Abstract
Sequence analysis of the internal transcribed spacer (ITS) region was employed as the gold standard method for yeast identification in the China Hospital Invasive Fungal Surveillance Net (CHIF-NET). It has subsequently been found that matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is potentially a more practical approach for this purpose. In the present study, the performance of the Vitek MS v2.0 system for the identification of yeast isolates collected from patients with invasive fungal infections in the 2011 CHIF-NET was evaluated. A total of 1,243 isolates representing 31 yeast species were analyzed, and the identification results by the Vitek MS v2.0 system were compared to those obtained by ITS sequence analysis. By the Vitek MS v2.0 system, 96.7% (n = 1,202) of the isolates were correctly assigned to the species level and 0.2% (n = 2) of the isolates were identified to the genus level, while 2.4% (n = 30) and 0.7% (n = 9) of the isolates were unidentified and misidentified, respectively. After retesting of the unidentified and misidentified strains, 97.3% (n = 1,209) of the isolates were correctly identified to the species level. Based on these results, a testing algorithm that combines the use of the Vitek MS system with selected supplementary ribosomal DNA (rDNA) sequencing was developed and validated for yeast identification purposes. By employing this algorithm, 99.7% (1,240/1,243) of the study isolates were accurately identified with the exception of two isolates of Candida fermentati and one isolate of Cryptococcus gattii. In conclusion, the proposed identification algorithm could be practically implemented in strategic programs of fungal infection surveillance.
INTRODUCTION
The global antifungal surveillance programs have provided important global epidemiology and susceptibility data for invasive yeast infections (1–6). In July 2009, the multicenter nationwide China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study was established, and since then it has provided important information on the epidemiology of fungal diseases in China (7). The importance of confirmatory identification in fungal surveillance programs has been highlighted by reports that the initial identifications by the participating laboratories may be inaccurate, particularly for isolates that are uncommon or have unusual biochemical or phenotypic profiles (7, 8). Currently, sequencing-based methods are recognized as the gold standard for identifying yeast species (9). In the 2010 CHIF-NET surveillance study, all 814 isolates were confirmed by internal transcribed spacer (ITS) region sequencing in a central laboratory (7). However, the subsequent expansion of the CHIF-NET program has led to a dramatic increase in the numbers of both the participating laboratories and the referred isolates, precluding the universal application of the high-cost and labor-intensive sequence-based identification methods.
Recently, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has emerged as an alternative for microbial identification (10–13). The Vitek MS system (bioMérieux, Marcy l'Etoile, France) is a relatively new commercialized MALDI-TOF-based method and is optimized for both bacterial and fungal identification utilizing single-spot inoculations without prior protein extraction (14–19). Several studies have evaluated the performance of the Vitek MS system with database version 1.0 in yeast identification (20–22). However, there is only one reported study that evaluated the performance of the Vitek MS system with database v2.0 (incorporating 755 species, including 645 bacterial and 110 fungal taxa) for yeast identification (23). In addition, there have been no studies that evaluated the application of the Vitek MS v2.0 system in a large fungal surveillance program such as CHIF-NET where a diverse range of yeasts are encountered.
Therefore, the aims of the present study were to (i) evaluate the performance of the Vitek MS v2.0 system for the identification of yeasts collected in the 2011 CHIF-NET with comparison to ITS sequence analysis, and (ii) develop a practical yeast identification strategy for future CHIF-NET and other fungal surveillance programs.
MATERIALS AND METHODS
Yeast isolates.
The 1,243 yeast isolates analyzed in this study were collected from 22 clinical microbiology laboratories situated in 15 provinces across China during the 2011 CHIF-NET program from August 2010 to July 2011. The invasive isolates were cultured from blood, cerebrospinal fluid, ascitic fluid, peritoneal fluid, or other sterile body fluids. Study strains were initially inoculated onto Brilliance Candida agar (Oxoid Ltd., Hampshire, United Kingdom) and then were subcultured on Sabouraud dextrose agar. Culture media were incubated for 24 to 48 h at 35°C.
Sequencing-based identification.
