilocus linkage disequilibrium analysis of allelic data at eight MLST loci (including gp60)
| Population | No. of specimens | Ha | ISA | PMC | VD | L | VD > L | LD or LE |
|---|---|---|---|---|---|---|---|---|
| Separate specimens | ||||||||
| All | 95 | 0.2895 ± 0.1075 | 0.1005 | <0.001 | 1.7013 | 1.0951 | Y | LD |
| IaA15R3 | 16 | 0.0917 ± 0.0754 | 0.3148 | 0.4325 | N | LE | ||
| IaA28R4 | 62 | 0.1639 ± 0.0919 | 0.0065 | 0.151 | 0.6514 | 0.6810 | N | LE |
| Ohio | 27 | 0.1275 ± 0.0757 | 0.639 | 0.5396 | 0.7110 | N | LE | |
| Non-Ohio | 35 | 0.0983 ± 0.0757 | 0.426 | 0.3736 | 0.4476 | N | LE | |
| IdA15G1 | 14 | 0.1470 ± 0.0875 | 0.4576 | 0.7687 | N | LE | ||
| IgA27 | 3 | 0.1250 ± 0.1250 | 0.0000 | 0.0000 | ||||
| MLG groupsb | ||||||||
| All | 24 | 0.3981 ± 0.1152 | 0.0738 | <0.001 | 1.7803 | 1.3948 | Y | LD |
| IaA15R3 | 4 | 0.1667 ± 0.1136 | 0.2667 | 0.6667 | N | LE | ||
| IaA28R4 | 11 | 0.2432 ± 0.1065 | 0.938 | 0.6822 | 1.1266 | N | LE | |
| Ohio | 8 | 0.2187 ± 0.1104 | 0.4907 | 0.9352 | N | LE | ||
| Non-Ohio | 5 | 0.2000 ± 0.1069 | 0.4889 | 1.1556 | N | LE | ||
| IdA15G1 | 6 | 0.2000 ± 0.1047 | 0.4000 | 1.1143 | N | LE | ||
| IgA27 | 3 | 0.1250 ± 0.1250 | 0.0000 | 0.0000 |
H, mean genetic diversity; ISA, standardized index of association; PMC, significance of obtaining this value in 1,000 Monte Carlo simulations; VD, variance of pairwise differences; L, 95% critical value for VD; VD > L indicates linkage disequilibrium; Y, yes; N, no.
Considering each group of specimens with the same multilocus genotype (MLG) as one individual.
TABLE 3.
Multilocus linkage disequilibrium analysis of allelic data at seven MLST loci (excluding gp60)
| Population | No. of specimens | Ha | ISA | PMC | VD | L | VD > L | LD or LE |
|---|---|---|---|---|---|---|---|---|
| Separate specimens | ||||||||
| All | 95 | 0.2553 ± 0.1177 | 0.0481 | <0.001 | 0.9659 | 0.8208 | Y | LD |
| IaA15R3 | 16 | 0.1048 ± 0.0858 | 0.3148 | 0.4325 | N | LE | ||
| IaA28R4 | 62 | 0.1873 ± 0.1026 | 0.0076 | 0.118 | 0.6514 | 0.6736 | N | LE |
| Ohio | 27 | 0.1457 ± 0.0849 | 0.628 | 0.5396 | 0.7110 | N | LE | |
| Non-Ohio | 35 | 0.1124 ± 0.0859 | 0.454 | 0.3736 | 0.4476 | N | LE | |
| IdA15G1 | 14 | 0.1680 ± 0.0980 | 0.4576 | 0.7687 | N | LE | ||
| IgA27 | 3 | 0.1429 ± 0.1429 | 0.0000 | 0.0000 | ||||
| MLG groupsb | ||||||||
| All | 24 | 0.3530 ± 0.1224 | 0.0375 | 0.041 | 1.1882 | 1.1737 | Y | LD |
| IaA15R3 | 4 | 0.1905 ± 0.1282 | 0.2667 | 0.6667 | N | LE | ||
| IaA28R4 | 11 | 0.2779 ± 0.1163 | 0.935 | 0.6822 | 1.1266 | N | LE | |
| Ohio | 8 | 0.2500 ± 0.1222 | 0.4907 | 1.0093 | N | LE | ||
| Non-Ohio | 5 | 0.2286 ± 0.1190 | 0.4889 | 1.1556 | N | LE | ||
| IdA15G1 | 6 | 0.2286 ± 0.1163 | 0.4000 | 1.1143 | N | LE | ||
| IgA27 | 3 | 0.1429 ± 0.1429 | 0.0000 | 0.0000 |
H, mean genetic diversity; ISA, standardized index of association; PMC, significance of obtaining this value in 1,000 Monte Carlo simulations; VD, variance of pairwise differences; L, 95% critical value for VD; VD > L indicates linkage disequilibrium; Y, yes; N, no.
