Abstract
We describe a nosocomial outbreak of diarrheal disease caused by extended-spectrum β-lactamase-producing multidrug-resistant Salmonella enterica serovar Typhimurium, focused on a pediatric ward in South Africa. The outbreak peaked between May 2012 and July 2012. Person-to-person transmission was the most likely mechanism of spread of the infection, expedited due to a breakdown in hand-washing and hygiene, suboptimal infection control practices, overcrowding of hospital wards, and an undesirable nurse-to-patient ratio.
TEXT
Salmonella is well recognized as an etiological agent of gastrointestinal and diarrheal disease (1). Outbreaks involving Salmonella have been extensively described, with food-borne disease outbreaks being very commonly reported (2, 3). To a lesser extent, nosocomial outbreaks of Salmonella have also been reported, with outbreaks occurring among hospitalized humans (4, 5) and outbreaks occurring among hospitalized animals (6). Worldwide, the epidemiology of human Salmonella diarrheal disease is dominated by only a few nontyphoidal serotypes. In Africa, Salmonella enterica serotype Typhimurium (Salmonella Typhimurium) and Salmonella enterica serotype Enteritidis (Salmonella Enteritidis) are the two most commonly reported serotypes of nontyphoidal Salmonella (NTS) (7). In the developed world, NTS disease is usually a self-limiting gastroenteritis with low mortality in humans; however, in sub-Saharan Africa, NTS also frequently cause invasive disease, which is associated with a substantial burden of illness and death (8). In the present study, we report on a nosocomial outbreak of extended-spectrum-β-lactamase (ESBL)-producing multidrug-resistant Salmonella Typhimurium which occurred among hospitalized humans in South Africa in 2012.
Names of provinces, districts, and hospitals have been concealed to maintain anonymity. Human strains of Salmonella Typhimurium were investigated between March 2011 to March 2013, from a particular district (part of a province) of South Africa. Bacterial isolations were performed at a regional clinical microbiology laboratory. The particular district includes several clinics and hospitals which are not in close proximity to a microbiology laboratory; consequently, human specimens are sent for analysis to the regional laboratory. Isolates were then forwarded to the Centre for Enteric Diseases (CED) of the National Institute for Communicable Diseases for extended characterization. The CED is the national reference center in South Africa for human infections caused by enteric pathogens, including Salmonella species, Shigella species, diarrheagenic Escherichia coli, and Vibrio cholerae. Isolates from across South Africa are voluntarily submitted to the CED through national laboratory-based surveillance from ∼200 clinical microbiology laboratories across the country. As part of routine activities, the CED receives all the above-mentioned enteric pathogens (surveillance isolates and outbreak isolates) and proceeds to confirm identification, perform serotyping, determine susceptibilities to antimicrobial agents, and perform molecular subtyping (if required, as in outbreak situations).
Laboratory methods included the following. Bacteria were cultured from stool specimens and identified using standard phenotypic microbiological identification and serotyping techniques. Susceptibility to antimicrobial agents (ampicillin, ceftriaxone, trimethoprim, sulfamethoxazole, chloramphenicol, nalidixic acid, ciprofloxacin, tetracycline, and imipenem) was determined by using Etests (bioMérieux, Marcy-l'Étoile, France). The presence of ESBL activity was investigated by combination disk diffusion-based screening tests using ceftazidime (30 μg), cefotaxime (30 μg), and cefpodoxime (30 μg) alone and in combination with clavulanic acid (10 μg), as described by the Clinical and Laboratory Standards Institute. Molecular typing of strains was performed using pulsed-field gel electrophoresis (PFGE) analysis, multilocus sequence typing (MLST), and multiple-locus variable-number tandem-repeats analysis (MLVA). For PFGE, XbaI-digested genomic DNA was analyzed with a Bio-Rad CHEF-DR III electrophoresis system (Bio-Rad Laboratories, Hercules, CA) using a PulseNet protocol (9). PFGE patterns were analyzed using BioNumerics (version 6.5) software (Applied Maths, Sint-Martens-Latem, Belgium), and dendrograms of the patterns were created using the unweighted pair group method with arithmetic averages, with analysis of banding patterns incorporating the Dice coefficient at an optimization setting of 1.5% and a position tolerance setting of 1.5%. MLST was performed as described at the Salmonella MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica), which included DNA sequencing analysis of the following seven housekeeping genes: aroC, dnaN, hemD, hisD, purE, sucA, and thrA. DNA sequencing was performed using the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and an Applied Biosystems 3500 genetic analyzer. DNA sequences were collated and analyzed using the DNASTAR Lasergene (version 8.0) software (DNASTAR, Inc., Madison, WI), followed by analysis at the Salmonella MLST database, where allele numbers and an MLST sequence type (ST) were assigned. For MLVA, repeat DNA sequences at five loci (STTR9, STTR5, STTR6, STTR10, and STTR3) were analyzed using methods, nomenclature, and reporting of MLVA profiles described previously (10). For MLVA, DNA fragments were analyzed using an Applied Biosystems 3500 genetic analyzer, followed by data analysis using GeneMapper (version 4.1) software (Applied Biosystems) and then by analysis at the Institut Pasteur MLVA database website (http://www.pasteur.fr/recherche/genopole/PF8/mlva/), where an MLVA repeat type (RT) was assigned; this database website has subsequently been shut down.