All isolates were identified by gold standard DNA sequencing of the fungal internal transcribed spacer (ITS) region. Primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) were used to amplify the ITS region (24). Amplification of the ITS region was carried out under the following conditions: denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 90 s, and elongation at 72°C for 75 s, with a final extension step of 10 min at 72°C. Strain species identification was performed as described by Wang et al. (7).
Vitek MS v2.0 system identification.
The Vitek MS analysis was performed according to the manufacturer's instructions. First, a small portion of a single colony after 24 or 48 h of incubation was smeared onto a target plate and covered with 0.5 μl formic acid (FA). Immediately after drying at room temperature, 1 μl α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution was applied and again allowed to dry prior to being loaded into the Vitek MS system. The identification control Escherichia coli ATCC 8739 strain was inoculated on the central spot of each acquisition group.
After the acquisition of spectra (mass range of 2 to 20 kDa in the linear mode), data were transferred from the Vitek MS acquisition station to the Vitek MS analysis server, which utilized software algorithms to compare the generated spectrum with the typical spectra within the database. These results were then exhibited in one of three forms, (i) a single identification (confidence value of 60.0% to 99.9%), (ii) a split identification for which a set of possible organisms is displayed, or (iii) no identification when no match is found.
Data analysis.
The Vitek MS result was considered accurate to the species level if a single identification was given and matched that obtained by the reference method. The Vitek MS result was considered correct to the genus level if multiple alternative identifications, all from the same genus, were reported and matched the genus obtained by the reference method. The Vitek MS result was considered a misidentification when a single identification that did not match the result obtained with the reference method was given, when multiple identifications of different genera were reported, or when multiple identifications of the same genera that did match the genus of the reference method were reported (25). In order to acquire more information, we reanalyzed all the isolates with the primary results of misidentification or no identification with a single spot.
RESULTS
Overall results.
The 1,243 study isolates represented 31 species, including 21 Candida species (1,137 isolates; 91.5%), 3 Cryptococcus species (87 isolates; 7.0%), and 7 other yeast species (19 isolates; 1.5%). The Vitek MS v2.0 system included reference spectra for only 22 of the 31 species encountered during the study period (Table 1). Among the 1,243 isolates analyzed by the Vitek MS v2.0 system, 1,202 (96.7%) isolates were correctly identified to the species level, with confidence values of 99.9% for 1,162 isolates and <99.9% for 40 isolates. Two isolates (0.2%) were correctly identified only to the genus level. Misidentification and no identification results were obtained for 0.7% and 2.4% of the isolates, respectively (Table 1). The confidence values for all the misidentified isolates were <99.9% with the exception of two Candida fermentati isolates misidentified as Candida guilliermondii and one Cryptococcus gattii isolate misidentified as Cryptococcus neoformans (Table 2). The results for the misidentified isolates after retesting are shown in Table 2. When isolates of species included in the v2.0 database that previously tested as no identification were retested, only a single isolate each of Candida parapsilosis sensu stricto and Trichosporon asahii remained unidentified. In total, 97.3% (n = 1,209) isolates were correctly identified to the species level after retesting of misidentified and unidentified strains.
TABLE 1.
