Abstract
Vitek 2 (bioMérieux, Inc., Durham, NC) is a widely used commercial antimicrobial susceptibility testing system. We compared MIC results obtained by Vitek 2 to those obtained by the Clinical and Laboratory Standards Institute (CLSI) broth microdilution (BMD) reference method for 134 staphylococcal and 84 enterococcal clinical isolates. Nineteen agents were evaluated, including all those available on Vitek 2 for testing staphylococci and enterococci. The resistance phenotypes tested included methicillin-resistant Staphylococcus aureus (MRSA) (n = 58), S. aureus with inducible clindamycin resistance (ICR) (n = 30), trimethoprim-sulfamethoxazole-resistant MRSA (n = 10), vancomycin-resistant Enterococcus (n = 37), high-level gentamicin-resistant Enterococcus (n = 15), linezolid-resistant Enterococcus (n = 5), and daptomycin-nonsusceptible Enterococcus faecalis (n = 6). For the staphylococci, there was 98.9% categorical agreement (CA). There was one very major error (VME) for gentamicin in a Staphylococcus hominis isolate, six VMEs for inducible clindamycin in S. aureus isolates, and two major errors (ME) for daptomycin in an S. aureus and a Staphylococcus epidermidis isolate. For enterococci, there was 97.3% CA. Two VMEs were observed for daptomycin in isolates of E. faecalis and 2 ME, 1 for high-level gentamicin resistance and 1 for nitrofurantoin, in E. faecium isolates. Overall, there was 98.3% CA and 99% essential agreement for the testing of staphylococci and enterococci by the Vitek 2. With the exception of detecting ICR in S. aureus, Vitek 2 performed reliably for antimicrobial susceptibility testing of staphylococci and enterococci.
INTRODUCTION
Clinical microbiology laboratories routinely perform antimicrobial susceptibility testing (AST) on all staphylococci and enterococci isolated from sterile body sites, as well as nonsterile sites, when they are the predominate pathogen. Both these genera are associated with important resistance phenotypes that are frequently encountered in clinical practice, including methicillin resistance in Staphylococcus aureus and vancomycin resistance in Enterococcus. As these phenotypes pose significant burdens on health care, reliable detection by the laboratory is imperative. In addition, less frequent but equally important resistance phenotypes, such as elevated vancomycin MICs in S. aureus and linezolid resistance and daptomycin nonsusceptibility in both staphylococci and enterococci, must be reliably detected by the laboratory. Prompt and accurate methods for susceptibility testing are needed for the laboratory to identify antimicrobial susceptibilities for these organisms for timely treatment decision making and implementation of infection control practices.
Commercial automated systems for identification and susceptibility testing of bacteria are used in most clinical microbiology laboratories in the United States. Due to their ease of use and cost-effectiveness they are often the preferred methods over the more labor-intensive Clinical and Laboratory Standards Institute (CLSI) reference methods of broth microdilution (BMD) and disk diffusion (DD). However, there have been reports that the commercial systems can produce inaccurate results for select antimicrobial agents when testing either staphylococci (1–3) or enterococci (4, 5). These reports have led to recommendations that laboratories confirm certain susceptibility phenotypes by a manual method (6–8). We evaluated the performance of the Vitek 2 (bioMérieux, Inc., Durham, NC) AST-GP71 and AST-GP72 cards for staphylococci and enterococci, respectively, compared to the CLSI BMD method.
(This work was presented in part at the 112th General Meeting of the American Society for Microbiology, San Francisco, CA, 16 to 19 June 2012.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 134 Staphylococcus and 84 Enterococcus single patient isolates were selected for testing, to represent various resistance phenotypes (Table 1). All isolates were obtained from specimens submitted between 2008 and 2012 to the UCLA clinical microbiology laboratory; 151 were freshly isolated (isolated from primary culture within 7 days and never frozen), and the remaining 67 were from frozen stock cultures. Isolates were recovered from a variety of sources, including blood, urine, respiratory secretions, wounds, tissues, fluids, and ocular specimens. Identification was performed with the Vitek 2 (bioMérieux Inc., Durham, NC) system using GP ID cards. Prior to testing, frozen isolates were subcultured twice and fresh isolates were subcultured once on tryptic soy agar plates containing 5% sheep blood (blood agar plate [BAP]) and incubated at 35°C for 18 to 24 h. The quality control (QC) strains tested with each run were S. aureus ATCC 29213 and E. faecalis ATCC 29212. Upon receipt of a new shipment of Vitek 2 cards or when changing lot numbers of cards, the following QC strains were tested according to the manufacturer's recommendations: S. aureus ATCC 29213 (AST-GP71 and AST-GP72), S. aureus ATCC BAA1026 (AST-GP71), S. aureus ATCC BAA976 (AST-GP71), S. aureus ATCC BAA977 (AST-GP71), E. faecalis ATCC 29212 (AST-GP71 and AST-GP72), E. faecalis ATCC 51299 (AST-GP72), and Escherichia coli ATCC 35318 (AST-GP72) (9, 10). QC procedures for in-house BMD panel preparation included testing 19 supplemental QC strains in addition to testing those recommended by CLSI for routine QC.
