Abstract
This study aimed to develop a loop-mediated isothermal amplification (LAMP) method for the rapid detection of Arcobacter species. Specific primers targeting the 23S ribosomal RNA gene were used to detect Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii. The specificity of the LAMP primer set was assessed using DNA samples from a panel of Arcobacter and Campylobacter species, and the sensitivity was determined using serial dilutions of Arcobacter species cultures. LAMP showed a 10- to 1,000-fold-higher sensitivity than multiplex PCR, with a detection limit of 2 to 20 CFU per reaction in vitro. Whereas multiplex PCR showed cross-reactivity with Campylobacter species, the LAMP method developed in this study was more sensitive and reliable than conventional PCR or multiplex PCR for the detection of Arcobacter species.
INTRODUCTION
Arcobacter species are Gram-negative, non-spore-forming, motile, spiral-shaped bacteria that require a low-oxygen environment and usually blood-based or complex media for their initial culture (1). Arcobacter organisms were first isolated as aerotolerant Campylobacter-like microorganisms from aborted bovine and pig fetuses in 1977 and were assigned to the family Campylobacteraceae (2, 3). In 1991, the aerotolerant campylobacters were reclassified, giving rise to the new genus Arcobacter, belonging to the class Epsilonproteobacteria (4, 5). Arcobacter species have high oxygen tolerance and can grow between 15 and 30°C, whereas Campylobacter species can grow only at 37°C and require a strict microaerophilic environment (5). Arcobacter nitrofigilis and Arcobacter cryaerophilus were previously included in the genus Campylobacter and later reassigned to the genus Arcobacter. In the past decade, the genus Arcobacter has been expanded to include 14 species with the following chronological order of discovery: Arcobacter butzleri, Arcobacter skirrowii, Arcobacter cibarius, Arcobacter halophilus, Arcobacter mytili, Arcobacter thereius, Arcobacter marinus, Arcobacter trophiarum, Arcobacter defluvii, Arcobacter molluscorum, Arcobacter bivalviorum, and Arcobacter venerupis (5–15).
Among the Arcobacter species described to date, A. butzleri, A. skirrowii, and A. cryaerophilus are considered human pathogens causing gastroenteritis or bacteremia (16). The clinical features of A. butzleri infection include watery and persistent diarrhea with abdominal pain (17). Presently, Arcobacter species are a serious potential concern in food safety because they can contaminate animal-origin foods and cause human diseases (17, 18). A. butzleri, A. cryaerophilus, and A. skirrowii were detected first in chicken and later in pork, beef, untreated water, and contaminated foods (19).
Selective culture methods for isolating Arcobacter species have been developed and compared in previous studies (20–23). PCR, multiplex PCR, real-time PCR, and multiplex real-time PCR for detecting Arcobacter species were developed, targeting the 16S or 23S rRNA, rpoB-rpoC, and gyrA genes (24–26). Loop-mediated isothermal amplification (LAMP) was recently developed for on-the-spot inspections (27). Importantly, this approach requires only simple equipment, such as a heating block and water bath. The method uses four or six different primers specifically designed to recognize six distinct regions in the target sequence (Eiken Chemical Co., Ltd.) and uses Bst DNA polymerase to interact with the template DNA during DNA replication. Bst DNA polymerase has a very high activity; thus, vast amounts of high-molecular-weight DNA can be produced within a short time (28), with the reaction proceeding at a constant temperature. LAMP was first introduced in 2000, and it has been successfully used for the detection of many pathogens (28–31).
Therefore, this study aimed to develop a rapid and sensitive LAMP technique for the detection of Arcobacter species and to compare the sensitivities of LAMP and multiplex PCR in the detection of Arcobacter in broth and chicken.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
Forty-two Arcobacter species and 27 food-borne pathogens were used in this study (Table 1). Arcobacter species were cultured using Arcobacter selection broth (Oxoid, Basingstoke, United Kingdom) or with added agar technical (Oxoid, United Kingdom) at 37°C for 48 h. Other pathogens were cultured in brain heart infusion broth at 37°C for 24 h. Bacterial DNA was extracted with an AccuPrep genomic DNA extraction kit (Bioneer, Daejeon, South Korea).
TABLE 1.
Reference strains for the sensitivity and specificity tests of LAMP used in this study
| Species | Strain(s)a |
|---|---|
| Arcobacter butzleri | ATCC 49616; CAU 076046, 076048, 076050, 080083, 080084, 080086, 080089, 080090, 080092, 080097, 080098, 080099, 080100, 080101, 080105, 080137, 080138, 080142, 080145, 080150, 080159, 080161, 080163, 080165, 080166, 080169, 080170, 080171, 080173, 080174, 080175, 080176, 080177, 080178, 080179, 080180 |
| Arcobacter cryaerophilus | ATCC 43158 |
| Arcobacter skirrowii | ATCC 51132; CAU 090017, 090019, 090023 |
| Escherichia coli O157:H7 | ATCC 43889, 43890 |
| Listeria monocytogenes | ATCC 15313, 19117, 19114 |
| Salmonella enterica serovar Typhimurium | ATCC 43971, 19585 |
| Staphylococcus aureus | KACC 10778, 10768, 10196; ATCC 49444,12600 |
| Vibrio parahaemolyticus | KCCM 4664, ATCC 43996, KCTC 2471 |
| Bacillus cereus | ATCC 13061, 10879 |
| Campylobacter jejuni | ATCC 33291; NCCP 10402, 10276, 11211, 10672 |
| Campylobacter coli | NCCP 11191 |
| Helicobacter pylori | ATCC 43504, 49503; KTCC B0233, B0322 |
ATCC, American Type Culture Collection; CAU, Chung-Ang University; KACC, Korean Agricultural Culture Collection; KCCM, Korean Culture Center of Microorganisms; KCTC, Korean Collection of Type Culture; NCCP, National Culture Collection for Pathogens.
