Abstract
We have developed a PCR–high-resolution melt (PCR-HRM) assay to discriminate nontypeable Haemophilus influenzae (NTHi) colonies from Haemophilus haemolyticus. This method is rapid and robust, with 96% sensitivity and 92% specificity compared to the hpd#3 assay. PCR-HRM is ideal for high-throughput screening for NTHi surveillance and clinical trials.
TEXT
Nontypeable Haemophilus influenzae (NTHi) is an important cause of respiratory system-related infections, including otitis media (OM), exacerbations of chronic obstructive pulmonary disease, chronic bronchitis, and bronchiectasis (1–3). There are also reports of its increasing importance as a cause of invasive infections in regions with H. influenzae type b immunization (4–6). NTHi isolates have colony phenotypes similar to those of nonhemolytic Haemophilus haemolyticus (7), a respiratory tract commensal that rarely causes disease (8, 9). Therefore, correct identification of NTHi requires novel methods of discrimination to complement traditional culture techniques (10, 11).
Clinical trials are under way to assess the effect of antibiotics and the 10-valent pneumococcal H. influenzae protein D-conjugate vaccine (PHiD-CV; Synflorix) on NTHi nasopharyngeal carriage and infection (for example, NCT01735084 and NCT01174849). An 11-valent precursor to PHiD-CV reduced NTHi-associated acute otitis media by 35% (12). Although a reduction in H. influenzae nasopharyngeal carriage was observed following primary and booster vaccination with the 11-valent vaccine (17), no substantial effect has been observed on NTHi carriage in subsequent studies with PHiD-CV, which has 8 of 10 serotypes conjugated to protein D (20, 21). Whether or not PHiD-CV will afford protection against OM or other NTHi infections, particularly in high-risk populations, is yet to be determined.
Given the high rates of misidentification of NTHi as the closely related commensal H. haemolyticus by standard culture-based techniques (10, 16), accurate molecular identification of NTHi is essential for surveillance of NTHi-targeted therapies. The development of a molecular tool for NTHi identification that can also identify H. haemolyticus will enable a greater understanding of the role of H. haemolyticus in carriage and disease. In addition, H. haemolyticus also expresses protein D (17), the NTHi antigen used in PHiD-CV. An assay that permits detection of H. haemolyticus is therefore important to monitor the impact of PHiD-CV on H. haemolyticus colonization. We have previously shown that the hpd#3 probe-based real-time PCR (RT-PCR) (13) provided superior NTHi identification compared to other PCR assays (14). We have utilized the discriminatory power of the hpd gene to develop a rapid PCR–high-resolution melt (PCR-HRM) assay for high-throughput identification of NTHi and discrimination from H. haemolyticus.
Two reference strains (H. haemolyticus ATCC 33390 and NTHi 86-028NP [15]), 60 nasopharyngeal carriage isolates from Western Australian children with and without a history of recurrent acute OM (14, 16) (20 NTHi, 19 H. haemolyticus, and 21 ambiguous Haemophilus isolates by 16S rRNA gene PCR), and 151 isolates from Northern Territory children with bronchiectasis (3) (49 nasopharyngeal, 52 bronchoalveolar lavage, and 50 throat isolates identified as NTHi or not by hpd#3 RT-PCR) were used to develop and validate the PCR-HRM assay. The Western Australian isolates were specifically chosen to represent NTHi, H. haemolyticus, and ambiguous strains, while the Northern Territory isolates were a sequential selection of clinical isolates selected to validate the method. Isolates were identified by colonial morphology, X and V growth factor dependence, and lack of reaction with capsular antisera using the Phadebact Haemophilus coagglutination test, and genomic DNA (gDNA) was isolated as previously described (16). gDNA was eluted in 100 μl sterile water and either used immediately or stored in aliquots at −20°C.
We assessed published DNA sequences for the gene encoding protein D, hpd, from NTHi and H. haemolyticus (13) and identified a hypervariable region (bp 580 to 1010) with potential discriminatory power. This region was amplified from the gDNA of 31 diverse strains using primers hpd1 fwd (5′-CAAAGTGTTGAAAAAATATGGCTATGA-3′) and hpd1 rev (5′-GTTGCACCTGATTTATTCAATAATGC-3′). Amplicons were then sequenced by the Australian Genome Research Facility using primers hpd2 fwd (5′-TTGCCTGGTTTAGATTGTTC-3′) and hpd2 rev (5′-GTTCAATTAGTGGCTTATACGG-3′) and standard DNA sequencing techniques (18). The hpd1 amplification primers were used for sequencing in cases where hpd2 sequencing reactions failed.