TABLE 1.
Numbers of isolates tested with various clinically significant resistant phenotypes
| Organism/resistance phenotypea | Total no. of isolates | Organism/resistance phenotype | Total no. of isolates |
|---|---|---|---|
| MRSA | 30 | E. faecalis/Vanco-S, HLSR | 9 |
| MRSA/ICR | 16 | E. faecalis/Vanco-S, HLGR | 8 |
| MRSA/T-S-R | 10 | E. faecalis/Vanco-S, Dap-NS | 6 |
| MRSA/Dap-NS/Vanco-S | 2 | E. faecium/VRE | 7 |
| MSSA/ICR | 14 | E. faecium/VRE, HLGR | 4 |
| CoNSb/Ox-R | 11 | E. faecium/VRE, LNZ-R | 5 |
| CoNS/Ox-R/ICR | 6 | E. faecium/VRE, HLSR | 22 |
| E. faecalis/VRE, HLSR | 3 | E. faecium/Vanco-S, HLGR | 1 |
| E. faecalis/VRE, HLGR | 3 | E. faecium/Vanco-S, HLSR | 3 |
MRSA, methicillin-resistant S. aureus; ICR, inducible clindamycin resistance; T-S, trimethoprim-sulfamethoxazole resistance; Dap-NS, daptomycin nonsusceptible; MSSA, methicillin-susceptible S. aureus; CoNS, coagulase-negative Staphylococcus; Ox-R, oxacillin resistance; VRE, vancomycin-resistant Enterococcus; HLGR, high-level gentamicin resistance; LNZ-R, linezolid resistance; HLSR, high-level streptomycin resistance; Vanco-S, vancomycin susceptible.
S. epidermidis (n = 17), S. lugdunensis (n = 7), S. haemolyticus (n = 6), S. capitis, (n = 4), S. hominis (n = 3), S. warneri (n = 1), S. caprae (n = 1), Staphylococcus spp. (n = 1).
Antimicrobial susceptibility testing.
BMD MIC testing was performed according to CLSI guidelines using in-house prepared panels (11). These panels were incubated at 35°C in ambient air, read manually at 16 to 20 h, and then reincubated at 35°C in ambient air to 24 h to obtain final results for oxacillin (staphylococci) and vancomycin (staphylococci and enterococci). For enterococci, high-level gentamicin and streptomycin were read at 24 h, and if high-level streptomycin screening was negative at 24 h, plates were reincubated at 35°C in ambient air and read at 48 h. Susceptibility tests with the Vitek 2 (bioMérieux, Inc., Durham, NC) system were performed using software version 5.01 and AST-GP71 (staphylococci) or AST-GP2 (enterococci) cards according to the manufacturer's instructions (9, 10). Each isolate was tested concurrently with both methods using isolated colonies from a single 18- to 24-h BAP. Purity plates were prepared following inoculation of each test by subculturing an aliquot of inoculum suspension onto a BAP and incubating for 18 to 24 h.
Testing for inducible clindamycin resistance.
BMD panels contained a single well with a combination of 0.5 μg/ml of clindamycin and 4 μg/ml of erythromycin. AST-GP71 cards contained two wells for inducible clindamycin resistance (ICR), one with 0.5 μg/ml of clindamycin and the other with a combination of 0.25 μg/ml of clindamycin and 0.5 μg/ml of erythromycin. For staphylococci that were erythromycin resistant and clindamycin susceptible or intermediate, ICR disk diffusion (D-zone test) was performed according to CLSI recommendations on BAP purity plates by placing a 15-μg erythromycin disk and a 2-μg clindamycin disk 15 mm apart on a heavily inoculated area of the purity plate. Flattening of the clindamycin zone adjacent to the erythromycin disk after 16 to 18 h of incubation at 35°C ambient air was interpreted as positive for inducible clindamycin resistance (11).
Confirmatory ICR testing.
S. aureus isolates that demonstrated discrepant results between Vitek 2 and the two CLSI methods (BMD and D-zone test) for ICR were sent to bioMérieux for further testing with two different lots of AST-GP71 cards. In addition, the D-zone test, ermA testing by PCR, and strain typing using the DiversiLab System were performed at bioMérieux.
Data analysis.
Essential agreement (EA), categorical agreement (CA), very major errors (VMEs), major errors (MEs), and minor errors (mEs) were calculated as previously described (12). The EA was defined as an MIC of ±1 doubling dilution of the reference BMD MIC. The CA was defined as a susceptible, intermediate, resistant, or nonsusceptible result that was the same with both methods. A VME was defined as a false susceptible result with the Vitek 2 system, whereas an ME was a false resistant or nonsusceptible result with the Vitek 2 system; an mE was identified when one method reported an intermediate result while the other method reported a susceptible or resistant (on nonsusceptible) result.
Discrepant resolution.