Multiplex PCR for Arcobacter species.
As multiplex PCR for Arcobacter species was widely used in previous studies (23, 26, 32, 33), the primers ARCO (5′-CCT GGA CTT GAC ATA GTA AGA ATG A-3′), BUTZ (5′-CGT ATT CAC CGT AGC ATA GC-3′), SKIR (5′-GGC GAT TTA CTG GAA CAC A-3′), CRY1 (5′-TGC TGG AGC GGA TAG AAG TA-3′), and CRY2 (5′-AAC AAC CTA CGT CCT TCG AC-3′) were used to detect Arcobacter species in this study. The multiplex PCR mixture contained 1 mM concentrations of deoxynucleoside triphosphate s (dNTPs) (Bioneer), 2 μl of 10× reaction buffer (Tris [pH 9.0], 15 mM MgCl2) (Bioneer), a 1.25 μM concentration of each primer, 0.05 U/μl Top polymerase (Bioneer), and 2 μl of DNA template, with deionized water added to bring the total reaction volume to 20 μl. Multiplex PCR was performed on an MJ mini personal thermal cycler (Bio-Rad, Mexico) with the following conditions: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 61°C for 45 s, extension at 72°C for 45 s, and final extension at 72°C for 10 min. PCR products were visualized on a 1.2% agarose gel using polyacrylamide gel electrophoresis (Bioneer).
LAMP for Arcobacter species.
LAMP primers specific for the A. butzleri 23S gene (GenBank no. FN600698.1) were designed by the online LAMP primer design software PrimerExplorer V4 (http://primerexplorer.jp) and were as follows: F3, 5′-ACT GTG ACA ACC AGG AGG TT-3′; B3, 5′-TCC AAC GCT CCT TAC CGG-3′; FIP, 5′-CGC GCA GAA TCA CTA GAC CAG TGG CTT AGA AGC AGC CAT CC-3′; and BIP, 5′-AAC GGG GCT AAG ATG TAC ACC GAC GCT GAA TAG AAC GCT CTC-3′. LAMP was carried out in a total reaction volume of 25 μl with a final concentration of 2 μM for both FIP and BIP and 0.2 μM for both F3 and B3, 0.8 mM concentrations of dNTPs (Bioneer), 2.5 μl of 1× ThermoPol reaction buffer [20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100] (New England BioLabs Inc., Ipswich, United Kingdom), 8 U of Bst DNA polymerase large fragment (New England BioLabs Inc.), 2 μl of DNA template, and diethylpyrocarbonate (DEPC)-treated deionized water to bring the reaction volume to 25 μl. The reaction mixture was incubated at 61°C for 50 min and heated at 80°C for 5 min to terminate the reaction. The LAMP reactions were conducted using an MJ mini personal thermal cycler (Bio-Rad) for temperature control.
Sensitivity and specificity of LAMP and multiplex PCR.
The specificity and sensitivity of the LAMP and multiplex PCR methods used in this study were determined using reference strains, including A. butzleri ATCC 49616, A. skirrowii ATCC 51132, and A. cryaerophilus ATCC 43158 (Table 1). To determine the detection limit of LAMP for Arcobacter species in chicken, 10 g of chicken skin and meat not containing Arcobacter species was spiked with 1 ml of A. butzleri, A. skirrowii, or A. cryaerophilus cultures at 104 to 108 CFU/ml. Each sample was homogenized for 2 min in a stomacher bag with 90 ml of 0.1% peptone water. One milliliter of homogenate was transferred to a 1.5-ml Eppendorf tube. The samples were centrifuged at 13,000 rpm for 15 min. The supernatant was discarded, and the bacterial pellet was resuspended in 100 μl of sterile water and boiled for 10 min to lyse the cells. The suspension was centrifuged at 8,000 rpm for 2 min, and the supernatant was used as a DNA template. From each sample, 2 μl of the supernatant was used for LAMP and PCR (26).
To confirm the LAMP amplification product specificity, restriction fragment length polymorphism (RFLP) analysis was performed with the restriction enzyme HindIII. The specific restriction sites in the LAMP products were analyzed and selected with NEB cutter V2.0 (http://tools.neb.com/NEBcutter2). Briefly, the DNA products amplified by LAMP were digested with 1 U/μl HindIII (Promega, Madison, WI) following the manufacturer's standard protocol. HindIII can specifically digest the DNA sequence 5′-AAGCTT-3′. The final digestion products were expected to be 147, 198, and 249 bp.
Experimental design.
Thirty fresh whole chickens were purchased from several local supermarkets in Gyeonggi province. The chickens were kept at 4°C and analyzed within 12 h of purchase. Five grams of skin from each chicken was collected in a stomacher bag with 45 ml of 0.1% peptone water and homogenized for 2 min. From each sample, bacterial DNA was extracted by boiling for 10 min. Another 1-ml homogenate of each sample was transferred to 9 ml of Arcobacter broth containing C.A.T. supplement (Oxoid) and incubated at 30°C in an incubator. After 3, 6, and 24 h of incubation, the DNA was extracted from 1 ml of culture broth. After 24 h of incubation, the cultured broth was filtered with a 0.45-μm sterile syringe filter and inoculated on an Arcobacter selective agar. After an additional 48 h of incubation at 30°C, single colonies were isolated for further analysis.