Performance of the Vitek MS v2.0 system for the identifications of 1,243 yeast isolates from CHIF-NET 2011
| Reference identification | No. (%) of isolates | No. (%) of isolates with Vitek MS results of: |
|||
|---|---|---|---|---|---|
| Correct identification to species level (single result)a | Correct identification to genus level (multiple results)b | Misidentification (single/multiple results)c | No identification | ||
| Candida species | 1,137 (91.5) | 1,101 (96.8) | 2 (0.2) | 8 (0.7) | 26 (2.3) |
| C. albicans | 556 (44.7) | 555 (99.8) | 0 (0) | 0 (0) | 1 (0.2) |
| C. tropicalis | 218 (17.5) | 214 (98.1) | 1 (0.5) | 0 (0) | 3 (1.4) |
| C. parapsilosis species complex | |||||
| C. parapsilosis sensu stricto | 161 (13.0) | 160 (99.4) | 0 (0) | 0 (0) | 1 (0.6) |
| C. metapsilosisd | 14 (1.1) | 0 (0) | 0 (0) | 3 (21.4) | 11 (78.6) |
| C. orthopsilosisd | 7 (0.6) | 0 (0) | 0 (0) | 2 (28.6) | 5 (71.4) |
| Lodderomyces elongisporusd | 2 (0.2) | 0 (0) | 0 (0) | 0 (0) | 2 (100) |
| C. glabrata sensu stricto | 115 (9.3) | 114 (99.1) | 0 (0) | 0 (0) | 1 (0.9) |
| C. krusei | 16 (1.3) | 16 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. guilliermondii | 16 (1.3) | 15 (93.7) | 0 (0) | 1 (6.3) | 0 (0) |
| C. pelliculosa | 10 (0.8) | 10 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. lusitaniae | 4 (0.3) | 4 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. haemulonii | 3 (0.2) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. intermedia | 3 (0.2) | 2 (66.7) | 1 (33.3) | 0 (0) | 0 (0) |
| C. lipolytica | 3 (0.2) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. inconspicua | 2 (0.2) | 2 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. catenulata | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. fermentatid | 2 (0.2) | 0 (0) | 0 (0) | 2 (100) | 0 (0) |
| C. freyschussii | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. opuntiaed | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| C. quercitrusad | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| C. rugosa | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Cryptococcus species | 87 (7.0) | 85 (97.8) | 0 (0) | 1 (1.1) | 1 (1.1) |
| C. neoformans | |||||
| C. neoformans var. grubii | 83 (6.7) | 83 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. neoformans var. neoformans | 2 (0.2) | 2 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. gattiid | 1 (0.1) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| C. laurentii | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| Other yeast species | 19 (1.5) | 16 (84.2) | 0 (0) | 0 (0) | 3 (15.8) |
| Kodamaea ohmeri | 4 (0.3) | 4 (100) | 0 (0) | 0 (0) | 0 (0) |
| Pseudozyma speciesd | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| Rhodotorula mucilaginosa | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Saccharomyces cerevisiae | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Sporobolomyces speciesd | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| Trichosporon asahii | 10 (0.8) | 9 (90) | 0 (0) | 0 (0) | 1 (10) |
| Trichosporon asteroides | 1 (0.1) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Total | 1,243 (100) | 1,202 (96.7) | 2 (0.2) | 9 (0.7) | 30 (2.4) |
The result was a single identification that matched the reference method to the species level.
The result was multiple possible identifications, all within the same genus, which matched (at the genus level) the identification obtained by the reference method.
The result was either a single identification that did not match the identification obtained by the reference method or multiple possible identifications that included more than one genus.
Nine species were not included in the Vitek MS v2.0 database.
TABLE 2.
Results of the misidentified isolates reanalyzed by the Vitek MS v2.0 system
| Reference identification | Sample no. | Vitek MS identification results (confidence value [%]) |
|
|---|---|---|---|
| Initial | Reanalyzed | ||
| C. guilliermondiia | 11H4034 | C. guilliermondii (93.6), Debaryomyces polymorphus (82.9) | C. guilliermondii (99.9) |
| C. fermentati | 10TJ113 | C. guilliermondii (99.9) | C. guilliermondii (98) |
| C. fermentati | 10TJ114 | C. guilliermondii (99.9) | C. guilliermondii (98.4) |
| C. metapsilosis | 11GH037 | C. laurentii (94.6) | Unidentified |
| C. metapsilosis | 11S1007 | C. laurentii (99.3) | Unidentified |
| C. metapsilosis | 10GZ105 | C. parapsilosis (74.5) | Unidentified |
| C. orthopsilosis | 11RJ039 | C. parapsilosis (92.1) | Unidentified |
| C. orthopsilosis | 11TJ188 | C. parapsilosis (93.5) | C. parapsilosis (97.3) |
| C. gattii | 11GZ074 | C. neoformans (99.9) | C. neoformans (99.9) |
C. guilliermondii is included in the v2.0 database.
The Vitek MS system performed well in identifying common Candida species (the species of isolates accounting for >1.0% of the overall isolates), including Candida albicans (99.8%), Candida tropicalis (98.1%), Candida parapsilosis sensu stricto (99.4%), Candida glabrata sensu stricto (99.1%), Candida krusei (100%), and Candida guilliermondii (93.7%) (Table 1). A total of 161 (87.5%) of 184 C. parapsilosis complex isolates were identified on sequencing as C. parapsilosis sensu stricto, and 160 (99.4%) isolates were correctly identified to the species level by the Vitek MS v2.0 system. The misidentifications of the less commonly encountered Candida species, including Candida metapsilosis, Candida orthopsilosis, and C. fermentati, are summarized in Table 2.