Isolates with a VME or ME were retested using both methods, as were select isolates with specific drug/organism combinations resulting in ≥10% mEs. Calculations of EA, CA, VMEs, MEs, and mEs were obtained following resolution of discrepant results after repeat testing.
RESULTS
Of 218 isolates tested, 8 (3.7%) terminated due to unacceptable growth in the control well of the Vitek 2 automated susceptibility testing (AST) card. These included 1 Staphylococcus capitis, 1 Staphylococcus spp. (not identified to species level by the GP ID card), 3 Enterococcus faecium, 2 Enterococcus faecalis, and 1 Enterococcus casseliflavus. One Staphylococcus caprae isolate was terminated because this species is not included in the Vitek 2 system AST database. For the remaining 209 isolates there were 88 discrepancies in categorical interpretation between Vitek 2 and BMD, out of a total of 2,950 organism-antimicrobial combinations. Upon repeat testing by both BMD and Vitek 2, 45 of the 88 (51.1%) discrepancies were resolved, resulting in EAs of 98.8% and 99.5% and CAs of 98.8% and 97.3% for staphylococci and enterococci, respectively. Of the 45 resolved discrepancies, 38 (84%) were due to BMD and 20 of the 38 (52%) were specifically due to linezolid. The discrepancies with linezolid were attributed to difficulty in manually reading the linezolid endpoint on BMD (5).
Results for staphylococci.
The Vitek 2 failed to identify ICR for 6 MRSA of 30 (20%) S. aureus (16 MRSA, 14 methicillin-susceptible S. aureus [MSSA]) isolates that were positive for ICR by the reference BMD single-well ICR test and the D-zone test (Table 2). The 6 isolates that were ICR negative by Vitek 2 but positive by reference BMD and D-zone tests were retested at bioMérieux and were confirmed to be ICR positive by the D-zone test and by presence of ermA by PCR. Two of 6 isolates gave a positive ICR reaction on one lot of Vitek AST-GP71 cards (Table 3). Strain typing was performed using the DiversiLab system to check for clonality of these isolates. The 6 isolates demonstrated 4 pattern types: two isolates belonged to pattern type one (P1) and two isolates belonged to pattern type two (P2). A single band difference was found between P1 and P2, indicating that these isolates were similar, but not indistinguishable. The remaining two isolates belonged to their own pattern types, P3 and P4 (Fig. 1).
TABLE 2.
Performance of AST-GP71 card for S. aureusa
| Antimicrobial | No. of isolatesb |
EAc (no. [%]) | CAd (no. [%]) | VMEe (no. [%]) | MEf (no. [%]) | mEg (no. [%]) | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | R | I | S | ||||||
| Cefoxitin screen | 94 | 58 | 0 | 36 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 94 | 56 | 4 | 34 | 93 (98.9) | 93 (98.9) | 0 (0) | 0 (0) | 1 (1.1) |
| Clindamycin | 94 | 41 | 0 | 53 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Inducible clindamycin resistance | 94 | 30 | 0 | 64 | 94 (100) | 88 (93.6) | 6 (20) | 0 (0) | 0 (0) |
| Daptomycin | 94 | 2 | 0 | 92 | 87 (92.5) | 93 (98.9) | 0 (0) | 1 (1.1) | 0 (0) |
| Erythromycin | 94 | 74 | 3 | 17 | 89 (94.7) | 91 (96.8) | 0 (0) | 0 (0) | 3 (3.2) |
| Gentamicin | 94 | 8 | 0 | 86 | 94 (100) | 93 (98.9) | 0 (0) | 0 (0) | 1 (1.1) |
| Linezolid | 94 | 0 | 0 | 94 | 93 (98.9) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Nitrofurantoin | 94 | 0 | 0 | 94 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Oxacillin | 94 | 58 | 0 | 36 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Penicillin | 94 | 89 | 0 | 5 | 93 (98.9) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Quinupristin-dalfopristin | 94 | 0 | 0 | 94 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Rifampin | 94 | 7 | 0 | 87 | 93 (98.9) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Tigecycline | 94 | 0 | 0 | 94 | 93 (98.9) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Trimethoprim-sulfamethoxazole | 94 | 11 | 0 | 83 | 94 (100) | 94 (100) | 0 (0) | 0 (0) | 0 (0) |
| Vancomycin | 94 | 0 | 0 | 94 | 94 (100) | 91 (96.8) | 0 (0) | 0 (0) | 3 (3.2) |
| Total | (98.7) | (98.9) | 6 | 1 | 8 | ||||
Results calculated following resolution of discrepancies after repeat testing.
R, resistant; I, intermediate; S, susceptible.
EA, essential agreement (MIC within ±1 doubling dilution).
CA, categorical agreement.
VME, very major error.
ME, major error.
mE, minor error.
TABLE 3.