Considering each group of specimens with the same multilocus genotype (MLG) as one individual.
TABLE 4.
Multilocus linkage disequilibrium analysis of concatenated sequences at seven MLST loci (excluding gp60)
| Population | No. of specimens | No. of segregating sites | No. of pairwise comparisons | No. of significant comparisons | No. of significant comparisons after Bonferroni correction | ZnSa | |D′| | LD | Minimum no. of recombination events |
|---|---|---|---|---|---|---|---|---|---|
| All | 95 | 8 | 28 | 11 | 11 | 0.3684 | y = 0.6618 + 0.0464x | Incomplete | 3 |
| IaA15R3 | 16 | 3 | 3 | 3 | 3 | 0.7229 | y = 1.0000 + 0.0000x | Complete | 0 |
| IaA28R4 | 62 | 4 | 6 | 3 | 1 | 0.0985 | y = 0.9397 − 0.1182x | Incomplete | 2 |
| Ohio | 27 | 4 | 6 | 1 | 1 | 0.1004 | y = 1.0147 − 0.1959x | Incomplete | 1 |
| Non-Ohio | 35 | 3 | 3 | 1 | 0 | 0.0982 | y = 0.5827 + 0.1516x | Incomplete | 1 |
| IdA15G1 | 14 | 2 | 1 | 0 | 0 | 0.4000 | 0 | ||
| IgA27 | 3 | 2 | 1 | 0 | 0 | 0.2500 | 0 |
ZnS, interlocus genetic association; |D′|, LD where y is the LD value and x is the nucleotide distance (in kb).
Two subpopulations of IaA28R4.
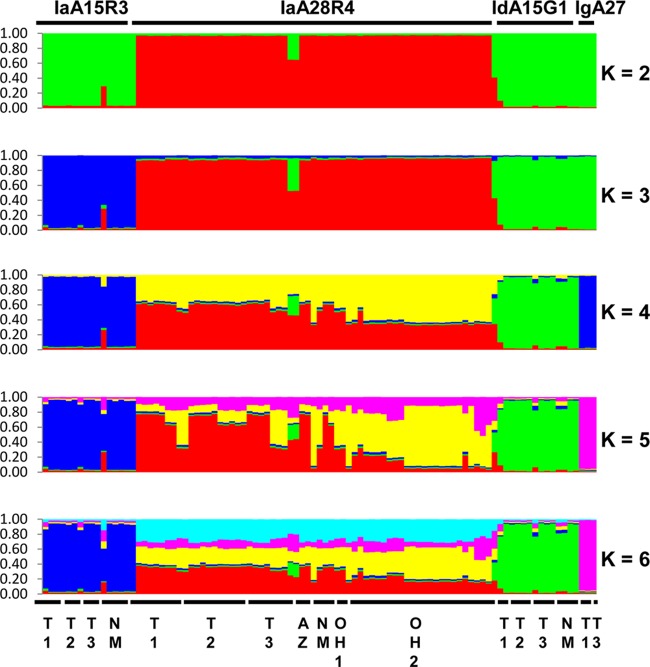
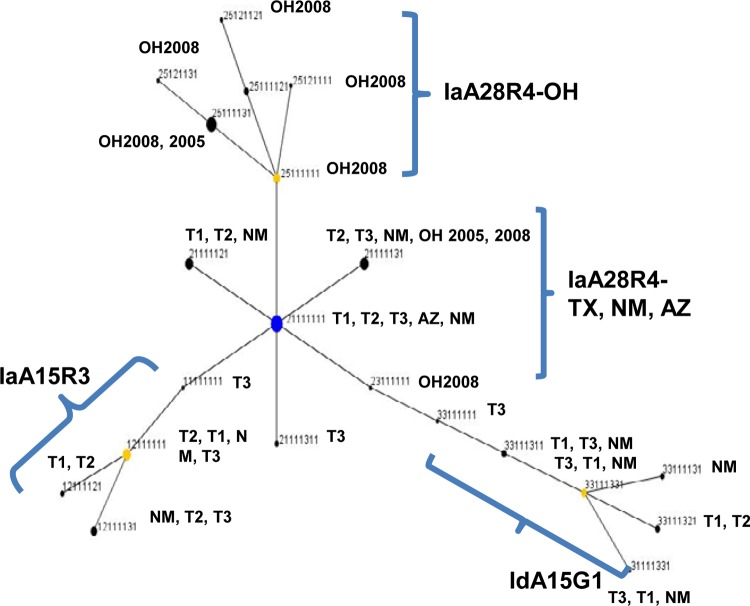
The genetic relationship between IaA28R4 and other C. hominis subtypes was inferred with Bayesian Structure analysis. At all K values used in the analysis, there was a clear separation of the 62 specimens of IaA28R4 from the 33 specimens of other C. hominis subtypes, including IaA15R3, which belongs to the same gp60 subtype family as IaA28R4 (Fig. 1). The best separation of gp60 subtype groups was seen at a K value of 5, with which all four C. hominis subtypes formed their own clusters. Pairwise FST analysis of these groups revealed the presence of highly significant population differentiation (FST = 0.172 to 0.300; P < 0.00001 for all comparisons) (Table 5). No genetic heterogeneity was seen in non-IaA28R4 subtypes at any of the K values (2 to 6) used. Admixed colors, however, were seen in IaA28R4 at K values of 4 to 6, and IaA28R4 specimens from Ohio showed a color pattern that was clearly different from that of IaA28R4 specimens from other areas, especially at a K value of 5 (Fig. 1). The admixed elements in IaA28R4 were not seen in any other gp60 subtypes. In eBURST analysis, there was also clear separation of IaA28R4 specimens from Ohio and IaA28R4 specimens from the southwestern states, with the IaA28R4 MLST subtypes from Ohio descending from the IaA28R4 MLST subtypes from the southwestern states (Fig. 2).