For accurate comparison, the sensitivities of multiplex PCR and LAMP were compared to that of a conventional PCR method developed in a previous study (34). The primers Arc1 (5′-AGA ACG GGT TAT AGC TTG CTA T-3′) and Arc2 (5′-GAT ACA ATA CAG GCT AAT CTC T-3′) were used for conventional PCR, which generated a 181-bp DNA product for A. butzleri, A. cryaerophilus, and A. skirrowii (34). Bacterial culture, conventional PCR, multiplex PCR, and LAMP were compared for the detection of Arcobacter spp. in the cultured samples at 0, 3, 6, and 24 h of incubation.
Statistical analysis.
Fisher's exact test was used to analyze the comparative detection of conventional PCR, multiplex PCR, and LAMP in experimentally A. butzleri-challenged samples at each time point. Analyses were carried out using SAS 9.2 software (SAS Institute Inc., Cary, NC). P values less than 0.05 were considered statistically significant.
RESULTS
Specificity of LAMP and multiplex PCR.
To evaluate the specificity of the primer set, we used multiplex PCR and LAMP to detect DNAs extracted from 42 bacterial strains of Arcobacter and 27 strains of other food-borne bacteria (Table 2). Multiplex PCR detected and differentiated A. butzleri, A. skirrowii, and A. cryaerophilus in the 42 Arcobacter strains used in this study. In addition, multiplex PCR detected 5 strains of Campylobacter jejuni and 1 strain of Campylobacter coli as false positives for Arcobacter species. Other food-borne pathogens were not detected by multiplex PCR. The size of the DNA product amplified from Campylobacter spp. was matched with the expected 641-bp amplicon from A. skirrowii (Fig. 1A).
TABLE 2.
LAMP and multiplex PCR detection with bacterial DNA isolated from reference strainsa
| Species | No. positive for Arcobacter DNA/no. tested by: |
|
|---|---|---|
| LAMP | Multiplex PCR | |
| A. butzleri | 37/37 | 37/37 |
| A. skirrowii | 4/4 | 4/4 |
| A. cryaerophilus | 1/1 | 1/1 |
| C. jejuni | 1/5 | 5/5 |
| C. coli | 0/1 | 1/1 |
| H. pylori | 0/4 | 0/4 |
| E. coli O157:H7 | 0/2 | 0/2 |
| L. monocytogenes | 0/3 | 0/3 |
| S. Typhimurium | 0/2 | 0/2 |
| S. aureus | 0/5 | 0/5 |
| V. parahaemolyticus | 0/3 | 0/3 |
| B. cereus | 0/2 | 0/2 |
The multiplex PCR developed by Houf et al. (26) to detect and differentiate Arcobacter species.
FIG 1.
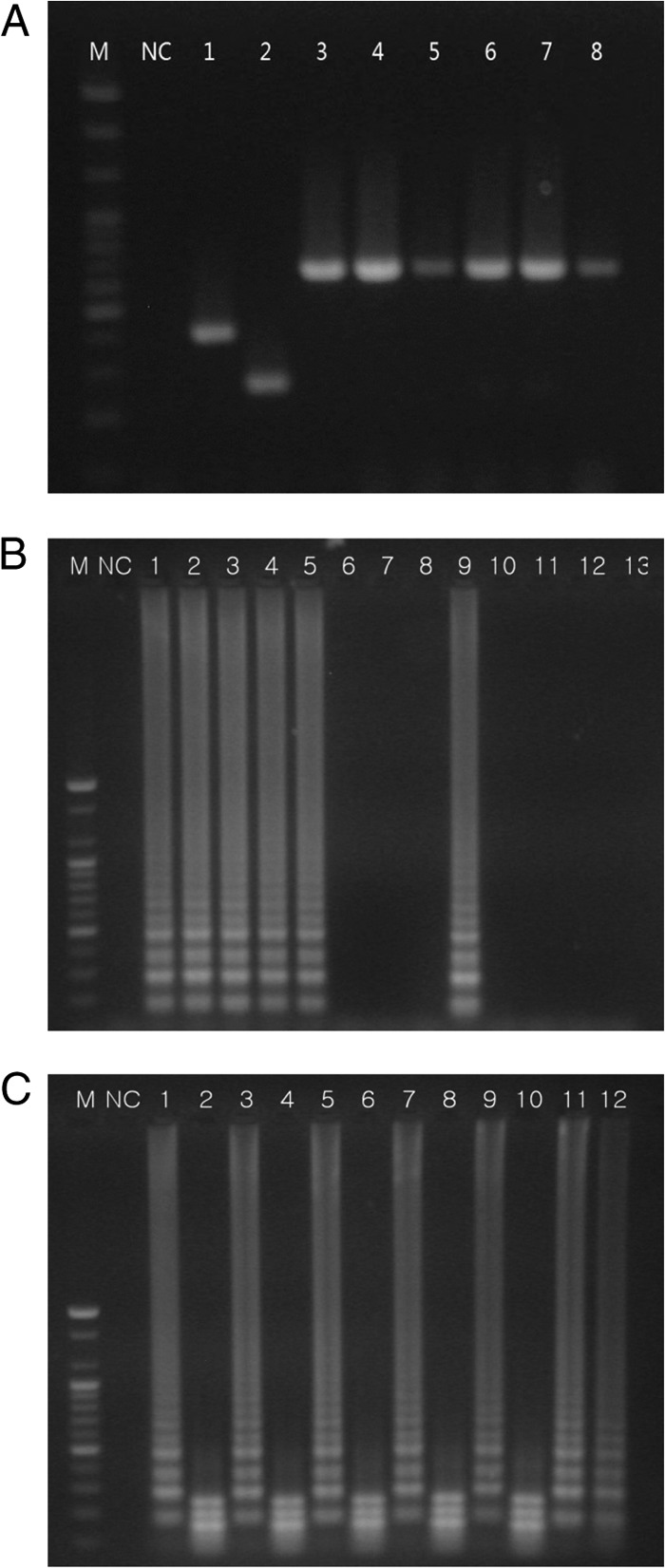
Specificity of multiplex PCR and LAMP. (A) PCR. Lane M, 100-bp DNA marker; lane NC, negative control; lane 1, A. butzleri ATCC 49616; lane 2, A. cryaerophilus ATCC 43152; lane 3, A. skirrowii ATCC 51132; lane 4, C. jejuni ATCC 33291; lane 5, C. jejuni NCCP 10402; lane 6, C. jejuni NCCP 10276; lane 7, C. jejuni NCCP 10672; lane 8, C. coli NCCP 11191. (B) LAMP. Lane M, 100-bp DNA marker; lane NC, negative control; lane 1, A. butzleri ATCC 49616; lane 2, A. butzleri CAU 076046; lane 3, A. skirrowii ATCC 51132; lane 4, A. skirrowii CAU 090029; lane 