Discrepant ICR results for MRSA
| Isolatea | UCLA |
bioMérieuxb |
||||
|---|---|---|---|---|---|---|
| AST-GP71 ICR lot 1 | BMD ICR test | D-zone test | AST-GP71 ICR lot 1/lot 2 | D-zone test | Molecular | |
| MRSA ICR no. 1 | Neg | Pos | Pos | Neg/Neg | Pos | ermA |
| MRSA ICR no. 2 | Neg | Pos | Pos | Neg/Neg | Pos | ermA |
| MRSA ICR no. 3 | Neg | Pos | Pos | Neg/Pos | Pos | ermA |
| MRSA ICR no. 4 | Neg | Pos | Pos | Neg/Neg | Pos | ermA |
| MRSA ICR no. 5 | Neg | Pos | Pos | Neg/Neg | Pos | ermA |
| MRSA ICR no. 6 | Neg | Pos | Pos | Neg/Pos | Pos | ermA |
MRSA, methicillin resistance in S. aureus; ICR, inducible clindamycin resistance.
Neg, negative for the ICR screen, clindamycin susceptible; Pos, positive for the ICR screen, clindamycin resistant.
FIG 1.
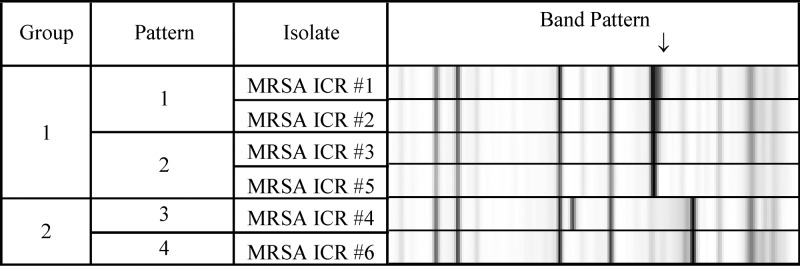
Strain typing by DiversiLab showing partial clonality. The results shown were produced from testing at bioMérieux. MRSA, methicillin-resistant S. aureus; ICR, inducible clindamycin resistance. The arrow points to an additional band that differentiates pattern 1 from pattern 2.
A VME was noted for gentamicin in a Staphylococcus hominis isolate (16.7% of 6 gentamicin-resistant coagulase-negative Staphylococcus). Two MEs were noted for daptomycin, one in an S. aureus isolate (1.1% of 92 daptomycin-susceptible S. aureus) and one in a Staphylococcus epidermidis isolate (2.7% of 37 daptomycin-susceptible coagulase-negative Staphylococcus) (Tables 2 and 4). mEs occurred with ciprofloxacin (n = 1), clindamycin (n = 2), erythromycin (n = 4), gentamicin (n = 3), and vancomycin (n = 4) (Tables 2 and 4).
TABLE 4.
Performance of AST-GP71 card for coagulase-negative Staphylococcusa
| Antimicrobial | No. of isolatesb |
EAc (no. [%]) | CAd (no. [%]) | VMEe (no. [%]) | MEf (no. [%]) | mEg (no. [%]) | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | R | I | S | ||||||
| Cefoxitin screen | 37 | 17 | 0 | 20 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 37 | 16 | 0 | 21 | 37 (100) | 36 (97.3) | 0 (0) | 0 (0) | 1 (2.7) |
| Clindamycin | 37 | 10 | 0 | 27 | 37 (100) | 36 (97.3) | 0 (0) | 0 (0) | 1 (2.7) |
| Daptomycin | 37 | 0 | 0 | 37 | 34 (91.9) | 36 (97.3) | 0 (0) | 1 (2.7) | 0 (0) |
| Erythromycin | 37 | 20 | 1 | 16 | 36 (97.3) | 36 (97.3) | 0 (0) | 0 (0) | 1 (2.7) |
| Gentamicin | 37 | 6 | 2 | 29 | 35 (94.6) | 34 (91.9) | 1 (16.7) | 0 (0) | 2 (5.4) |
| Inducible clindamycin resistance | 37 | 12 | 0 | 25 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Linezolid | 36 | 0 | 0 | 36 | 36 (100) | 36 (100) | 0 (0) | 0 (0) | 0 (0) |
| Nitrofurantoin | 37 | 0 | 0 | 37 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Oxacillin | 37 | 17 | 0 | 20 | 36 (97.3) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Penicillin | 35 | 27 | 0 | 8 | 35 (100) | 35 (100) | 0 (0) | 0 (0) | 0 (0) |
| Quinupristin-dalfopristin | 37 | 0 | 0 | 37 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Rifampin | 37 | 0 | 0 | 37 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Tigecycline | 37 | 0 | 0 | 37 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Trimethoprim-sulfamethoxazole | 37 | 15 | 0 | 22 | 37 (100) | 37 (100) | 0 (0) | 0 (0) | 0 (0) |
| Vancomycin | 37 | 0 | 0 | 37 | 37 (100) | 36 (97.3) | 0 (0) | 0 (0) | 1 (2.7) |
| Total | (98.8) | (98.7) | 1 | 1 | 6 | ||||
Results calculated following resolution of discrepancies after repeat testing.