Alignment of our and all published hpd sequences identified polymorphic regions suitable for differentiating NTHi from H. haemolyticus. PCR primers were designed to amplify a 50-bp product containing two nucleotide polymorphisms for differentiation of NTHi (A and T) from H. haemolyticus (G and G) using high-resolution melt (HRM) technology (Table 1). Degeneracies in the reverse primer were necessary to ensure hpd amplification in both NTHi and H. haemolyticus. The BLAST function (NCBI) was used to rule out primer sequence similarity with other species of respiratory bacteria.
TABLE 1.
Variant NTHi and H. haemolyticus sequences amplified by hpd HRM primers
| Identifier | Species | 50-bp sequence (5′ to 3′) | % GC | Curve |
|---|---|---|---|---|
| Hi86 | NTHi | ATGGATTTGAAATTAGTTCAATTAATTGCTTATACAGATTGGAAAGAAAC | 26 | A |
| H17 | NTHi | ATGGATTTGAAATTAGTTCAATTAATTGCTTATACAGACTGGAAAGAAAC | 28 | B |
| H4 | H. haemolyticus | ATGGATTTGAAATTAGTTCAATTAGTGGCTTATACAGATTGGAAAGAAAC | 30 | C |
| 33390 | H. haemolyticus | ATGGATTTGAAATTAGTTCAATTAGTGGCTTATACGGATTGGAAAGAAAC | 32 | D |
| H9 | H. haemolyticus | ATGGATTTGAAATTAGTTCAATTAGTGGCTTATACTGACTGGCAAGAAAC | 34 | E |
| H12 | H. haemolyticus | ATGGATTTGAAACTAGTTCAATTAGTGGCTTATACGGATTGGAAAGAAAC | 34 | E |
| H47 | H. haemolyticus | ATGGATTTGAAATTAGTTCAATTAGTGGCTTATACTGACTGGCATGAAAC | 34 | E |
| H16 | H. haemolyticus | ATGGATTTGAAACTAGTTCAATTAGTGGCTTATACTGACTGGCATGAAAC | 36 | F |
| Forward primer | hpd HRM FWD | ATGGATTTGAAATTAGTTCAATTA | ||
| Reverse primer | hpd HRM REV | CGAATATGHCTRACCKTWCTTTG |
Real time-PCR and the HRM assay were performed on the Rotorgene 6000 system (Corbett Life Science). Reaction mixtures contained 5 μl of 2× SensiMix SYBR green (Bioline), 100 nM (each) primer, and 1 μl of gDNA in a total volume of 10 μl. To reduce sample preparation time, the PCR-HRM method was also tested using 1 μl of a colony boil preparation rather than gDNA. Two to three Haemophilus colonies from a subset of isolates were placed into 200 μl sterile water, heated at 100°C for 10 min, placed on ice, and centrifuged, and 1 μl of supernatant was used in the PCR-HRM assay. Cycling conditions were 50°C for 2 min, 95°C for 2 min, and then 40 cycles of 95°C for 15 s, 66°C for 15 s, and 70°C for 30 s. The HRM assay was carried out from 65°C to 80°C in 0.1°C increments for 2 s each. Raw HRM curves were normalized in the region of 64.5 to 79°C prior to analysis using the Rotorgene 6000 software v1.7. All samples were run in duplicate and were required to be within 0.5 cycles.