5, A. cryaerophilus ATCC 43152; lane 6, C. jejuni ATCC 33291; lane 7, C. jejuni NCCP 10402; lane 8, C. jejuni NCCP 10276; lane 9, C. jejuni NCCP 10672; lane 10, C. jejuni NCCP 11211; lane 11, C. coli NCCP 11191; lane 12, H. pylori ATCC 43504; lane 13, E. coli O157:H7 ATCC 43889. (C) Restriction fragment length polymorphism of LAMP products. Lane M, 100-bp DNA marker; lane NC, negative control; lane 1, A. butzleri ATCC 49616 LAMP; lane 2, A. butzleri ATCC 49616 LAMP product digested with HindIII; lane 3, A. butzleri CAU 076046 LAMP; lane 4, A. butzleri CAU 076046 LAMP product digested with HindIII; lane 5, A. skirrowii ATCC 51132 LAMP; lane 6, A. skirrowii ATCC 51132 LAMP product digested with HindIII; lane 7, A. skirrowii CAU 090017 LAMP; lane 8, A. skirrowii CAU 090017 LAMP product digested with HindIII; lane 9, A. cryaerophilus ATCC 43152 LAMP; lane 10, A. cryaerophilus ATCC 43152 LAMP product digested with HindIII; lane 11, C. jejuni NCCP 10672 LAMP; lane 12, C. jejuni NCCP 10672 LAMP product digested with HindIII.
The LAMP method developed in this study detected 37 A. butzleri, 4 A. skirrowii, and 1 A. cryaerophilus strains. The other food-borne pathogens, except C. jejuni NCCP 10672, were not amplified by LAMP (Table 2) (Fig. 1B). RFLP analysis of the LAMP products amplified from 42 Arcobacter strains, including 3 reference strains, yielded three fragment sizes: 147, 198, and 249 bp (Fig. 1C). The LAMP products of C. jejuni NCCP 10672 were not digested with HindIII (Fig. 1C). The specificities of multiplex PCR and LAMP are summarized in Table 2.
Sensitivity of LAMP and multiplex PCR.
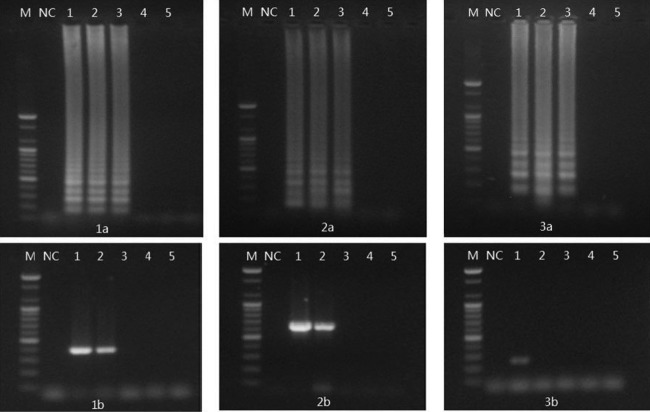
The sensitivities of LAMP and multiplex PCR were determined by serial 10-fold dilutions of Arcobacter reference strains (Fig. 2). The detection limit of LAMP in cultured broth was 2 CFU per reaction for A. butzleri ATCC 49616 and A. skirrowii ATCC 51132, and 20 CFU per reaction for A. cryaerophilus ATCC 43158. However, the detection limit of multiplex PCR in cultured broth was 20 CFU per reaction for A. butzleri, 20 CFU per reaction for A. skirrowii, and 2 × 104 CFU per reaction for A. cryaerophilus. The detection sensitivity of LAMP in cultured broth was 10-fold higher for A. butzleri and A. skirrowii and 1,000-fold higher for A. cryaerophilus than that of multiplex PCR.
FIG 2.
Comparison of LAMP and multiplex PCR for the detection of Arcobacter species in broth. (Panels 1a and 1b) A. butzleri ATCC 49616; (panels 2a and 2b) A. skirrowii ATCC 51132; (panels 3a and 3b) A. cryaerophilus ATCC 43152. Lane M, 100-bp DNA marker; lane NC, negative control; lane 1, 2 × 105 CFU per reaction; lane 2, 2 × 104 CFU per reaction; lane 3, 2 × 103 CFU per reaction; lane 4, 2 × 102 CFU per reaction; lane 5, 2 × 101 CFU per reaction; lane 6, 2 CFU per reaction; lane 7, 2 × 10−1 CFU per reaction; lane 8, 2 × 10−2 CFU per reaction.