In June 2012, routine laboratory-based surveillance detected an unusual increase in the number of human Salmonella Typhimurium isolates identified in a particular district of South Africa. On average, this district usually reports fewer than 5 isolations of Salmonella Typhimurium per month, so the sudden identification of more than 5 isolates within a few days signaled a potential outbreak. The peak of the outbreak occurred between May 2012 and July 2012 (dates are based on patient specimen collection dates), and further discussion is focused on this peak period. During this peak in the outbreak, 22 cases (patients) were involved. Available information showed that 4 patients were HIV positive, while a further 11 patients had one or more of the following background problems suggesting an immunocompromised state as a potential risk factor for Salmonella infection: chronic organ disease, malnutrition, tuberculosis infection, or perinatal HIV exposure. Three deaths were recorded; all three of these patients had multiple background medical conditions, and Salmonella Typhimurium infection was thought to have been a contributory but not final or singular cause of death. Patients were all hospitalized in a particular hospital; most patients were children between the ages of 1 and 22 months (n = 19). For all patients, the median age was 11 months and the interquartile range of age was 15 months. Child patients were from a pediatric ward, while adult patients (n = 3) were from multiple wards. One adult was an infected adult staff member (see below), but no apparent links between the other 2 adults and the pediatric ward could be determined. Patients presented with diarrhea and stool specimens were collected for laboratory analysis, with most specimens being collected ≥48 h after hospital admission.
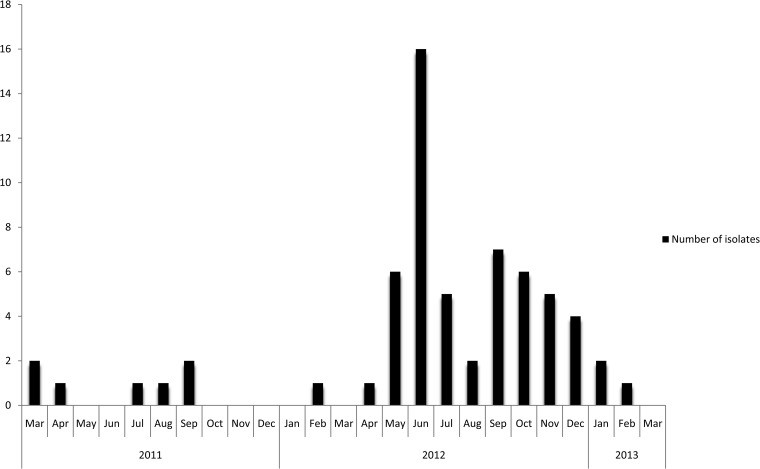
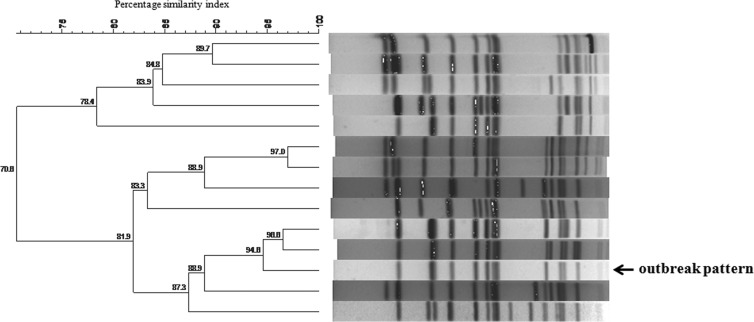
At the regional clinical microbiology laboratory, a Salmonella species was cultured from stool specimens and identified as the probable etiological agent of the diarrheal outbreak. Most isolates were then forwarded to the CED for extended characterization. From the 22 cases which resulted in 32 isolates being identified during the peak of the outbreak, 23 (72%) viable isolates were received by the CED; it must be noted that the number of isolates received and analyzed by the CED is often less than that identified at a district/regional laboratory, as some isolates become lost in the system or die in transit. The CED determined that the outbreak strain was multidrug-resistant Salmonella Typhimurium (multidrug resistance is defined as resistance to 3 or more classes of antimicrobial agents). The outbreak strain showed resistance to ampicillin, ceftriaxone, trimethoprim, sulfamethoxazole, chloramphenicol, and tetracycline and susceptibility to nalidixic acid, ciprofloxacin, and imipenem. The outbreak strain also showed ESBL activity. ESBL-producing Salmonella strains are not uncommon in South Africa, and these have been previously described, including previous descriptions of ESBL production in Salmonella Typhimurium (11, 12). A graph (Fig. 1) of the numbers of human isolates of Salmonella Typhimurium with an antimicrobial susceptibility profile (including ESBL production) identical to that of the current outbreak strain, for strains isolated in the same district as the outbreak strain, showed a peak of isolates in June 2012, which coincided with the peak of the outbreak. Importantly, this graph illustrates only the isolates investigated by the CED; as explained above, the number of strains received and analyzed by the CED is often less than that identified at district level. Molecular typing of the outbreak strains revealed indistinguishable PFGE patterns, indistinguishable MLVA profiles, and indistinguishable MLST sequence types. Figure 2 shows the PFGE outbreak pattern compared to other/background PFGE patterns identified in the province. The outbreak strain had the MLVA profile 3-12-10-NA-0211, which was assigned MLVA molecular subtype RTd61 