FIG 1.
Population substructure of 95 specimens of C. hominis from Ohio, Texas, New Mexico, and Arizona in the United States by Bayesian analyses of the MLST allelic data. Subpopulation patterns are shown for K values of 2 to 6 used in the analysis. Colored regions indicate major ancestral contributions. Mixed subtypes are indicated by the pattern of color combinations. Subtype identities of the specimens at the gp60 locus are shown above the color patterns, and the geographic origins of the specimens are shown below the color patterns. T1 and T2, specimens from an outbreak in two neighboring northeastern counties in Texas; T3, specimens from sporadic cases in a central Texas city; NM, specimens from sporadic cases (except for two IaA15R3 specimens) from New Mexico; AZ, specimens from an outbreak in Arizona; OH1 and OH2, specimens from two outbreaks in central Ohio. All specimens are from 2008, except for OH1, which represents two specimens from 2005.
TABLE 5.
Population differentiation among gp60 subtypes of C. hominis in pairwise FST analysis
| Parameter and subtype | IaA15R3 | IaA28R4 | IdA15G1 |
|---|---|---|---|
| Population pairwise FST | |||
| IaA28R4 | 0.21403 | ||
| IdA15G1 | 0.24477 | 0.17231 | |
| IgA27 | 0.30047 | 0.20569 | 0.22100 |
| FST P values | |||
| IaA28R4 | 0.00000 ± 0.0000 | ||
| IdA15G1 | 0.00000 ± 0.0000 | 0.00000 ± 0.0000 | |
| IgA27 | 0.00000 ± 0.0000 | 0.00000 ± 0.0000 | 0.00000 ± 0.0000 |
FIG 2.
Patterns of evolutionary descent among multilocus sequence typing (MLST) types of Cryptosporidium hominis, as revealed by eBURST analysis of allelic data. The numbers for each MLST type represent the sequence type at each of the eight MLST loci, in the following order: gp60, CP47, Mucin1, CP56, RPGR, MSC6-7, DZ-HRGP, and HSP70. Blue dot, primary founder; yellow dots, subgroup founders. The size of each dot is proportional to the number of specimens of the sequence type. The three MLST types of the gp60 IgA27 subtype did not cluster in this diagram. See Fig. 1 legend for the origins of the specimens.