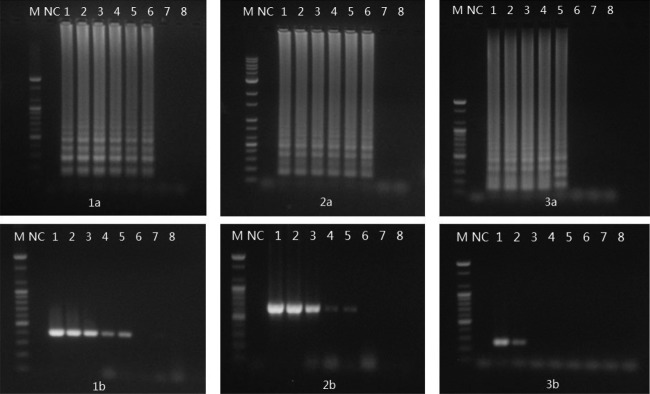
The detection limits of LAMP and multiplex PCR were also determined in chicken spiked with an Arcobacter reference strains (Fig. 3). The detection limit of LAMP in chicken was 2 × 102 CFU per reaction for A. butzleri ATCC 49616, A. skirrowii ATCC 51132, and A. cryaerophilus ATCC 43158. However, the detection limit of multiplex PCR in chicken was 2 × 103 CFU per reaction for A. butzleri and A. skirrowii and 2 × 104 CFU for A. cryaerophilus. The detection sensitivity of LAMP for Arcobacter species was 10- to 100-fold higher than that of multiplex PCR.
FIG 3.
Comparison of LAMP and multiplex PCR for the detection of Arcobacter species in chicken. (Panels 1a and 1b) A. butzleri ATCC 49616; (panels 2a and 2b) A. skirrowii ATCC 51132; (panels 3a and 3b) A. cryaerophilus ATCC 43152. Lane M, 100-bp DNA marker; lane NC, negative control; lane 1, 2 × 104 CFU per reaction; lane 2, 2 × 103 CFU per reaction; lane 3, 2 × 102 CFU per reaction; lane 4, 2 × 101 CFU per reaction; lane 5, 2 CFU per reaction.
Detection of Arcobacter species in chickens from retail markets.
Ten (33.3%) Arcobacter butzleri and three (10.0%) A. cryaerophilus strains were isolated by bacterial culture using Arcobacter selective medium from 30 chickens purchased from retail markets. Culture homogenates from the 30 chicken samples were analyzed by conventional PCR, multiplex PCR, and LAMP at 0, 3, 6, and 24 h of incubation (Table 3). Although Arcobacter species were not detected in chicken homogenates without incubation by conventional PCR, homogenates incubated for 3, 6, and 24 h showed positivity for 6, 18, and 27 of 30 chickens, respectively. Whereas 9 of 30 chicken homogenates were positive for Arcobacter species by multiplex PCR at 0 h of incubation, homogenates incubated for 3, 6, and 24 h showed positivity for 19, 22, and 28 of 30 chickens, respectively. In LAMP detection, homogenates incubated for 0, 3, 6, and 24 h showed positivity for 20, 24, 27, and 28 of 30 chickens, respectively. The detection rate of LAMP was significantly higher than rates of conventional PCR and multiplex PCR at 0, 3, and 6 h incubation (P < 0.05). However, the detection rates of conventional PCR, multiplex PCR, and LAMP at 24 h incubations were not significantly different.
TABLE 3.
Comparative detection of Arcobacter species in 30 chickens purchased from retail markets
In homogenates cultured for 24 h, multiplex PCR detected both A. butzleri and A. cryaerophilus in 10 samples, and simultaneously detected A. butzleri, A. cryaerophilus, and A. cryaerophilus in five chickens. Five chickens were positive only for A. butzleri, and four chickens were positive for both A. butzleri and A. skirrowii. Two chickens were positive for either A. skirrowii or A. cryaerophilus.
DISCUSSION
Arcobacter species are considered a major public health concern (18). In severe chronic human diseases caused by Arcobacter spp., the rapid detection and treatment of Arcobacter infection are required (35). Nine A. butzleri isolates were cultured from diarrheal stool samples in Turkish hospitals, although A. butzleri was first isolated from the blood of a Korean patient with liver cirrhosis (36–38). Bacterial culture is not practical for the rapid detection of Arcobacter species because of their slow growth in Campylobacter blood-free selective medium; therefore, conventional PCR, multiplex PCR, and enterobacterial repetitive intergenic consensus (ERIC)-PCR were used in previous studies for the rapid detection of Arcobacter spp. (23, 35, 36). In this study, five C. jejuni strains and one C. coli strain were detected as false positives by multiplex PCR described previously (26). The Campylobacter species amplicons were matched with the expected size of the A. cryaerophilus amplicon. Because of a lack of a simple identification system for Arcobacter spp., multiplex PCR was widely used for the detection of Arcobacter spp. in previous studies (19, 23, 25, 26). However, the cross-reactivity of multiplex PCR with Campylobacter spp. has not been addressed thus far. A high rate of contamination with Campylobacter in chicken was also reported in a previous study (39). Thus, it is possible that the prevalence data obtained from multiplex PCR reflected Campylobacter contamination. Whereas Campylobacter spp. could be cultured only under anaerobic conditions, Arcobacter species were differentiated by bacterial culture under aerobic conditions. Further, given that bacterial isolation takes 2 to 4 days and anaerobic chamber culture requires skilled microbiologists, bacterial culture is not practical for on-the-spot inspection of Arcobacter contamination.
In the present study, whereas Campylobacter strains were detected by multiplex PCR, LAMP specifically detected only Arcobacter species except C. jejuni NCCP 10672. Because Arcobacter and Helicobacter also belong to the genus Campylobacter of the rRNA superfamily VI, 23S ribosomal RNA gene sequences show high homology between Campylobacter and Arcobacter (15). As LAMP primers were designed in 23S rRNA genes of Arcobacter species, LAMP in this study did not show the cross-reactivity with Campylobacter or Helicobacter species. Of all the reference strains tested, only C. jejuni NCCP 10672 showed a false-positive reaction by LAMP; however, the LAMP product of this strain was not digested with restriction enzyme HindIII. Thus, RFLP analysis of LAMP products clearly differentiated Arcobacter spp. and Campylobacter spp.