by the Institut Pasteur MLVA database. Subtype RTd61 has previously been described in the MLVA database and is associated with Salmonella Typhimurium strains isolated in Europe. The outbreak strain had MLST molecular subtype ST34. This was not unexpected, as ST34 is a conventional ST and commonly found in Salmonella Typhimurium; ST34 and ST19 are the 2 most commonly described STs in Salmonella Typhimurium worldwide, including Africa (13, 14). It was noteworthy that our investigation found no occurrence of ST313; this was also not unexpected, as ST313 is more commonly associated with invasive disease, and there are reports of ST313 causing epidemic invasive disease in sub-Saharan Africa (15, 16).
FIG 1.
Number of human isolates of Salmonella Typhimurium with an antimicrobial susceptibility profile (including ESBL production) identical to that of the current outbreak strain, for isolates from the same district as the outbreak strain, March 2011 to March 2013. Importantly, these data show only the number of isolates received and investigated by the CED.
FIG 2.
PFGE patterns (XbaI digestion) associated with Salmonella Typhimurium strains isolated in the province in which the nosocomial outbreak occurred. The outbreak pattern is indicated for comparison to other/background patterns identified in the province.
Additional molecular typing was performed on randomly selected Salmonella Typhimurium isolates from the population in the district served by the regional laboratory associated with the particular hospital in question, with isolation dates preceding and following the peak of the outbreak. For the period March 2011 to April 2012, 28 isolates (∼2 for each month) were selected, of which 2 (1 from February 2012 and 1 from April 2012) showed the molecular subtype of the outbreak strain. For the period August to November 2012, 16 isolates (∼4 for each month) were selected, of which the majority (10 isolates) showed the molecular subtype of the outbreak strain. This suggests the presence and circulation of the outbreak strain within the community, before and after the peak (May 2012 to July 2012) of the nosocomial outbreak. Therefore, we hypothesize that the outbreak strain was circulating in the community prior to its introduction into the pediatric ward of the hospital by a patient(s) or patient contacts. The pediatric ward consists of 25 beds, including 8 beds devoted to patients with gastroenteritis. Further examination of the structural facilities and resource availability revealed several shortcomings which hampered the institution of effective contact precautions and isolation of cases during the outbreak. First and most importantly, inadequate availability of alcohol-based hand sanitizer, soap, and paper towels and the lack of a basin in the child toilet during the outbreak period impaired basic hand hygiene practices. Second, although each cubicle has its own hand-washing basin, access to the basin was interpreted as inconvenient. Third, small cubicles holding 1 to 6 beds resulted in close physical proximity between patients. Fourth, it was observed that mothers do not exclusively handle their own children, thus potentially facilitating the transmission of organisms between patients. Finally, an increased influx of patients (a June 2012 bed occupancy rate of 81% compared to a mean of 65% for 2012; June 2012 gastroenteritis admission of 61 cases compared to a mean of 32 cases per month for 2012) was observed over the outbreak period. The majority of cases admitted with gastroenteritis during the 3-month period were not thought to be associated with infection by Salmonella Typhimurium. It has been suggested (although there is no laboratory evidence to substantiate this) that an initial wave of community-acquired, possibly viral gastroenteritis acted as a precursor event which resulted in increased admissions and pressure on staff, without a concomitant increase in staff numbers. Decompensation in the nursing services would be expected to exacerbate the factors favoring disease transmission within the ward.