The results of the FST analysis supported the presence of significant population differentiation between the IaA28R4 specimens from Ohio and the IaA28R4 specimens from other areas (FST = 0.271, P < 0.00001). The two IaA28R4 subpopulations had different sequences mostly at the CP47 locus.
Genetic recombination within IaA28R4.
We analyzed the gp60 subtype-specific population structure. In multilocus LD analyses of allelic data including and excluding gp60, we observed less LD within each of the gp60 subtypes than in C. hominis as a whole. In most cases, no ISA values were obtained, the Monte-Carlo P value was >0.05, and the VD was less than L regardless of whether the analysis included all allelic data (Table 2) or data from each unique multilocus subtype (considering each group of specimens with the same multilocus subtype as one individual) (Table 3). In multilocus LD analysis of concatenated nucleotide sequences excluding gp60, however, we obtained complete LD for IaA15R3 and incomplete LD for IaA28R4. The numbers of segregation sites in the sequences for IdA15G1 and IgA27 were too few for LD analysis. The negative slopes in the LD equations for all IaA28R4 specimens and the IaA28R4 specimens from Ohio indicated decreased linkage with increased nucleotide distance (Table 4), evidence of the occurrence of some genetic recombination. A recombination test revealed a minimum of 2 potential recombination events within IaA28R4. This genetic recombination was seen in both Ohio and non-Ohio IaA28R4 specimens (Table 4).
DISCUSSION
Data from this study clearly show that, despite the genetic identity of the IaA28R4 subtype at the gp60 locus, specimens of this subtype differed from each other at other genetic loci, especially DZ-HRGP, which showed discordant subtyping results with respect to gp60. Although CP47 generally produced concordant subtyping results with respect to the nearby gp60 locus, it did further divide IaA28R4 into two geographic subpopulations of IaA28R4, i.e., Ohio and southwestern states. This separation was supported by the results of Structure, eBURST, and FST analyses. Because all three DZ-HRGP subtypes were found within each subpopulation, each subpopulation had multiple MLST subtypes. In fact, the three DZ-HRGP subtypes were also found in the three other gp60 subtypes in the MLST analysis, even though only a small number of specimens with these gp60 subtypes were used in the study. Because the two IaA28R4 subpopulations had sequences as divergent at other genetic loci as other gp60 subtypes, IaA28R4 at gp60 apparently represents a fitness marker for C. hominis; the widespread distribution of this gp60 subtype in the United States in recent years is independent of the sequence characteristics at other genetic loci examined in this study. The common detection of this subtype in the United States, where cryptosporidiosis is severely underdiagnosed (3), and its propensity to cause statewide outbreaks (18) suggest its high virulence in humans. Recently, in an outbreak investigation in China, it was shown that the gp60 subtype IaA14R4, a subtype almost identical to IaA28R4 at the gp60 locus, was significantly more virulent than the IdA19 subtype (25).
There are at least two origins of IaA28R4 in the United States. Although this gp60 subtype was first identified in a cryptosporidiosis outbreak in Ohio in 2005 and was the cause of another outbreak in the same area in 2008, Structure analysis clearly indicated that IaA28R4 circulating in Ohio was genetically different from IaA28R4 in southwestern states in the 2008 outbreak season. The results of eBURST analysis further suggested that the Ohio IaA28R4 subpopulation was actually a descendant of IaA28R4 in southwestern states. Therefore, IaA28R4 was probably present in the United States before 2005. Both Structure and eBURST analyses have failed to provide any clue regarding the initial source of IaA28R4 in the United States.