R, resistant; I, intermediate; S, susceptible.
EA, essential agreement (MIC within ±1 doubling dilution).
CA, categorical agreement.
VME, very major error.
ME, major error.
mE, minor error.
Vancomycin MICs determined by Vitek 2 and BMD for 45 of 58 (78%) MRSA were identical. Of the 13 isolates for which vancomycin MICs were not identical, 4 had vancomycin MICs of 1 μg/ml by BMD and 2 μg/ml by Vitek 2. The remaining 9 MRSA isolates had a vancomycin MIC of either 0.5 μg/ml or 1 μg/ml with either BMD or Vitek 2. No consistent trend toward higher or lower vancomycin MICs was observed for Vitek 2 compared to BMD. All MRSA isolates had vancomycin MICs within the susceptible range of ≤2 μg/ml, but 3 mEs were observed when the Vitek 2 Advanced Expert System (AES) software edited a vancomycin-susceptible interpretation to intermediate for 3 MRSA isolates that had daptomycin MICs in the nonsusceptible range (MIC 4 μg/ml).
Results for enterococci.
Two VMEs were observed for daptomycin, both in E. faecalis isolates (33.3% of 6 daptomycin-nonsusceptible E. faecalis). Both VMEs were confirmed to be daptomycin-nonsusceptible by Etest (data not shown). Two MEs occurred in E. faecium, one for high-level gentamicin (1.6% of 64 high-level gentamicin-susceptible Enterococcus) and one for nitrofurantoin (2.4% of 42 nitrofurantoin-susceptible Enterococcus) (Table 5). mEs among Enterococcus included ciprofloxacin (n = 5), erythromycin (n = 3), linezolid (n = 2), and nitrofurantoin (n = 6).
TABLE 5.
Performance of AST-GP72 card for Enterococcus spp.a
| Antimicrobial | No. of isolatesb |
EAc (no. [%]) | CAd (no. [%]) | VMEe (no. [%]) | MEf (no. [%]) | mEg (no. [%]) | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | R | I | S | ||||||
| Ampicillin | 78 | 36 | 0 | 42 | 78 (100) | 78 (100) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 78 | 48 | 6 | 24 | 78 (100) | 73 (93.6) | 0 (0) | 0 (0) | 5 (6.4) |
| Daptomycin | 32h | 6 | 0 | 26 | 31 (96.9) | 30 (93.8) | 2 (33.3) | 0 (0) | 0 (0) |
| Erythromycin | 78 | 58 | 15 | 5 | 78 (100) | 75 (96.2) | 0 (0) | 0 (0) | 3 (3.8) |
| Gentamicin (high level) | 78 | 16 | 0 | 62 | 78 (100) | 77 (98.7) | 0 (0) | 1 (1.6) | 0 (0) |
| Linezolid | 78 | 5 | 4 | 69 | 78 (100) | 76 (97.4) | 0 (0) | 0 (0) | 2 (2.6) |
| Nitrofurantoin | 78 | 19 | 17 | 42 | 76 (97.4) | 71 (91) | 0 (0) | 1 (2.4) | 6 (7.7) |
| Quinupristin-dalfopristin | 73 | 25 | 5 | 43 | 73 (100) | 73 (100) | 0 (0) | 0 (0) | 0 (0) |
| Streptomycin (high level) | 78 | 37 | 0 | 41 | 78 (100) | 78 (100) | 0 (0) | 0 (0) | 0 (0) |
| Tigecycline | 78 | 0 | 0 | 78 | 78 (100) | 78 (100) | 0 (0) | 0 (0) | 0 (0) |
| Vancomycin | 73 | 35 | 0 | 38 | 73 (100) | 73 (100) | 0 (0) | 0 (0) | 0 (0) |
| Total | (99.5) | (97.3) | 2 | 2 | 16 | ||||
Results calculated following resolution of discrepancies after repeat testing.
R, resistant; I, intermediate; S, susceptible.
EA, essential agreement (within ±1 doubling dilution).
CA, categorical agreement.
VME, very major error.
ME, major error.
mE, minor error.
E. faecalis only.
DISCUSSION
Vitek 2 is a widely used system for identification and antimicrobial susceptibility testing. Recent publications that describe the performance of Vitek 2 with staphylococci or enterococci focused on an individual antimicrobial agent such as vancomycin (7), cefoxitin (1), clindamycin (ICR test) (2, 3, 8), or linezolid (5). Noteworthy discrepancies occurred in S. aureus with clindamycin (ICR test), which had a reported sensitivity of 91 to 95% (2, 3, 8), and with vancomycin, which had a reported sensitivity of 91% (7). It has been over 8 years since a comprehensive evaluation of Vitek 2 performance for staphylococci and enterococci was reported in the peer-reviewed literature (13). Here, we report the performance of Vitek 2 for both staphylococci and enterococci for testing contemporary isolates with currently used antimicrobial agents and the most up-to-date software and AST cards available from bioMérieux.