Development of a yeast identification algorithm using Vitek MS and rDNA sequence analysis.
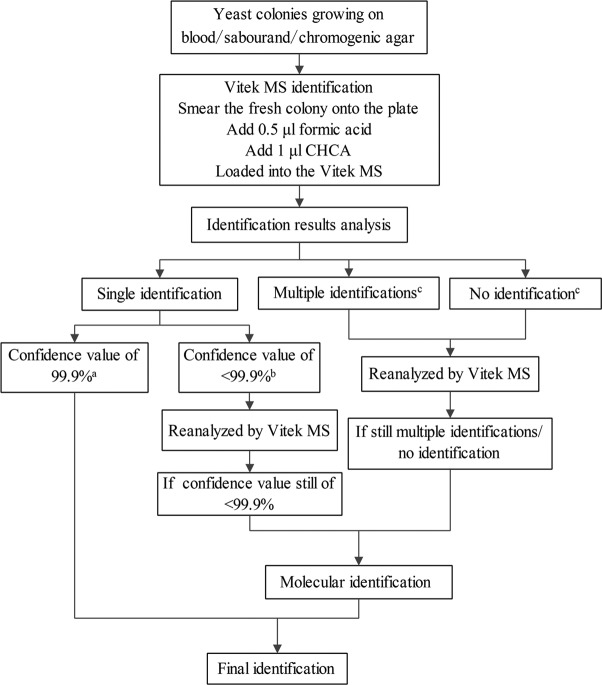
A testing algorithm for yeast was developed based on the analysis of the Vitek MS system and ribosomal DNA (rDNA) sequencing results from the present study (Fig. 1). Under this proposal, strains with identification confidence values of <99.9% are initially retested by the Vitek MS system. Isolates with confidence values of <99.9% on retesting should be confirmed by molecular methods. Uncommon species (species accounting for <1% of total yeast isolates as determined by existing epidemiological data) are confirmed by molecular identification, even if the confidence value is 99.9% (Fig. 1). When the initial Vitek MS analysis reports multiple identifications or when no identification is provided, strains should be initially retested by the Vitek MS system. If this repeat analysis fails to provide a high confidence score, these isolates should be identified by molecular methods (Fig. 1).
FIG 1.
An identification testing algorithm for yeast based on the Vitek MS system and selective molecular identification in a large surveillance program. a, Identification results of the uncommon yeast species should be confirmed by the molecular identification; b, the strain identification results with confidence values of <99.9% should be reanalyzed by the Vitek MS system; c, The strains with multiple identifications or no identification should be reanalyzed by the Vitek MS system.
Applying the proposed testing algorithm in the 2011 CHIF-NET, we found that 73 isolates required reanalysis by the Vitek MS system (40 isolates with confidence values of <99.9%, 3 isolates with multiple identifications, and 30 isolates with no identification), and only 65 isolates met the criteria for molecular confirmation. Of the 1,243 study isolates, a total of 1,240 (99.7%) isolates could be accurately identified when we employed the proposed algorithm, with the three errors relating to two isolates of C. fermentati, which are indistinguishable from those of C. guilliermondii by phenotypic methods, including the Vitek MS system, and one isolate of C. gattii, which was indistinguishable from C. neoformans using the Vitek MS system.