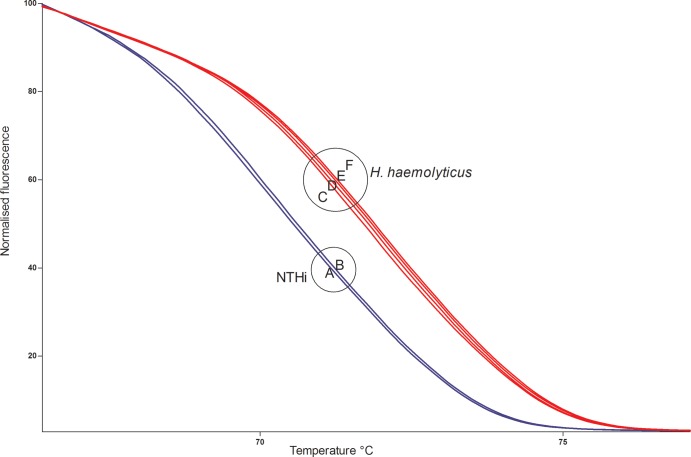
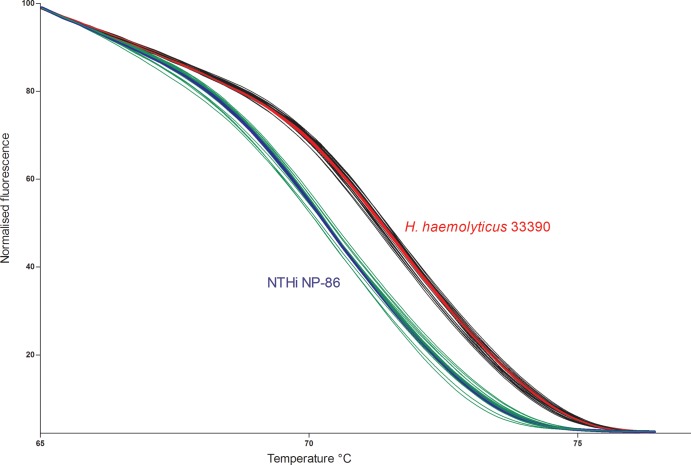
Based on all available sequences for the 50-bp amplicon of the hpd gene and the assumption that GC content defines the melting temperature (19), 6 different melt curves were predicted (Table 1, curves A to F). The PCR-HRM assay was conducted on all 31 sequenced isolates, and curves representing each of the 6 unique sequence types are shown in Fig. 1. The largest temperature demarcation occurred between NTHi and H. haemolyticus isolates, as would be predicted by the central A-to-G and T-to-G single nucleotide polymorphisms (SNPs). The PCR-HRM assay was then conducted on gDNA of the remaining 180 isolates (29 Western Australian and 151 Northern Territory isolates). Figure 2 shows the melt curves from 19 representative isolates tested (including the 2 reference strains). One hundred sixty-one of the 180 isolates clustered with the melt curves for either NTHi or H. haemolyticus reference strains. Isolates were classified as NTHi or H. haemolyticus based on the reference strain with which they clustered. Compared with the hpd#3 RT-PCR assay, the PCR-HRM assay had 96% sensitivity and 92% specificity (Table 2). This was based on the assumption that for microbiologically identified Haemophilus isolates, a positive hpd#3 RT-PCR assay confirmed H. influenzae and that an hpd#3 RT-PCR-negative result identified presumptive H. haemolyticus. The PCR-HRM assay also allowed the clear classification of strains that were previously ambiguous by 16S rRNA gene PCR. A comparison of the PCR-HRM assay on gDNA versus colony boils demonstrated that both DNA preparation methods worked equally well; the colony boil method is now standard for our studies.
FIG 1.
Normalized PCR-HRM curves of NTHi and H. haemolyticus variants. NTHi amplicons encompass an internal ATT and melt earlier (blue) than do H. haemolyticus amplicons, which encompass an internal GTG (red). A to F relate to curve types in Table 1.
FIG 2.
Separation of PCR-HRM curves from 8 H. haemolyticus (black) and 9 NTHi (green) isolates. All H. haemolyticus isolates align with the reference H. haemolyticus strain ATCC 33390, shown in red, and NTHi isolates align with NTHi reference strain 86-028NP, shown in blue. Duplicate reactions are displayed.
TABLE 2.
Comparison of sensitivity and specificity for molecular detection of suspected H. influenzae coloniesa
| Assay for H. influenzae detection | TP/TP + TN | % sensitivity | 95% CI | TN/TN + FP | % specificity | 95% CI |
|---|---|---|---|---|---|---|
| hpd#3 assay | 134/138 | 97.1 | 92–99 | 48/54 | 88.8 | 77–95 |
| hpd HRM assay | 134/140 | 95.7 | 90–98 | 48/52 | 92.3 | 81–97 |
Abbreviations: TP, number of true positives; TN, number of true negatives; FP, number of false positives; CI, confidence interval.
Nineteen Haemophilus isolates (4 ambiguous Western Australian and 15 Northern Territory isolates that were all X and V factor dependent) could not be amplified and were also negative by the hpd#3 RT-PCR assay. The failure to amplify suggests significant variation or absence of the hpd gene. While this is a limitation of the current assay, the 4 ambiguous Western Australian isolates could not be classified using other PCR targets (ompP2, ompP6, lgtC, 16S rRNA, fucK, and iga [11]) and are a reminder that complete species differentiation by PCR will likely remain elusive.