With a few exceptions, Vitek 2 performed reliably for staphylococci and enterococci for all antimicrobials tested, with overall EAs of 98.8% and 99.5% and CAs of 98.8% and 97.3%, respectively. Categorical error rates for staphylococci (n = 131) tested with all antimicrobial agents (n = 16) were 1.7% VMEs, 0.2% MEs, and 0.8% mEs. Categorical error rates for enterococci (n = 78) and all antimicrobial agents (n = 11) were 0.7% VMEs, 0.5% MEs, and 2% mEs. Performances of the Vitek 2 AST-GP71 (staphylococci) and AST-GP-72 (enterococci) cards met the criteria suggested for acceptable performance of an automated susceptibility test system outlined in Cumitech 31A for AST systems (e.g., >89.9% EA and CA) (12).
The most notable deficiency of the Vitek 2 performance was the detection of ICR in S. aureus, for which 20% VME were observed among 30 S. aureus with the ICR phenotype. All 6 isolates that tested negative for ICR by Vitek 2 were confirmed to harbor an ermA gene by PCR. Strain typing performed by bioMérieux showed that only 2 of the 6 isolates were clonal (Fig. 1). Several groups have reported discrepant results for ICR and Vitek 2. Buchan and colleagues published a sensitivity of 91% when 51 of 56 ICR-positive isolates were tested (3). Lavallee et al. (2) and Gardiner et al. (8) published similar sensitivities of 93% and 95% when 124 of 134 ICR-positive and 191 of 201 ICR-positive isolates were tested, respectively. All three groups used the D-zone test as confirmation and included a mix of MRSA and MSSA isolates. The CLSI BMD recommendation for detection of ICR involves a single well containing 4 μg/ml erythromycin and 0.5 μg/ml clindamycin (11). It has been suggested that a possible reason for the failed detection of ICR with Vitek 2 is due to an insufficient concentration of erythromycin to induce the erm gene (8). The concentration of erythromycin in the ICR well on Vitek 2 cards is 0.5 μg/ml and the concentration of clindamycin is 0.25 μ/ml (9). The shortened incubation time in the Vitek 2 (∼8 h) might also contribute to failure to detect ICR in some strains of MRSA.
Treatment failures have been reported for infections caused by MRSA isolates with ICR when clindamycin was used for therapy (14–17). A solution to the ICR problem for laboratories using Vitek 2 would be the addition of clindamycin and erythromycin disks placed 15 mm apart on the BAP purity plate inoculated at the time of AST-GP71 card setup (6, 11). Some investigators have presented an alternative solution and report all erythromycin-resistant Staphylococcus spp. as resistant to clindamycin, without confirming the inducible resistance phenotype (2, 6, 8). However, the prevalence of ICR is variable depending upon whether the infection is hospital or community acquired and whether the organism is MRSA or MSSA (18–21), suggesting this solution may not be applicable to all settings.
Some discrepancies noted between Vitek 2 and BMD were a result of the Vitek 2 AES. The AES is designed to analyze the antimicrobial susceptibility pattern of each organism to determine biologic validity. Results are reviewed and categorized based on consistency with previously defined wild or resistant phenotypes and inconsistent results are flagged for further review. Antimicrobials are grouped together to create resistant phenotype profiles. The AES groups vancomycin and daptomycin together in a phenotypic group for S. aureus. When the daptomycin MIC is ≥2 μg/ml (nonsusceptible) and the vancomycin MIC is 1 or 2 μg/ml, the AES edits the vancomycin interpretation from susceptible to intermediate. Three mEs occurred for vancomycin and MRSA as a result of AES editing a vancomycin MIC of 2 μg/ml to “intermediate” when the daptomycin MIC was 4 μg/ml (nonsusceptible). This AES correction may be misleading because not all daptomycin-nonsusceptible MRSA are vancomycin intermediate (22, 23). Further, reporting vancomycin as intermediate and daptomycin as nonsusceptible leaves very few treatment options, creating an unnecessary challenge for physicians. Laboratories may consider manually overriding this rule in their laboratory information system.