The yeast identification methods utilizing only rDNA sequencing, only the Vitek MS system, and a combination of these methods are compared by the proposed algorithm in Table 3. Molecular identification of all 1,243 2011 CHIF-NET isolates took two technicians nearly 30 weeks with a consumable cost of US$6,215. Utilizing the Vitek MS system alone, two technicians could complete identification of the same isolates within 4 weeks with a consumable cost of approximately $4,100. When the proposed testing algorithm was applied to the study isolates, 1,170 (94.1%) of the isolates were identified by initial Vitek MS analysis, 73 (5.9%) isolates needed reanalysis by Vitek MS, and 65 required further confirmation by rDNA sequencing. Overall, the algorithm led to the accurate identification of 99.7% isolates. Two technicians were able to complete the task in 6 weeks with an approximate cost of $4,668.
TABLE 3.
Comparison of the three methods for yeast identification in CHIF-NET 2011
| Item | rDNA sequencing | Vitek MS | Vitek MS plus rDNA sequencing |
|---|---|---|---|
| No. of analyzed isolates | 1,243 | 1,243 | 1,243 (Vitek MS), 73 (reanalyzed), 65 (rDNA sequencing) |
| Total time (wk) for 2 technicians | 30 | 4 | 6 |
| Total cost (US$)a | 6,215 | 4,100 | 4668 |
| Accuracy (%) | 100 | 96.7 | 99.7 |
| Results analysis | BLAST with GenBank/CBS database | Identification results could be read directly |
The expenses of the equipment and technicians were not included.
DISCUSSION
Accurate identification of strains in surveillance programs is essential for providing reliable epidemiological data (8). In the first year of study in the 2010 CHIF-NET, a total of 814 isolates were confirmed in a central laboratory by ITS sequence analysis, providing clinically useful data on yeast infections in China (7). With the rapid expansion of the CHIF-NET program, more laboratories (∼100) will participate, and more isolates will be collected. DNA sequence-based analysis is impractical for identification on such a scale.
Many previous studies have evaluated the performance of the Bruker Biotyper system (Bruker Daltonics, Germany) for identification of bacteria or fungi. However, there have been limited reports relating to the Vitek MS system, especially for the identification of fungi (20–23). Iriart et al. showed that the Vitek MS v1.0 system performed well for the identification of yeasts and Aspergillus fungi (93.2% of correct identifications) (20). Mancini et al. compared the performance of the Vitek MS v1.0 system to that of the Bruker Biotyper. While the Vitek MS system proved to be as accurate as the Bruker Biotyper for the identification of the most common medically important yeasts, it yielded a lower rate of correct identification for rarer Candida and non-Candida species. This difference may be partially attributed to its nonexpandable database (22). The present study is the first to evaluate the performance of the Vitek MS v2.0 system for the identification of invasive yeast isolates in China and only the second reported study of the Vitek MS v2.0 system for yeast identification. Our results are comparable to findings from the previous single Vitek MS v2.0 system study, where >96% of the 852 isolates were correctly identified to the species level, 2.8% were not identified, and 0.6% were misidentified (23).
In the present study, 22 isolates representing seven species not included in the Vitek MS v2.0 database obtained results of no identification, highlighting the specificity of the Vitek MS system. Of the eight isolates unidentified by the Vitek MS v2.0 system despite being included in the database, six (75%) were correctly identified when a second Vitek MS analysis was performed. Therefore, laboratories should consider routinely repeating testing of all isolates initially identified as no identification. C. parapsilosis sensu stricto could be identified, while C. orthopsilosis and C. metapsilosis species could not be identified by the Vitek MS v2.0 system, a result attributable to the absence of reference spectra in the v2.0 database. Given that the Bruker Biotyper can identify C. orthopsilosis and C. metapsilosis, enhancements to the current Vitek MS v2.0 database may be warranted (26). Some closely related species that are difficult to identify by conventional methods, such as C. dubliniensis, which is closely related to C. albicans, were not included in this study. However, the Vitek MS system has been successful in separating these species when encountered in a previous study (23).