Design of a conserved reverse primer downstream from the chosen discriminatory SNPs was not possible for amplification of both Haemophilus species. Therefore, a degenerate primer which could add complexity to the amplicon melt profile was designed; that is, amplicon variation may widen the area at which species-specific curves fall, potentially obscuring species differentiation. To gauge this potential effect, we conducted the PCR-HRM assay on representatives of each amplicon variant identified from the sequencing. There was 1°C separation between the highest melt curve for the NTHi variants (74.6°C) and the lowest melt curve for H. haemolyticus variants (75.6°C). Furthermore, as shown in Fig. 1 and 2, the dichotomous separation of isolates by HRM was almost completely explained by the GC differences of the internal SNPs with seemingly little influence by GC variation in the primer region. In short, variation in the primer region had little effect on the melting temperature and no effect on assay outcome. This may be the result of a primer binding bias within the degenerate primer pool despite the stringency of the relatively high annealing temperature used for this assay. Furthermore, despite the potential for overlap in GC content theoretically afforded by the degenerate primer, this has not been observed.
NTHi has a significant role in morbidity, and thus, accurate surveillance is required for determining vaccine efficacy (i.e., the impact of Synflorix) and monitoring antibiotic resistance. The hpd#3 RT-PCR assay (13) is useful for discriminating H. influenzae from closely related Haemophilus species and can be applied to swabs directly. However, in the context of routine microbiological identification of NTHi, the PCR-HRM assay is rapid (colony boil preparations), accurate, and inexpensive (no probe required) and has potential for high-throughput analysis. It also has the added advantage of identifying H. haemolyticus. In summary, our PCR-HRM assay allows discrimination of NTHi from H. haemolyticus and has excellent concordance with the hpd#3 RT-PCR assay.
Nucleotide sequence accession numbers.
The hpd sequences were deposited in GenBank under accession numbers KF048057 to KF048087.
ACKNOWLEDGMENTS
We thank the families who participated in these studies; staff from the Child Health Respiratory Group (Menzies School of Health Research); the Vaccine Trials Group (Telethon Institute for Child Health Research, Centre for Child Health Research); McCourt Street Day surgery, Subiaco; Colin Street Day surgery, West Perth; St. John of God Hospital, Subiaco and Murdoch; and Jacinta Bowman at PathWest Diagnostics for provision of clinical specimens.
This study was supported by NHMRC grants 1023781, 1011172, and NHMRC CRE in Lung Health of Aboriginal and Torres Strait Islander Children (1040830). J.P. is supported by an NHMRC Australian Postgraduate Scholarship, a University of Western Australia Top-Up Scholarship, and a Princess Margaret Hospital Foundation PhD Supplementary Scholarship. M.J.B. is supported by NHMRC Dora Lush Postgraduate Scholarship 1017225. K.M.H. is supported by NHMRC Gustav Nossal Postgraduate Scholarship 1038072. K.M.H. and M.J.B. are also supported by Douglas and Lola Douglas Scholarships. H.S.-V. is supported by NHMRC Career Development Fellowship 1024175.
Footnotes
Published ahead of print 4 December 2013
REFERENCES
- 1.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr. Infect. Dis. J. 28:43–48. 10.1097/INF.0b013e318184dba2 [DOI] [PubMed] [Google Scholar]
- 2.Thanavala Y, Lugade AA. 2011. Role of nontypeable Haemophilus influenzae in otitis media and chronic obstructive pulmonary disease. Adv. Otorhinolaryngol. 72:170–175. 10.1159/000324785 [DOI] [PubMed] [Google Scholar]
- 3.Hare KM, Binks MJ, Grimwood K, Chang AB, Leach AJ, Smith-Vaughan H. 2012. Culture and PCR detection of Haemophilus influenzae and Haemophilus haemolyticus in Australian indigenous children with bronchiectasis. J. Clin. Microbiol. 50:2444–2445. 10.1128/JCM.00566-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Wessel K, Rodenburg GD, Veenhoven RH, Spanjaard L, van der Ende A, Sanders EA. 2011. Nontypeable Haemophilus influenzae invasive disease in The Netherlands: a retrospective surveillance study 2001–2008. Clin. Infect. Dis. 53:e1–e7. 10.1093/cid/cir268 [DOI] [PubMed] [Google Scholar]