Thus, a breakdown in hand-hygiene and infection control practices, secondary to overwhelmed (but also inadequate) ward infrastructural capabilities, combined with an undesirable nurse-to-patient ratio, resulted in nosocomial transmission of the strain and amplification of diarrheal cases in the hospital. Available supporting data included the following: 4 cases did not have diarrhea on admission, a further 3 cases had initial cultures on admission which did not yield Salmonella Typhimurium, a further 2 patients had a previous (within 12 days) admission to the same pediatric ward, and the time from admission to specimen collection was prolonged, with a median time of 4 days (range 1 to 31 days). Also, investigation determined that patients were from diverse geographic regions in the district (mostly living in informal settlements) and that there were no common community exposures, such as at child day care centers, common food sources, or common social gatherings. For this nosocomial outbreak, food as a source was excluded; it was unlikely that the source of the infection was food prepared by the hospital, because a large majority of patients were infants who did not consume hospital food but rather were fed on ready-made, commercial, prepackaged milk foods, which does not require further preparation prior to consumption. In addition, the kitchen prepared and served food to many wards throughout the hospital, yet the outbreak clustered in only a few wards. Testing of a representative sample from the commercially prepared milk foods failed to culture any pathogens. In terms of identification of a common meeting place in the hospital where patients (and patient contacts) could have gathered together and socialized, no such common area or mechanism was identified. All staff members (n = 17) from the pediatric ward were screened for infection by culturing rectal swabs. Salmonella Typhimurium was cultured from 1 staff member, with the isolate showing the same phenotypic and molecular subtyping characteristics as the outbreak strain; further investigation determined that the staff member had a chronic, untreated medical condition and reported chronic diarrhea since 2011. Directional causality relating to the whether the staff member was a recipient of or participated in the propagation of the outbreak cannot be determined. Sixteen environmental swabs taken from feeding cups, hand basins, baby cots, and resuscitation equipment present in the pediatric ward failed to culture any bacterial pathogens.
The outbreak was eventually contained by institution of the following interventions. The first, emphasized as being the most important intervention, is the uninterrupted supply of hand-washing consumables and the improvement of hand-washing facilities. Second, patients and staff were (re)educated regarding contact precautions. Third, sick-staff protocols were improved, and the infected staff member was counseled. Fourth, appropriate use of antimicrobial therapy was reinforced. Last, the communication of critical microbial results and patterns by the laboratory was enhanced, and the ability of the hospital to mount an appropriate outbreak response was strengthened.
Nosocomial outbreaks of Salmonella (including Salmonella Typhimurium) are occasionally reported in published literature. In developing countries, nosocomial outbreaks mostly occur in pediatric wards, due to additional risk factors (including malnutrition) associated with NTS infection. Recent reports of nosocomial outbreaks include that of Salmonella Typhimurium in a neonatal unit in Turkey (17) and that of Salmonella Isangi in a pediatric ward in South Africa (5). In Africa, the source and mode of transmission of Salmonella are unclear; transmission could include animals, animal products, water, and infected humans. Kariuki and coworkers (7) described a population of asymptomatic human carriers of NTS in Kenya and suggested that in Africa, asymptomatic carriers represent an important reservoir of NTS, which may drive person-to-person transmission (anthroponotic transmission) of NTS in Africa.
The outbreak investigation described here has its limitations and shortcomings. The investigation may not have been conducted in the same systematic way as an investigation in the United States or Europe. In South Africa, there are acute resource constraints and a shortage of adequately trained personnel to conduct proper outbreak investigations. As a result, most outbreaks are not investigated or inadequately investigated. This is exacerbated by the regular lack of complete data, including the lack of availability of exhaustive clinical data. For the current outbreak, we have done our best given the trying circumstances and limited resources available; reporting and publication of limited data are better than no reporting of data.
In conclusion, due to the limitations described above, a final and unifying theory of the pathogenesis of the outbreak cannot be offered at this time. The outbreak most likely involved person-to-person transmission, expedited due to a breakdown in hand-washing and hygiene, suboptimal infection control practices, overcrowding of hospital wards, and an undesirable nurse-to-patient ratio. Due to the persistence of the immunocompromised niche population of patients within this hospital and indeed other similar hospitals in South Africa, and the transmission dynamics exhibited during the outbreak, the threat of a similar outbreak persists where factors facilitating the transmission remain.