Our study has demonstrated that the Vitek MS system performs well for the routine laboratory identification of the commonly encountered yeast species. However, when identification of rarer species is considered critical for clinical or epidemiological studies, the gold standard for identification is a molecular method. Our proposed algorithm offers a clear and structured approach for yeast identification that can be applied to large epidemiological studies, including future CHIF-NET reports. The benefits of the algorithm include the high degree of accurate identification (99.7%) as well as the significant reduction in labor and material costs associated with the lower number of isolates requiring molecular identification. However, due to the inability of the Vitek MS database to distinguish species with high phenotypic homogeneity, two isolates of C. fermentati were indistinguishable from C. guilliermondii, and a single C. gattii isolate was indistinguishable from C. neoformans when the testing algorithm was applied (27). We envisage that with future revisions of the Vitek MS reference spectra database, closely related species may be able to be separated.
This study has several strengths. First, the 1,243 yeast strains were isolated from patients with invasive fungal infections in laboratories of 22 hospitals across China, with wide geographic representation and strain heterogeneity. Second, the isolates represented 31 species, including both common and rarely encountered yeast species, nine of which were not included in the v2.0 database. The large number of well-characterized strains collected across China constitutes a valuable resource for enhancement of the Vitek MS comprehensive database. In addition, all isolates were analyzed by the Vitek MS v2.0 system and ITS sequence analysis, which is the first time that the performance of the Vitek MS v2.0 system has been evaluated using molecular identification as a reference method in a developing country. Finally, this is the first time that Vitek MS v2.0 evaluation has been performed on yeast isolates collected from a large national surveillance program, providing valuable insights for formulating a strategy for yeast identification in similar surveillance programs.
In summary, the present evaluation involving 1,243 yeast isolates from the 2011 CHIF-NET demonstrated that the Vitek MS system is a valuable method for yeast identification. The implementation of this technology in clinical microbiology laboratories in China will improve the diagnostic accuracy of fungal infection. More importantly, a practical and cost-effective algorithm for yeast identification incorporating the Vitek MS system with rDNA sequencing has been proposed for large surveillance programs such as the CHIF-NET. This algorithm will be an important foundation for the establishment of protocols for laboratory diagnosis of fungal infections in developing countries. With further enhancements to the Vitek MS system, the testing algorithm and subsequently laboratory workflow will improve and lead to further efficiencies.
ACKNOWLEDGMENTS
We appreciate the anonymous reviewers' very helpful comments, which helped improve the paper significantly.
This study was supported by the Innovation Fund of Peking Union Medical College (grant 2012-1002-023) and the Young Foundation of Peking Union Medical College Hospital (grant 2010145).
We thank all the laboratories participating in 2011 CHIF-NET for making this large project possible.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Pfaller MA, Diekema DJ, Messer SA, Boyken L, Hollis RJ. 2003. Activities of fluconazole and voriconazole against 1,586 recent clinical isolates of Candida species determined by Broth microdilution, disk diffusion, and Etest methods: report from the ARTEMIS Global Antifungal Susceptibility Program, 2001. J. Clin. Microbiol. 41:1440–1446. 10.1128/JCM.41.4.1440-1446.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377. 10.1128/JCM.02117-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Bijie H, Dzierzanowska D, Klimko NN, Letscher-Bru V, Lisalova M, Muehlethaler K, Rennison C, Zaidi M. 2009. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: 10.5-year analysis of susceptibilities of noncandidal yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 47:117–123. 10.1128/JCM.01747-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortorano AM, Kibbler C, Peman J, Bernhardt H, Klingspor L, Grillot R. 2006. Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 27:359–366. 10.1016/j.ijantimicag.2006.01.002 [DOI] [PubMed] [Google Scholar]
- 5.Messer SA, Kirby JT, Sader HS, Fritsche TR, Jones RN. 2004. Initial results from a longitudinal international surveillance programme for anidulafungin (2003). J. Antimicrob. Chemother. 54:1051–1056. 10.1093/jac/dkh504 [DOI] [PubMed] [Google Scholar]