Regardless of the initial source of IaA28R4, it appears that genetic recombination has played a major role in the emergence and transmission of this gp60 subtype in the United States. Although the overall population of C. hominis specimens had a clonal population structure, genetic recombination was seen in IaA28R4 and probably played an important role in the emergence of the dominant IaA28R4 subtype at the gp60 locus. The occurrence of genetic recombination was identified by LD analysis of both allelic data and concatenated multilocus sequences and by Bayesian Structure analysis. The gp60 locus appears to be the site of genetic recombination, as many MLST subtypes differed from each other only at gp60 and the neighboring CP47, adding support for the theory that genetic recombination is a driving force in the emergence of the gp60 IaA28R4 subtype. Genetic exchange within an overall clonal population of Cryptosporidium parvum has been reported in Italy recently (26), and genetic recombination has been identified as a driving force in the emergence of the virulent epidemic C. hominis gp60 subtype IbA10G2 and the C. parvum subtype IIaA15G2R1 (27, 28). Comparative genomic studies are needed to confirm the role of genetic recombination in the evolution of hyperinfectious or virulent Cryptosporidium subtypes.
Results of this study have been useful for understanding the transmission of Cryptosporidium in the United States. Texas, New Mexico, Arizona, and Ohio all experienced increased cryptosporidiosis incidence in the summer of 2008 (7), and the increases in Texas and Ohio were largely due to two large prolonged outbreaks of cryptosporidiosis in major metropolitan areas, each associated with multiple recreational water venues. Although IaA28R4 was the dominant gp60 subtype in the two outbreaks, MLST analysis clearly indicated that the outbreaks were not related to each other. In fact, each outbreak was caused by multiple MLST subtypes, although they all belonged to the IaA28R4 subtype at the gp60 locus. As these two outbreaks were each associated with multiple recreational water venues, the MLST subtype results point to the complicated nature of these outbreaks and the complex epidemiology of IaA28R4 in the United States. Further studies are needed to understand the evolution and transmission of this virulent parasite and its role in the recent increase in the reported incidence of cryptosporidiosis in the United States.
ACKNOWLEDGMENTS
This work was supported by the Centers for Disease Control and Prevention and the National Natural Science Foundation of China (grants 31229005 and 31110103901). N.T. was the recipient of an APHL/CDC Emerging Infectious Disease Fellowship.
We are grateful to the state and local public health epidemiologists and laboratory staff members for providing specimens used in this study.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 4 December 2013
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 2.Baldursson S, Karanis P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks: an update 2004–2010. Water Res. 45:6603–6614. 10.1016/j.watres.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States: major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady MT, Oleske JM, Williams PL, Elgie C, Mofenson LM, Dankner WM, Van Dyke RB, Pediatric AIDS Clinical Trials Group 219/219C Team 2010. Declines in mortality rates and changes in causes of death in HIV-1-infected children during the HAART era. J. Acquir. Immune Defic. Syndr. 53:86–94. 10.1097/QAI.0b013e3181b9869f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoder JS, Beach MJ. 2007. Cryptosporidiosis surveillance—United States, 2003–2005. MMWR Surveill. Summ. 56(SS-7):1–10 [PubMed] [Google Scholar]
- 6.Hlavsa MC, Watson JC, Beach MJ. 2005. Cryptosporidiosis surveillance—United States 1999–2002. MMWR Surveill. Summ. 54(SS-1):1–8 [PubMed] [Google Scholar]
- 7.Yoder JS, Harral C, Beach MJ. 2010. Cryptosporidiosis surveillance—United States, 2006–2008. MMWR Surveill. Summ. 59(SS-6):1–14 [PubMed] [Google Scholar]
- 8.Yoder JS, Wallace RM, Collier SA, Beach MJ, Hlavsa MC. 2012. Cryptosporidiosis surveillance—United States, 2009–2010. MMWR Surveill. Summ. 61(SS-5):1–12 [PubMed] [Google Scholar]
- 9.Yoder JS, Beach MJ. 2010. Cryptosporidium surveillance and risk factors in the United States. Exp. Parasitol. 124:31–39. 10.1016/j.exppara.2009.09.020 [DOI] [PubMed] [Google Scholar]