The detection limit of LAMP was determined both in vitro and in food. With DNA isolated from cultured bacteria, LAMP could detect as little as 2 CFU per reaction. This value is similar to those reported in previous studies, which detected 1, 20, 7.9, and 3.8 CFU per reaction for Escherichia coli O157:H7, Plesiomonas shigelloides, C. jejuni, and C. coli, respectively (30, 31, 40). The sensitivity of the LAMP method developed in this study was 10- to 1,000-fold higher than that of conventional PCR or multiplex PCR (30, 40). In concordance with the results of previous studies, LAMP showed 100-fold-greater sensitivity than conventional PCR or multiplex PCR.
Because of the high prevalence of Arcobacter in chicken meat (19), the sensitivity of LAMP for the detection of Arcobacter spp. in chicken was assessed. The detection sensitivity of LAMP was 100-fold higher than that of multiplex PCR for A. cryaerophilus and 10-fold higher for A. butzleri and A. skirrowii. Its detection limit was as low as 200 CFU per reaction in chicken spiked with Arcobacter species. According to previous studies, food components can inhibit or interfere with DNA amplification in LAMP and PCR assays for the detection of Campylobacter spp. (41). In the present study, the blood components or nucleic acids from the chicken samples were considered PCR inhibitors; thus, they may explain the difference in LAMP or PCR sensitivity between the bacterial culture and chicken samples.
This study showed that 93.3% of the chickens sampled were contaminated with Arcobacter species, with A. butzleri being the most prevalent species. Although the Arcobacter contamination rates obtained in this study were consistent with those in previous reports (23, 42), studies in the United States and Japan showed much lower rates than those reported herein (43, 44). In contrast to molecular detection, bacterial culture for Arcobacter spp. took at least 3 to 4 days and had low sensitivity. Although conventional PCR could not detect Arcobacter spp. in all samples without enrichment, the combination of bacterial culture with multiplex PCR and LAMP showed a higher detection rate. Compared with multiplex PCR, the detection rate of LAMP was significantly increased by 3 to 6 h of enrichment (P < 0.05). Thus, the LAMP assay developed in this study proved to be more rapid, specific, and sensitive than conventional PCR or multiplex PCR.
In contrast to PCR and real-time PCR, the detection time of LAMP is less than 1 h because the target gene is amplified under isothermal conditions. LAMP is more specific than PCR, because 4 or 6 primers used in LAMP increase specificity (45). Although short detection time and specificity of LAMP are the major advantages, continuous amplification of the target gene by LAMP may be confused with nonspecific PCR products on gel electrophoresis. In order to overcome this disadvantage of LAMP, alternative LAMP-based detection techniques were recently proposed (45–48). In one variation that does not use gel electrophoresis, a positive reaction was determined by Ca2+ precipitation and SYBR green fluorescent or color dye with naked-eye inspection (45, 47). In addition, real-time LAMP and probe-based LAMP have also been reported (46, 48). To enhance the specificity of LAMP, probe-based LAMP may be developed for the detection of Arcobacter in future research.
In conclusion, the LAMP assay developed in this research was able to detect Arcobacter spp. rapidly and reliably in vitro and in chicken samples. This method could be used for the rapid diagnosis of Arcobacter spp. infections in food poisoning cases or for on-the-spot inspection of slaughterhouses.
ACKNOWLEDGMENT
This research was supported by the Fishery Commercialization Technology Development Program (112089-3) of the Ministry for Food, Agriculture, Forestry, and Fisheries, Republic of Korea.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Vandamme P, De Ley J. 1991. Proposal for a new family, Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451–455. 10.1099/00207713-41-3-451 [DOI] [Google Scholar]
- 2.Ellis WA, Neill SD, O'Brien JJ, Hanna J. 1978. Isolation of Spirillum-like organisms from pig fetuses. Vet. Rec. 102:106. [DOI] [PubMed] [Google Scholar]
- 3.Ellis WA, Neill SD, O'Brien JJ, Ferguson HW, Hanna J. 1977. Isolation of Spirillum/Vibrio-like organisms from bovine fetuses. Vet. Rec. 100:451–452. 10.1136/vr.100.21.451 [DOI] [PubMed] [Google Scholar]
- 4.Neill SD, Ellis WA, O'Brien JJ. 1979. Designation of aerotolerant Campylobacter-like organisms from porcine and bovine abortions to the genus Campylobacter. Res. Vet. Sci. 27:180–186 [PubMed] [Google Scholar]
- 5.Vandamme P, Falsen E, Rossau R, Hoste B, Seqers P, Tytqat R, De Lev J. 1991. Revision of Campylobacter, Helicobacter and Wolinella taxonomy: emendation of generic description and proposal for Arcobacter gen. nov.. Int. J. Syst. Bacteriol. 41:88–103 [DOI] [PubMed] [Google Scholar]
- 6.Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ. 2009. Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int. J. Syst. Evol. Microbiol. 59:1391–1396. 10.1099/ijs.0.003749-0 [DOI] [PubMed] [Google Scholar]