- 5.Gkentzi D, Slack MP, Ladhani SN. 2012. The burden of nonencapsulated Haemophilus influenzae in children and potential for prevention. Curr. Opin. Infect. Dis. 25:266–272. 10.1097/QCO.0b013e32835310a4 [DOI] [PubMed] [Google Scholar]
- 6.MacNeil JR, Cohn AC, Farley M, Mair R, Baumbach J, Bennett N, Gershman K, Harrison LH, Lynfield R, Petit S, Reingold A, Schaffner W, Thomas A, Coronado F, Zell ER, Mayer LW, Clark TA, Messonnier NE. 2011. Current epidemiology and trends in invasive Haemophilus influenzae disease—United States, 1989–2008. Clin. Infect. Dis. 53:1230–1236. 10.1093/cid/cir735 [DOI] [PubMed] [Google Scholar]
- 7.Sandstedt SA, Zhang L, Patel M, McCrea KW, Qin Z, Marrs CF, Gilsdorf JR. 2008. Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. J. Microbiol. Methods 75:369–371. 10.1016/j.mimet.2008.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morton DJ, Hempel RJ, Whitby PW, Seale TW, Stull TL. 2012. An invasive Haemophilus haemolyticus isolate. J. Clin. Microbiol. 50:1502–1503. 10.1128/JCM.06688-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson R, Wang X, Briere EC, Katz LS, Cohn AC, Clark TA, Messonnier NE, Mayer LW. 2012. Haemophilus haemolyticus isolates causing clinical disease. J. Clin. Microbiol. 50:2462–2465. 10.1128/JCM.06575-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81–89. 10.1086/509824 [DOI] [PubMed] [Google Scholar]
- 11.Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, Unrath WC, Campos J, Crook DW, Farley MM, Henderson FW, Jacobs RF, Muhlemann K, Satola SW, van Alphen L, Golomb M, Smith AL. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 73:5853–5863. 10.1128/IAI.73.9.5853-5863.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prymula R, Peeters P, Chrobok V, Kriz P, Novakova E, Kaliskova E, Kohl I, Lommel P, Poolman J, Prieels JP, Schuerman L. 2006. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study. Lancet 367:740–748. 10.1016/S0140-6736(06)68304-9 [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Mair R, Hatcher C, Theodore MJ, Edmond K, Wu HM, Harcourt BH, Carvalho MDGS, Pimenta F, Nymadawa P, Altantsetseg D, Kirsch M, Satola SW, Cohn A, Messonnier NE, Mayer LW. 2011. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int. J. Med. Microbiol. 301:303–309. 10.1016/j.ijmm.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 14.Binks MJ, Temple B, Kirkham LA, Wiertsema SP, Dunne EM, Richmond PC, Marsh RL, Leach AJ, Smith-Vaughan HC. 2012. Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS One 7:e34083. 10.1371/journal.pone.0034083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakaletz LO, Tallan BM, Hoepf T, DeMaria TF, Birck HG, Lim DJ. 1988. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkham LA, Wiertsema SP, Mowe EN, Bowman JM, Riley TV, Richmond PC. 2010. Nasopharyngeal carriage of Haemophilus haemolyticus in otitis-prone and healthy children. J. Clin. Microbiol. 48:2557–2559. 10.1128/JCM.00069-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prymula R, Kriz P, Kaliskova E, Pascal T, Poolman J, Schuerman L. 2009. Effect of vaccination with pneumococcal capsular polysaccharides conjugated to Haemophilus influenzae-derived protein D on nasopharyngeal carriage of Streptococcus pneumoniae and H. influenzae in children under 2 years of age. Vaccine 28:71–78. 10.1016/j.vaccine.2009.09.113 [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467. 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson P, Tong SY, Bell JM, Turnidge JD, Giffard PM. 2012. Minim typing—a rapid and low cost MLST based typing tool for Klebsiella pneumoniae. PLoS One 7:e33530. 10.1371/journal.pone.0033530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prymula R, Hanovcova I, Splino M, Kriz P, Motlova J, Lebedova V, Lommel P, Kaliskova E, Pascal T, Borys D, Schuerman L. 2011. Impact of the 10-valent pneumococcal non-typeable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) on bacterial nasopharyngeal carriage. Vaccine 29:1959–1967. 10.1016/j.vaccine.2010.12.086 [DOI] [PubMed] [Google Scholar]
- 21.Van den Bergh MR, Spijkerman J, Swinnen KM, Francois NA, Pascal TG, Borys D, Schuerman L, Ijzerman EP, Bruin JP, van der Ende A, Veenhoven RH, Sanders EA. 2013. Effects of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugate vaccine on nasopharyngeal bacterial colonization in young children: a randomized controlled trial. Clin. Infect. Dis. 56:e30–e39. 10.1093/cid/cis922 [DOI] [PMC free article] [PubMed] [Google Scholar]