- 10.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 11.Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Thomas E, Watkins J, Casemore D, Kay D. 2010. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J. Water Health 8:311–325. 10.2166/wh.2009.185 [DOI] [PubMed] [Google Scholar]
- 12.Mayne DJ, Ressler KA, Smith D, Hockey G, Botham SJ, Ferson MJ. 2011. A community outbreak of cryptosporidiosis in Sydney associated with a public swimming facility: a case-control study. Interdiscip. Perspect. Infect. Dis. 2011:341065. 10.1155/2011/341065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng JS, Pingault N, Gibbs R, Koehler A, Ryan U. 2010. Molecular characterisation of Cryptosporidium outbreaks in western and south Australia. Exp. Parasitol. 125:325–328. 10.1016/j.exppara.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 14.Waldron LS, Ferrari BC, Cheung-Kwok-Sang C, Beggs PJ, Stephens N, Power ML. 2011. Molecular epidemiology and spatial distribution of a waterborne cryptosporidiosis outbreak in Australia. Appl. Environ. Microbiol. 77:7766–7771. 10.1128/AEM.00616-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournet N, Deege MP, Urbanus AT, Nichols G, Rosner BM, Chalmers RM, Gorton R, Pollock KG, van der Giessen JW, Wever PC, Dorigo-Zetsma JW, Mulder B, Mank TG, Overdevest I, Kusters JG, van Pelt W, Kortbeek LM. 2013. Simultaneous increase of Cryptosporidium infections in the Netherlands, the United Kingdom and Germany in late summer season, 2012. Euro Surveill. 18:pii=20348 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20348 [PubMed] [Google Scholar]
- 16.Xiao L, Hlavsa MC, Yoder J, Ewers C, Dearen T, Yang W, Nett R, Harris S, Brend SM, Harris M, Onischuk L, Valderrama AL, Cosgrove S, Xavier K, Hall N, Romero S, Young S, Johnston SP, Arrowood M, Roy S, Beach MJ. 2009. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: widespread occurrence of one Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium horse genotype. J. Clin. Microbiol. 47:3017–3020. 10.1128/JCM.00226-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantey PT, Kurian AK, Jefferson D, Moerbe MM, Marshall K, Blankenship WR, Rothbarth GR, Hwang J, Hall R, Yoder J, Brunkard J, Johnston S, Xiao L, Hill VR, Sarisky J, Zarate-Bermudez MA, Otto C, Hlavsa MC. 2012. Outbreak of cryptosporidiosis associated with a man-made chlorinated lake—Tarrant County, Texas, 2008. J. Environ. Health 75:14–19 [PubMed] [Google Scholar]
- 18.Valderrama AL, Hlavsa MC, Cronquist A, Cosgrove S, Johnston SP, Roberts JM, Stock ML, Xiao L, Xavier K, Beach MJ. 2009. Multiple risk factors associated with a large statewide increase in cryptosporidiosis. Epidemiol. Infect. 137:1781–1788. 10.1017/S0950268809002842 [DOI] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention 2009. Outbreak of cryptosporidiosis associated with a splash park—Idaho, 2007. MMWR Morb. Mortal. Wkly. Rep. 58:615–618 [PubMed] [Google Scholar]
- 20.Xiao L, Ryan UM. 2008. Molecular epidemiology, p 119–171 In Fayer R, Xiao L. (ed), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL [Google Scholar]
- 21.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744–2747. 10.1128/JCM.41.6.2744-2747.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatei W, Das P, Dutta P, Sen A, Cama V, Lal AA, Xiao L. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197–205. 10.1016/j.meegid.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 23.Gatei W, Hart CA, Gilman RH, Das P, Cama V, Xiao L. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53(Suppl 1):S43–S48. 10.1111/j.1550-7408.2006.00169.x [DOI] [PubMed] [Google Scholar]
- 24.Cama VA, Arrowood MJ, Ortega YR, Xiao L. 2006. Molecular characterization of the Cryptosporidium parvum IOWA isolate kept in different laboratories. J. Eukaryot. Microbiol. 53(Suppl 1):S40–S42. 10.1111/j.1550-7408.2006.00168.x [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, Zhao X, Hu J, Zhang N, Xiao L. 2012. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg. Infect. Dis. 18:312–314. 10.3201/eid1802.110666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drumo R, Widmer G, Morrison LJ, Tait A, Grelloni V, D'Avino N, Pozio E, Caccio SM. 2012. Evidence of host-associated populations of Cryptosporidium parvum in Italy. Appl. Environ. Microbiol. 78:3523–3529. 10.1128/AEM.07686-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, Feng Y. 2013. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg. Infect. Dis. 19:1573–1582. 10.3201/eid1910.121361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. 2013. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int. J. Parasitol. 43:1141–1147. 10.1016/j.ijpara.2013.09.002 [DOI] [PubMed] [Google Scholar]