- 7.Collado L, Levicán A, Perez J, Figueras MJ. 2011. Arcobacter defluvii sp. nov., isolated from sewage samples. Int. J. Syst. Evol. Microbiol. 61:2155–2161. 10.1099/ijs.0.025668-0 [DOI] [PubMed] [Google Scholar]
- 8.De Smet S, Vandamme P, De Zutter L, On S, Douidah L, Houf K. 2011. Arcobacter trophiarum sp. nov. isolated from fattening pigs. Int. J. Syst. Evol. Microbiol. 61:356–361. 10.1099/ijs.0.022665-0 [DOI] [PubMed] [Google Scholar]
- 9.Donachie SP, Bowman JP, On SL, Alam M. 2005. Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55:1271–1277. 10.1099/ijs.0.63581-0 [DOI] [PubMed] [Google Scholar]
- 10.Figueras MJ, Collado L, Levican A, Perez J, Solsona MJ, Yustes C. 2011. Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Syst. Appl. Microbiol. 34:105–109. 10.1016/j.syapm.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 11.Houf K, On SL, Coenye T, Debruyne L, De Smet S, Vandamme P. 2009. Arcobacter thereius sp. nov., isolated from pigs and ducks. Int. J. Syst. Evol. Microbiol. 59:2599–2604. 10.1099/ijs.0.006650-0 [DOI] [PubMed] [Google Scholar]
- 12.Houf K, On SL, Coenye T, Mast J, Van Hoof J, Vandamme P. 2005. Arcobacter cibarius sp. nov., isolated from broiler carcasses. Int. J. Syst. Evol. Microbiol. 55:713–717. 10.1099/ijs.0.63103-0 [DOI] [PubMed] [Google Scholar]
- 13.Kim HM, Hwang CY, Cho BC. 2010. Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60:531–536. 10.1099/ijs.0.007740-0 [DOI] [PubMed] [Google Scholar]
- 14.Levican A, Collado L, Aguilar C, Yustes C, Diéguez AL, Romalde JL, Figueras MJ. 2012. Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35:133–138. 10.1016/j.syapm.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P, Vancanneyt M, Pot B, Mels L, Hoste B, Dewettinck D, Vlaes L, van den Borre C, Higgins R, Hommez J. 1992. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42:344–356. 10.1099/00207713-42-3-344 [DOI] [PubMed] [Google Scholar]
- 16.Hsueh PR, Teng LJ, Yang PC, Wang SK, Chang SC, Ho SW, Hsieh WC, Luh KT. 1997. Bacteremia caused by Arcobacter cryaerophilus 1B. J. Clin. Microbiol. 35:489–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, Douat N, Zissis G, Butzler JP, Vandamme P. 2004. Arcobacter species in humans. Emerg Infect. Dis. 10:1863–1867. 10.3201/eid1010.040241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme P, Pugina P, Benzi G, Van Etterijck R, Vlaes L, Kesters K, Butzler JP, Lior H, Lauwers S. 1992. Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J. Clin. Microbiol. 30:2335–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho HT, Lipman LJ, Gaastra W. 2006. Arcobacter, what is known about a potential foodborne zoonotic agent. Vet. Microbiol. 115:1–13. 10.1016/j.vetmic.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 20.Atabay HI, Waino M, Madsen M. 2006. Detection and diversity of various Arcobacter species in Danish poultry. Int. J. Food Microbiol. 109:139–145. 10.1016/j.ijfoodmicro.2006.01.020 [DOI] [PubMed] [Google Scholar]
- 21.de Boer E, Tilburg JJ, Woodward DL, Lior H, Johnson WM. 1996. A selective medium for the isolation of Arcobacter from meats. Lett. Appl. Microbiol. 23:64–66. 10.1111/j.1472-765X.1996.tb00030.x [DOI] [PubMed] [Google Scholar]
- 22.Johnson LG, Murano EA. 1999. Comparison of three protocols for the isolation of Arcobacter from poultry. J. Food Prot. 62:610–614 [DOI] [PubMed] [Google Scholar]
- 23.Lee MH, Cheon DS, Choi S, Lee BH, Jung JY, Choi C. 2010. Prevalence of Arcobacter species isolated from retail meats in Korea. J. Food Prot. 73:1313–1316 http://www.ingentaconnect.com/content/iafp/jfp/2010/00000073/00000007/art00013?token=004f14c95b1f405847447b49796c5f316a6f7c4763474833757e6f4f2858592f3f3b57b863b24fd [DOI] [PubMed] [Google Scholar]
- 24.Abdelbaqi K, Ménard A, Prouzet-Mauleon V, Bringaud F, Lehours P, Mégraud F. 2007. Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol. Med. Microbiol. 49:337–345. 10.1111/j.1574-695X.2006.00208.x [DOI] [PubMed] [Google Scholar]
- 25.Brightwell G, Mowat E, Clemens R, Boerema J, Pulford DJ, On S. 2007. Development of a multiplex and real time PCR assay for the specific detection of Arcobacter butzleri and Arcobacter cryaerophilus. J. Microbiol. Methods 68:318–325. 10.1016/j.mimet.2006.09.008 [DOI] [PubMed] [Google Scholar]
- 26.Houf K, Tutenel A, De Zutter L, Van Hoof J, Vandamme P. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89–94. 10.1111/j.1574-6968.2000.tb09407.x [DOI] [PubMed] [Google Scholar]
- 27.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:63e. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niessen L, Gräfenhan T, Vogel RF. 2012. ATP citrate lyase 1 (acl1) gene-based loop-mediated amplification assay for the detection of the Fusarium tricinctum species complex in pure cultures and in cereal samples. Int. J. Food Microbiol. 158:171–185. 10.1016/j.ijfoodmicro.2012.06.021 [DOI] [PubMed] [Google Scholar]