For enterococci, most errors in this study were for antimicrobial agents that are generally not considered primary agents for treating enterococcal infections. Two VMEs for daptomycin were found in vancomycin-susceptible E. faecalis isolates. Another major limitation with the Vitek 2 for daptomycin testing is that reporting of daptomycin susceptibility results is limited to vancomycin-susceptible E. faecalis, as is required by the FDA to be consistent with the FDA clinical indications for daptomycin. However, daptomycin is frequently used to treat difficult infections caused by vancomycin-resistant E. faecalis and E. faecium, such as endocarditis, as there are very limited FDA-approved agents for the treatment of such infections (e.g., linezolid and quinupristin-dalfopristin). Numerous reports have documented treatment-emergent daptomycin nonsusceptibility in Enterococcus, primarily in vancomycin-resistant E. faecium (24–26). Clinical laboratories need reliable methods for testing daptomycin for all enterococcal species, but Vitek 2 is at the present not a viable option for this, due to both limitations in reporting and poor detection of nonsusceptibility in E. faecalis, as we document herein. The disk diffusion method is not reliable for daptomycin (11), leaving laboratories with few options for testing this agent. This issue is not unique to Vitek 2, Bryant and colleagues recently reported that MicroScan detected only 9 of 30 daptomycin-nonsusceptible enterococci with a VME rate of 70% for the MicroScan prompt inoculation method (27). They attributed this high percentage of error to inherent error in the system, as all errors occurred within 1 dilution of the susceptible breakpoint; nevertheless, they too advocate the use of an alternative method for testing enterococci with daptomycin. Although there is a limitation included in the Vitek 2 package insert, which states that the ability to detect linezolid resistance in Enterococcus spp. is unknown due to lack of resistant strains at the time of comparative testing (10), Vitek 2 detected all 5 linezolid-resistant E. faecium isolates tested (Table 5). For the 2 mEs that occurred with linezolid, one occurred with an intermediate BMD interpretation and a susceptible Vitek 2 and one was with a susceptible BMD interpretation and an intermediate Vitek 2. Tenover and colleagues published similar results of no VMEs or MEs and only 3 mEs, when 10 linezolid-resistant E. faecium isolates and 5 linezolid-resistant E. faecalis isolates were tested by Vitek 2 (5).
A limitation of this study was the small sample number of some clinically important phenotypes, including daptomycin-nonsusceptible E. faecalis (n = 6), daptomycin-nonsusceptible S. aureus (n = 2), linezolid-resistant E. faecium (n = 5), and ICR staphylococci (n = 30). The percentages of errors for ICR and daptomycin-nonsusceptible isolates were high due to the low number of resistant isolates tested. However, results were consistent with previously published reports (2, 3, 8), and worth noting because of the clinical significance of this observation. No VMEs were observed for linezolid-resistant E. faecium, but the sample size may have been too small to detect any problem. Additionally, no VMEs were detected for vancomycin and linezolid in staphylococci, which may be misleading because no resistant isolates were tested. Future testing is warranted as the prevalences of these resistant organisms increase.
Overall, the Vitek 2 AST-GP71 and GP72 performed comparably to BMD. Performance was reliable for organisms with significant resistant phenotypes, such as MRSA, high-level gentamicin-resistant enterococci, and vancomycin-resistant Enterococcus. Discrepancies were observed for ICR in S. aureus and daptomycin nonsusceptibility in E. faecalis. Based on our findings and others, we advise supplemental testing for S. aureus ICR and for Enterococcus spp. and daptomycin. We also urge users to be aware of the AES correction of daptomycin-nonsusceptible S. aureus isolates with vancomycin MICs of 1 or 2 μg/ml.
ACKNOWLEDGMENTS
This study was funded by bioMérieux, Inc.
We thank Jennifer Freie from the customer response lab at bioMérieux for performing confirmatory AST, PCR, and DiversiLab Systems testing on MRSA with ICR discrepancies. We also thank Farzaneh Sooudipour, Myra Maldonado, Marissa Carvalho, and Maria Tagarao for their technical assistance.
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Junkins AD, Lockhart SR, Heilmann KP, Dohrn CL, Von Stein DL, Winokur PL, Doern GV, Richter SS. 2009. BD Phoenix and Vitek 2 detection of mecA-mediated resistance in Staphylococcus aureus with cefoxitin. J. Clin. Microbiol. 47:2879–2882. 10.1128/JCM.01109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavallée C, Rouleau D, Gaudreau C, Roger M, Tsimiklis C, Locas MC, Gagnon S, Delorme J, Labbé A. 2010. Performance of an agar dilution method and a Vitek 2 card for detection of inducible clindamycin resistance in Staphylococcus spp. J. Clin. Microbiol. 48:1354–1357. 10.1128/JCM.01751-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan BW, Anderson NW, Ledeboer NA. 2012. Comparison of BD Phoenix and bioMérieux Vitek 2 automated systems for the detection of macrolide-lincosamide-streptogramin B resistance among clinical isolates of Staphylococcus. Diagn. Microbiol. Infect. Dis. 72:291–294. 10.1016/j.diagmicrobio.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 4.Raponi G, Ghezzi MC, Gherardi G, Lorino G, Dicuonzo G. 2010. Analysis of methods commonly used for glycopeptide and oxazolidinone susceptibility testing in Enterococcus faecium isolates. J. Med. Microbiol. 59:672–678. 10.1099/jmm.0.016444-0 [DOI] [PubMed] [Google Scholar]