- 6.Messer SA, Moet GJ, Kirby JT, Jones RN. 2009. Activity of contemporary antifungal agents, including the novel echinocandin anidulafungin, tested against Candida spp., Cryptococcus spp., and Aspergillus spp.: report from the SENTRY Antimicrobial Surveillance Program (2006 to 2007). J. Clin. Microbiol. 47:1942–1946. 10.1128/JCM.02434-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Xiao M, Chen SC, Kong F, Sun ZY, Liao K, Lu J, Shao HF, Yan Y, Fan H, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Xu YC. 2012. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J. Clin. Microbiol. 50:3952–3959. 10.1128/JCM.01130-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller MA, Woosley LN, Messer SA, Jones RN, Castanheira M. 2012. Significance of molecular identification and antifungal susceptibility of clinically significant yeasts and moulds in a global antifungal surveillance programme. Mycopathologia 174:259–271. 10.1007/s11046-012-9551-x [DOI] [PubMed] [Google Scholar]
- 9.Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. 1999. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49(Part 1):329–337. 10.1099/00207713-49-1-329 [DOI] [PubMed] [Google Scholar]
- 10.Hsieh SY, Tseng CL, Lee YS, Kuo AJ, Sun CF, Lin YH, Chen JK. 2008. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol. Cell. Proteomics 7:448–456. 10.1074/mcp.M700339-MCP200 [DOI] [PubMed] [Google Scholar]
- 11.Qian J, Cutler JE, Cole RB, Cai Y. 2008. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal. Bioanal. Chem. 392:439–449. 10.1007/s00216-008-2288-1 [DOI] [PubMed] [Google Scholar]
- 12.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551. 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 13.Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr AM. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 14.Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J. Clin. Microbiol. 50:2568–2576. 10.1128/JCM.00343-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang H, Ohlsson AK, Ullberg M, Ozenci V. 2012. Evaluation of species-specific PCR, Bruker MS, Vitek MS and the Vitek 2 system for the identification of clinical Enterococcus isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31:3073–3077. 10.1007/s10096-012-1667-x [DOI] [PubMed] [Google Scholar]
- 16.Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, Howard W, Pruessner J, Safwat N, Cockerill FR, Bossler AD, Patel R, Richter SS. 2012. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol. 50:2034–2039. 10.1128/JCM.00330-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917. 10.1128/JCM.00389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175. 10.1128/JCM.01881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616. 10.1128/JCM.02381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. 2012. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization-time of flight system with a new time-effective strategy. J. Clin. Microbiol. 50:2107–2110. 10.1128/JCM.06713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavergne RA, Chauvin P, Valentin A, Fillaux J, Roques-Malecaze C, Arnaud S, Menard S, Magnaval JF, Berry A, Cassaing S, Iriart X. 2013. An extraction method of positive blood cultures for direct identification of Candida species by Vitek MS matrix-assisted laser desorption ionization time of flight mass spectrometry. Med. Mycol. 51:652–656. 10.3109/13693786.2012.762607 [DOI] [PubMed] [Google Scholar]
- 22.Mancini N, De Carolis E, Infurnari L, Vella A, Clementi N, Vaccaro L, Ruggeri A, Posteraro B, Burioni R, Clementi M, Sanguinetti M. 2013. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry systems for identification of yeasts of medical importance. J. Clin. Microbiol. 51:2453–2457. 10.1128/JCM.00841-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Burnham CA. 2013. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J. Clin. Microbiol. 51:2267–2272. 10.1128/JCM.00680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo LN, Xiao M, Kong F, Chen SC, Wang H, Sorrell TC, Jiang W, Dou HT, Li RY, Xu YC. 2011. Three-locus identification, genotyping, and antifungal susceptibilities of medically important Trichosporon species from China. J. Clin. Microbiol. 49:3805–3811. 10.1128/JCM.00937-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia L, Westblade LF, Ferraro MJ, Branda JA. 2013. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J. Clin. Microbiol. 51:2225–2231. 10.1128/JCM.00682-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quiles-Melero I, García-Rodriguez J, Gómez-López A, Mingorance J. 2012. Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:67–71. 10.1007/s10096-011-1277-z [DOI] [PubMed] [Google Scholar]
- 27.San Millán R, Wu LC, Salkin IF, Lehmann PF. 1997. Clinical isolates of Candida guilliermondii include Candida fermentati. Int. J. Syst. Bacteriol. 47:385–393. 10.1099/00207713-47-2-385 [DOI] [PubMed] [Google Scholar]