- 29.Zhang LQ, Zhao FR, Liu ZG, Kong WL, Wang H, Ouyang Y, Liang HB, Zhang CY, Qi HT, Huang CL, Guo SH, Zhang GH. 2012. Simple and rapid detection of swine hepatitis E virus by reverse transcription loop mediated isothermal amplification. Arch. Virol. 157:2383–2388. 10.1007/s00705-012-1425-5 [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki W. 2013. Sensitive and rapid detection of Campylobacter jejuni and Campylobacter coli using loop-mediated isothermal amplification. Methods Mol. Biol. 943:267–277. 10.1007/978-1-60327-353-4_18 [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Jiang L, Ge B. 2012. Loop-mediated isothermal amplification assays for detecting Shiga toxin-producing Escherichia coli in ground beef and human stools. J. Clin. Microbiol. 50:91–97. 10.1128/JCM.05612-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amare LB, Saleha AA, Zunita Z, Jalila A, Hassan L. 2011. Prevalence of Arcobacter spp. on chicken meat at retail markets and in farm chickens in Selangor, Malaysia. Food Control 22:732–736. 10.1016/j.foodcont.2010.11.004 [DOI] [Google Scholar]
- 33.Hamill S, Neill SD, Madden RH. 2008. A comparison of media for the isolation of Arcobacter spp. from retail packs of beef. J. Food Prot. 71:850–854 http://www.ingentaconnect.com/content/iafp/jfp/2008/00000071/00000004/art00028?token=004b110876e5865462431516f3b6b672123663b25705e4e26634a492f253033297628dc7ba6 [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez I, Garcia T, Antolin A, Hernandez PE, Martin R. 2000. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Lett. Appl. Microbiol. 30:207–212. 10.1046/j.1472-765x.2000.00696.x [DOI] [PubMed] [Google Scholar]
- 35.Shah AH, Saleha AA, Zunita Z, Murugaiyah M, Aliyu AB. 2012. Antimicrobial susceptibility of an emergent zoonotic pathogen, Arcobacter butzleri. Int. J. Antimicrob. Agents 40:569–570. 10.1016/j.ijantimicag.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 36.Kayman T, Abay S, Hizlisoy H, Atabay HI, Diker KS, Aydin F. 2012. Emerging pathogen Arcobacter spp. in acute gastroenteritis: molecular identification, antibiotic susceptibilities and genotyping of the isolated arcobacters. J. Med. Microbiol. 61:1439–1444. 10.1099/jmm.0.044594-0 [DOI] [PubMed] [Google Scholar]
- 37.Ko KS, Oh WS, Lee MY, Peck KR, Lee NY. 2007. A new Microbacterium species isolated from the blood of a patient with fever: Microbacterium pyrexiae sp. nov. Diagn. Microbiol. Infect. Dis. 57:393–397. 10.1016/j.diagmicrobio.2006.10.010 [DOI] [PubMed] [Google Scholar]
- 38.Shin KS, Son BR, Hong SB. 2005. A case of bacteremia caused by Arcobacter butzleri. Korean J. Lab. Med. 25:259–261 http://pdf.medrang.co.kr/Kjlm/2005/259.pdf [Google Scholar]
- 39.Kudra LL, Sebranek JG, Dickson JS, Mendonca AF, Zhang Q, Jackson-Davis A, Prusa KJ. 2012. Control of Campylobacter jejuni in chicken breast meat by irradiation combined with modified atmosphere packaging including carbon monoxide. J. Food Prot. 75:1728–1733. 10.4315/0362-028X.JFP-12-178 [DOI] [PubMed] [Google Scholar]
- 40.Meng S, Xu J, Xiong Y, Ye C. 2012. Rapid and sensitive detection of Plesiomonas shigelloides by loop-mediated isothermal amplification of the hugA gene. PLoS One 7:e41978. 10.1371/journal.pone.0041978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki W, Taguchi M, Kawai T, Kawatsu K, Sakata J, Inoue K, Misawa N. 2009. Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol. 75:1597–1603. 10.1128/AEM.02004-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang JS, Lee YD, Park ZH. 2003. Growth inhibition of newly emerging Arcobacter butzleri by organic acids and trisodium phosphate. Korean J. Food Sci. Technol. 35:1169–1173 [Google Scholar]
- 43.Wesley IV, Baetz AL. 1999. Natural and experimental infections of Arcobacter in poultry. Poultry Sci. 78:536–545 [DOI] [PubMed] [Google Scholar]
- 44.Kabeya H, Maruyama S, Morita Y, Ohsuga T, Ozawa S, Kobayashi Y, Abe M, Katsube Y, Mikami T. 2004. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int. J. Food Microbiol. 90:303–308. 10.1016/S0168-1605(03)00322-2 [DOI] [PubMed] [Google Scholar]
- 45.Li Q, Yue Z, Liu H, Liang C, Zheng X, Zhao Y, Chen X, Xiao X, Chen C. 2010. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of lymphocystis disease virus. J. Virol. Methods 163:378–384. 10.1016/j.jviromet.2009.10.028 [DOI] [PubMed] [Google Scholar]
- 46.Patel JC, Oberstaller J, Xayavong M, Narayanan J, Debarry JD, Srinivasamoorthy G, Villegas L, Escalante AA, Dasilva A, Peterson DS, Barnwell JW, Kissinger JC, Udhayakumar V, Lucchi NW. 2013. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS One 8:e54986. 10.1371/journal.pone.0054986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravindran A, Levy J, Pierson E, Gross DC. 2012. Development of a loop-mediated isothermal amplification procedure as a sensitive and rapid method for detection of ‘Candidatus Liberibacter solanacearum' in potatoes and psyllids. Phytopathology 102:899–907. 10.1094/PHYTO-03-12-0055-R [DOI] [PubMed] [Google Scholar]
- 48.Seetang-Nun Y, Jaroenram W, Sriurairatana S, Suebsing R, Kiatpathomchai W. 2013. Visual detection of white spot syndrome virus using DNA-functionalized gold nanoparticles as probes combined with loop-mediated isothermal amplification. Mol. Cell. Probes 27:71–79. 10.1016/j.mcp.2012.11.005 [DOI] [PubMed] [Google Scholar]