- 5.Tenover FC, Williams PP, Stocker S, Thompson A, Clark LA, Limbago B, Carey RB, Poppe SM, Shinabarger D, McGowan JE. 2007. Accuracy of six antimicrobial susceptibility methods for testing linezolid against staphylococci and enterococci. J. Clin. Microbiol. 45:2917–2922. 10.1128/JCM.00913-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen JH, Crawford SA, McElmeel ML, Fiebelkorn KR. 2004. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J. Clin. Microbiol. 42:1800–1802. 10.1128/JCM.42.4.1800-1802.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swenson JM, Anderson KF, Lonsway DR, Thompson A, McAllister SK, Limbago BM, Carey RB, Tenover FC, Patel JB. 2009. Accuracy of commercial and reference susceptibility testing methods for detecting vancomycin-intermediate Staphylococcus aureus. J. Clin. Microbiol. 47:2013–2017. 10.1128/JCM.00221-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner BJ, Grayson ML, Wood GM. 2013. Inducible resistance to clindamycin in Staphylococcus aureus: validation of Vitek-2 against CLSI D-test. Pathology 45:181184. 10.1097/PAT.0b013e32835cccda [DOI] [PubMed] [Google Scholar]
- 9.bioMérieux 2013. VITEK 2 AST-GP71 product information. bioMérieux, Inc., Durham, NC [Google Scholar]
- 10.bioMérieux 2013. VITEK 2 AST-GP72 product information. bioMérieux, Inc., Durham, NC [Google Scholar]
- 11.Clinical Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23 Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12.Clark RB, Lewinski MA, Loeffelholz MJ, Thibbetts RJ. 2009. Cumitech 31A, Verification and validation of procedures in the clinical microbiology laboratory. Coordinating ed, Sharp SE. ASM Press, Washington, DC [Google Scholar]
- 13.Eigner U, Schmid A, Wild U, Bertsch D, Fahr AM. 2005. Analysis of the comparative workflow and performance characteristics of the VITEK 2 and Phoenix systems. J. Clin. Microbiol. 43:3829–3834. 10.1128/JCM.43.8.3829-3834.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drinkovic D, Fuller ER, Shore KP, Holland DJ, Ellis-Pegler R. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315–316. 10.1093/jac/48.2.315 [DOI] [PubMed] [Google Scholar]
- 15.Siberry GK, Tekle T, Carroll K, Dick J. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257–1260. 10.1086/377501 [DOI] [PubMed] [Google Scholar]
- 16.Frank AL, Marcinak JF, Mangat PD, Tjhio JT, Kelkar S, Schreckenberger PC, Quinn P. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530–534. 10.1097/00006454-200206000-00010 [DOI] [PubMed] [Google Scholar]
- 17.Patra KP, Vanchiere JA, Bocchini JA. 2011. Adherence to CLSI recommendations for testing of Staphylococcus aureus isolates in Louisiana hospitals: report of a clinical failure and results of a questionnaire study. J. Clin. Microbiol. 49:3019–3020. 10.1128/JCM.00944-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fokas S, Tsironi M, Kalkani M, Dionysopouloy M. 2005. Prevalence of inducible clindamycin resistance in macrolide-resistant Staphylococcus spp. Clin. Microbiol. Infect. 11:337–340. 10.1111/j.1469-0691.2005.01101.x [DOI] [PubMed] [Google Scholar]
- 19.Patel M, Waites KB, Moser SA, Cloud GA, Hoesley CJ. 2006. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates. J. Clin. Microbiol. 44:2481–2484. 10.1128/JCM.02582-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreckenberger PC, Ilendo E, Ristow KL. 2004. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J. Clin. Microbiol. 42:2777–2779. 10.1128/JCM.42.6.2777-2779.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I. 2007. Detection and prevalence of inducible clindamycin resistance in staphylococci. J. Med. Microbiol. 56:342–345. 10.1099/jmm.0.46761-0 [DOI] [PubMed] [Google Scholar]
- 22.Humphries RM, Hindler JA. 2012. Should laboratories test methicillin-resistant Staphylococcus aureus for elevated vancomycin minimum inhibitory concentrations by Etest as a driver of treatment changes? Clin. Infect. Dis. 55:612–613. 10.1093/cid/cis469 [DOI] [PubMed] [Google Scholar]
- 23.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56:92–102. 10.1128/AAC.00432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelesidis T, Humphries R, Uslan DZ, Pegues DA. 2011. Daptomycin nonsusceptible enterococci: an emerging challenge for clinicians. Clin. Infect. Dis. 52:228–234. 10.1093/cid/ciq113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphries RM, Kelesidis T, Tewhey R, Rose WE, Schork N, Nizet V, Sakoulas G. 2012. Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 56:6051–6053. 10.1128/AAC.01318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelesidis T, Tewhey R, Humphries RM. 2013. Evolution of high-level daptomycin resistance in Enterococcus faecium during daptomycin therapy is associated with limited mutations in the bacterial genome. J. Antimicrob. Chemother. 68:1926–1928. 10.1093/jac/dkt117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant KA, Roberts AL, Rupp ME, Anderson JR, Lyden ER, Fey PD, Van Schooneveld TC. 2013. Susceptibility of enterococci to daptomycin is dependent upon testing methodology. Diagn. Microbiol. Infect. Dis. 76:497–501. 10.1016/j.diagmicrobio.2013.04.019 [DOI] [PubMed] [Google Scholar]