ACKNOWLEDGMENTS
This work was supported by a Cooperative Agreement (5U19GH000571-02) from the Centers for Disease Control and Prevention, Atlanta, GA.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 2.Niehaus AJ, Apalata T, Coovadia YM, Smith AM, Moodley P. 2011. An outbreak of foodborne salmonellosis in rural KwaZulu-Natal, South Africa. Foodborne Pathog. Dis. 8:693–697. 10.1089/fpd.2010.0749 [DOI] [PubMed] [Google Scholar]
- 3.Maki DG. 2009. Coming to grips with foodborne infection—peanut butter, peppers, and nationwide salmonella outbreaks. N. Engl. J. Med. 360:949–953. 10.1056/NEJMp0806575 [DOI] [PubMed] [Google Scholar]
- 4.Lee MB, Greig JD. 2013. A review of nosocomial Salmonella outbreaks: infection control interventions found effective. Public Health 127:199–206. 10.1016/j.puhe.2012.12.013 [DOI] [PubMed] [Google Scholar]
- 5.Wadula J, von Gotteberg A, Kilner D, de Jong G, Cohen C, Khoosal M, Keddy K, Crewe-Brown H. 2006. Nosocomial outbreak of extended-spectrum beta-lactamase-producing Salmonella Isangi in pediatric wards. Pediatr. Infect. Dis. J. 25:843–844. 10.1097/01.inf.0000233543.78070.a2 [DOI] [PubMed] [Google Scholar]
- 6.Steneroden KK, Van Metre DC, Jackson C, Morley PS. 2010. Detection and control of a nosocomial outbreak caused by Salmonella Newport at a large animal hospital. J. Vet. Intern. Med. 24:606–616. 10.1111/j.1939-1676.2010.0484.x [DOI] [PubMed] [Google Scholar]
- 7.Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Muyodi J, Githinji JW, Kagendo D, Munyalo A, Hart CA. 2006. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J. Med. Microbiol. 55:585–591. 10.1099/jmm.0.46375-0 [DOI] [PubMed] [Google Scholar]
- 8.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 10.Larsson JT, Torpdahl M, Petersen RF, Sorensen G, Lindstedt BA, Nielsen EM. 2009. Development of a new nomenclature for Salmonella typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro. Surveill. 14:19174 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19174 [PubMed] [Google Scholar]
- 11.Usha G, Chunderika M, Prashini M, Willem SA, Yusuf ES. 2008. Characterization of extended-spectrum beta-lactamases in Salmonella spp. at a tertiary hospital in Durban, South Africa. Diagn. Microbiol. Infect. Dis. 62:86–91. 10.1016/j.diagmicrobio.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 12.Kruger T, Szabo D, Keddy KH, Deeley K, Marsh JW, Hujer AM, Bonomo RA, Paterson DL. 2004. Infections with nontyphoidal Salmonella species producing TEM-63 or a novel TEM enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 48:4263–4270. 10.1128/AAC.48.11.4263-4270.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikumapayi UN, Antonio M, Sonne-Hansen J, Biney E, Enwere G, Okoko B, Oluwalana C, Vaughan A, Zaman SM, Greenwood BM, Cutts FT, Adegbola RA. 2007. Molecular epidemiology of community-acquired invasive non-typhoidal Salmonella among children aged 2–29 months in rural Gambia and discovery of a new serovar, Salmonella enterica Dingiri. J. Med. Microbiol. 56:1479–1484. 10.1099/jmm.0.47416-0 [DOI] [PubMed] [Google Scholar]
- 14.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 68:771–777. 10.1093/jac/dks496 [DOI] [PubMed] [Google Scholar]
- 15.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, MacLennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G. 2012. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 44:1215–1221. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leekitcharoenphon P, Friis C, Zankari E, Svendsen CA, Price LB, Rahmani M, Herrero-Fresno A, Fashae K, Vandenberg O, Aarestrup FM, Hendriksen RS. 2013. Genomics of an emerging clone of Salmonella serovar Typhimurium ST313 from Nigeria and the Democratic Republic of Congo. J. Infect. Dev. Ctries. 7:696–706. 10.3855/jidc.3328 [DOI] [PubMed] [Google Scholar]
- 17.Anil M, Helvaci M, Ozkalay N, Toprak E, Anil AB, Dilek M, Agus N. 2009. Salmonella typhimurium outbreak in a neonatal unit in Turkey. Indian J. Pediatr. 76:629–633. 10.1007/s12098-009-0083-4 [DOI] [PubMed] [Google Scholar]